Abstract
Human papillomavirus (HPV) infection is the most common cause of sexually transmitted viral infection and is the main cause of cervical cancer. Identification of HPV T-cell epitopes would be instrumental not only in our understanding of the protective immune response but also in the development of vaccines and immunotherapies. In contrast to viruses which cause systemic infection, identification of HPV epitopes is technically challenging because HPV causes a localized mucosal infection and the frequency of pathogen-specific T lymphocytes in peripheral blood is expected to be low. Here we describe three new antigenic epitopes (E7 7-15 [TLHEYMLDL], E6 52-61 [FAFRDLCIVY], and E7 79-87 [LEDLLMGTL]) of HPV 16 E6 and E7 proteins which have oncogenic activities. E7 7-15 was identified among peptides previously shown to bind to human leukocyte antigen (HLA)-A2.1 molecule, but it was found likely to be restricted by the HLA-B48 molecule. E6 52-61 (likely to be restricted by HLA-B57) and E7 79-87 (likely to be restricted by HLA-B60) were detected, based on the magnitude of the T-cell immune responses, in another individual. In particular, T-cell clones specific for the E6 52-61 epitope were isolated effectively by magnetically selecting them based on gamma interferon secretion. This is an efficient method of identifying new epitopes of antigens for which the number of specific T lymphocytes in the circulation is expected to be small, and it should be widely applicable in identifying new T-cell epitopes.
Worldwide, cervical cancer is the second most common malignancy in women, and an estimated 470,000 new cases are diagnosed annually (4). Although the association between human papillomavirus (HPV) infection and the development of squamous intraepithelial lesions (SIL) is well described, the exact mechanisms that lead from HPV infection to cancer are not well understood. Studies suggest that cell-mediated immune responses are critical for HPV control. Unfortunately, the antigenic epitopes involved in HPV control have not been well characterized. Studying the immune response to the HPV early genes E6 and E7 has been of central interest since these genes are required for the maintenance of cellular transformation and since they are constitutively expressed in the majority of precancer tumor cells. In addition, our earlier work has demonstrated more frequent cell-mediated immune responses to E6 and E7 in HPV 16-positive women who had not developed SIL than in HPV 16-positive women with SIL (9). These data suggest that identifying immunogenic and immunodominant epitopes of the E6 and E7 genes may be of considerable importance in developing therapeutic approaches to prevent the progression of precancers. To date, several T-cell epitopes of HPV 16 have been described, mostly by identifying peptides capable of stimulating T-cell responses among those shown to bind to common HLA class I molecules (2, 5, 10). Here we identify three novel antigenic epitopes of HPV 16 in women who were able to clear their HPV 16 infection, suggesting that these epitopes may have played a direct role in infection control.
MATERIALS AND METHODS
Subjects.
The 10 subjects in the present study were participants in a longitudinal study of HPV infection initiated in 1991 (8). As a part of the parent study, the subjects were being monitored by cervical HPV DNA testing by PCR (13), cytology, and colposcopy every 4 months. These 10 subjects were selected for being HLA-A2 positive and having had HPV 16 infection which subsequently became undetectable for a minimum of two consecutive visits. Established T-cell lines stimulated with recombinant vaccinia virus-infected autologous dendritic cells were used from seven subjects (subjects 1 to 7) to assess the immunogenicity of HLA-A2.1 binding E7 peptides (see Fig. 1); an established CD8 T-cell line was used for one subject (subject 10) (see Fig. 4) in a similar manner. Established peptide-stimulated T-cell lines were used for five subjects (subjects 1, 2, 6, 8, and 9). Informed consent was obtained as specified in the guidelines of the Committee on Human Research at the University of California, San Francisco.
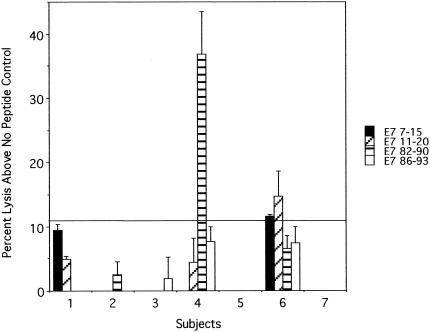
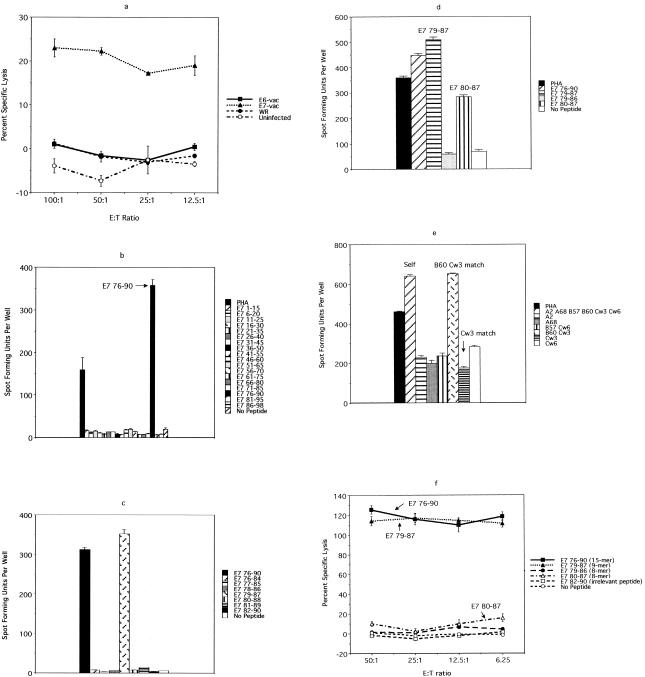
FIG. 1.
Chromium release assay of T-cell lines established from HLA-A2.1-positive women with past HPV 16 infection who have not developed SIL. A positive response to E7 7-15, E7 11-20, or E7 82-90 was demonstrated once each (above the cutoff line).
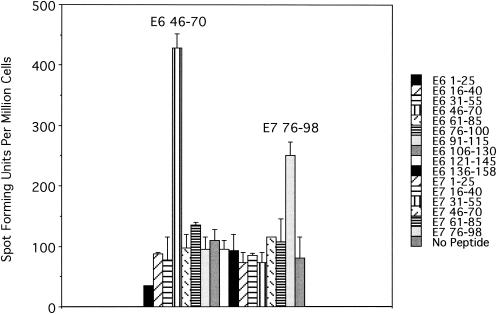
FIG. 4.
An ELISPOT assay performed using a CD8 T-cell line from subject 10, with overlapping 15-mer peptides, in pools of three, of the HPV 16 E6 and E7 protein, suggesting the presence of one antigenic epitope in the E6 46-70 region and another one in the E7 76-98 region. Each pool contained three 15-mer peptide which overlap by 10 central amino acids. For example, the first pool, which covered the E6 1-25 region, contained E6 1-15, E6 6-20, and E6 11- 25. A total of 2 × 105 cells from the CD8 T-cell line were plated per well.
HLA typing.
Peripheral blood mononuclear cells (PBMC) were serologically typed for HLA class I molecules by the University of California, San Francisco, Immunogenetics Laboratory. High-resolution HLA typing by sequencing was also performed whenever necessary to identify the alleles (i.e., to identify HLA-A2.1 positive subjects).
Peptides.
The overlapping 9-mer peptides (overlapping by 8 amino acids) covering the entire HPV 16 E6 and E7 proteins (12) were synthesized by SynPep (Dublin, Calif.). All other peptides, including the overlapping 15-mer peptides (overlapping by 10 amino acids), covering the entire HPV 16 E6 and E7 proteins were synthesized by the Biomolecular Resource Center at the University of California, San Francisco.
Generation of T-cell lines.
HPV 16 E6- and E7-specific T-cell lines were established by in vitro stimulation of PBMC using autologous dendritic cells infected with recombinant vaccinia viruses expressing the E6 and E7 proteins (E6-vac and E7-vac [9]) for subjects 1 through 7. Autologous dendritic cells were established by isolating monocytes from PBMC by using CD14 antibody coupled to magnetic beads (Miltenyi Biotec), by growing them in the presence of granulocyte-macrophage colony-stimulating factor (50 ng/ml) and recombinant interleukin-4 (rIL-4) (100 U/ml) for 7 days. Half of these autologous dendritic cells were cryopreserved so that they would be available for the second round of in vitro stimulation. They were matured by being cultured in wells containing irradiated L cells expressing CD40 ligand for the last 24 h while they were infected with E6-vac and E7-vac. PBMC were stimulated for 7 days in vitro by using these mature autologous dendritic cells infected with E6-vac and E7-vac. The stimulation was repeated for an additional 7 days by using thawed autologous dendritic cells.
Peptide-stimulated CD8 T-cell lines were established from subjects 1, 2, 6, 8, and 9 by using E7 7-15 (TLHEYMLDL), E7 11-20 (YMLDLQPETT), and E7 82-90 (LLMGTLGIV) to generate T-cell lines with a high enough frequency of T cells specific to these peptides in order to isolate T cell clones. They were generated by in vitro stimulation of magnetically selected CD8 cells, using autologous immature dendritic cells pulsed with 40 μg each of peptides per ml in the presence of 4 μg of β2-microglobulin per ml, 20 U of rIL-2 per ml, and 10 ng of rIL-7 per ml (see Fig. 6). Two cycles of stimulation were performed, with each cycle lasting for 7 days.
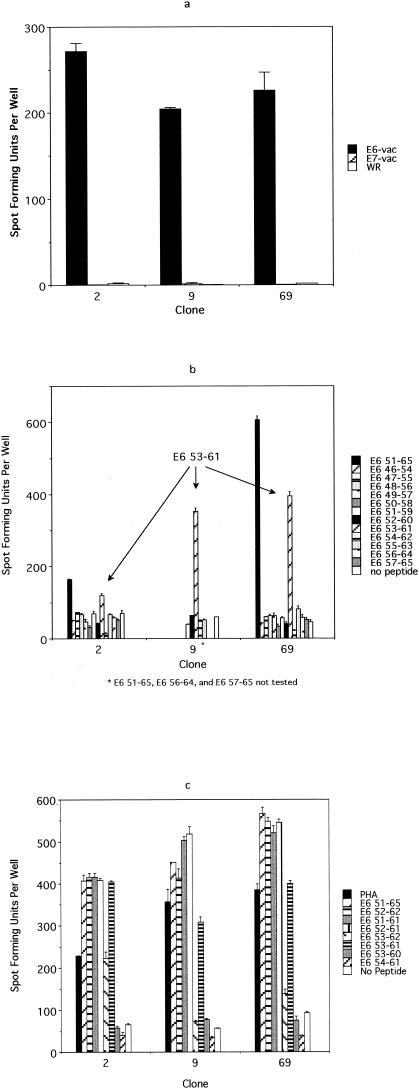
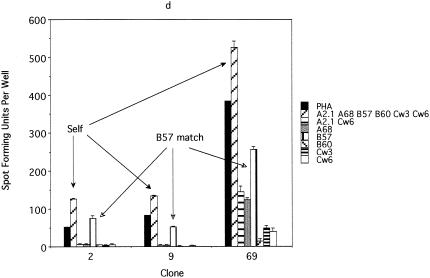
FIG. 6.
An ELISPOT assay performed to characterize the E6 epitope from subject 10. (a) An ELISPOT assay performed using E6-vac-, E7-vac-, and WR-infected autologous EBV-LCL as antigen-presenting cells revealed that the T-cell clones (no. 2, 9, and 69) recognize an E6 epitope, which is likely to be naturally processed. A total of 103 T-cell clones were plated along with 105 autologous EBV-LCL infected with E6-vac, E7-vac, and WR, respectively. (b) An ELISPOT assay using overlapping 9-mer peptides within the E6 46-70 region demonstrated a response with the E6 53-61 peptide. A total of 103 T-cell clones were plated along with 105 autologous EBV-LCL. (c) The comparison of E6 51-65 (15-mer), E6 52-62 (11-mer), E6 51-61 (11-mer), E6 52-61 (10-mer), E6 53-62 (10-mer), E6 53-61 (9-mer), E6 53-60 (8-mer), and E6 54-61 (8-mer) using an ELISPOT assay revealed that the optimal peptide of minimum length was E6 52-61 (10-mer). A total of 103 T-cell clones were plated along with 105 autologous EBV-LCL. (d) An ELISPOT assay examining the restriction element for the E6 52-61 epitope. The result suggests that the HLA-B57 molecule is the restriction element. A total of 103 T-cell clones were plated along with 105 EBV-LCL matching at the designated HLA types.
For subject 10, CD8 cells were selected magnetically before the T-cell line was established, using autologous mature dendritic cells infected with E6-vac and E7-vac (see Fig. 4 and 5). Otherwise, the methods were the same as those described for subjects 1 to 7. Because not enough cells were available from this CD8 cell line for isolation of T-cell clones recognizing the newly described E6 epitope, a second CD8 T-cell line was established from this subject by using the three 15-mer peptides contained in a pool to which the first T-cell line showed positivity (see Fig. 4) (E6 46-60 [RREVYDFAFRDLCIV], E6 51-65 [DFAFRDLCIVYRDGN], and E6 56-70 [DLCIVYRDGNPYAVC]). The methods were the same as described for establishing peptide-stimulated line for subjects 1, 2, 6, 8, and 9 above.
FIG. 5.
Chromium release and ELISPOT assays performed to characterize the E7 epitope from subject 10. (a) A chromium release assay demonstrated the specificity of the 27G6 clone for an epitope from the E7 protein. (b) An ELISPOT assay using overlapping 15-mer peptides of E7 demonstrated that the antigenic peptide is contained within the E7 76-90 region. A total of 103 T-cell clones were plated along with 105 autologous EBV-LCL. (c) An ELISPOT assay using overlapping 9-mer peptides within the E7 76-90 region demonstrated a response only with the E7 79-87 peptide. A total of 103 T-cell clones were plated along with 105 autologous EBV-LCL. (d) The comparison of E7 76-90 (15-mer), E7 79-87 (9-mer), E7 79- 86 (8-mer), and E7 80-87 (8-mer) by an ELISPOT assay showed an optimal response with E7 79-98 (9-mer). A total of 103 T-cell clones were plated along with 105 autologous EBV-LCL. (e) An ELISPOT assay examining the restriction element for the 27G6 clone. The result suggests that the HLA-B60 molecule is the restriction element. A total of 103 T-cell clones were plated along with 105 EBV-LCL matching at designated HLA types. (f) A chromium release assay demonstrated that the E7 76-90 (15-mer) and E7 79-87 (9-mer) peptides were much more vigorously recognized than were the E7 79-86 (8-mer) and E7 80-87 (8-mer) peptides. E:T ratio, effector-to-target-cell ratio.
Chromium release assay.
Either the cells of the autologous Epstein-Barr virus (EBV)-transformed B-lymphoblastoid cell line were infected with recombinant vaccinia viruses (E6-vac, E7-vac, or WR) at multiplicity of infection of 1, or they were pulsed with 10 μM peptide antigens. They were radiolabeled using 200 μCi of sodium chromate (Na251CrO4; specific activity 5 mCi/ml). The cells were incubated with peptides at this time if applicable. After being washed, the cells were plated in triplicate in 96-well plates at 3 × 103 cells per well. The peptides were added to the medium in appropriate wells. Effector cells were added at four different effector-to-target-cell ratios or at a single effector-to-target-cell ratio of 50:1. The plated cells were pelleted by centrifugation and then incubated for 5 h at 37°C in a humidified 5% CO2 incubator. The supernatants were harvested using a Skatron harvesting press and the 51Cr was counted using a gamma counter (Packard Instruments, Meriden, Conn.). The percent specific lysis was calculated as described previously (9).
To establish a cutoff defining a positive response for the E7 peptide assay (see Fig. 1), the standard deviations of the triplicate of the no-peptide negative controls were averaged for all assays performed. Any response greater than 3 standard deviations or 10.8% above the corresponding negative control was considered positive.
Magnetic selection of IFN-γ-secreting cells.
At 8 h prior to performing limiting-dilution assays, 2.5 × 106 cells from each T-cell line were stimulated with the appropriate peptide(s) at 10 μM. The peptide-specific T cells were positively selected using the gamma interferon (IFN-γ) secretion assay enrichment kit (Miltenyi Biotec).
Limiting dilution.
Briefly, the cells were plated at concentration of 0.5 cell per well in the presence of a 0.5× feeder cell mixture (Yssel's medium containing 1% pooled human serum, penicillin G [100 U/ml], streptomycin [100 μg/ml], 5 × 105 irradiated allogeneic PBMC per ml, 5 × 104 irradiated JY cells per ml, 0.1 μg of phytohemagglutinin per ml) with or without 10 U of rIL-4 per ml. Control wells for growth contained 1 to 1,000 cells per well. On day 5, 100 μl of Yssel's medium containing 20 U of rIL-2 per ml was added to each well. Growing microcultures were transferred to wells of 24-well plates containing 1 ml of a 1× feeder cell mixture (Yssel's medium containing 1% pooled human serum, penicillin G [100 U/ml], streptomycin [100 μg/ml], 1 × 106 irradiated allogeneic PBMC per ml, 1 × 105 irradiated JY cells per ml, 0.1 μg of phytohemogglutinin per ml).
IFN-γ enzyme-linked immunospot (ELISPOT) assay.
A method described by Larsson et al. was used with the following modifications (7). A total of 2 × 105 cells of a T-cell line (see Fig. 4) or 1 × 103 T-cell clone cells with 1 × 105 EBV-transformed lymphoblastoid cell line cells (LCL) (see Fig. 3, 5, and 6) were plated in each well. For screening of T-cell clones, 50 μl of culture medium containing the T-cell clones was added with 105 autologous EBV-LCL. E6-vac, E7-vac, and WR were added at a multiplicity of infection of 10 (see Fig. 2a, 3a, and 6a). Peptide antigens were added at 10 μM, along with 20 U of rIL-2 per ml (except for screening, when no rIL-2 was added).
FIG. 3.
Confirmation of the E7 11-20 specificity of the screen-positive T-cell clones and identification of their restriction element. (a) The E7 11-20-specific T-cell clones recognize the E7 11-20 peptide but not the E7 11-19 peptide, the E7 12-20 peptide, or E7-vac-infected autologous EBV-LCL. A total of 103 T-cell clones were plated along with 105 autologous EBV-LCL. (b) The restriction element for E7 11-20 appears to be the HLA-A2.1 molecule. A total of 103 T-cell clones were plated along with 105 EBV-LCL matching at the designated HLA types.
FIG. 2.
Confirmation of the E7 7-15 specificity of the screen-positive T-cell clones and identification of their restriction element. (a) The E7 7-15-specific T-cell clones recognize the E7 7-15 peptide but not E7-vac-infected autologous EBV-LCL. A total of 103 T-cell clones were plated along with 105 autologous EBV-LCL. (b) The results suggest that the restriction element for the E7 7-15 peptide is likely to be the HLA-B48 molecule. A total of 103 T-cell clones were plated along with 105 EBV-LCL matching at the designated HLA types. Only the HLA class I molecules matching the subject being studied are indicated.
To determine the frequencies of peptide-specific T cells, 5 × 104 cells of T-cell lines per well were plated in triplicate for each peptide being examined and for the no-peptide control. The frequencies were determined by subtracting the number of spot-forming units in the no-peptide control wells from the number in the peptide-containing wells and dividing the difference by the number of cells plated.
Fluorescence-activated cell sorter analysis.
The T-cell clones were stained for surface markers with anti-CD4 anti-CD8 and anti-CD3 anti-CD16 (Caltag, Burlingame, Calif.) and were analyzed (FACScan; Beckton Dickinson Immunocytometry Systems, San Jose, Calif.).
RESULTS
Identifying antigenic peptides among those shown to bind to the HLA-A2.1 molecule.
To examine the immunogenicity of HPV 16 E6 and E7 peptides (5), cytotoxic T-lymphocyte lines were derived from PBMC of women who were participants in a cohort study investigating the natural history of HPV infection. Seven women (subjects 1 to 7) were selected for being HLA-A2 positive and having had a history of cleared HPV 16 infection. The mean age at infection was 24 years. The mean duration since the last HPV 16-positive result was 25.0 months (range, 8.5 to 38.9 months), which means that on average, the subjects had five consecutive visits with negative PCR results. The cytotoxic T-lymphocyte lines were established by stimulating PBMC using autologous dendritic cells infected with E6-vac and E7-vac. Chromium release assays were performed to assess the lysis of peptide (E7 7-15, E7 11-20, E7 82-90, or E7 86-93 [TLGIVCPI]-pulsed target cells) (only E7 peptides were studied because of low cell yield). These peptides were previously shown to bind the HLA-A2.1 molecule by Kast et al. (5). Of the seven subjects, two demonstrated positive responses to one (E7 82-90 in subject 4) or two (E7 7-15 and E7 11-20 in subject 6) (Fig. 1) peptides. The antigenicity of E7 11-20 and E7 82-90 has been previously described (1, 3, 10, 14), but the antigenicity of the E7 7-15 peptide has never been described, to our knowledge. To fully characterize these epitopes, attempts were made to isolate T-cell clones. First, T-cell lines were established from five subjects by serially stimulating CD8 T lymphocytes with autologous dendritic cells pulsed with E7 7-15, E7 11-20, or E7 82-90 peptides. IFN-γ ELISPOT assays were performed to determine the frequency of T lymphocytes specific to these peptides. Only subject 6 demonstrated the presence of E7 7-15-specific T lymphocytes (data not shown). This T-cell line was also positive for E7 11-20. To isolate T-cell clones, the cell line was stimulated for two additional cycles. The frequencies of E7 7-15- and E7 11-20-specific T lymphocytes, determined by an ELISPOT assay, were 0.20 and 0.47%, respectively.
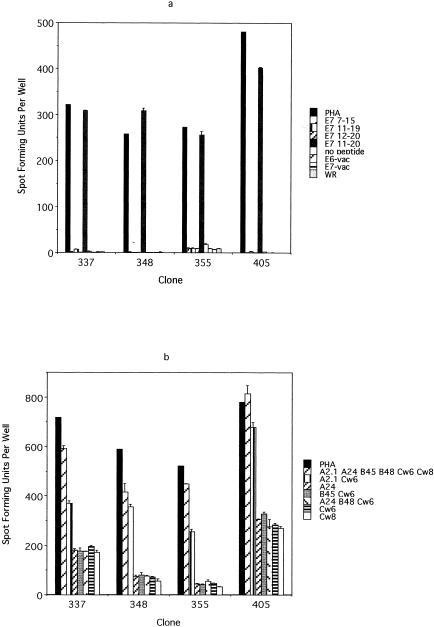
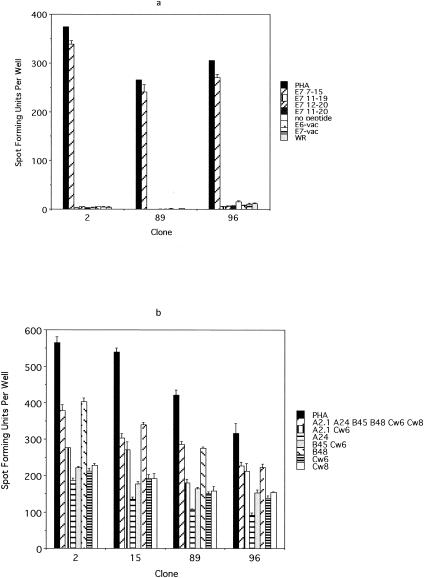
Approximately 1.5 × 103 E7 7-15-specific T cells and 2.5 × 104 E7 11-20-specific T cells from the T-cell line from subject 6 were isolated magnetically on the basis of IFN-γ secretion. The limiting-dilution experiments were performed separately for each peptide by plating cells at a frequency of 0.5 cell per well. A total of 288 clones for E7 7-15 and 157 clones for E7 11-20 were expanded. Then 96 randomly selected clones were screened for specificity to the E7 7-15 peptide by using ELISPOT (data not shown). Of these clones, 81 (84.3%) were positive for E7 7-15 peptide (i.e., positive IFN-γ secretion in the presence of the peptide but not in its absence) and 2 (2.1%) were positive for EBV antigens (i.e., positive IFN-γ secretion in the presence and absence of the peptide). Likewise, 94 randomly selected clones were screened for E7 11-20; 89 (94.7%) were positive for E7 11-20 and 2 (2.1%) clones were positive for EBV antigens. The results of restriction mapping of E7 7-15 suggested that the HLA-B48 molecule was the restriction element, which was unexpected (Fig. 2b). On the other hand, the restriction element of the E7 11-20 peptide appeared to be the HLA-A2.1 molecule, as expected (Fig. 3b). The fact that the E7 7-15 peptide was likely to be restricted by the HLA-B48 molecule demonstrates a limitation of an approach in which peptide binding to only common HLA types was studied. The T-cell clones specific for the E7 7-15 and E7 11-20 epitopes did not recognized E7-vac-infected autologous EBV-LCL (Fig. 2a and 3a). Fluorescence-activated cell sorter analysis demonstrated that all T-cell clones examined have the surface phenotype CD3+ CD4− CD8+ CD16−.
Identifying epitopes within the HPV 16 E6 and E7 proteins based on the magnitude of the immune response.
Demonstration of T-cell responses to peptides known to bind HLA molecules can lead to the identification of important T-cell epitopes; however, this method does not take into consideration the magnitude of the immune response to the peptide. We developed a method which would allow us to identify antigenic epitopes based on the magnitude of the immune response by first quantifying the number of peptide-specific CD8 T cells in an established T-cell line from one of the subjects (subject 10). This subject was HLA-A2.1 positive and had an HPV 16 infection, at the age of 26 years, which was cleared 29 months prior to the blood draw. Overlapping 15-mer peptides pooled in groups of three spanning all the E6 and E7 proteins were used in a IFN-γ ELISPOT assay. Responses to the E6 46-70 and E7 76-98 regions were demonstrated, with the response to the E6 epitope being larger than that to the E7 epitope (Fig. 4).
A limiting-dilution assay was performed to isolate T-cell clones recognizing these antigenic epitopes by using a classical approach in which the cells of the CD8 T-cell line were plated at a frequency of 0.5 cell per well without selection for IFN-γ secretion. A total of 3 × 103 were plated, and 348 of 586 clones that grew were screened by a chromium release assay, using E6-vac- or E7-vac-infected autologous EBV-LCL (data not shown). Of the 348 T-cell clones tested, 1 (the 27G6 clone) had an E7 specificity, as shown by a chromium release assay (Fig. 5a). By using overlapping 15-mer peptides covering the E7 protein, the epitope recognized by this T-cell clone was shown to be in the E7 76-90 (IRTLEDLLMGTLGIV) peptide (Fig. 5b). The epitope was further defined using an overlapping 9-mer within the 15-amino-acid region to the E7 79-87 peptide (LEDLLMGTL) (Fig. 5c). Examination of shorter 8-mer peptides (E7 79-86 [LEDLLMGT] and E7 80-87 [EDLLMGTL]) demonstrated a smaller response compared to that for the E7 79-87 (9-mer) (Fig. 5d). Using allogeneic EBV-LCL sharing HLA class I molecules with subject 10, the restriction element for the E7 79-87 epitope was investigated. The results suggest that the HLA-B60 molecule is the restriction element (Fig. 5e). The 9-mer peptide E7 79-87 does contain the known HLA-B60 motif, which includes a glutamic acid residue at position 2 and a leucine residue at position 9. This explained why the E7 80-87 (8-mer) peptide, which contains both anchor residues, exhibited partial IFN-γ secretion while E7 79-86 (8-mer), which contains only one of the two anchor residues, did not show any activity above the background (Fig. 5d). These results were similar to those of the same analysis performed using a chromium release assay; however, the quantitative difference between the strength of the immunogenicity of E7 76-90 (15-mer) and E7 79-87 (9-mer) was not apparent in the chromium release assay (Fig. 5f). The difference between the activity of E7 79-86 (8-mer) and E7 80-87 (8-mer) was less obvious as well. Nevertheless, the major advantage of using the ELISPOT assay lies in the fact that only 103 T-cell clones were needed per well, which translates to needing only 1/600 of the number of T-cell clones used in the chromium release assay.
Using the approach described above, we were unable to isolate any T-cell clones recognizing the E6 epitope, most probably because the frequency of this clone was too low (1 in 2,500) (Fig. 4). Indeed, it was only by sheer luck that we were able to isolate the T-cell clone recognizing the E7 epitope, since its frequency was even lower (1 in 4,000), and we screened 348 T-cell clones which grew from approximately 3,000 that were plated in a limiting-dilution experiment. A second CD8 T-cell line was generated from the same individual (subject 10), because the cells from the first CD8 T-cell line had run out, using autologous dendritic cells pulsed with the peptide pool to which this subject demonstrated a response (E6 46-60, E6 51-65, and E6 56-70 peptides). Stimulation was done using peptides because we observed more robust proliferation of T cells than occurred after stimulation with recombinant vaccinia virus-infected antigen-presenting cells. The frequencies of the peptide-specific T cells were 0.09, 0.11, and 0.04%, respectively. To circumvent the problem of the low frequency of the T-cell clone, we attempted to isolate T-cell clones by using a more selective strategy. The T cells with specificity to three peptides were isolated magnetically for IFN-γ production before the limiting-dilution assay was performed. One thousand cells selected on the basis of IFN-γ secretion were plated. An ELISPOT assay was performed on 94 randomly selected clones of the 480 clones which grew after the limiting-dilution experiment. Each clone was plated in quadruplicate and was tested separately with E6 46-60, E6 51-65, E6 56-70, and the no-peptide control. A total of 18 T-cell clones were positive for the E6 46-60 peptide, 6 clones were positive for E6 51-65, 18 clones were positive for both E6 46-60 and E6 51-65, and 1 clone was positive for E6 56-70. To further define which peptide contained the E6 epitope, eight representative clones (two positive for E6 46-60, two positive for E6 51-65, three positive for E6 46-60 and E6 51-65, and one positive for E6 56-70) were examined to determine if they recognize naturally processed epitopes by testing their recognition of E6-vac-infected autologous EBV-LCL. This criterion, whether naturally processed E6 antigen was recognized or not, was used to distinguish T-cell clones of interest from irrelevant T-cell clones because the first CD8 T-cell line was established using autologous dendritic cells infected with recombinant vaccinia viruses expressing the E6 protein. Three of the eight clones positively recognized E6-vac-infected autologous EBV-LCL. These three clones recognized E6-vac-infected autologous EBV-LCL but not those infected with E7-vac or WR (wild type), suggesting that these clones are specific for a naturally processed epitope of the E6 protein (Fig. 6a). Using the overlapping 9-mer peptides within this region, the epitope was determined to be E6 53-61 (AFRDLCIVY) (Fig. 6b). However, the response to the 15-mer E6 51-65 was greater than that to the 9-mer E6 53-51, suggesting that the optimal sequence of this epitope was longer than 9 amino acids. We next tested for peptide sequences, E6 53-60 (AFRDLCIV) (8-mer), E6 54-61 (FRDLCIVY) (8-mer), E6 53-61 (9-mer), E6 53-62 (10-mer), E6 52-61 (FAFRDLCIVY) (10-mer), E6 51-61 (DFAFRDLCIVY) (11-mer), E6 52-62 (FAFRDLCIVYR) (11-mer), and E6 51-65 (15-mer) (Fig. 6c). We determined the shortest optimal peptide sequence to be the 10-mer E6 52-61 (Fig. 6c). The restriction element for the E6 52-61 was likely to be the HLA-B57 molecule (Fig. 6d). The surface phenotypes of all three T-cell clones were CD3+ CD4− CD8+ CD16−.
DISCUSSION
This is the first study that we are aware of which has identified antigenic peptides of HPV 16 E6 and E7 proteins in women with documented HPV clearance. Although this task seems straightforward in theory, it is technically challenging because HPV does not cause a systemic infection. Therefore, only a few circulating HPV-specific circulating memory cells are expected to be present. We utilized three different technical approaches to identify these peptides. The first approach, which led to the description of the novel E7 7-15 epitope, required first having the information about which peptides bind to an HLA-A2.1 molecule. Although this method should have identified an HLA-A2.1-restricted epitope, the restriction element was found likely to be the HLA-B48 molecule, underscoring the limitation of this approach in identifying HLA-specific epitopes. The second approach, which led to the description of the E7 79-87 epitope, involved generating a T-cell line by in vitro stimulation of CD8 cells with autologous dendritic cells infected with E6-vac and E7-vac and performing a limiting-dilution assay to isolate a T-cell clone before defining the peptide sequence and its associated restriction element. The latter technique is a more classical approach, and many viral epitopes have been described as a result of its use (6, 11). However, this approach is impractical for identifying antigenic epitopes of pathogens which are expected to generate a small number of circulating T lymphocytes. Although we were able to isolate a T-cell clone recognizing the E7 epitope by chance, this method failed to identify the E6 epitope due to a low frequency of T-cell clones. The last approach, with which we described the E6 51-62 epitope, incorporated key technical advances which should make it feasible to identify new epitopes even when the particular T lymphocytes with the specificity may be scarce. These advances included (i) using overlapping 15-mer peptides to identify the region in which the epitope is contained, (ii) magnetically selecting for IFN-γ-secreting peptide-specific T lymphocytes, and (iii) seeding autologous and allogeneic EBV-LCL cells for the ELISPOT assay, reducing the number of T-cell clone cells needed to 103 cells per well.
Unexpected observations in our study were the lack of recognition of E7-vac-infected autologous EBV-LCL by the E7 7-15 and E7 11-20 specific T-cell clones (Fig. 2a and 3a), especially since Ressing et al. (10) have demonstrated cross-reactive lysis of CaSki cells (an HLA A2.1-positive cervical carcinoma cell line expressing the HPV 16 E6 and E7 proteins) by an E7 11-20-specific T-cell clone. This may be a reflection of the low affinity of our E7 7-15- and E7 11-20-specific T-cell clones, since they were isolated from a peptide-stimulated T-cell line. Alternatively, it is possible that these epitopes are not naturally processed in EBV-LCL, although they are processed in dendritic cells used to stimulate the T-cell lines.
For subject 10, the magnitude of the immune responses to the E6 and E7 proteins was measured using a CD8 T-cell line stimulated in vitro (Fig. 4). Despite the in vitro manipulation, the repertoire of the resulting HPV 16 E6- and E7-specific T cells is likely to be representative of that in natural infection, since antigenic epitopes in E6-vac- and E7-vac-infected cells would be processed in the same manner as in cells infected with the whole virus; i.e., proteins are synthesized by the cellular machinery of the host, cleaved by proteosomes or proteases, and transported by the transporter-associated protein to the endoplasmic reticulum, where antigenic peptides would bind to HLA molecules.
Unique to this study was the identification of novel epitopes (Table 1) in women with a documented history of HPV 16 clearance or control. Consequently, we think that these epitopes are likely to be important in HPV control. Therefore, they may be key in the development of vaccines to enhance immunity to HPV, whether preventive or therapeutic.
TABLE 1.
Amino acid sequences and the suggested restriction elements of the novel HPV 16 T-cell epitopes described
| Epitope | Sequence | Suggested restriction element |
|---|---|---|
| E6 52-61 | FAFRDLCIVY | B57 |
| E7 7-15 | TLHEYMLDL | B48 |
| E7 79-87 | LEDLLMGTL | B60 |
Since many women in this study had cleared their HPV 16 infection many months prior to analysis, it is possible that some key epitopes were undetected. Also, we examined only women whose HPV 16 infection had cleared but not women whose HPV 16-positive SIL had regressed. It would be important to study the latter group because the T-cell epitopes to be described should be at least as important as the ones described from the former group.
Although all of the women studied were HLA-A2 positive, new epitopes described were likely to be restricted by other, less common HLA types (B48, B57, and B60), underscoring the limitation of studying only common HLA types. The new and more efficient method of identifying antigenic epitopes described here should lead to a description of many more key epitopes not only for HPV but also for many other pathogens and even for autoantigens.
Acknowledgments
This work was supported by the National Institutes of Health (NCI CA51323, NCI K07 CA75974, and M01 RR01271), the Cancer Research Institute (Pre-Clinical Grant-HPV), and the UCSF Cancer Center Clinical Investigator Research Program (CI-02-3).
REFERENCES
- 1.Alexander, M., M. L. Salgaller, E. Celis, A. Sette, W. A. Barnes, S. A. Rosenberg, and M. A. Steller. 1996. Generation of tumor-specific cytolytic T lymphocytes from peripheral blood of cervical cancer patients by in vitro stimulation with a synthetic human papillomavirus type 16 E7 epitope. Am. J. Obstet. Gynecol. 175:1586-1593. [DOI] [PubMed] [Google Scholar]
- 2.Bourgault Villada, I., N. Beneton, C. Bony, F. Connan, J. Monsonego, A. Bianchi, P. Saiag, J. P. Levy, J. G. Guillet, and J. Choppin. 2000. Identification in humans of HPV-16 E6 and E7 protein epitopes recognized by cytolytic T lymphocytes in association with HLA-B18 and determination of the HLA-B18-specific binding motif. Eur. J. Immunol. 30:2281-2289. [DOI] [PubMed] [Google Scholar]
- 3.Evans, E. M., S. Man, A. S. Evans, and L. K. Borysiewicz. 1997. Infiltration of cervical cancer tissue with human papillomavirus-specific cytotoxic T-lymphocytes. Cancer Res. 57:2943-2950. [PubMed] [Google Scholar]
- 4.Ferlay, J., F. Bray, P. Pisani, and D. Parkin. 2001. Globocan 2000: cancer incidence, mortality and prevalence worldwide, version 1.0. IARC CancerBase no.5. IARC Press, Lyons, France.
- 5.Kast, W. M., R. M. Brandt, J. Sidney, J. W. Drijfhout, R. T. Kubo, H. M. Grey, C. J. Melief, and A. Sette. 1994. Role of HLA-A motifs in identification of potential CTL epitopes in human papillomavirus type 16 E6 and E7 proteins. J. Immunol. 152:3904-3912. [PubMed] [Google Scholar]
- 6.Kaul, R., and S. L. Rowland-Jones. 2000. Methods of detection of HPV-specific CTL and their role in protection against HIV infection, p. 35-44. In B. Korber, C. Brander, B. Haynes, R. Koup, C. Kuiken, J. P. Moore, B. D. Walker, and D. I. Watkins (ed.), HIV molecular immunology 2000. Los Alamos National Laboratory, Los Alamos, N.M.
- 7.Larsson, M., D. T. Wilkens, J. F. Fonteneau, T. J. Beadle, M. J. Merritt, R. G. Kost, P. A. Haslett, S. Cu-Uvin, N. Bhardwaj, D. F. Nixon, and B. L. Shacklett. 2002. Amplification of low-frequency antiviral CD8 T cell responses using autologous dendritic cells. Aids 16:171-180. [DOI] [PubMed] [Google Scholar]
- 8.Moscicki, A. B., S. Shiboski, J. Broering, K. Powell, L. Clayton, N. Jay, T. M. Darragh, R. Brescia, S. Kanowitz, S. B. Miller, J. Stone, E. Hanson, and J. Palefsky. 1998. The natural history of human papillomavirus infection as measured by repeated DNA testing in adolescent and young women. J. Pediatr. 132:277-284. [DOI] [PubMed] [Google Scholar]
- 9.Nakagawa, M., D. P. Stites, S. Farhat, J. R. Sisler, B. Moss, F. Kong, A. B. Moscicki, and J. M. Palefsky. 1997. Cytotoxic T lymphocyte responses to E6 and E7 proteins of human papillomavirus type 16: relationship to cervical intraepithelial neoplasia. J. Infect. Dis. 175:927-931. [DOI] [PubMed] [Google Scholar]
- 10.Ressing, M. E., A. Sette, R. M. Brandt, J. Ruppert, P. A. Wentworth, M. Hartman, C. Oseroff, H. M. Grey, C. J. Melief, and W. M. Kast. 1995. Human CTL epitopes encoded by human papillomavirus type 16 E6 and E7 identified through in vivo and in vitro immunogenicity studies of HLA-A*0201-binding peptides. J. Immunol. 154:5934-5943. [PubMed] [Google Scholar]
- 11.Rickinson, A. B., and D. J. Moss. 1997. Human cytotoxic T lymphocyte responses to Epstein-Barr virus infection. Annu. Rev. Immunol. 15:405-431. [DOI] [PubMed] [Google Scholar]
- 12.Seedorf, K., G. Krammer, M. Durst, S. Suhai, and W. G. Rowekamp. 1985. Human papillomavirus type 16 DNA sequence. Virology 145:181-185. [DOI] [PubMed] [Google Scholar]
- 13.Ting, Y., and M. M. Manos. 1990. Detection and typing of genital human papillomaviruses, p. 356-367. In, M. A. Innis, D. H. Gelfand, J. J. Sninsky, and T. J. White (ed.), PCR protocols: a guide to methods and applications. Academic Press, Inc., San Diego, Calif.
- 14.Youde, S. J., P. R. Dunbar, E. M. Evans, A. N. Fiander, L. K. Borysiewicz, V. Cerundolo, and S. Man. 2000. Use of fluorogenic histocompatibility leukocyte antigen- A*0201/HPV 16 E7 peptide complexes to isolate rare human cytotoxic T-lymphocyte- recognizing endogenous human papillomavirus antigens. Cancer Res. 60:365-371. [PubMed] [Google Scholar]