Supplemental Digital Content is available in the text.
Keywords: ADCK1, antipsychotics, genome-wide association study, heritability, paliperidone, pharmacogenetics, pharmacogenomics, polygenic, schizophrenia
Abstract
Objective
Clinical response to the atypical antipsychotic paliperidone is known to vary among schizophrenic patients. We carried out a genome-wide association study to identify common genetic variants predictive of paliperidone efficacy.
Methods
We leveraged a collection of 1390 samples from individuals of European ancestry enrolled in 12 clinical studies investigating the efficacy of the extended-release tablet paliperidone ER (n1=490) and the once-monthly injection paliperidone palmitate (n2=550 and n3=350). We carried out a genome-wide association study using a general linear model (GLM) analysis on three separate cohorts, followed by meta-analysis and using a mixed linear model analysis on all samples. The variations in response explained by each single nucleotide polymorphism (h2SNP) were estimated.
Results
No SNP passed genome-wide significance in the GLM-based analyses with suggestive signals from rs56240334 [P=7.97×10−8 for change in the Clinical Global Impression Scale-Severity (CGI-S); P=8.72×10−7 for change in the total Positive and Negative Syndrome Scale (PANSS)] in the intron of ADCK1. The mixed linear model-based association P-values for rs56240334 were consistent with the results from GLM-based analyses and the association with change in CGI-S (P=4.26×10−8) reached genome-wide significance (i.e. P<5×10−8). We also found suggestive evidence for a polygenic contribution toward paliperidone treatment response with estimates of heritability, h2SNP, ranging from 0.31 to 0.43 for change in the total PANSS score, the PANSS positive Marder factor score, and CGI-S.
Conclusion
Genetic variations in the ADCK1 gene may differentially predict paliperidone efficacy in schizophrenic patients. However, this finding should be replicated in additional samples.
Introduction
Identification of the factors influencing variation in response to pharmacological therapy has important health and economic implications. Predicting which individuals will respond positively to a medication before the administration of a therapeutic compound, or those who can avoid adverse effects, can lead to more targeted treatment and thereby potentially improve health outcomes and reduce healthcare costs.
Genomic sequence variation has been shown to influence response to particular pharmacotherapies. For example, warfarin and clopidogrel are two widely prescribed drugs that exert considerable pharmacogenetic effects 1–3. In the case of warfarin, variants at seven different genetic loci affect optimal dosing 4. Thus, the determination of an individual’s genetic profile before warfarin administration is essential as improper dosing can lead to thrombosis or bleeding. Similarly, genetic variants in the CYP2C19 gene are associated with rare, yet potentially catastrophic clotting and stent closure in individuals prescribed clopidogrel. Despite these and similar observations for other medications, responses to currently prescribed pharmaceuticals have rarely been subjected to rigorous genetic analyses and for those that have, the results are generally less conclusive than those found for warfarin and clopidogrel.
In the field of psychiatric genetics, few pharmacogenetic genome-wide association studies (GWAS) have been published on antipsychotic efficacy using clinical scales as outcome measurements. Lavedan et al. 5 examined the total Positive and Negative Syndrome Scale (PANSS) score in 407 patients from an iloperidone phase 3 clinical trial. Their top finding identified rs11851892 (14q12–q13) in the neuronal PAS domain protein 3 gene (NPAS3) as a potential mediator of the pharmacologic effect on PANSS treatment response (discovery P=8.6×10−5; confirmatory P=0.099, n=210). This locus was in close proximity to a translocation breakpoint site observed previously in a family with schizophrenia. McClay et al. 6 examined PANSS and the PANSS five-factor structure 7 in schizophrenic patients enrolled in the Clinical Antipsychotic Trial of Intervention Effectiveness (CATIE) and identified the intergenic marker rs17390445 (4p15) as a potential mediator of the effect of ziprasidone on the PANSS positive factor (P=9.82×10−8, n=160). In addition, the study by Clark et al. 8, which also used data generated from the CATIE study, identified PDE4D (5q12.1) as a potential mediator of the effect of quetiapine on patient-reported severity (P=4.2×10−8, n=238) as measured by a patient global impression of illness. A recent study by Sacchetti et al. 9 identified GRM7 as a potential predictor for risperidone treatment response using Emsley’s positive domain derived from PANSS (n=86; replicated in CATIE, n=97). Another recent study by Stevenson et al. 10 identified GRM7 and GRID2 as potential predictors for risperidone treatment response using the Brief Psychiatric Rating Scale total score in first-episode psychotic patients (P=1.10×10−8 for GRID2 variant rs9307122, n=86). However, in each of these studies, the sample size and effects were relatively small partially because of stratification by compound. In the last example (10), it is unclear whether these results would generalize beyond a first-episode population.
One reason why genetic analyses of pharmacotherapeutic responses may not yield findings as striking as those for warfarin and clopidogrel is that such responses might be influenced by the combined effect of many loci and not necessarily by a large effect from a single locus. Such polygenic or multifactorial influences on pharmaceutical responses are consistent with a growing appreciation among geneticists that an emphasis on the ‘common disease/complex trait’ hypothesis is limited and flawed. This has motivated a shift toward the discovery and interpretation of the likely polygenic nature of phenotypic variation 11,12. Some proponents of polygenic modeling hypothesize that assumptions on selective pressures on various human diseases and traits are most consistent with a polygenic origin. They argue that singular deleterious genetic factors that contribute toward a disease are likely related to survival and are thus unlikely to persist in the population. However, genetic variants that contribute toward pharmacotherapeutic efficacy and adverse drug reactions are less likely to be under similar selective forces as the introduction and exposure to pharmaceuticals in the population at large has been recent, and therefore, responses to them are likely not to have been shaped by selection simply because not enough time has elapsed since their introduction for selection to have shown its effects. It remains an important open question whether genetically associated pharmaceutical-response traits are, in part, because of a small number of genetic variants that contribute a major effect toward patient outcome or whether they are attributable to a large number of genetic variants, each contributing a minor effect toward the overall observed phenotype. Importantly, each therapeutic and each outcome (e.g. efficacy versus adverse response) may have its own unique characteristics.
To address this question, we explored the pharmacogenetic determinants of response to paliperidone in schizophrenic patients of European ancestry. The sample studied represented data pooled from 12 clinical studies. Paliperidone is an atypical antipsychotic medication used to treat symptoms in patients with schizophrenia. The compound can be administered once-daily through the OROS extended-release tablet or once-monthly through paliperidone palmitate long-acting injectable formulation. Unlike many psychiatric medications, paliperidone is mostly excreted unchanged and is thus not extensively metabolized in the liver. We focused on the change in the severity of symptoms of schizophrenia between baseline measurements before therapeutic administration and those at the end of the clinical trial. To explore different hypotheses surrounding the genetic basis of response to paliperidone, we implemented a two-step approach. This consisted of (a) a traditional GWAS analysis that identified individual loci with an appreciable singular effect on response and (b) estimation of the outcome variation explained by all genetic markers by polygenic modeling. Our results identify a number of loci that may harbor variants that influence paliperidone response. It was anticipated that treatment response was polygenic in origin. Our polygenic modeling analyses suggest a polygenic basis for paliperidone response, but are inconclusive because of our limited sample size. We discuss the overall limitations of our findings as well as potential ways of designing future clinical trials on the basis of these results.
Methods
Characteristics of study participants
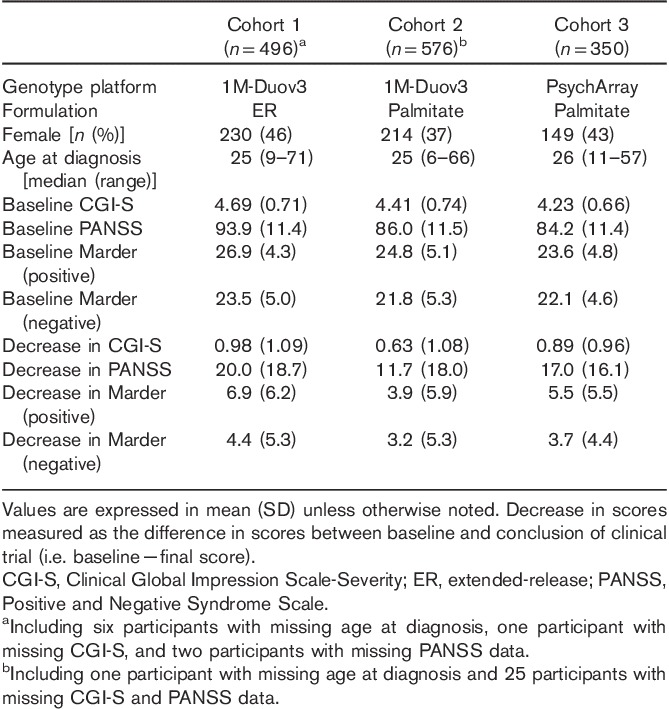
Study participants were enrolled in one of 12 trials 13–26 (Supplementary Table 1, Supplemental digital content 1, http://links.lww.com/FPC/B114) who fulfilled all of the following inclusion criteria: have a Diagnostic and Statistical Manual of Mental Disorders, 4th ed. (DSM-IV) diagnosis of schizophrenia (295.10, 295.20, 295.30, 295.60, or 295.90) for at least 1 year and experienced an acute episode of schizophrenia with a total PANSS score between 60 and 120 or 70 and 120 at screening depending on the trial. Additional inclusion and exclusion criteria can be found at http://clinicaltrials.gov. The present study only included participants who were genetically determined to be of European ancestry (methods described below) and who received treatment with paliperidone ER or paliperidone palmitate. Baseline clinical and demographic characteristics were collected on each participant (Table 1). At the end of the acute treatment phase of each trial (except NCT00210717, where response at day 92 last observation carried forward rather than at the end of 53 weeks treatment was used to ensure a treatment duration more comparable with the other studies that were short-term treatment studies), endpoint scores were obtained for the primary outcomes in the present study: total PANSS, positive and negative Marder factor scores, and Clinical Global Impression-Severity (CGI-S). The clinical studies were carried out in accordance with the ethical principles outlined in the Declaration of Helsinki, Good Clinical Practices guidelines, and applicable regulatory requirements. Participation in the genetic study was optional and all participants provided written informed consent before enrollment.
Table 1.
Characteristics of the study participants
Genotyping
Samples from 11 out of the 12 clinical studies were included previously in a candidate gene study using a custom-designed CNS chip 27. Genotyping in this study was performed using either the Human1M-Duov3 or PsychArray (Illumina Inc., San Diego, California, USA). Genetic markers that showed high missingness (>0.05), failed Hardy–Weinberg equilibrium (P<10−6), or had exceedingly rare alternative alleles [minor allele frequency (MAF)<0.01] were excluded. For samples genotyped using Human1M-Duov3, the degree of European ancestry for each study participant was calculated using a supervised clustering analysis 28 from publicly available European reference panels (n=1335). Participants estimated to be of less than 90% ancestrally European were excluded from downstream analyses. Among the remaining participants, relatedness was assessed using pairwise identity by descent estimation in PLINK 29. Participants were excluded such that the estimated proportion of identity by descent between any two remaining individuals was less than 0.1. In addition, individuals enrolled in more than one clinical study and with genetically inferred sex discrepant from sex from case report form were excluded. Following these procedures, 1072 unrelated participants of European ancestry remained for analysis. For samples genotyped using PsychArray, the same QC criteria were applied, except that smartpca 30,31 was used to remove outliers from self-reported European ancestry participants. Using these criteria, 350 participants genotyped using PsychArray were retained for further analysis.
Imputation
Genomic data were prephased 32 and genome-wide imputation was performed on the resulting haplotypes separately for samples genotyped using the Human1M-Duov3 or PsychArray beadchip and the default parameters in IMPUTE v2.3 33–35. The 1000 Genomes Phase 1 integrated variant set haplotypes were used as the reference panel 36. Genomes were divided into approximately 5 Mbp segments (avoiding chromosome and centromere boundaries), with phasing and imputation performed on each. GTOOL v0.7.5 was used to convert imputed genotype posterior probabilities into calls. Genotypes were considered to be missing if the posterior probability of any genotype was not greater than 0.90. The best-guessed imputed genotypes for all samples were merged into a single dataset and additional QC criteria [such as single nucleotide polymorphism (SNP)-wise missing rate>0.05, Hardy–Weinberg equilibrium P<10−6 and MAF<0.01] were applied.
GWAS
GLM-based genome-wide association analyses were carried out on three cohorts of study participants: patients receiving paliperidone ER (n=496 genotyped using Illumina Human1M-Duov3, effective sample size n=488 for PANSS score endpoints and 489 for CGI-S) and patients receiving paliperidone palmitate (n1=576 genotyped using Illumina Human1M-Duov3, effective sample size n=550; and n2=350 genotyped using PsychArray, effective sample size n=350). A meta-analysis across the three cohorts was used to summarize the association statistics. Four primary outcomes were analyzed: the change in the total PANSS score from baseline, the change in the positive and negative Marder factor scores of the PANSS from baseline, and the change in the CGI-S scale from baseline. On the basis of stepwise regression procedures, the following covariates were included as fixed-effects covariates: the respective baseline measurement, sex, and age at diagnosis. The first 10 principal components were calculated within each grouping, and were likewise included as fixed-effects covariates in GLM. Genetic association was performed using allelic dosage probability at each common (MAF>0.01) autosomal marker that passed genotype and imputation quality control greater than 0.3 using an additive model in PLINK. Haplotype markers based on directly genotyped markers from the top hit locus were also associated with the endpoint.
In addition to the respective baseline measurement, sex, and age at diagnosis, drug received (paliperidone ER or paliperidone palmitate) was also included in the MLM analysis using all samples (three cohorts combined). An MLM-based genome-wide analysis (both univariate and multivariate) using best-guessed imputed genotypes with imputation quality control greater than 0.5 was carried out using studentized residuals for each endpoint after adjusting the clinical covariates in GEMMA 37.
Estimation of h2SNP
The proportion of phenotypic variance explained by autosomal markers (h2SNP) was calculated for each of the primary outcomes using GCTA 12 and GEMMA in all samples. Like the MLM analysis, studentized residuals for each endpoint and best-guessed imputed genotype were used as the phenotype and genotype, respectively, in both heritability estimation analyses. Genetic relationship matrices were calculated using genotype data on all patients making up the cohorts. No pairs of participants showed an estimated relatedness greater than 0.1. The variance explained by markers used to estimate the genetic relationship matrices was calculated using restricted maximum likelihood. Twenty principal components derived from GCTA were also included as covariates in the GCTA analysis.
Gene set enrichment analysis
INRICH is a pathway analysis tool for GWAS, designed for detecting enriched association signals of linkage disequilibrium (LD)-independent genomic regions within biologically relevant gene sets 38. Reference gene sets used in the INRICH analysis include KEGG, Gene Ontology, and Molecular Signature Database (v5.0). Top variants from GLM-based change in the total PANSS score analysis with a nominal association P-values less than 0.0005, 0.0001, 0.00005, and 0.00001 were separately fed into PLINK to clump the variants into LD-independent genomic intervals (r2 threshold using 0.2, 0.3, and 0.5, respectively), and then LD-independent genomic regions were used for INRICH (version 1.0) analyses. No multiple testing corrections were applied for running INRICH against multiple reference gene sets or for using multiple parameters (P-value cutoff and LD threshold).
Cross-reference between the top association results with GTEx, BRAINEAC, and EnigmaVis findings
To examine tissue expression patterns and explore possible functional evidence for the associated variants, such as their being a known eQTL or having been found to be associated with neural imaging-derived phenotypes, the top associated genes and variants were queried in the GTEx portal (http://www.gtexportal.org/), Data Source: GTEx Analysis Release V6 (dbGaP Accession phs000424.v6.p1) 39, BRAINEAC (http://braineac.org/) 40, and EnigmaVis (http://enigma.ini.usc.edu/) 41. These resources include GWAS results for a few imaging genetics studies for human hippocampal and intracranial volumes (ICVs) 42, lentiform nucleus volume 43, temporal lobe 44, and caudate 45. Furthermore, the GWAS results from ENIGMA2 (seven subcortical regions and the ICV derived from magnetic resonance images of 30 717 individuals from 50 cohorts) 46 were downloaded and used to intersect with the association results from this study.
Results
The study participants from 12 clinical trials designed to elucidate efficacy of paliperidone ER and paliperidone palmitate are characterized in Table 1. All participants included in the present study were determined to be of European ancestry through the use of genetic markers. For GLM-based association analysis, participants were stratified into three cohorts on the basis of the therapeutic treatment received during the clinical trials and the chip used for the genotyping: extended-release paliperidone genotyped using Illumina Human1M-Duov3 (n=490), paliperidone palmitate genotyped using Illumina Human1M-Duov3 (n=550), or paliperidone palmitate genotyped using PsychArray (n=350). All analyses were carried out independently on each cohort using changes from baseline in each of the primary outcomes as the phenotypic endpoint or the dependent variable in our analyses: total PANSS score, positive and negative Marder factor scores, and CGI-S score.
GLM-based association analyses
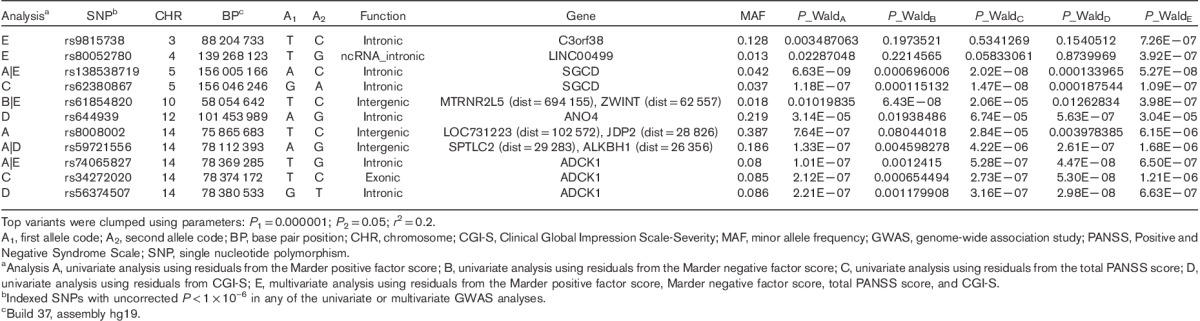
Following quality control procedures, 913 899 and 288 712 genotyped markers were used to impute unobserved genotypes and ∼9M polymorphic imputed markers remained for downstream analyses. Controlling for covariates, genome-wide association analyses of these markers followed by meta-analysis across three cohorts showed four regions with somewhat rare variants (MAF between 1 and 5%) reaching genome-wide significance (P<5×10−8, Supplementary Fig. 1, Supplemental digital content 2, http://links.lww.com/FPC/B115 and Supplementary Fig. 2, Supplemental digital content 3, http://links.lww.com/FPC/B116 for Manhattan and quantile–quantile plots for the total PANSS score, the positive Marder factor score, the negative Marder factor score, and CGI-S, respectively) and 20+ regions including aarF domain containing kinase 1 (ADCK1) that showed a trend toward association (P<1.0×10−6). Table 2 provides only those indexed variants with MAF greater than or equal to 10% in the 1000 Genomes Project. Rs56240334 (P=7.97×10−8 for CGI-S; P=8.72×10−7 for total PANSS; P=2.09×10−7 for the PANSS positive Marder factor; and P=0.003 for the PANSS negative Marder factor) in the intron of ADCK1 (14q24.3) was identified as a potential associated variant of paliperidone efficacy. Other suggestive association signals included rs12915820 (P=1.28×10−7 for total PANSS score) in the intergenic region (15q25.1–25.2) between transmembrane channel-like 3 (TMC3) and mex-3 RNA-binding family member B (MEX3B). The full list of variants with P<1×10−5 in either the meta-analysis or any of the three cohorts is available in Supplementary Table 2 (Supplemental digital content 4, http://links.lww.com/FPC/B117). The association of relatively rare variants must be interpreted with caution as the sample sizes for rare genotype groups are small and imputation tends to be less accurate. Among the common variants showing suggestive associations, the strongest signal came from variants in ADCK1, which showed largely a consistent trend for symptom severity measurements across the three cohorts (Table 3). Overall, each additional copy of the rs56240334-G allele was associated with an average of a 5.9 point reduction in the total PANSS score, a 2.1 point reduction in the positive Marder factor score, a 1.0 point reduction in the negative Marder factor score, and a 0.38 point reduction in CGI-S. The common homozygote genotype group was associated with greater therapeutic efficacy (i.e. a greater decrease in the symptom severity score from baseline; Fig. 1). Rs56240334 was imputed and not genotyped directly in both Human1M-Duov3and PsychArray platforms, although the imputation confidence info scores from Impute2 are high (0.959 and 0.894, respectively, for Human1M-Duov3 and PsychArray platforms, respectively). A nearby intronic SNP rs10147707 in LD with rs56240334 (r2=0.86, D′=0.99) was genotyped directly in Human1M-Duov3, but not in PsychArray (Imputation confidence info score is 0.816). In cohort 1, the common homozygote rs10147707 C/C (n=402), heterozygote C/T (n=89), and the rare homozygote T/T (n=3) had an average of 21.22, 14.47, and 27.67 point reductions in the total PANSS score, respectively. Similarly, the common homozygote C/C (n=461), heterozygote C/T (n=84), and the rare homozygote T/T (n=6) had an average of 12.76, 7.43, and −6.17 point reductions in the total PANSS score, respectively, in cohort 2. The sample size for the rare homozygote T/T genotype group was too rare to have a robust estimate of group mean. There was also a synonymous variant rs34272020 (S188S) in ADCK1 in strong LD (r2=0.87, D′=1) with rs56240334. The LD in the associated regions spanned a few neighboring genes, although the strongest association signal came from ADCK1 (Fig. 2). Haplotype association for markers from ADCK1 region was also available from Supplementary Text S1 (Supplemental digital content 5, http://links.lww.com/FPC/B118). If we relax the criteria by requiring variants with P-value less than 0.01 in all three primary cohorts and the effect size is consistent across all three cohorts, the only variant surviving is rs2532643 (first variant in Table 2) for the CGI-S endpoint (P1=0.001259, P2=0.009209, P3=0.009618, β1=−2.5666, β2=−2.2791 and β3=−2.3803).
Table 2.
Summary of GLM GWAS meta-analysis – SNPs with uncorrected P<1×10–6 and MAF≥10%
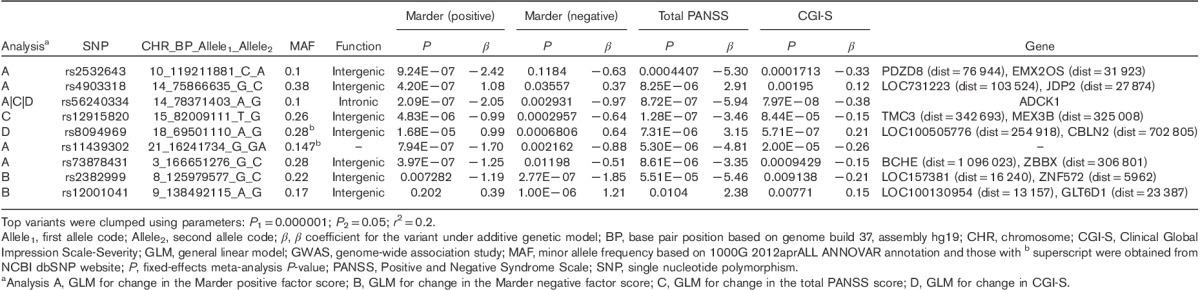
Table 3.
GWAS results of the top common variant finding among all cohorts
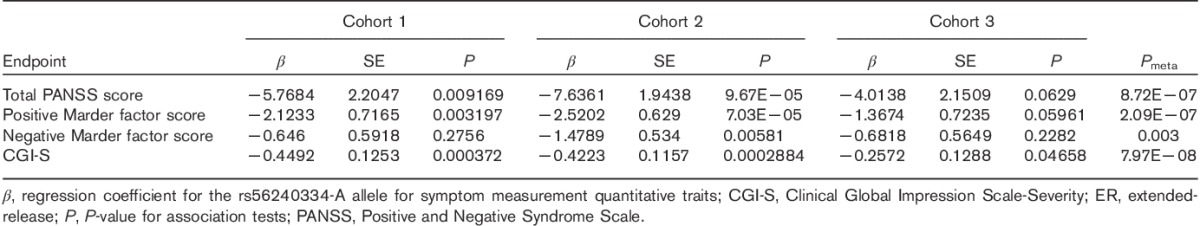
Fig. 1.
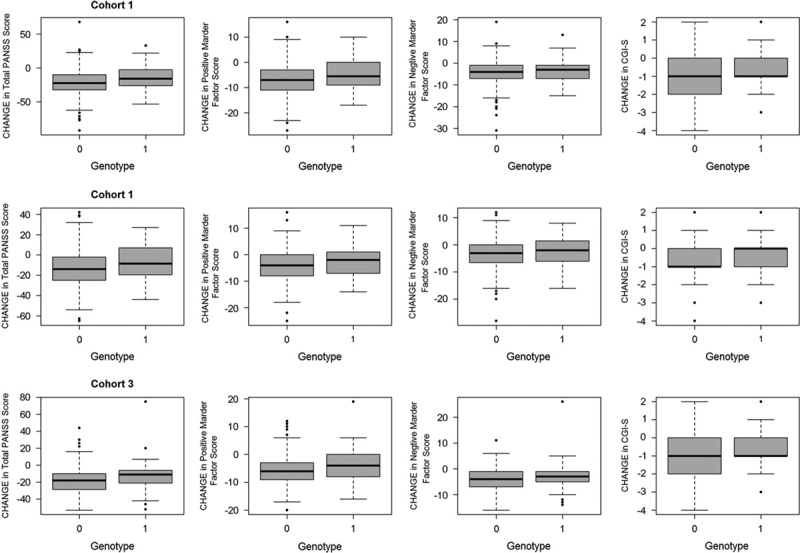
Changes in outcome measurements stratified by genotype for rs56240334 in ADCK1, G/G homozygotes (i.e. A-noncarriers), and A-carriers correspond to 0 and 1 genotypes in the graph. CGI-S, Clinical Global Impression Scale-Severity; PANSS, Positive and Negative Syndrome Scale.
Fig. 2.
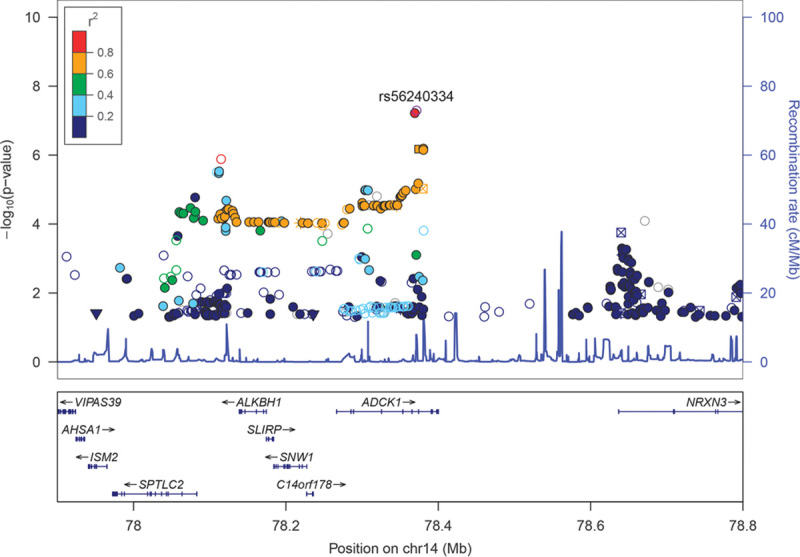
Regional plot for ADCK1. Association results [−log10(P)] are plotted for all single nucleotide polymorphisms (SNPs) passing quality control. Chromosome position is plotted with reference to the NCBI build 37. Recombination rate as estimated from the HapMap Project is plotted in light blue. SNPs are color coded according to the linkage disequilibrium measure (r2) with reference SNP based on the reference panel of CEU population from the 1000 Genome Project (March 2012 release). SNP annotation for all 1000GP SNPs are represented by the annotation categories: nonsynonymous (inverted triangle), synonymous (square), UTR (square), TFBScons (star), MCS44 Placental (square with diagonal lines), and none of the above (filled circle).
As paliperidone is an active metabolite of risperidone, we took a close look for replication evidence among variants reported to be associated with risperidone in the literature. Variants from GRM7 9,10, GRID2 10, DRD2 (including rs2514218, a genome-wide significant variant for schizophrenia disease susceptibility reported by PGC 47 and subsequently reported to be related to treatment response 48 and others implicated in candidate gene studies 49), and DRD3 were not replicated (P>0.05) in the meta-analysis of paliperidone response (Supplementary Table 3, Supplemental digital content 6, http://links.lww.com/FPC/B119), except rs6314 of HTR2A, where it was nominally associated with changes in CGI-S (P=0.01), Marder negative factor score (P=0.08), and total PANSS score (P=0.1), with G allele being the better response allele.
MLM-based association analyses
Both univariate and multivariate MLM were fitted using GEMMA (Supplementary Fig. 3, Supplemental digital content 7, http://links.lww.com/FPC/B120 and Supplementary Fig. 4, Supplemental digital content 8, http://links.lww.com/FPC/B121 for Manhattan and quantile–quantile plots for the total PANSS score, the positive Marder factor score, the negative Marder factor score, and CGI-S univariate analyses and multivariate analysis, respectively) and SNPs with unadjusted P-value less than 1×10−6 are shown in Table 4. The GEMMA univariate mixed linear model results were largely consistent with the GLM-based results, although association P-values could still differ by 1–2 orders of magnitude. Using the same P-value threshold cutoff (i.e. 10−6), there were few independent groups of associated variants from the MLM-based analysis compared with the GLM-based analysis, primarily because of a few rare variants showing stronger associations in the MLM-based analysis. MLM-based association P-values for rs56240334 from ADCK1 (in LD with rs56374507, r2=0.86 and rs34272020, r2=0.87) for univariate CGI-S, total PANSS, PANSS positive Marder factor, PANSS negative Marder factor, and multivariate analysis are 4.26×10−8, 9.56×10−7, 4.74×10−7, 1.12×10−3, and 1.39×10−6, respectively, which are consistent with the results from general linear model-based analyses, whereas the association with change in CGI-S reached genome-wide significance. The full list of variants with association P-values less than 1×10−5 in any of univariate or multivariate MLM analyses is shown in Supplementary Table 4 (Supplemental digital content 9, http://links.lww.com/FPC/B122).
Table 4.
Summary of MLM GWAS analyses – SNPs with an uncorrected P<1×10–6
Estimation of h2SNP
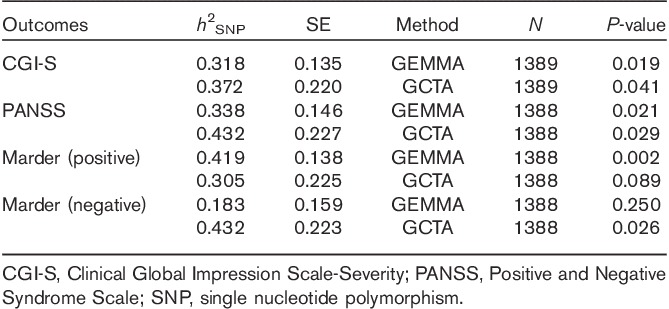
The proportion of variance explained by autosomal markers, or marker-based heritability (h2SNP), in the combined therapeutic grouping was calculated for each of the primary outcomes using the best-guessed imputed markers (Table 5). The proportion of variance explained by genotyped markers was the largest in the PANSS positive Marder factor score (h2SNP=0.42, SE=0.14), followed by the total PANSS score (h2SNP=0.34, SE=0.15) and CGI-S (h2SNP=0.32, SE=0.14) on the basis of estimates from GEMMA. The SE estimates from GCTA were larger in general compared with GEMMA, making the probabilistic significance of the polygenic heritability estimates inconclusive.
Table 5.
Proportion of variation explained by genotyped, autosomal markers (h2SNP)
Gene set enrichment results
INRICH enrichment analysis using total PANSS score GLM analysis showed nominal enrichments of immune-related Biocarta IL-2 pathway (Pcorr=0.1), hallmark tumor necrosis factor α (TNFα) signaling by NFκβ (Pcorr=0.09), and the Biocarta TCR pathway (Pcorr=0.03–0.08). The genes from the genomic intervals driving the enrichment and common to at least two of three gene sets above were lymphocyte-specific protein tyrosine kinase (LCK), mitogen-activated protein kinase 8 (MAPK8), and mitogen-activated protein kinase kinase 1 (MAP2K1), and Jun_proto-oncogene (JUN). The complete list of nominally enriched pathways with Pcorr less than 0.1 is shown in Supplementary Table 5 (Supplemental digital content 10, http://links.lww.com/FPC/B123) and the genes driving the enriched pathways are listed in Supplementary Table 6 (Supplemental digital content 11, http://links.lww.com/FPC/B124).
Cross-reference results
ADCK1 is expressed in multiple brain regions including the cortex and pituitary. Intriguingly, variant rs144082574 in the same genomic region, not in LD with the top associated variant rs56240334 from this study (r2=0.004, D′=1), had a significant eQTL relationship with expression levels of ADCK1 in blood (P=1.2×10−15). Another variant rs9323656 in ADCK1 in LD with the top variant rs56240334 (r2=0.83, D′=0.99) had a nominal association (P=0.007) with ICV 42 in healthy individuals. The association P-values for rs9323656 with total PANSS, Marder positive factor score, Marder negative factor score, and CGI-S were 0.0002, 3.9×10−5, 0.037, and 5.3×10−5, respectively (in GLM-based models, Supplementary Fig. 5, Supplemental digital content 12, http://links.lww.com/FPC/B125), whereas rs56240334 was not captured in the Stein et al. association study. Other candidate genes with subtle eQTL relationships with rs56240334 in the ADCK1 genomic region shown by BRAINEAC are listed in Supplementary Table 7 (Supplemental digital content 13, http://links.lww.com/FPC/B126) and their tissue expression patterns are described in Supplementary Fig. 6 (Supplemental digital content 14, http://links.lww.com/FPC/B127). None of these candidate genes neighboring ADCK1 is a more likely candidate than ADCK1 to explain the association of rs56240334 with paliperidone association, although we cannot rule them out completely.
Discussion
Our results suggest that candidate genomic regions that harbor variants that influence paliperidone efficacy among patients with schizophrenia may exist, although there is no genetic locus with large effect size predictive of paliperidone efficacy. The most strongly associated SNP, rs56240334, from ADCK1 was consistently associated with each of the three cohorts and efficacy outcome measurements [all consistent direction-wise; most are significant at the P<0.1 in individual cohorts, except for cohort 1 (P=0.28) and cohort 3 (P=0.23) with respect to the negative Marder factor score]. This suggests that these markers contribute toward paliperidone efficacy irrespective of the mode of administration (i.e. oral extended-release or long-acting injectable palmitate), which is expected if the polymorphism reflects the underlying mechanism of action of paliperidone. The association signal is stronger with respect to the positive Marder factor score and the total PANSS score compared with the negative Marder factor score, which probably reflects the fact that antipsychotics in general are known to be more effective in treating positive symptoms rather than negative symptoms and that change in positive symptoms tends to drive the change in the overall PANSS scores in antipsychotic medication trials. The association signals were also independent of the statistical models used (i.e. GLM-based or MLM-based models). The biological relationship between variants in ADCK1 and paliperidone is unknown, except for the imaging genetics relationship with ICV. On the basis of data from 2028 individuals with schizophrenia and 2540 healthy controls from the ENIGMA consortium, patients with schizophrenia had smaller hippocampus (Cohen’s d=−0.46), amygdala (d=−0.31), thalamus (d=−0.31), accumbens (d=−0.25), and ICVs (d=−0.12), as well as larger pallidum (d=0.21) and lateral ventricle volumes (d=0.37) compared with healthy controls. No group differences were identified for putamen and caudate volumes 50. The volumes of the subcortical regions included in the basal ganglia were reported to be predictive of/associated with treatment response to antipsychotic drugs (APDs), although those studies were small in sample size, did not control for other factors that could also be used to explain variance in treatment response 51–53, and some did not normalize the brain region volumes by ICV 51,53. However, APDs could also impact the volumes of brain regions such as the caudate nucleus and putamen in patients with schizophrenia 53,54 and could complicate interpretation of results unless studying APD-naive patients. In a small (n=23) yet well-controlled 6-week risperidone treatment study in unmedicated schizophrenic patients, basal ganglia volumes including the bilateral caudate, putamen, and pallidum were normalized by ICV and the caudate volume showed the strongest correlation with treatment response even when controlling for baseline symptom severity and duration of illness 55. The relationships between rs9323656 and other brain regions are unknown, except those examined in EnigmaVis and ENIGMA2 46. Given the results with risperidone, it is noteworthy that the ADCK1 variant influences both ICV and paliperidone response, especially as paliperidone (9-hydroxyrisperidone) is the active metabolite of risperidone. Risperidone is metabolized by cytochrome p450 (CYP) enzymes and polymorphisms in the CYP genes may influence risperidone efficacy, whereas paliperidone is not further metabolized by CYP enzymes. Variants reported to be associated with risperidone treatment response were largely not replicated in our paliperidone response analysis.
It is intriguing to observe the nominal enrichment of several immune-related gene sets in the INRICH analysis. Although the hypothesis of the interplay between immune and nervous system was postulated a century ago, there has been increasing evidence to support this hypothesis in the past 20 years 56,57. Notably, cytokines could affect the CNS by modulating the activity of several monoaminergic systems, which are the primary targets of psychotropic therapies. Elevation in cytokine levels could inhibit dopamine synthesis and reduce dopaminergic signaling and serotonin bioavailability 58,59. Antipsychotics including risperidone were reported to decrease TNFα 60,61. It is therefore of interest to observe the nominal enrichment of ‘Hallmark TNFα Signaling via NFκβ’ and other immune-related gene sets in the gene set enrichment analysis. Other variants with a suggestive association signal for paliperidone response are discussed in the Supplementary Text S2 (Supplemental digital content 15, http://links.lww.com/FPC/B128).
Our main finding, that a common homozygous genotype for ADCK1 was associated with greater therapeutic efficacy, is of interest, although it is important to note that the candidate regions that we identified each explain only a small fraction of the variance in therapeutic response. Therefore, the clinical utility of these markers for use as classifiers to guide treatment decisions is limited. However, these results seem to rule out the existence of common variants with very large genetic effect sizes as such variants should have been detectable in this study.
Our discovery of multiple candidate markers provides some evidence that efficacy may be influenced by polygenic factors. This observation was partially supported in our estimations of h2SNP, the proportion of therapeutic response attributable to the combined effects of genetic variants throughout the genome. For at least some of the outcome measurements (CGI-S, positive Marder factor score, and total PANSS score), our GEMMA results suggested that a modest proportion of phenotypic variance can be explained by imputed genetic markers. This relationship is less evident in negative Marder factor responses, which tend to respond less to antipsychotic medication treatment. However, the estimates remain uncertain because of sample size limitations. It is unclear why the SE estimates from GCTA and GEMMA varied considerably, with GCTA having larger SEs than GEMMA. Although not described in this manuscript, we also estimated h2SNP in a larger cohort that combined the White patients with schizophrenia with non-Caucasians and/or bipolar patients receiving paliperidone. The resulting estimates of h2SNP were 0.217 in total PANSS and 0.274 in CGI-S (SE=0.130 and 0.166, respectively; positive and negative Marder factor scores were not analyzed) using GCTA. These results are consistent with a polygenic model of therapeutic efficacy, and yet population stratification and dissimilar disease etiology may be a confounding factor in these analyses. Nevertheless, these estimates of h2SNP present a potential upper bound on the predictive capacity of the collective effect of genetic markers that could be imputed using common genotyping platforms. Ideally, findings and interpretations from pharmacogenetic studies can be applied to guide clinical decision-making by identifying patients who will most benefit from a particular treatment or avoid an adverse reaction before administration. This observation may be explained by the fact that although attempts were made to control for heterogeneity in this cohort (e.g. by taking into consideration ethnicity, compound, diagnosis, etc.), other factors could not be controlled. Variation in the dosage of the drugs administered to the participants presents a particularly challenging form of heterogeneity. Although some study participants received a set dosage, a large proportion of the study population was enrolled into ‘flexible’ dosage arms designed to optimize treatment response. Hence, dosage was nonuniformly altered to reach a target response, although there was a lack of a clear PK–PD relationship on the basis of unpublished PK modeling and the dose–response relationship was not always observable from the fixed-dose studies. Other likely sources of heterogeneity include suboptimal dosing potentially because of a suboptimal injection site and needle length (with possible injection into fatty tissue rather than muscle) in the case of paliperidone palmitate, unknown adherence over time to oral paliperidone ER, and variable trial length across the clinical studies. The variable length of studies was somewhat mitigated by the fact that therapeutic response reached a plateau as early as 6 weeks after the initiation of paliperidone treatment 17. Such heterogeneity may have limited estimation of pharmacogenetic effects in a cohort that is small for genetic association evaluations.
The issues of heterogeneity and limitation of the study sample size identified here are not unique to this study. To address this, the Psychiatric Genomic Consortium involving academic and pharmaceutical companies has been formed to address these challenges by sharing data to increase sample size and contribute analytic methods 47,62,63. However, at present, only examination of disease susceptibility is within the scope of consortium activities perhaps because of the scarcity of controlled clinical studies. The field has witnessed growing success in identifying safety-related genetic associations derived from clinical trial studies, but less success in attempts to identify efficacy biomarkers. Finding efficacy genetic associations for psychiatric disease has proven to be particularly challenging, even with access to large number of samples in clinical trials. For greater success in finding genetic markers for efficacy in complex disorders such as schizophrenia, pharmaceutical companies, regulatory bodies, and academic groups should continue to share data. In addition, clinical trial study design must consider disease heterogeneity. Finally, in our clinical studies described in the paper, DNA sampling was optional among study participants. After selecting samples of European ancestry and patients treated with study drug, the sample size became small. Future clinical studies might consider willingness to consent for DNA sampling as a potential inclusion criterion to increase the sample size for pharmacogenomics study.
Supplementary Material
Supplemental digital content is available for this article. Direct URL citations appear in the printed text and are provided in the HTML and PDF versions of this article on the journal's website (www.pharmacogeneticsandgenomics.com).
Acknowledgements
The authors are grateful to the study volunteers for participating in the research studies and to the clinicians and support staff for enabling patient recruitment and blood sample collection. They also thank the staff at the Neuroscience Biomarkers Genomic Lab at Janssen for sample processing and the staff at Illumina for genotyping these DNA samples. They thank the Neuroscience Biomarkers operational team of Janssen Research & Development for operational support.
This study was funded by Janssen Research & Development, LLC (formerly known as Johnson & Johnson Pharmaceutical Research & Development, LLC), Raritan, New Jersey, USA, including funding to The Scripps Research Institute.
Conflicts of interest
Dr Cohen is a former employee of Janssen Research & Development. Drs Li, Fu, Savitz, and Gopal are employees of Janssen Research & Development, LLC. Dr Alphs is employee of Janssen Scientific Affairs. Drs Li, Fu, Alphs, Savitz, and Gopal may be shareholders in Johnson & Johnson, which is the parent company of the Janssen companies. Drs Libiger and Schork are former employees of The Scripps Research Institute. Dr Libiger can now be contacted at MD Revolution and Dr Schork can now be contacted at J. Craig Venter Institute. For the remaining authors there are no conflicts of interest.
Footnotes
Qingqin Li and Nathan E. Wineinger contributed equally to the writing of this article.
References
- 1.Cooper GM, Johnson JA, Langaee TY, Feng H, Stanaway IB, Schwarz UI, et al. A genome-wide scan for common genetic variants with a large influence on warfarin maintenance dose. Blood 2008; 112:1022–1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Furuya H, Fernandez-Salguero P, Gregory W, Taber H, Steward A, Gonzalez FJ, et al. Genetic polymorphism of CYP2C9 and its effect on warfarin maintenance dose requirement in patients undergoing anticoagulation therapy. Pharmacogenetics 1995; 5:389–392. [DOI] [PubMed] [Google Scholar]
- 3.Hulot JS, Bura A, Villard E, Azizi M, Remones V, Goyenvalle C, et al. Cytochrome P450 2C19 loss-of-function polymorphism is a major determinant of clopidogrel responsiveness in healthy subjects. Blood 2006; 108:2244–2247. [DOI] [PubMed] [Google Scholar]
- 4.International Warfarin Pharmacogenetics Consortium, Klein TE, Altman RB, Eriksson N, Gage BF, Kimmel SE, et al. Estimation of the warfarin dose with clinical and pharmacogenetic data. N Engl J Med 2009; 360:753–764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lavedan C, Licamele L, Volpi S, Hamilton J, Heaton C, Mack K, et al. Association of the NPAS3 gene and five other loci with response to the antipsychotic iloperidone identified in a whole genome association study. Mol Psychiatry 2009; 14:804–819. [DOI] [PubMed] [Google Scholar]
- 6.McClay JL, Adkins DE, Aberg K, Stroup S, Perkins DO, Vladimirov VI, et al. Genome-wide pharmacogenomic analysis of response to treatment with antipsychotics. Mol Psychiatry, 16:76–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Van Der Gaag M, Hoffman T, Remijsen M, Hijman R, de Haan L, van Meijel B, et al. The five-factor model of the Positive and Negative Syndrome Scale II: a ten-fold cross-validation of a revised model. Schizophr Res 2006; 85:280–287. [DOI] [PubMed] [Google Scholar]
- 8.Clark SL, Souza RP, Adkins DE, Aberg K, Bukszar J, McClay JL, et al. Genome-wide association study of patient-rated and clinician-rated global impression of severity during antipsychotic treatment. Pharmacogenet Genomics, 23:69–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sacchetti E, Magri C, Minelli A, Valsecchi P, Traversa M, Calza S, et al. The GRM7 gene, early response to risperidone, and schizophrenia: a genome-wide association study and a confirmatory pharmacogenetic analysis. Pharmacogenomics J 2016. [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- 10.Stevenson JM, Reilly JL, Harris MS, Patel SR, Weiden PJ, Prasad KM, et al. Antipsychotic pharmacogenomics in first episode psychosis: a role for glutamate genes. Transl Psychiatry 2016; 6:e739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chatterjee N, Wheeler B, Sampson J, Hartge P, Chanock SJ, Park JH. Projecting the performance of risk prediction based on polygenic analyses of genome-wide association studies. Nat Genet 2013; 45:400–405. 5e1–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang J, Lee SH, Goddard ME, Visscher PM. GCTA: a tool for genome-wide complex trait analysis. Am J Hum Genet 2011; 88:76–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bossie CA, Sliwa JK, Ma YW, Fu DJ, Alphs L. Onset of efficacy and tolerability following the initiation dosing of long-acting paliperidone palmitate: post-hoc analyses of a randomized, double-blind clinical trial. BMC psychiatry 2011; 11:79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Canuso CM, Dirks B, Carothers J, Kosik-Gonzalez C, Bossie CA, Zhu Y, et al. Randomized, double-blind, placebo-controlled study of paliperidone extended-release and quetiapine in inpatients with recently exacerbated schizophrenia. Am J Psychiatry 2009; 166:691–701. [DOI] [PubMed] [Google Scholar]
- 15.Davidson M, Emsley R, Kramer M, Ford L, Pan G, Lim P, et al. Efficacy, safety and early response of paliperidone extended-release tablets (paliperidone ER): results of a 6-week, randomized, placebo-controlled study. Schizophr Res 2007; 93 (1–3):117–130. [DOI] [PubMed] [Google Scholar]
- 16.Fleischhacker WW, Gopal S, Lane R, Gassmann-Mayer C, Lim P, Hough D, et al. A randomized trial of paliperidone palmitate and risperidone long-acting injectable in schizophrenia. Int J Neuropsychopharmacol 2012; 15:107–118. [DOI] [PubMed] [Google Scholar]
- 17.Gopal S, Hough DW, Xu H, Lull JM, Gassmann-Mayer C, Remmerie BM, et al. Efficacy and safety of paliperidone palmitate in adult patients with acutely symptomatic schizophrenia: a randomized, double-blind, placebo-controlled, dose–response study. Int Clin Psychopharmacol 2010; 25:247–256. [DOI] [PubMed] [Google Scholar]
- 18.Kane J, Canas F, Kramer M, Ford L, Gassmann-Mayer C, Lim P, et al. Treatment of schizophrenia with paliperidone extended-release tablets: a 6-week placebo-controlled trial. Schizophr Res 2007; 90 (1–3):147–161. [DOI] [PubMed] [Google Scholar]
- 19.Kramer M, Litman R, Hough D, Lane R, Lim P, Liu Y, et al. Paliperidone palmitate, a potential long-acting treatment for patients with schizophrenia. Results of a randomized, double-blind, placebo-controlled efficacy and safety study. Int J Neuropsychopharmacol 2010; 13:635–647. [DOI] [PubMed] [Google Scholar]
- 20.Kramer M, Simpson G, Maciulis V, Kushner S, Vijapurkar U, Lim P, et al. Paliperidone extended-release tablets for prevention of symptom recurrence in patients with schizophrenia: a randomized, double-blind, placebo-controlled study. J Clin Psychopharmacol 2007; 27:6–14. [DOI] [PubMed] [Google Scholar]
- 21.Marder SR, Kramer M, Ford L, Eerdekens E, Lim P, Eerdekens M, et al. Efficacy and safety of paliperidone extended-release tablets: results of a 6-week, randomized, placebo-controlled study. Biol Psychiatry 2007; 62:1363–1370. [DOI] [PubMed] [Google Scholar]
- 22.Meltzer HY, Bobo WV, Nuamah IF, Lane R, Hough D, Kramer M, et al. Efficacy and tolerability of oral paliperidone extended-release tablets in the treatment of acute schizophrenia: pooled data from three 6-week, placebo-controlled studies. J Clin Psychiatry 2008; 69:817–829. [DOI] [PubMed] [Google Scholar]
- 23.Nasrallah HA, Gopal S, Gassmann-Mayer C, Quiroz JA, Lim P, Eerdekens M, et al. A controlled, evidence-based trial of paliperidone palmitate, a long-acting injectable antipsychotic, in schizophrenia. Neuropsychopharmacology 2010; 35:2072–2082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pandina G, Lane R, Gopal S, Gassmann-Mayer C, Hough D, Remmerie B, et al. A double-blind study of paliperidone palmitate and risperidone long-acting injectable in adults with schizophrenia. Prog Neuropsychopharmacol Biol Psychiatry 2011; 35:218–226. [DOI] [PubMed] [Google Scholar]
- 25.Pandina GJ, Lindenmayer JP, Lull J, Lim P, Gopal S, Herben V, et al. A randomized, placebo-controlled study to assess the efficacy and safety of 3 doses of paliperidone palmitate in adults with acutely exacerbated schizophrenia. J Clin Psychopharmacol 2010; 30:235–244. [DOI] [PubMed] [Google Scholar]
- 26.Tzimos A, Samokhvalov V, Kramer M, Ford L, Gassmann-Mayer C, Lim P, et al. Safety and tolerability of oral paliperidone extended-release tablets in elderly patients with schizophrenia: a double-blind, placebo-controlled study with six-month open-label extension. Am J Geriatr Psychiatry 2008; 16:31–43. [DOI] [PubMed] [Google Scholar]
- 27.Wang D, Li Q, Favis R, Jadwin A, Chung H, Fu DJ, et al. SULT4A1 haplotype: conflicting results on its role as a biomarker of antipsychotic response. Pharmacogenomics 2014; 15:1557–1564. [DOI] [PubMed] [Google Scholar]
- 28.Libiger O, Schork NJ. A method for inferring an individual’s genetic ancestry and degree of admixture associated with six major continental populations. Front Genet 2012; 3:322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 2007; 81:559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Patterson N, Price AL, Reich D. Population structure and eigenanalysis. PLoS Genet 2006; 2:e190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet 2006; 38:904–909. [DOI] [PubMed] [Google Scholar]
- 32.Howie B, Fuchsberger C, Stephens M, Marchini J, Abecasis GR. Fast and accurate genotype imputation in genome-wide association studies through pre-phasing. Nat Genet 2012; 44:955–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Howie B, Marchini J, Stephens M. Genotype imputation with thousands of genomes. G3 (Bethesda) 2011; 1:457–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Howie BN, Donnelly P, Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet 2009; 5:e1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Marchini J, Howie B. Genotype imputation for genome-wide association studies. Nat Rev Genet 2010; 11:499–511. [DOI] [PubMed] [Google Scholar]
- 36.1000 Genomes Project Consortium, Abecasis GR, Altshuler D, Auton A, Brooks LD, Durbin RM, et al. A map of human genome variation from population-scale sequencing. Nature 2010; 467:1061–1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhou X, Stephens M. Genome-wide efficient mixed-model analysis for association studies. Nat Genet 2012; 44:821–824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee PH, O’Dushlaine C, Thomas B, Purcell SM. INRICH: interval-based enrichment analysis for genome-wide association studies. Bioinformatics 2012; 28:1797–1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.GTEx Consortium. The genotype-tissue expression (GTEx) project. Nat Genet 2013; 45:580–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ramasamy A, Trabzuni D, Guelfi S, Varghese V, Smith C, Walker R, et al. Genetic variability in the regulation of gene expression in ten regions of the human brain. Nat Neurosci 2014; 17:1418–1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Novak NM, Stein JL, Medland SE, Hibar DP, Thompson PM, Toga AW. EnigmaVis: online interactive visualization of genome-wide association studies of the Enhancing NeuroImaging Genetics through Meta-Analysis (ENIGMA) Consortium. Twin Res Hum Genet 2012; 15:414–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stein JL, Medland SE, Vasquez AA, Hibar DP, Senstad RE, Winkler AM, et al. Identification of common variants associated with human hippocampal and intracranial volumes. Nat Genet 2012; 44:552–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hibar DP, Stein JL, Ryles AB, Kohannim O, Jahanshad N, Medland SE, et al. Genome-wide association identifies genetic variants associated with lentiform nucleus volume in N=1345 young and elderly subjects. Brain Imaging Behav 2013; 7:102–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stein JL, Hua X, Morra JH, Lee S, Hibar DP, Ho AJ, et al. Genome-wide analysis reveals novel genes influencing temporal lobe structure with relevance to neurodegeneration in Alzheimer’s disease. NeuroImage 2010; 51:542–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stein JL, Hibar DP, Madsen SK, Khamis M, McMahon KL, de Zubicaray GI, et al. Discovery and replication of dopamine-related gene effects on caudate volume in young and elderly populations (N=1198) using genome-wide search. Mol Psychiatry 2011; 16:927–937. 881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hibar DP, Stein JL, Renteria ME, Arias-Vasquez A, Desrivieres S, Jahanshad N, et al. Common genetic variants influence human subcortical brain structures. Nature 2015; 520:224–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schizophrenia Working Group of the Psychiatric Genomics Consortium. Biological insights from 108 schizophrenia-associated genetic loci. Nature 2014; 511:421–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang JP, Robinson DG, Gallego JA, John M, Yu J, Addington J, et al. Association of a schizophrenia risk variant at the DRD2 locus with antipsychotic treatment response in first-episode psychosis. Schizophr Bull 2015; 41:1248–1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Llerena A, Berecz R, Penas-Lledo E, Suveges A, Farinas H. Pharmacogenetics of clinical response to risperidone. Pharmacogenomics 2013; 14:177–194. [DOI] [PubMed] [Google Scholar]
- 50.Van Erp TG, Hibar DP, Rasmussen JM, Glahn DC, Pearlson GD, Andreassen OA, et al. Subcortical brain volume abnormalities in 2028 individuals with schizophrenia and 2540 healthy controls via the ENIGMA consortium. Mol Psychiatry 2015; 21:547–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Buchsbaum MS, Shihabuddin L, Brickman AM, Miozzo R, Prikryl R, Shaw R, et al. Caudate and putamen volumes in good and poor outcome patients with schizophrenia. Schizophr Res 2003; 64:53–62. [DOI] [PubMed] [Google Scholar]
- 52.Molina V, Martin C, Ballesteros A, de Herrera AG, Hernandez-Tamames JA. Optimized voxel brain morphometry: association between brain volumes and the response to atypical antipsychotics. Eur Arch Psychiatry Clin Neurosci 2011; 261:407–416. [DOI] [PubMed] [Google Scholar]
- 53.Li M, Chen Z, Deng W, He Z, Wang Q, Jiang L, et al. Volume increases in putamen associated with positive symptom reduction in previously drug-naive schizophrenia after 6 weeks antipsychotic treatment. Psychol Med 2012; 42:1475–1483. [DOI] [PubMed] [Google Scholar]
- 54.Okugawa G, Nobuhara K, Takase K, Saito Y, Yoshimura M, Kinoshita T. Olanzapine increases grey and white matter volumes in the caudate nucleus of patients with schizophrenia. Neuropsychobiology 2007; 55:43–46. [DOI] [PubMed] [Google Scholar]
- 55.Hutcheson NL, Clark DG, Bolding MS, White DM, Lahti AC. Basal ganglia volume in unmedicated patients with schizophrenia is associated with treatment response to antipsychotic medication. Psychiatry Res 2014; 221:6–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Baumeister D, Ciufolini S, Mondelli V. Effects of psychotropic drugs on inflammation: consequence or mediator of therapeutic effects in psychiatric treatment? Psychopharmacology (Berl) 2015; 233:1575–1589. [DOI] [PubMed] [Google Scholar]
- 57.Khandaker GM, Cousins L, Deakin J, Lennox BR, Yolken R, Jones PB. Inflammation and immunity in schizophrenia: implications for pathophysiology and treatment. Lancet Psychiatry 2015; 2:258–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Felger JC, Lotrich FE. Inflammatory cytokines in depression: neurobiological mechanisms and therapeutic implications. Neuroscience 2013; 246:199–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Baumeister D, Russell A, Pariante CM, Mondelli V. Inflammatory biomarker profiles of mental disorders and their relation to clinical, social and lifestyle factors. Soc Psychiatry Psychiatr Epidemiol 2014; 49:841–849. [DOI] [PubMed] [Google Scholar]
- 60.Chen ML, Wu S, Tsai TC, Wang LK, Tsai FM. Regulation of macrophage immune responses by antipsychotic drugs. Immunopharmacol Immunotoxicol 2013; 35:573–580. [DOI] [PubMed] [Google Scholar]
- 61.Chen ML, Tsai TC, Wang LK, Lin YY, Tsai YM, Lee MC, et al. Risperidone modulates the cytokine and chemokine release of dendritic cells and induces TNF-alpha-directed cell apoptosis in neutrophils. Int Immunopharmacol 2012; 12:197–204. [DOI] [PubMed] [Google Scholar]
- 62.Cross-Disorder Group of the Psychiatric Genomics Consortium. Identification of risk loci with shared effects on five major psychiatric disorders: a genome-wide analysis. Lancet 2013; 381:1371–1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Schizophrenia Psychiatric Genome-Wide Association Study Consortium. Genome-wide association study identifies five new schizophrenia loci. Nat Genet 2011; 43:969–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental digital content is available for this article. Direct URL citations appear in the printed text and are provided in the HTML and PDF versions of this article on the journal's website (www.pharmacogeneticsandgenomics.com).


