Abstract
Membrane interfaces formed at cell-cell junctions are associated with characteristic patterns of membrane protein organization, such as E-cadherin enrichment in epithelial junctional complexes and CD45 exclusion from the signaling foci of immunological synapses. To isolate the role of protein size in these processes, we reconstituted membrane interfaces in vitro using giant unilamellar vesicles decorated with synthetic binding and non-binding proteins. We show that size differences between binding and non-binding proteins can dramatically alter their organization at membrane interfaces in the absence of active contributions from the cytoskeleton, with as little as a ~5 nm increase in non-binding protein size driving its exclusion from the interface. Combining in vitro measurements with Monte Carlo simulations, we find that non-binding protein exclusion is also influenced by lateral crowding, binding protein affinity, and thermally-driven membrane height fluctuations that transiently limit access to the interface. This simple, sensitive, and highly effective means of passively segregating proteins has implications for signaling at cell-cell junctions and protein sorting at intracellular contact points between membrane-bound organelles.
Direct physical contact between cells forms membrane interfaces that are important for cell-cell communication; for example, in epithelia and endothelia, initiation of cell-cell fusion during muscle formation, and T-cell activation in the immune system1–3. These membrane interfaces are comprised of two closely apposed plasma membranes that are densely packed with binding proteins that form adhesions and non-binding proteins that occupy space in the interface but do not form adhesions. Specific binding proteins, such as E-cadherins, are known to be enriched at membrane interfaces4 and to hold the membranes together. The membrane separation distances at these junctions are determined, at least in part, by size of the binding proteins that form the interface and could, in principle, influence which non-binding proteins are permitted at the interface. However, there is no quantitative understanding of the fundamental interplay of protein size and membrane properties on the segregation of binding and non-binding proteins at membrane interfaces.
Recently, spatial organization of binding and non-binding proteins at membrane interfaces has been found to be critical for function5. One well-studied example is the immunological synapse (IS) formed during initiation of the adaptive immune response, during which peptide-bound Major Histocompatibility Complexes (pMHCs) on the surface of an antigen-presenting cell interact with T-Cell Receptors (TCRs) on the apposing T-cell membrane6–8. Subsequent activation of the T-cell relies on spatial segregation of proteins that make up a kinase–phosphatase system, wherein the transmembrane phosphatase CD45, which has a large extracellular domain, is excluded from pMHC/TCR clusters9–12, permitting stable TCR phosphorylation and a downstream signaling cascade leading to activation13–15.
Multiple mechanisms have been implicated in the organization of proteins at membrane interfaces, including receptor-ligand clustering by diffusion and trapping, lipid raft formation, intracellular protein-protein interactions, protein displacement based on size of the extracellular domain, and reorganization driven by the underlying cortical cytoskeleton16–18. Modeling of binding proteins at membrane interfaces has revealed that binding affinities, membrane fluctuations, and mixing entropy can produce phase transitions that segregate different binding proteins19. In the immunological synapse, it has been suggested that exclusion of the non-binding protein CD45 is driven by a size-dependent mechanism11,20,21, since CD45 isoforms can have extended conformations that are 15–40 nm larger than the space between apposing membranes imposed by pMHC/TCR binding at the membrane interface15,22. Detailed simulations of membrane interfaces have provided important insight into the organization and affinity of binding proteins23–25, but little is known about how the interplay between binding proteins and non-binding proteins contributes to protein segregation at membrane interfaces.
Here we show that size alone is sufficient to titrate non-binding protein exclusion from membrane interfaces, absent the underlying dynamics of the actin cortex, receptor-ligand clustering, and complex lipid composition of the plasma membrane. We find that nanometer-scale increases in non-binding protein height beyond that of the binding protein will monotonically increase non-binding protein exclusion. We also show that lateral crowding of binding proteins at the membrane interface can exclude non-binding proteins regardless of their height within an interface. Since lateral crowding depends on surface area coverage of the binding protein, this demonstrates a direct connection between lateral footprint and binding affinity of one protein species and exclusion of another. We use Monte Carlo simulations to generalize and extend these results to show that changes in binding protein affinity govern the maximum inclusion of non-binding proteins due to lateral crowding and to reveal that changes in membrane bending rigidity have only a modest effect on protein segregation. The experiments and simulations presented here provide a framework for predicting the size-dependent organization of both binding and non-binding proteins at membrane interfaces – patterns that can either directly contribute to functional cell-cell interactions or must be modified by active processes in the cell for productive signaling.
In vitro membrane interface system
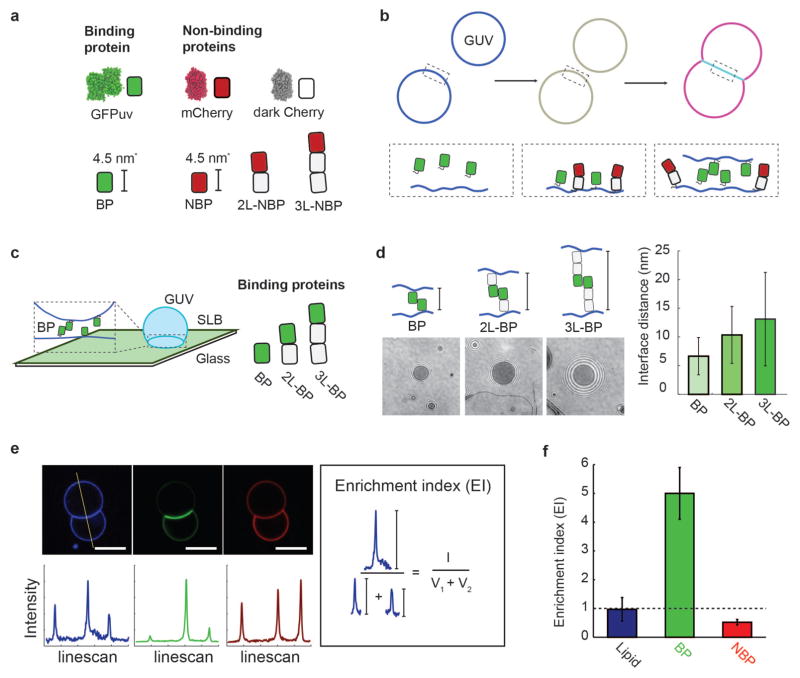
To experimentally isolate the role of protein size on segregation, we developed a simplified membrane interface system using giant unilamellar vesicles (GUVs) decorated with synthetic binding and non-binding proteins (Fig. 1a and 1b). We chose Green Fluorescent Protein (GFP)uv, an anti-parallel homodimer, as a homophilic binding protein (BP) because its binding affinity (Kd = 20–100 μM26,27) is in the range of TCR/pMHC interactions (Kd = 0.1–500 μM28, and references therein). Since GFPuv is not spherical (~4.5 nm height, ~4 nm width, ~ (4 nm)2 cross-sectional area; PDB: 1GFL), we refer to protein size in two distinct ways: (i) height above the membrane and (ii) lateral footprint on the membrane. We used mCherry, which has nearly identical dimensions to GFPuv, as the modular building block for a set of non-binding proteins of different heights above the membrane but constant lateral footprints (see Methods): single length (NBP), double length (2L-NBP), and triple length (3L-NBP). We similarly constructed a set of binding proteins of increasing height above the membrane using dark mCherry as the modular spacer (see Methods): single length (BP), double length (2L-BP), and triple length (3L-BP). All binding and non-binding proteins had a single fluorescent molecule (either mCherry or GFPuv) and used non-fluorescent mCherry for additional length.
Figure 1.
In vitro membrane interface system. (a) Synthetic binding proteins (GFPuv, BP) and non-binding proteins (mCherry variants, NBP, 2L-NBP, 3L-NBP) are used to investigate protein segregation at membrane interfaces. (b) Membrane interfaces are formed after the synthetic proteins bind to giant unilamellar vesicles (GUVs) that then come into contact. His-tagged binding proteins and non-binding proteins first attach to DOGS-Ni-NTA containing GUV membranes, and then binding protein dimerization leads to interface formation and protein segregation, which can be monitored fluorescently. (c) Membrane interfaces with different gap sizes were formed using binding proteins of different lengths (BP, 2L-BP, 3L-BP) and quantified using reflection interference contrast microscopy (RICM) at an interface formed between a supported lipid bilayer (SLB) and GUV. (d) Representative RICM images at the interface between GUV and SLB for interfaces formed with BP, 2L-BP, and 3L-BP. Image contrast and refractive index differences were used to extract the axial distance between the two membranes. Interface distance increased monotonically with the addition of spacer modules, forming interfaces of 6.2 +/− 3 nm (BP), 10.3 +/− 5.0 nm (2L-BP) or 13.1 +/− 8.1 nm (3L-BP). Error bars are standard deviation across N = 10 vesicles. (e) Representative confocal fluorescence images of GUVs (composition: 97.7 DOPC, 2.5 DOGS-Ni-NTA, 0.3 Atto 390-DOPE) incubated with 100 nM BP and 100 nM 2L-NBP in solution for 10 min. Scale bar is 5 μm long (blue channel: Atto 390-DOPE, green channel: BP, red channel: NBP). Linescans through vesicles dimers allow for quantification of fluorescence intensity at outside vesicle membranes (V1 and V2) and interface (I). We calculate an ‘Enrichment index’ (EI) by taking the ratio between (I) and the sum of (V1 and V2) and reveal uniform distribution of the fluorescently-labeled lipid (EI=1), enrichment of BP (EI>1) and exclusion of 2L-NBP (EI<1) at the interface. Error bars are standard errors of the mean from three independent experiments on separate vesicle batches, each with ~50 vesicles quantified.
The binding proteins and non-binding proteins were expressed and purified with an N-terminal deca-His tag, enabling fluid protein attachment to Ni-chelating lipids (DOGS-Ni-NTA) on the synthetic GUV membranes. GUVs were prepared with typical protein densities on the vesicle surface of ~2,300 molecules/μm2 as measured by fluorescence correlation spectroscopy (FCS) (see Methods and Supplementary Figure S1). For proteins with lateral footprints of (4 nm)2, this corresponds to a protein surface area coverage of approximately 3.7 , which is lower than the total membrane protein coverage on cellular membranes (~20,000 to 130 000 molecules/μm2) 28,29. GUVs containing only binding proteins showed protein enrichment at the interface, while GUVs containing only non-binding proteins showed no interface formation, as expected (Supplementary Figure S2).
We used reflection interference contrast microscopy (RICM) between a GUV and a supported lipid bilayer30 to determine the average distance between membranes at interfaces formed by the synthetic binding proteins (Fig. 1c; see Methods). Single length binding protein BP on both membranes formed membrane interfaces separated by 6.2 +/− 3.0 nm, consistent with size estimates from crystal structures of GFP-dimers (PDB: 1GFL31). Double and triple length binding proteins (2L-BP and 3L-BP) on both membranes formed membrane interfaces separated by 10.3 +/− 5.0 nm for 2L-BP and 13.1 +/− 8.1 nm for 3L-BP (Fig. 1d). As expected for thermally-driven proteins that may tilt at the membrane interface, the measured RICM distance is somewhat shorter than the estimated extended structure. The size of the non-binding proteins could not be directly measured with this approach because of the lack of a second interface needed for RICM.
To quantify the relative proportion of proteins at membrane interfaces containing both binding and non-binding proteins, we measured fluorescence intensity along a line bisecting the GUV-GUV pair and calculated an “Enrichment Index” (EI) – the intensity ratio between the interface (I) and the sum of the individual vesicle intensities (V1+V2) (Fig. 1e). EI values greater than 1 indicate an enrichment of molecules (increased surface density, #/μm2) at the membrane interface, while EI values that are less than 1 indicate an exclusion of molecules (decreased surface density) at the membrane interface (Fig. 1f). An EI of 1 is expected for a fluorescent molecule that is distributed homogenously across each vesicle’s surface.
Effect of protein height on segregation
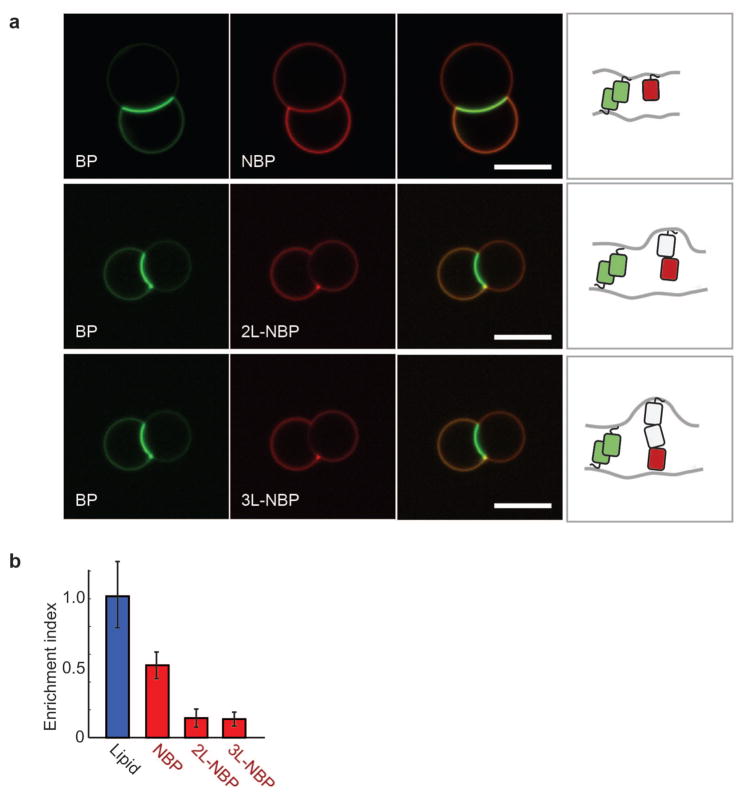
We began quantifying size-dependent segregation by separately incorporating the three non-binding proteins of different heights (NBP, 1L-NBP, 2L-NBP) with the single-length (shortest) binding protein (Fig. 2a). Pairs of GUVs containing both binding proteins and non-binding proteins showed significant protein reorganization at the membrane interface (Fig. 1c), with the binding protein BP significantly enriched (EI = 3.54 +/− 1.25), leading to an estimated binding protein density of ~8,200 molecules/μm2 at the interface, or 13.2 membrane area coverage (see Methods). In contrast, we found that all non-binding proteins were excluded, though to varying extent (Fig. 2b). Single length non-binding protein (NBP) was partially excluded (EI = 0.52 +/− 0.17), while double length non-binding protein (2L-BP) was significantly excluded (EI = 0.14 +/− 0.11). Triple length non-binding protein (3L-BP) was similarly excluded (EI = 0.13 +/− 0.13), suggesting that protein exclusion has reached a limit. To control for any changes in membrane morphology at the interface, we included a fluorescently-labeled lipid (Atto 390-DOPE) and confirmed that it was neither enriched nor excluded (EI = 0.97 +/− 0.41) (Fig. 2b).
Figure 2.
Effect of non-binding protein height on segregation at membrane interfaces. (a) Representative confocal fluorescence images of non-binding protein exclusion from GUV interfaces formed with the single length binding protein (BP). GUVs (composition: 97.7 DOPC, 2.5 DOGS-Ni-NTA, 0.3 Atto 390-DOPE) were incubated with 100 nM BP and 100 nM NBP, 2L-NBP, or 3L-NBP in solution for 10 min. Scale bar is 10 μm long (green channel: BP, red channel: NBP, 2L-NBP, or 3L-NBP). (b) Quantification of NBP, 2L-NBP and 3L-NBP exclusion from BP interfaces in comparison to a fluorescently-labeled lipid (Atto 390-DOPE), which should not exhibit size-dependent exclusion. Error bars are standard errors of the mean from three independent experiments on separate vesicle batches, each with ~50 vesicles quantified.
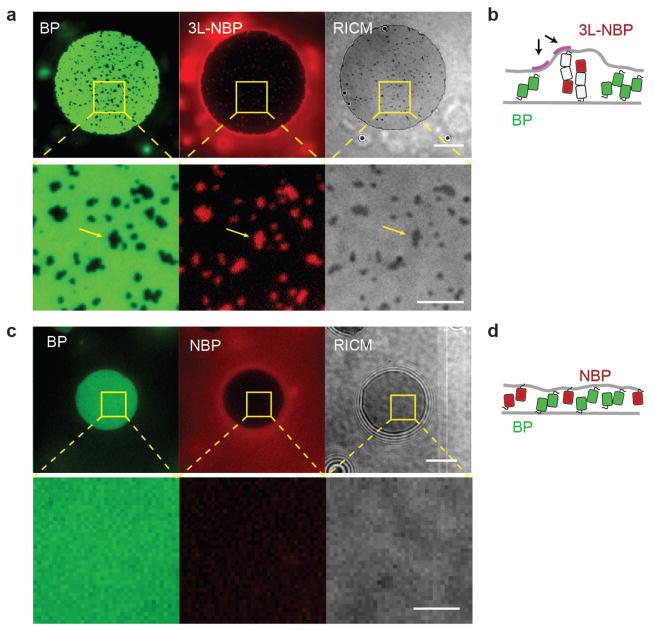
A single mCherry is approximately 4.5 nm (PDB: 2H5Q) in height, so the 2L-NBP and 3L-NBP are expected to be ~9.0 nm and ~13.5 nm, respectively, in their extended configuration. Since the measured distance of the membrane interface created by the single length binding protein (BP) is only 6.2 +/− 3 nm, the membrane interface must bend in order to accommodate the 2L-NBP and 3L-NBP. To test whether membrane bending is associated with exclusion of long non-binding proteins from the membrane interface, we captured multi-channel fluorescence and RICM images of membrane interfaces formed between SLBs and GUVs. When single length binding protein (BP) interfaces were formed in the presence of triple length non-binding proteins (3L-NBP), small clusters of the tall non-binding proteins were observed that excluded the short binding protein (Fig. 3a). RICM imaging of the membrane interface revealed regions of increased membrane separation that were co-localized with 3L-NBP clusters (yellow arrows in Fig. 3a lower panel), which coarsened over time to minimize total bending across the membrane interface (Fig. 3b). This behavior is consistent with an elastic model that attributes exclusion to the minimization of membrane bending energy32.
Figure 3.
Membrane bending by long non-binding proteins at membrane interfaces. (a) Representative multi-channel fluorescence and RICM images of membrane interface formation between SLBs and GUVs showing cluster formation of the triple-length non-binding protein (3L-NBP, red). The 3L-NBP clusters exclude the single length binding protein (BP, green) and co-localize with increased membrane distance (darker contrast, RICM), indicating there is a bending energy penalty for inclusion of long non-binding proteins at the membrane interface. Lower panels show zoomed regions of images in the upper panel. At long times, the long non-binding protein clusters are fully excluded from the interface. Scale bars 10 μm. (b) Cartoon representation of long non-binding protein cluster formation and associated membrane bending stress (arrows). (c) Representative multi-channel fluorescence and RICM images of membrane interface formation between SLBs and GUVs show no cluster formation of the single-length non-binding protein (NBP, red), no exclusion of the single-length binding protein, and no membrane bending in RICM. Scale bars 10 μm. (d) Cartoon representation of single-length non-binding proteins and single-length binding proteins at a membrane interface, where protein crowding could influence exclusion.
If long non-binding proteins are excluded from membrane interfaces due to membrane bending, proteins shorter than the interface distance should not be excluded. However, the single length non-binding protein (NBP), which is shorter (~4.5 nm) than the measured single length binding protein (BP) membrane interface distance (6.2 +/− 3 nm), was still partially excluded (EI = 0.52 +/− 0.17) (Fig. 2a and 2b). Using a combination of fluorescence microscopy and RICM, we again looked for localized membrane deformation in interfaces formed with BP and NBP but found only uniform fluorescence and no clusters or RICM contrast changes indicating deformation (Fig. 3c), suggesting that exclusion of a non-binding protein shorter than the interface cannot be explained by a simple elastic membrane model of protein segregation. Instead, we hypothesized that lateral protein crowding may be involved (Fig. 3d).
Effect of protein crowding on segregation
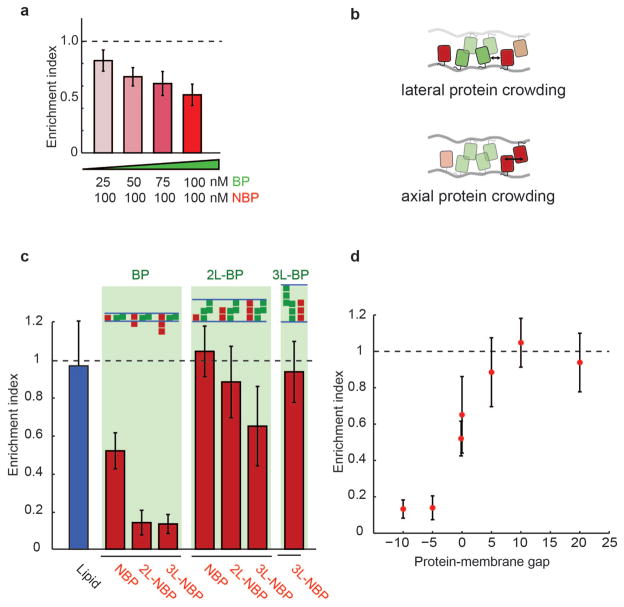
To test whether lateral protein crowding at the interface contributes to exclusion of non-binding proteins from the membrane interface, we lowered the concentration of the binding protein (BP) in solution from 100 nM to 25 nM, resulting in a proportional decrease in protein density at the interface from 11,585 / μm2 (or 18.5 coverage) at 100 nM to 1,930 molecules / μm2 (or 3 coverage) at 25 nM (Supplementary Figure S4). This reduced binding protein density resulted in a decrease in single length non-binding protein (NBP) exclusion (Fig. 4a, EI = 0.52 +/− 0.1, 0.62 +/− 0.1, 0.68 +/− 0.08, and 0.8 +/− 0.9 at 100 nM, 75 nM, 50 nM, and 25 nM respectively), indicating that lateral space taken up by the binding protein due to its footprint on the membrane, as well as its tilt and flexibility, reduces the space available for non-binding proteins. Proteins in the interface have been suggested to experience reduced entropy due to alignment and a reduction in tilt states33, which could additionally influence the crowding-induced NBP exclusion.
Figure 4.
Effect of protein crowding on segregation at membrane interfaces. (a) Single-length non-binding protein exclusion increases with increasing single-length binding protein concentration in solution. Error bars are standard error of the mean from three independent experiments on separate vesicle batches, each with ~50 vesicles quantified. (b) Cartoon representation of lateral protein crowding (due to steric conflict with proteins on the same membrane) and axial protein crowding (due to steric conflict with proteins present on opposite membranes). Lateral crowding of binding proteins, which depends on both protein footprint on the membrane and binding affinity, limits space available for non-binding proteins. (c) Summary of measured non-binding protein exclusion for each pair of binding protein (BP, 1L-BP, 2L-BP) and non-binding protein (NBP, 1L-NBP, 2L-NBP), showing increases in exclusion with increases in non-binding protein size. (d) Non-binding protein exclusion data is plotted as a function of the space between non-binding proteins and the apposing membrane (protein-membrane gap). A negative gap indicates the non-binding protein is larger than the average measured distance of the interface, while a positive gap indicates there is space between the non-binding protein and the apposing membrane. Error bars are standard error of the mean from three independent experiments on separate vesicle batches, each with ~50 vesicles quantified.
Since lateral space at membrane interfaces is taken up not only by binding proteins but also by non-binding proteins on opposite membranes when the gap is small (Fig. 4b), we next investigated whether steric exclusion across membranes contributes to non-binding protein exclusion. To test for this, we increased the height of the binding protein to the double length binding protein (2L-BP, gap = 10.3 +/− 5.0 nm) while continuing to use the single length non-binding protein (NBP) on the two membranes. We found that NBP was no longer excluded from the interface (EI = 1.05 +/− 0.23) (quantification in Fig. 4b and confocal images in Supplementary Figure S5), consistent with reduced crowding from the single length non-binding protein on opposite membranes. Interestingly, we found that enrichment of the double length binding protein (2L-BP) in the membrane interface (EI = 1.28 +/− 0.28) was significantly less than that of the single length binding protein (BP) (EI = 3.55 +/− 1.7) (Fig. 4c; Supplementary Figure S4 and Supplementary Figure S5), likely because 2D affinity decreases with protein length20,33, indicating that reduced crowding of the 2L-BP (from ~11,000 to ~3,000 molecules per μm2 at the interface, Supplementary Table) could also contribute to the lack of NBP exclusion.
To directly test whether steric interference of non-binding proteins from opposite membranes contribute to exclusion, we held the double length binding protein (2L-BP) constant and doubled the length of the non-binding protein to 2L-NBP (~9 nm), which is shorter than the measured membrane gap but tall enough to interact with non-binding proteins on the opposite membrane. Indeed, we found that the double length non-binding protein (2L-NBP) was excluded to a small but statistically significant amount (EI = 0.89 +/− 0.33), consistent with limited interaction between non-binding proteins in the opposite membrane (Fig. 4c). Increasing the length of the non-binding protein by ~4.5 nm to the triple length non-binding protein increased the exclusion (EI = 0.65 +/− 0.36) (Fig. 4c).
Overall, our experimental data show that a combination of changes in protein height and lateral crowding can titrate exclusion of non-binding proteins from membrane interfaces with only small changes in protein size. This can be seen more explicitly by re-plotting the data as a function of the difference between non-binding protein length and measured membrane gap size (set by the binding protein), which we refer to as protein-membrane gap (Fig. 4d, where 0 corresponds to non-binding protein size = membrane gap). To test in detail size-based mechanisms of protein exclusion and investigate the role of membrane properties, we turned to Monte Carlo simulations.
Monte Carlo simulations of size-dependent protein segregation
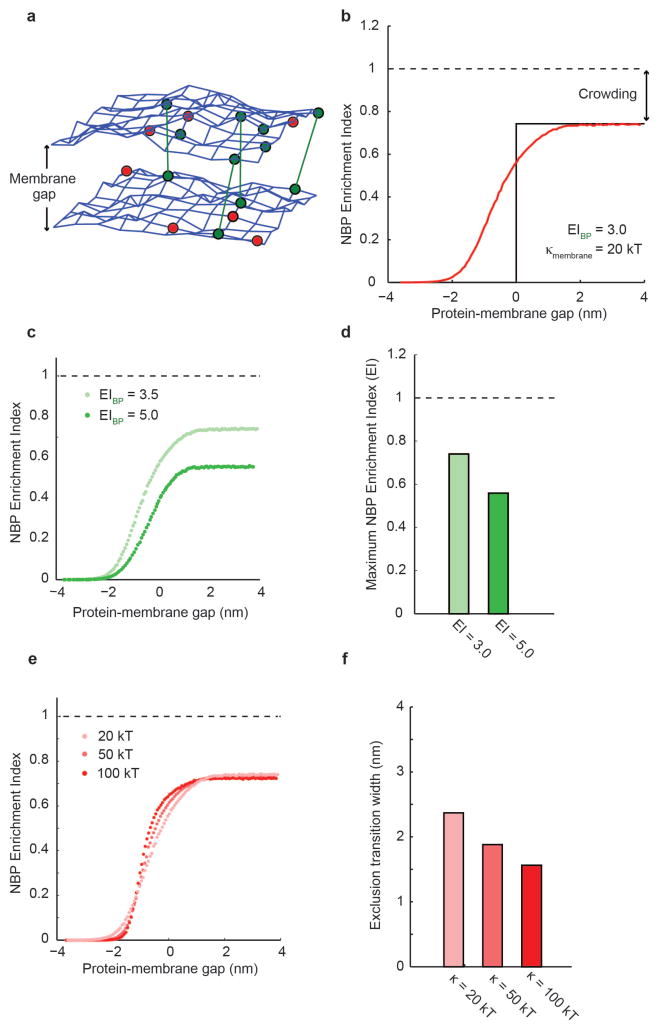
Monte Carlo models of membranes have been productively used to capture both membrane dynamics and spatial organization of adhesion proteins19,35. To explore the interplay between protein height, membrane gap, and lateral crowding at the interface, we developed a simulation of binding and non-binding protein segregation at membrane interfaces that includes as parameters the properties identified as important in our experimental work (non-binding protein height, binding protein height, protein lateral footprint, and protein density) and those that were not easily accessible experimentally (membrane physical properties, binding protein affinity). To that end, we constructed a Monte Carlo simulation of a deformable, fluid membrane interface in which a fluctuating triangulated mesh represents each of the two membrane surfaces that form the interface, and nodes of the mesh can be occupied by diffusing proteins of defined height and binding potentials36,37 (Fig. 5a, details in Methods). We seeded the membranes with a binding protein that defines a membrane gap size of 10 nm, comparable to the double length binding protein (2L-BP) interface, together with non-binding proteins of heights between 1 nm (protein-membrane gap = 9 nm) and 20 nm (protein-membrane gap = −10 nm).
Figure 5.
Monte Carlo simulations of size-dependent protein segregation. (a) A fluctuating triangulated mesh represents each of the two membrane surfaces that form a membrane interface in the simulations. Nodes of the mesh can be occupied by non-binding proteins (red) and by binding proteins (green) that may form bonds across the interface (green lines). (b) Simulation of protein enrichment for a non-binding protein as a function of the protein-membrane gap. At positive protein-membrane gap (right), non-binding proteins are smaller than the membrane interface, and lateral crowding in the plane of the membrane results in a maximum enrichment index, or maximum inclusion, below 1.0. At negative protein-membrane gap (left), non-binding proteins are completely excluded to minimize bending of the membrane. Near a protein-membrane gap of 0, fluctuations of the membrane surface and steric interactions between apposite membranes and proteins result in a smooth transition between these two extremes. (c & d) The maximum enrichment index, or maximum inclusion, at large protein-membrane gaps decreases with increasing binding protein enrichment, since increased binding protein density within the interface crowds out non-binding proteins. (e & f) Increasing the bending rigidity of the membrane makes the transition between maximum inclusion and maximum exclusion of non-binding proteins sharper. If the exclusion transition width is defined as the width along the protein-membrane gap axis between 10 and 90 of maximum inclusion, the change in transition width is less than 1 nm for a factor of five increase in bending rigidity, suggesting that size-dependent segregation is only modestly affected by bending rigidity.
Using parameters based on our experimental system, we found that non-binding protein exclusion varies sigmoidally as a function of the protein-membrane gap (Fig. 5b), consistent with our experimental observations (Fig. 4d). The simulations predict that proteins more than 2 nm taller than the membrane gap are fully excluded (EI = 0) from the interface, while proteins more than 2 nm shorter than the membrane gap are included to a level below full mixing (EI = 1) due to crowding, comparable to the effect of a ~4.5 nm change in protein height seen experimentally. Decreasing the binding protein density at the membrane interface by reducing its binding potential decreases the level of exclusion of non-binding proteins in the simulation (Fig. 5c, quantification in Fig. 5d), which is also consistent with our experiments (Fig. 4c and Supplementary Figure S4). Increases in membrane bending rigidity from 20 kT to 100 kT in the simulation sharpened the transition between maximum exclusion and maximum inclusion of non-binding proteins (Fig. 5e, quantification in Fig. 5f), although this effect was over a very small protein-membrane gap that was not resolved in our experiments (Supplementary Figure S4). These results indicate that thermal fluctuations both make room for proteins taller than the membrane gap and exclude proteins shorter than the membrane gap, to a degree that is reduced with increasing bending rigidity. Interestingly, the small magnitude of membrane thermal fluctuations at even low membrane bending rigidities suggests that passive thermal fluctuations at crowded membrane interfaces may not be sufficient to bring a second species of short binding proteins into contact during cell-cell signaling, such as engagement of the peptide MHC-TCR (~13 nm) within a membrane interface formed by the comparatively longer LFA-ICAM adhesion (~40 nm)2.
Conclusions and implications
Our experiments and simulations show that protein size is a simple, sensitive, and highly effective means of altering local protein concentrations at membrane interfaces. Nanometer-scale changes in the height of non-binding proteins can dramatically change their densities at membrane interfaces, while high surface area coverage of binding proteins, which is dependent on both protein lateral footprint and binding affinity, can exclude non-binding proteins even when membrane gap size is not limiting. Our data and simulations show that increasing non-binding protein height has a large effect when it is comparable to membrane gap size (e.g. NBP to 2L-NBP in a BP interface) and a small effect when the protein height already exceeds the membrane gap size (e.g. 2L-NBP to 3L-NBP in a BP interface).
The large difference between the reported height of CD45 isoforms (28 to 53 nm 22) and the membrane interface gap size between T-cells and antigen presenting cells (~13 nm 15) is surprising given our finding that only ~4.5 nm increase in non-binding protein size is necessary for significant protein exclusion. However, the active cytoskeletal processes that are involved in T-cell membrane reorganization and receptor engagement likely create a highly dynamic membrane interface with a variable protein-membrane gap size, for which extra non-binding protein length is needed to ensure exclusion. While T-cell receptors are at low abundance on a T-cell prior to cell-cell junction formation (100 molecules per μm2), signaling occurs in micron-scale TCR micro-clusters that are highly enriched in TCRs and other membrane proteins, suggesting that lateral crowding may also be relevant for CD45 exclusion in the immunological synapse. More broadly, we believe that size-dependent protein segregation may be a general organizing mechanism at membrane interfaces. In fact, the dramatic size-diversity of membrane proteins may have evolved in order to influence cell-cell signaling. It is notable that many proteins on the extracellular surface of cells of multicellular eukaryotes, including CD45, are members of the immunoglobulin superfamily, and are extended in height above the membrane from 5 to greater than 50 nm by addition of a modular building block – the Ig domain38,39. Interestingly, this domain has a size of ~4 nm, which in our experiments is the size difference needed to lead to complete exclusion from the interface. Therefore, adding or removing a single Ig domain may be sufficient to completely re-localize a protein from a membrane interface in the absence of active processes, a potentially powerful evolutionary tool.
We expect that the basic principles of protein segregation at membrane interfaces reported here are at work not only at cell-cell adhesions but also at intracellular membrane contact sites40–42, where size-dependent enrichment and exclusion of proteins at interfaces can drive key steps in molecular sorting and signaling.
Methods
Chemical reagents
Hepes (4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid), MOPS (3-(N-morpholino)propanesulfonic acid), TCEP (tris(2-carboxyethyl)phosphine), KCl, glucose, sucrose, β-Mercaptoethanol, and imidazole were purchased from Fisher Scientific. EDTA (Ethylenediaminetetraacetic acid), was purchased from Acros Organics. Imidazole was purchased from Sigma Aldrich. Atto 390-DOPE was purchased from ATTO-TEC. DOGS-Ni-NTA (1,2-dioleoyl-sn-glycero-3-[(N-(5-amino-1-carboxypentyl)iminodiacetic acid)succinyl], with nickel salt) and DOPC (1,2-dioleoyl-sn-glycero-3-phosphocholine), were purchased from Avanti Polar Lipids (Alabaster, AL). All purchased chemical reagents were used without further purification.
Cloning, protein expression and purification
Standard methods of molecular biology, including reagents from Quiagen, Thermo Scientific and Zymo Research, were used to generate the following constructs. BP (GFPuv) and NBP (mCherry) variants were cloned into petMz backbone vector (a kind gift from Dr. Peter Bieling). We cloned combinations of dark mCherry (Y72S), mCherry and GFPuv fusions to generate our synthetic toolset of binding or non-binding membrane bound proteins of different sizes. For a list of all used proteins please see domain structures in Supplementary Figure S2. Multimers of mCherry/GFPuv were created by linking C to N-termini via a 7 amino acid (GGGGSTS) linker. To render mCherry into a non-fluorescent dark mCherry we mutated tyrosine 72 into a serine by site-directed mutagenesis.
All proteins were expressed as a Deca-His-tag fusion proteins in BL21 (DE3) pLysS cells (Stratagene). Cells were grown at 37 °C until OD600 of 0.3 – 0.5, induced with 0.3 mM IPTG and grown for 14 – 16 h at 18 °C. Cells were harvested and resuspended in 25 mM Hepes, pH 7.4, 150 mM KCl, 1 mM TCEP and 10 mM Imidazole, and lysed by freeze thawing and sonication. The lysate was centrifuged for 45 min at 20,000 × g, 4 °C and affinity purified over a His-Trap HP or a Hi-Trap Chelating column HP (GE Healthcare), including extensive washing with 25 mM Hepes, pH 7.4, 150 mM KCl, 1 mM TCEP and 10–50 mM Imidazole, followed by Imidazole gradient elution on an AKTA Pure (GE Healthcare) system. Peak fractions were pooled and proteins were cleaved in elution buffer by incubation for 2 hours at room temperature and overnight at 4°C with His6-TEV protease (purchased from Macrolab at UC Berkeley). Proteins were buffer exchanged over PD-10 columns (GE Healthcare) into 25 mM Hepes, pH 7.4, 150 mM KCl, 1 mM TCEP. Protein solution was spun at 16,000 × g at 4deg C to remove aggregated His-6 TEV protease and supernatant was rebound to the chelating column to remove the non-his-tagged z-tag. After imidazole gradient elution on the AKTA PURE system, protein was concentrated and gelfiltered via a Superdex 200 column in 25 mM Hepes, pH 7.4, 150 mM KCl, 1 mM TCEP. Proteins were concentrated, aliquoted and snap frozen. Purity was assessed via SDS PAGE.
GUV preparation
Electroformation of giant unilamellar vesicles (GUVs) was performed according to published protocols43. To ensure mixing of all lipid components we performed electroformation at approximately 55 °C. Vesicles were electroformed in solution containing approximately 350 mM sucrose (~350 mOsm). Lipid composition was kept constant in all our experiments with 97.5 DOPC and 2.5 DOGS-Ni-NTA. 0.3 Atto 390-DOPE dye was used as a membrane where indicated.
Preparation and characterization of protein-coated GUVs
After electroformation, vesicles were diluted, exposed to proteins, and prepared for imaging. 1 μl GUVs were gently transferred to a home-made Polydimethylsiloxane (PDMS, Sylgard) chamber containing 100 μl of protein solution (in 25 mM Hepes buffer with 150 mM KCl and 1mM TCEP, osmotically balanced to 320 mOsm). The mixture was incubated on room temperature for 10 minutes and imaged immediately thereafter.
To avoid complications of dimer formation in solution we used protein concentrations at least 200 fold below the dimer Kd. Using fluorescence correlation spectroscopy (FCS) of supported bilayers having the same composition (for FCS and supported bilayers see Methods sections below and Supplementary Figure S1), we measured a lateral protein density of 2,317 +/− 370 molecules per μm2 for these conditions. Given an estimated footprint of ~ (4 nm)2 for GFPuv, this protein density corresponds to 3.8 membrane area coverage by protein. For each GFPuv at this density there are ten DOGS-Ni-NTA lipids available, leading to nearly irreversible protein-to-membrane attachment32. Interface size and shape was equilibrated rapidly (within 2 minutes), consistent with fast protein diffusion on the GUV membranes. No nonspecific binding of GFPuv to membranes lacking DOGS-Ni-NTA was observed (see Supplementary Figure S2).
Imaging
GUVs were imaged on a spinning disc confocal microscope (AxioObserverZI, Zeiss, with motorized nosepiece and spinning disk confocal, Yokogawa CSU-10) for confocal microscopy with a cooled EMCCD camera (Cascade II, Photometrics). Images were acquired using a 63× objective (Zeiss, Plan Apochromat 1.4 NA, oil), and analyzed using ImageJ (National Institutes of Health) and Matlab (Mathworks).
Image analysis
Vesicle dimers were localized within spinning disk confocal images and line scans were drawn to cross both vesicles and the vesicle interface using ImageJ (NIH). An intensity trace across the length of the line scan was extracted from the confocal images after background subtraction using a custom Matlab script (Mathworks). The three intensity peaks, corresponding to each of the two vesicles and the vesicle interface, were localized using a peak finding algorithm, and the intensity values from these peaks were used to compute the EI.
Supported Lipid Bilayer (SLB) Preparation
SLBs were formed by fusion of small unilamellar vesicles (SUVs) on RCA cleaned glass coverslips. SUVs were prepared from a lipid film composed of 97.5 DOPC and 2.5 DOGS-Ni-NTA rehydrated in buffer containing 25 mM MOPS and 125 mM NaCl at pH 7.4. The rehydrated solution was freeze-thawed in liquid nitrogen and agitated using a high-power bath sonicator (Avanti), then filtered through a 200 nm filter (Millipore) to select for SUVs. A solution of SUVs was incubated in a home-made Polydimethylsiloxane (PDMS, Sylgard) well chamber above a clean glass coverslip for 10 minutes, then washed with MOPS buffer 10 times to remove excess SUVs from solution. Lipid bilayers were tested for fluidity after binding proteins to DOGS-Ni-NTA lipid by photo-bleaching a small region and monitoring for rapid recovery.
Distance measurement preparation
SLBs were incubated with 100 nM protein (BP, 2L-BP, 3L-BP) in the presence of GUVs to coat both the planar and vesicle membranes. GUVs settled onto the planar membrane, and binding between proteins on the SLB and GUV results in the formation of a flattened membrane-membrane interface at the base of the GUV.
Quantitative distance measurement by Reflection Interference Contrast Microscopy (RICM)
Narrow-band illumination light (546 nm) selected from a mercury arc lamp (Excite) was directed through an anti-flex (EC-Plan Neofluor 63x, NA 1.25) through a crossed linear polarizing cube. A reflected light interference-contrast image was formed, such that image contrast was a function of the distance between the SLB and the bottom surface of the GUV as well as the refractive index difference at the glass-media and media-GUV interfaces. The RICM images were processed using a method previously described to extract the axial distance between the two membranes with a precision of a few nanometers30. Briefly, the SLB-GUV interface was modeled as a multilayer film consisting of the following layers: glass (N=5.0), lipid (N=4.2), buffer (N=3.0, h=variable), lipid (N=4.2), sucrose (N=2.0), where the height of each lipid layer was the thickness of a typical bilayer (5.0 nm) and the buffer layer (N=3.0) was of variable axial extent. The theoretical contrast as a function of the height of the variable buffer layer, which corresponds to the distance between SLB and the bottom surface of the GUV, was calculated. RICM contrast images of the SLB-GUV interface were processed using custom Matlab scripts to extract the radially averaged intensity as a function of distance from the center of the interface. This function was used to determine the minimum and maximum of the first periodic interference fringe, and the average contrast of the flattened GUV-SLB interface area was normalized with these extrema. This normalized interface contrast was mapped to the theoretical contrast function to determine the best-fit height of the buffer layer, and thus the separation between SLB and GUV imposed by the adhesive proteins (Supplementary Figure S3).
FCS
Fluorescence correlation spectroscopy experiments are performed on a custom-built setup based around a Nikon Eclipse TE2000 microscope body (Nikon). A 488 nm CW laser was used as an excitation source (Sapphire, Coherent). Photons counts were detected with an avalanche photo diode (SPCM-AQR-14, Excelitas), and photon arrival times were registered by the counter module of an NI DAQ board (PCI-6321, National Instruments). Autocorrelation analysis and data fitting were performed by custom written Matlab scripts (Mathworks). Experiments were carried out with a 100x oil immersion objective (NA 1.49). On each measurement day, an autocorrelation measurement from a 50nM solution of Fluorescein was acquired to calibrate the focal volume of the system. SLBs were incubated with a mixture of GFPuv and a mutated dark GFPuv (Y66S) at a 7 ratio, such that the concentration of fluorescent molecules on the SLB was within the range needed for FCS. To measure the density of molecules on the SLB, photon arrival times were recorded for periods of 10 seconds, with 6 independent measurements performed per bilayer (for a total acquisition time of 60 seconds per bilayer). An autocorrelation was performed on the photon arrival record, and the resulting curve was fit to a model of single-component two-dimensional diffusion45. The value of G(0) was extrapolated from the fit, and the concentration of fluorescent molecules on the bilayer was expressed as (G(0)*Aeff)−1, where Aeff is the area defined by the confocal observation volume. Concentration measurements were repeated for 5 independent SLBs.
Estimates of protein density and membrane coverage
To estimate the density of molecules on the membrane for each BP species, we used the FCS measurements of protein density for GFPuv (BP, in molecules/um2) along with the fluorescence intensity (in arbitrary units) from confocal images of GUVs to compute a scaling factor between arbitrary fluorescence units and molecular density (AU to molecules/um2). The fluorescence emission from each (BP, 2L-BP, 3L-BP) molecule is expected to be equivalent because each species contains a single fluorescent GFPuv module. This scaling factor is used to compute molecular density for both 2L-BP and 3L-BP. The molecular density at the interface for BP, 2L-BP, and 3L-BP is estimated by multiplying the density on the vesicle with the enrichment index of the binding protein. The area of membrane covered by protein is estimated using a footprint of (4 nm)2 for both binding proteins and non-binding proteins.
Generation of protein-membrane gap curve in Fig. 4b
To compute the protein-membrane gap, we used estimates of non-binding protein height based on crystal structure (NBP = 5 nm, 2L-NBP = 10 nm and 3L-NBP = 15 nm) and equivalent estimates of interface distance imposed by BPs (BP dimer = 5.0 nm, 2L-BP dimer = 15.0 nm and 3L-dimer 25.0 nm). These values are in the range of interface distance measured by RICM. The protein-membrane gap is the difference between the interface distance imposed by BPs and the size of the NBP.
Monte Carlo simulation model
We are simulating two adjacent quasi-flat periodic membrane interfaces employing a Metropolis Monte Carlo (MC) method based on a dynamically triangulated surface model for fluctuating fluid membranes. The bending energy of the membrane is calculated using a discretized version of the Laplacian on the triangulated lattice at constant bending rigidity46. During the initialization procedure, the area (i.e. number of vertices in the triangulation) of a single bare membrane patch is equilibrated using a grand canonical MC move. Afterwards, a vertically translated copy of the resulting membrane patch is created and the number of membrane vertices is kept constant throughout the simulation. Initially, the two membrane surfaces are placed at a relative height that corresponds to the linker rest length l of the bound BP complex in simulation and all membrane vertices are populated by BP and linked pairwise. An additional MC move attempts to (un-)bind pairs of BP across the two lattices incorporating a constant binding energy 4.5kT and a harmonic linker potential of given rest length l=10nm and spring constant 20kT/nm² between the two. The density of unbound BP and NBP is equilibrated using a grand canonical MC move that inserts and subtracts these particle species within the membrane lattice at a constant chemical potential that corresponds to a protein-membrane lattice coverage of 0.1 within the implicit reservoir outside the bound membrane interface. After equilibration, fluctuating protein densities of BP (bound and unbound) and NBP are monitored. The linear length of the projected quadratic membrane area is 200nm, while the maximum length of the purely entropic membrane-internal linkers is 9.5 nm. Membrane vertices interact with themselves and other protein species on apposing membranes via steric interactions. The hard-sphere node diameter is constant at dmem=5nm for the bare membrane and at BP = 10nm for vertices occupied by BP. The NBP diameter is varied in the range 5nm ≤ dNBP ≤ 20nm. The corresponding gap size for plotting NBP exclusion curves is calculated as the average distance between membranes in simulation.
Supplementary Material
Supplementary Figure S1. BP density measurement by FCS
(a) Glass supported lipid bilayers (SLBs, composition: 94.47 DOPC, 2.5 DOGS-Ni-NTA) are incubated 100 nM BP solution for 10 minutes. (b) The density of BP protein on the SLB is measured by FCS. 7 fluorescent GFPuv (BP) was mixed with 93 mutated non-fluorescent GFPuv (BP-Y66S) (7 bright) is used to achieve the correct concentration range for FCS measurements. Normalized autocorrelation curves are fit with a single component two-dimensional diffusion model to extract the diffusion coefficient and protein density of the BP (molecules/μm2) (c) Measurements on 5 independent SLBs reveal an BP density of 2,317 +/− 370 molecules/um2 for an SLB with 2.5 DOGS-Ni-NTA incubated with 100 nM BP protein.
Supplementary Figure S2.
(a) Representative field of view of BP (green) and NBP (red) bound to DOGS-Ni-NTA containing GUV membranes (composition: 97.47 DOPC, 2.5 DOGS-Ni-NTA incubated with 100 nM protein solution for 10 min). Note how BP molecules form GUV interfaces while NBP molecules do not. Scale bar is 20 μm long. (b) A mixture of GUVs with His-tagged protein binding capability (97.5 DOPC, 2.5 DOGS-Ni-NTA) and GUVs lacking DOGS-Ni-NTA (99.7 DOPC, 0.3 Atto647-DOPE) were incubated with 100 nM GFPuv (BP). BP only bound to DOGS-Ni-NTA containing vesicles, confirming that there is no non-specific binding of BP from solution to membranes. Furthermore, we could not detect unspecific interface formation between BP-covered GUVs and protein-free GUVs. All scale bars are 10 μm. (c) Protein domain architectures illustrate proteins used in this study.
Supplementary Figure S3. Distance measurements by RICM
(left) Radially averaged RICM contrast as a function of the distance from the center of the GUV. (right) Theoretical RICM contrast as a function of the height of the variable buffer layer, which corresponds to the distance between SLB and the bottom surface of the GUV (see Methods).
Supplementary Figure S4
(a) Increasing concentrations of BP in solution leads to increased fluorescence intensities on GUVs. Vesicle (V) fluorescence intensities are plotted versus interface (I) intensities. (b) EIs for BP at different protein concentrations in solution. We note that while the intensities in a) scale with concentration, protein distribution over the vesicles stays remarkably constant at different protein densities. (c) EI values for different lengths adhesion molecules averaged over all experiments. Single lengths BP enrich more at the interface possibly due to higher 2D affinity than the longer 2L-BP and 3L-BP. (d) Exclusion of NBP or 2L-NBP from BP interfaces does not change significantly with increased membrane rigidity. GUVs from 80 Sphingomyelin and 20 Cholesterol mixtures (SM + Chol) have a approximately 6 fold higher bending rigidity than the DOPC GUVs. SM+Chol (77.5 Brain Sphingomyelin (Avanti), 20 Cholesterol (Avanti) 2.5 DOGS Ni-NTA) GUVs were incubated with 100 nM BP and 100 nM NBP or 2L-NBP. Interface formation and NBP and 2L NBP exclusion did not differ significantly from the same experiment performed on DOPC (97.5 DOPC, 2.5 DOGS-NiNTA) vesicles.
Supplementary Figure S5
Representative confocal images for data in Fig. 4a. Increasing the size of the adhesive protein (2L-BP, 3L-BP) leads to decreased exclusion of NBP variants from interfaces. Representative confocal fluorescence images of GUVs (composition: 97.47 DOPC, 2.5 DOGS-Ni-NTA) incubated with 100 nM BP variants and 100 nM NBP variants in solution for 10 min. Scale bars are 5 μm long (green channel: BP, red channel: NBP). Error bars are standard error of the mean from three independent experiments on separate vesicle batches, each with ~50 vesicles categorized.
Supplementary Table. BP densities
Acknowledgments
We acknowledge Ron Vale and Christopher Peel for helpful discussions. This work was supported by a Graduate Fellows Research Program grant from the National Science Foundation (NSF) for MHB; a Cancer Research Institute Post-Doctoral Fellowship and a K99 grant from the National Institute of Health (NIH, K99AI093884 and R00AI093884) for KC; a Forschungsstipendium of the Deutsche Forschungsgemeinschaft (DFG grant no. We 5004/2) for JW; a NIH grant (R37AI043542), a NIGMS Nanomedicine Development Center grant (PN2EY016586) and a Wellcome Trust Principal Research Fellowship to MLD; and a NIH Nanomedicine Development Center grant (PN2EY016546) and an NIH R01 grant (GM1145091) to DAF.
Footnotes
Author Contributions
All authors contributed to design of the experiments. EMS and MB performed experiments. EMS, HSA, and MB created unique materials. JW, MB and PLG performed simulations and modeling. EMS, MB and DAF wrote the manuscript. All authors discussed the results and commented on the manuscript.
References
- 1.Bell GI. Models for the specific adhesion of cells to cells. Sci N Y NY. 1978;200:618–627. doi: 10.1126/science.347575. [DOI] [PubMed] [Google Scholar]
- 2.Dustin ML. The immunological synapse. Arthritis Res Ther. 2002;4:S119–25. doi: 10.1186/ar559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rochlin K, Yu S, Roy S, Baylies MK. Myoblast fusion: When it takes more to make one. Dev Biol. 2010;341:66–83. doi: 10.1016/j.ydbio.2009.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Adams CL, Chen YT, Smith SJ, Nelson WJ. Mechanisms of epithelial cellW cell adhesion and cell compaction revealed by highWresolution tracking of EW cadherinWgreen fluorescent protein. J Cell Biol. 1998;142:1105–1119. doi: 10.1083/jcb.142.4.1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Goodridge HS, et al. Activation of the innate immune receptor DectinW1 upon formation of a ‘phagocytic synapse’. Nature. 2011;472:471–475. doi: 10.1038/nature10071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Monks CR, Freiberg BA, Kupfer H, Sciaky N, Kupfer A. ThreeWdimensional segregation of supramolecular activation clusters in T cells. Nature. 1998;395:82–86. doi: 10.1038/25764. [DOI] [PubMed] [Google Scholar]
- 7.Dustin ML. Making a little affinity go a long way: a topological view of LFAW1 regulation. Cell Adhes Commun. 1998;6:255–262. doi: 10.3109/15419069809004481. [DOI] [PubMed] [Google Scholar]
- 8.Grakoui A, et al. The immunological synapse: a molecular machine controlling T cell activation. Sci N Y NY. 1999;285:221–227. doi: 10.1126/science.285.5425.221. [DOI] [PubMed] [Google Scholar]
- 9.Bunnell SC, et al. T cell receptor ligation induces the formation of dynamically regulated signaling assemblies. J Cell Biol. 2002;158:1263–1275. doi: 10.1083/jcb.200203043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Varma R, Campi G, Yokosuka T, Saito T, Dustin ML. T cell receptorWproximal signals are sustained in peripheral microclusters and terminated in the central supramolecular activation cluster. Immunity. 2006;25:117–127. doi: 10.1016/j.immuni.2006.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.James JR, Vale RD. Biophysical mechanism of TWcell receptor triggering in a reconstituted system. Nature. 2012;487:64–69. doi: 10.1038/nature11220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cordoba SWP, et al. The large ectodomains of CD45 and CD148 regulate their segregation from and inhibition of ligated TWcell receptor. Blood. 2013;121:4295–4302. doi: 10.1182/blood-2012-07-442251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Siu G, Springer EA, Hedrick SM. The biology of the TWcell antigen receptor and its role in the skin immune system. J Invest Dermatol. 1990;94:91S–100S. doi: 10.1111/1523-1747.ep12876046. [DOI] [PubMed] [Google Scholar]
- 14.Davis SJ, van der Merwe PA. CD2: an exception to the immunoglobulin superfamily concept? Sci N Y NY. 1996;273:1241–1242. doi: 10.1126/science.273.5279.1241. [DOI] [PubMed] [Google Scholar]
- 15.Choudhuri K, Wiseman D, Brown MH, Gould K, van der Merwe PA. TWcell receptor triggering is critically dependent on the dimensions of its peptideWMHC ligand. Nature. 2005;436:578–582. doi: 10.1038/nature03843. [DOI] [PubMed] [Google Scholar]
- 16.Bethani I, Skånland SS, Dikic I, AckerWPalmer A. Spatial organization of transmembrane receptor signalling. EMBO J. 2010;29:2677–2688. doi: 10.1038/emboj.2010.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li P, et al. Phase transitions in the assembly of multivalent signalling proteins. Nature. 2012;483:336–340. doi: 10.1038/nature10879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sheetz MP, Sable JE, Döbereiner HWG. CONTINUOUS MEMBRANEW CYTOSKELETON ADHESION REQUIRES CONTINUOUS ACCOMMODATION TO LIPID AND CYTOSKELETON DYNAMICS. Annu Rev Biophys Biomol Struct. 2006;35:417–434. doi: 10.1146/annurev.biophys.35.040405.102017. [DOI] [PubMed] [Google Scholar]
- 19.Weikl TR, Asfaw M, Krobath H, Rózycki B, Lipowsky R. Adhesion of membranes via receptor–ligand complexes: Domain formation, binding cooperativity, and active processes. Soft Matter. 2009;5:3213–12. [Google Scholar]
- 20.Milstein O, et al. Nanoscale increases in CD2WCD48Wmediated intermembrane spacing decrease adhesion and reorganize the immunological synapse. J Biol Chem. 2008;283:34414–34422. doi: 10.1074/jbc.M804756200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Alakoskela JWM, et al. Mechanisms for SizeWDependent Protein Segregation at Immune Synapses Assessed with Molecular Rulers. Biophys J. 2011;100:2865–2874. doi: 10.1016/j.bpj.2011.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McCall MN, Shotton DM, Barclay AN. Expression of soluble isoforms of rat CD45. Analysis by electron microscopy and use in epitope mapping of antiWCD45R monoclonal antibodies. Immunology. 1992;76:310–317. [PMC free article] [PubMed] [Google Scholar]
- 23.Rózycki B, Lipowsky R, Weikl TR. Segregation of receptor–ligand complexes in cell adhesion zones: phase diagrams and the role of thermal membrane roughness. New J Phys. 2010;12:095003–22. [Google Scholar]
- 24.Burroughs NJ, et al. Boltzmann EnergyWbased Image Analysis Demonstrates that Extracellular Domain Size Differences Explain Protein Segregation at Immune Synapses. PLoS Comput Biol. 2011;7:e1002076–11. doi: 10.1371/journal.pcbi.1002076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hu J, Lipowsky R, Weikl TR. Binding constants of membraneWanchored receptors and ligands depend strongly on the nanoscale roughness of membranes. Proc Natl Acad Sci U S A. 2013;110:15283–15288. doi: 10.1073/pnas.1305766110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Phillips GN. Structure and dynamics of green fluorescent protein. Curr Opin Struct Biol. 1997;7:821–827. doi: 10.1016/s0959-440x(97)80153-4. [DOI] [PubMed] [Google Scholar]
- 27.CLONTECH Laboratories, Inc. Living Colors, Clontech User Manual. 1998. pp. 1–51. [Google Scholar]
- 28.Rudolph MG, Luz JG, Wilson IA. Structural and thermodynamic correlates of T cell signaling. Annu Rev Biophys Biomol Struct. 2002;31:121–149. doi: 10.1146/annurev.biophys.31.082901.134423. [DOI] [PubMed] [Google Scholar]
- 29.Quinn P, Griffiths G, Warren G. Density of newly synthesized plasma membrane proteins in intracellular membranes II. Biochemical studies J Cell Biol. 1984;98:2142–2147. doi: 10.1083/jcb.98.6.2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Takamori S, et al. Molecular anatomy of a trafficking organelle. Cell. 2006;127:831–846. doi: 10.1016/j.cell.2006.10.030. [DOI] [PubMed] [Google Scholar]
- 31.Theodoly O, Huang ZWH, Valignat MWP. New modeling of reflection interference contrast microscopy including polarization and numerical aperture effects: application to nanometric distance measurements and object profile reconstruction. Langmuir ACS J Surf Colloids. 2010;26:1940–1948. doi: 10.1021/la902504y. [DOI] [PubMed] [Google Scholar]
- 32.Yang F, Moss LG, Phillips GN. The molecular structure of green fluorescent protein. Nat Biotechnol. 1996;14:1246–1251. doi: 10.1038/nbt1096-1246. [DOI] [PubMed] [Google Scholar]
- 33.Krobath H, Rózycki B, Lipowsky R, Weikl TR. Line Tension and Stability of Domains in CellWAdhesion Zones Mediated by Long and Short ReceptorWLigand Complexes. PLoS ONE. 2011;6:e23284. doi: 10.1371/journal.pone.0023284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu Y, Vendome J, Shapiro L, BenWShaul A, Honig B. Transforming binding affinities from three dimensions to two with application to cadherin clustering. Nature. 2011;475:510–513. doi: 10.1038/nature10183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Müller M, Katsov K, Schick M. A new mechanism of model membrane fusion determined from Monte Carlo simulation. Biophys J. 2003;85:1611–1623. doi: 10.1016/S0006-3495(03)74592-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ho JWS, BAUMGARTNER A. Simulations of Fluid SelfWAvoiding Membranes. Europhys Lett. 1990;12:295–300. [Google Scholar]
- 37.Gompper G, Kroll DM. Network models of fluid, hexatic and polymerized membranes. J Phys Condens Matter Inst Phys J. 1997:8795–8834. [Google Scholar]
- 38.Teichmann SA, Chothia C. Immunoglobulin superfamily proteins in Caenorhabditis elegans. J Mol Biol. 2000;296:1367–1383. doi: 10.1006/jmbi.1999.3497. [DOI] [PubMed] [Google Scholar]
- 39.Vogel C. The immunoglobulin superfamily in Drosophila melanogaster and Caenorhabditis elegans and the evolution of complexity. Dev Camb Engl. 2003;130:6317–6328. doi: 10.1242/dev.00848. [DOI] [PubMed] [Google Scholar]
- 40.Helle SCJ, et al. Organization and function of membrane contact sites. BBA I Mol Cell Res. 2013;1833:2526–2541. doi: 10.1016/j.bbamcr.2013.01.028. [DOI] [PubMed] [Google Scholar]
- 41.Kornmann B. The molecular hug between the ER and the mitochondria. Curr Opin Cell Biol. 2013;25:443–448. doi: 10.1016/j.ceb.2013.02.010. [DOI] [PubMed] [Google Scholar]
- 42.Martens S, McMahon HT. Mechanisms of membrane fusion: disparate players and common principles. Nat Rev Mol Cell Biol. 2008;9:543–556. doi: 10.1038/nrm2417. [DOI] [PubMed] [Google Scholar]
- 43.Angelova MI, Dimitrov DS. Liposome electroformation. Faraday Discuss Chem Soc. 1986;81:303. [Google Scholar]
- 44.Nye JA, Groves JT. Kinetic control of histidineWtagged protein surface density on supported lipid bilayers. Langmuir ACS J Surf Colloids. 2008;24:4145–4149. doi: 10.1021/la703788h. [DOI] [PubMed] [Google Scholar]
- 45.Krichevsky O, Bonnet G. Fluorescence correlation spectroscopy: the technique and its applications. Rep Prog Phys. 2002;65:251–297. [Google Scholar]
- 46.Gompper DMKG. Random Surface Discetizations and the Renormalization of the Bending Rigidity. J Phys Fr. 1996;6:1305–1320. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure S1. BP density measurement by FCS
(a) Glass supported lipid bilayers (SLBs, composition: 94.47 DOPC, 2.5 DOGS-Ni-NTA) are incubated 100 nM BP solution for 10 minutes. (b) The density of BP protein on the SLB is measured by FCS. 7 fluorescent GFPuv (BP) was mixed with 93 mutated non-fluorescent GFPuv (BP-Y66S) (7 bright) is used to achieve the correct concentration range for FCS measurements. Normalized autocorrelation curves are fit with a single component two-dimensional diffusion model to extract the diffusion coefficient and protein density of the BP (molecules/μm2) (c) Measurements on 5 independent SLBs reveal an BP density of 2,317 +/− 370 molecules/um2 for an SLB with 2.5 DOGS-Ni-NTA incubated with 100 nM BP protein.
Supplementary Figure S2.
(a) Representative field of view of BP (green) and NBP (red) bound to DOGS-Ni-NTA containing GUV membranes (composition: 97.47 DOPC, 2.5 DOGS-Ni-NTA incubated with 100 nM protein solution for 10 min). Note how BP molecules form GUV interfaces while NBP molecules do not. Scale bar is 20 μm long. (b) A mixture of GUVs with His-tagged protein binding capability (97.5 DOPC, 2.5 DOGS-Ni-NTA) and GUVs lacking DOGS-Ni-NTA (99.7 DOPC, 0.3 Atto647-DOPE) were incubated with 100 nM GFPuv (BP). BP only bound to DOGS-Ni-NTA containing vesicles, confirming that there is no non-specific binding of BP from solution to membranes. Furthermore, we could not detect unspecific interface formation between BP-covered GUVs and protein-free GUVs. All scale bars are 10 μm. (c) Protein domain architectures illustrate proteins used in this study.
Supplementary Figure S3. Distance measurements by RICM
(left) Radially averaged RICM contrast as a function of the distance from the center of the GUV. (right) Theoretical RICM contrast as a function of the height of the variable buffer layer, which corresponds to the distance between SLB and the bottom surface of the GUV (see Methods).
Supplementary Figure S4
(a) Increasing concentrations of BP in solution leads to increased fluorescence intensities on GUVs. Vesicle (V) fluorescence intensities are plotted versus interface (I) intensities. (b) EIs for BP at different protein concentrations in solution. We note that while the intensities in a) scale with concentration, protein distribution over the vesicles stays remarkably constant at different protein densities. (c) EI values for different lengths adhesion molecules averaged over all experiments. Single lengths BP enrich more at the interface possibly due to higher 2D affinity than the longer 2L-BP and 3L-BP. (d) Exclusion of NBP or 2L-NBP from BP interfaces does not change significantly with increased membrane rigidity. GUVs from 80 Sphingomyelin and 20 Cholesterol mixtures (SM + Chol) have a approximately 6 fold higher bending rigidity than the DOPC GUVs. SM+Chol (77.5 Brain Sphingomyelin (Avanti), 20 Cholesterol (Avanti) 2.5 DOGS Ni-NTA) GUVs were incubated with 100 nM BP and 100 nM NBP or 2L-NBP. Interface formation and NBP and 2L NBP exclusion did not differ significantly from the same experiment performed on DOPC (97.5 DOPC, 2.5 DOGS-NiNTA) vesicles.
Supplementary Figure S5
Representative confocal images for data in Fig. 4a. Increasing the size of the adhesive protein (2L-BP, 3L-BP) leads to decreased exclusion of NBP variants from interfaces. Representative confocal fluorescence images of GUVs (composition: 97.47 DOPC, 2.5 DOGS-Ni-NTA) incubated with 100 nM BP variants and 100 nM NBP variants in solution for 10 min. Scale bars are 5 μm long (green channel: BP, red channel: NBP). Error bars are standard error of the mean from three independent experiments on separate vesicle batches, each with ~50 vesicles categorized.
Supplementary Table. BP densities