Supplemental Digital Content is available in the text.
Keywords: antidepressants, desvenlafaxine, major depression, personalized medicine, pharmacogenetics, pharmacogenomics, precision medicine
Abstract
Background
Pharmacogenetic-based dosing support tools have been developed to personalize antidepressant-prescribing practice. However, the clinical validity of these tools has not been adequately tested, particularly for specific antidepressants.
Objective
To examine the concordance between the actual dose and a polygene pharmacogenetic predicted dose of desvenlafaxine needed to achieve symptom remission.
Materials and methods
A 10-week, open-label, prospective trial of desvenlafaxine among Caucasian adults with major depressive disorder (n=119) was conducted. Dose was clinically adjusted and at the completion of the trial, the clinical dose needed to achieve remission was compared with the predicted dose needed to achieve remission.
Results
Among remitters (n=95), there was a strong concordance (Kendall’s τ-b=0.84, P=0.0001; Cohen’s κ=0.82, P=0.0001) between the actual and the predicted dose need to achieve symptom remission, showing high sensitivity (≥85%), specificity (≥86%), and accuracy (≥89%) of the tool.
Conclusion
Findings provide initial evidence for the clinical validity of a polygene pharmacogenetic-based tool for desvenlafaxine dosing.
Introduction
The effectiveness of antidepressants and their dosing in practice is variable. This variability, in part, can be attributed to genetic polymorphisms that influence antidepressant bioavailability – phase I and II metabolism, and the active efflux at the blood–brain barrier 1. The most studied of these are the CYP2D6 and CYP2C19 genetic polymorphisms, which encode enzymes involved in phase I metabolism of most second-generation antidepressants, and commonly show functional variance between individuals 2,3. In fact, independent expert groups such as the Clinical Pharmacogenetics Implementation Consortium have developed dosing guidelines for serotonin selective reuptake inhibitors and tricyclic antidepressants exclusively on the basis of CYP2D6 and CYP2C19 genetic variation 4–6. These guidelines have contributed toward the ‘personalized psychiatry’ movement and have stimulated the development of several commercial pharmacogenetic-based decision support tools, all of which contain CYP2D6 and CYP2C19 to aid in the optimization of antidepressant prescribing practices 7. However, not all antidepressants are metabolized by CYP2D6 and CYP2C19, suggesting that pharmacogenetic-based decision support tools may need to include additional pharmacokinetic genes.
One such commercial pharmacogenetic-based decision support tool is CNSDose. In addition to genetic variation in CYP2D6 and CYP2C19, CNSDose also measures genetic variation in the UGT1A1 (UDP-glucuronosyltransferase 1A1) gene that encodes a phase II metabolism enzyme as well as two ATP-binding cassette (ABC) genes (ABCB1 and ABCC1) that encode efflux transporters that restrict permeability of drugs at the blood–brain barrier 8. These additional genes are particularly relevant to the pharmacokinetics of desvenlafaxine, the active metabolite of venlafaxine and a serotonin norepinephrine reuptake inhibitor. Desvenlafaxine is not subject to CYP450 metabolism 9, but is subject to UGT1A1 metabolism 10, and genetic variation in UGT1A1’s promoter region has been shown to affect its function 11. Furthermore, there is some evidence suggesting that the ABCB1 (also known as P-glycoprotein) efflux transporter moderates desvenlafaxine concentrations in the brain 12, and functional genetic variants in both ABCB1 and ABCC1 (also known as multidrug resistance-associated protein 1, MRP1) have been associated with antidepressants’ efficacy 13–21.
The clinical utility of CNSDose was examined recently in a 12-week double-blind randomized clinical trial. Individuals diagnosed with major depressive disorder (MDD) who received CNSDose-guided prescribing were 2.5 times more likely to achieve symptom remission compared with those receiving unguided prescribing 22. However, a limited proportion (6%) of participants in that trial were prescribed desvenlafaxine and as such the usefulness of the CNSDose tool for guiding desvenlafaxine dosing is unclear. Therefore, we carried out a 10-week, open-label, prospective cohort study of desvenlafaxine in MDD and compared the CNSDose predicted dose with the actual dose required for symptom remission to estimate the clinical validity and performance of the CNSDose for guiding desvenlafaxine dosing.
Materials and methods
Participants
Participants were antidepressant-naive, self-identified Caucasian outpatients aged 18 years and older with a principal Diagnostic and Statistical Manual of Mental Disorders, 5th ed. (DSM-5) diagnosis of MDD (semistructured psychiatrist assessment) and a 17-item Hamilton Depression Rating Scale (HDRS) score greater than or equal to 18. Participants with a history of childhood trauma or active psychiatric diagnoses other than MDD were excluded, specifically those with anxiety disorder, adjustment disorder with depressed mood, persistent depressive disorder, and patients with a principal clinical diagnosis of a personality disorder. Additional exclusion criteria included pregnancy or breastfeeding, hepatic or renal impairments, coprescription of commonly prescribed UGT1A1 or ABCB1 inducers/inhibitors in the mood disorder care setting (i.e. valproate, carbamazepine, and lamotrigine) as well as St Johns wort, regular grapefruit juice consumption, and current smoking as these may influence appropriate dosing 23–27. Participants were allowed to have hypnotics (i.e. temazepam or zolpidem CR), but no other psychotropic medications were permitted. A total of 131 individuals were screened for eligibility criteria. Seven individuals did not fulfill the inclusion/exclusion criteria and an additional five failed to return for inclusion in the study, resulting in a final study sample of 119 participants.
Study procedures
All participants received desvenlafaxine in an open-label manner during the 10-week study period. At baseline, age, sex, duration of the current depressive episode, and number of depressive episodes was recorded. Desvenlafaxine dose was increased, decreased, or left unchanged every 2 weeks (from baseline) on the basis of subjectively reported tolerability and clinical assessment of symptom improvement. Dose increases were limited to 50 mg increments every 2 weeks to mitigate potential adverse events (e.g. orthostatic hypotension) and the dosing range followed Australian pharmaceutical prescribing recommendations (50–200 mg/day) 28. Symptom severity was assessed with the HDRS at baseline and weeks 2, 4, 6, 8, and 10 postbaseline. Remission was defined as an HDRS score of 7 or less 29 by week 10 of the study. Physicians and the symptom rater were blinded to genotypes. All participants provided written informed consent and procedures were in accordance with the Declaration of Helsinki and were approved by an ethics committee at Deakin University, Australia.
Pharmacogenetic interpretive report
A commercially available pharmacogenetic interpretive report (CNSDose; Baycrest Biotechnology Pty Ltd, Melbourne, Victoria, Australia) was ordered at the conclusion of the trial (week 10) for each participant using a proprietary algorithm described previously 22 (Fig. 1). The interpretive report predicted each participant’s optimal desvenlafaxine dose range as low (≤50 mg), medium (>50 and <150 mg), or high (≥150 mg) on the basis of genetic variation in ABCB1 (rs1045642), ABCC1 (rs212090), CYP2C19, CYP2D6, and UGT1A1 (rs8175347), albeit for this study, CYP2C19 and CYP2D6 genetic information was not used because of its lack of relevance to desvenlafaxine pharmacokinetics. DNA was extracted from participant self-administered buccal brush samples using the QIAamp DNA Mini Kit (QIAGEN Inc., Chadstone, Victoria, Australia). Genotyping was performed by PCR, followed by single primer extension and analysis on a Sequenom Matrix-assisted laser desorption/ionization time-of-flight mass spectrometry 384-well genetic analysis system by Healthscope Molecular (Clayton, Victoria, Australia).
Fig. 1.
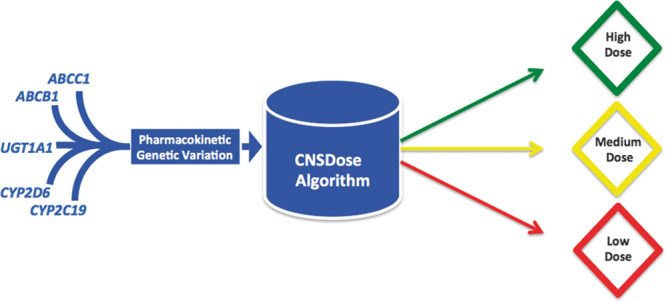
Overview of the CNSDose dosing support tool. Dosing predictions are derived from genetic variants in ABCC1, ABCB1, UGT1A1, CYP2D6, and CYP2C19 by a pharmacogenetic evidence-based algorithm. Clinical information is not included in the algorithm.
Analysis
Among remitters, performance of the CNSDose tool was estimated by comparing the predicted desvenlafaxine dosing range derived from the interpretive report with the actual desvenlafaxine dose required to achieve symptom remission. Concordance between received and predicted dose was estimated using two approaches: (a) the nonparametric Kendall’s τ-b (Tb) correlation coefficient was used to compare the actual dose in milligrams with the dose range predicted by CNSDose and (b) the Cohen’s κ was used to compare the actual dose range with the CNSDose predicted dose range. Sensitivity, specificity, false positive, and false negative rates, as well as accuracy of the CNSDose predicted dose range relative to the actual dose range were also calculated. In addition, individual genes/variants comprising the CNSDose tool were compared with the actual dose required for remission to determine whether any one gene/variant performed better than the CNSDose tool. Among nonremitters who showed a 50% reduction in the HDRS score from baseline, exploratory analyses were carried out using the same analytical methods as those used in the remitted sample.
Results
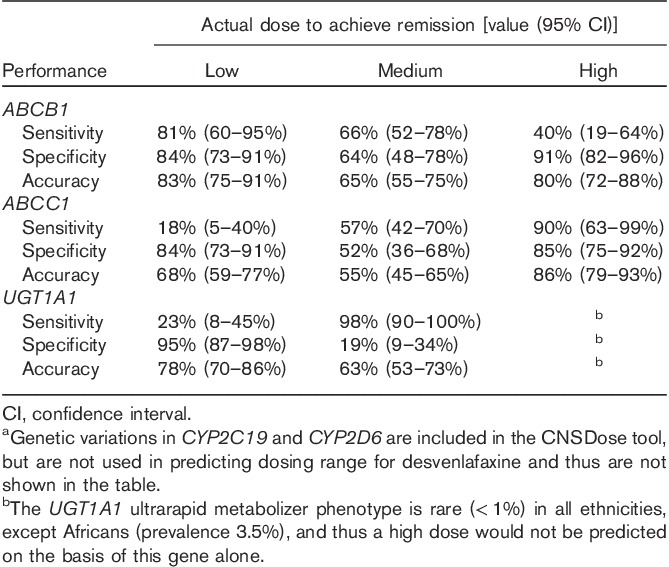
After 10 weeks of desvenlafaxine treatment, 80% (n=95) of participants achieved symptom remission (Table 1). The average time to remission was 8.4 (SD=1.4) weeks. Those predicted to required a high dose had significant longer times (mean=9.3, SD=1.0 weeks) to remission compared with those in the low (mean=8.1, SD=1.3 weeks; Bonferroni’s P=0.01) and medium (mean=8.2, SD=1.4 weeks; Bonferroni’s P=0.006) predicted dose groups. Of the 95 participants who achieved symptom remission, 22 (23%) received a low dose; 53 (56%) received a medium dose; and 20 (21%) received a high dose. The CNSDose tool predicted that 22 (23%) required a low dose, 55 (58%) required a medium dose, and 18 (19%) required a high dose to achieve remission. Comparison of the actual and CNSDose predicted doses required for remission indicated strong concordance (Tb=0.84, P=0.0001; κ=0.82, P=0.0001) (Fig. 2). In addition, the CNSDose predicted dose showed high sensitivity (85–92%), specificity (86–92%), and accuracy (89–96%) relative to the actual dose required for symptom remission (Table 2). Examination of the individual genes/variants included in CNSDose showed moderate concordance between the actual dose and predicted dose for ABCB1 (Tb=0.54, P=0.0001; κ=0.40, P=0.0001) and ABCC1 (Tb=0.48, P=0.0001; κ=0.25, P=0.001), but weak concordance for UGT1A1 (Tb=0.14, P=0.149; κ=0.15, P=0.03). Sensitivity and specificity of each individual gene/variant were more variable and accuracy estimates were lower than observed for CNSDose (Table 3).
Table 1.
Participant characteristics by remission status and actual dose to achieve remission
Fig. 2.
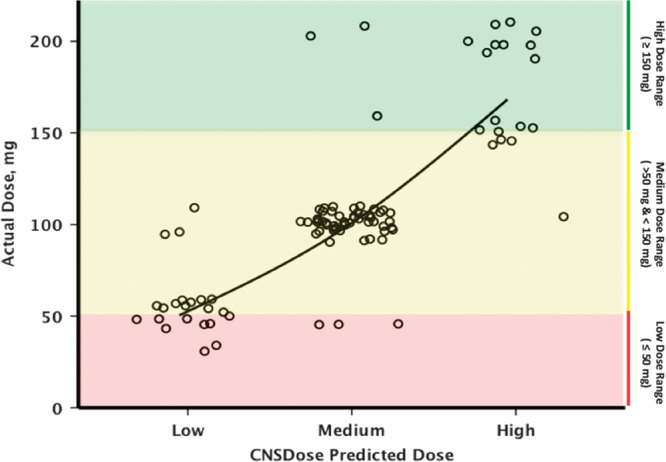
Concordance between actual desvenlafaxine dose and CNSDose predicted desvenlafaxine dose required for symptom remission. Each point represents a patient who achieved symptom remission.
Table 2.
CNSDose performance in predicting required desvenlafaxine dose needed to achieve remission among 95 major depressive disorder remitters
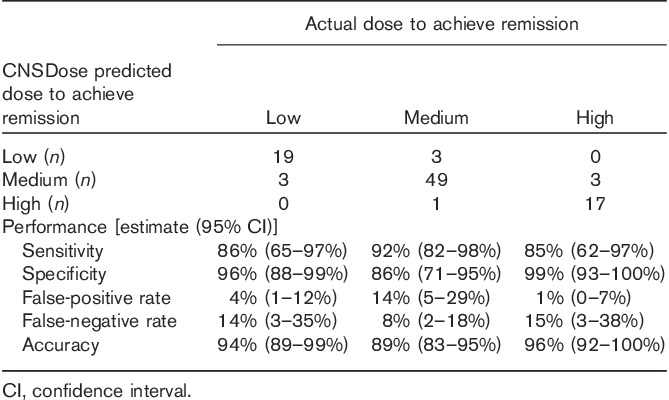
Table 3.
Individual genea performance in predicting required desvenlafaxine dose needed to achieve remission among 95 major depressive disorder remitters
Among the 24 participants who did not achieve symptom remission by week 10, 42% (n=10) had a greater than 50% reduction in HDRS from the baseline. Among these nonremitted responders, two (20%) received a low dose, six (60%) received a medium dose, and two (20%) received a high dose. The CNSDose tool predicted that three (30%) patients would require a low dose, five (50%) patients would require a medium dose, and two (20%) patients would require a high dose. Similar to the remitter analysis, comparison of the actual and CNSDose predicted doses required for response indicated strong concordance (Tb=0.87, P=0.004; κ=0.83, P=0.0001; Supplementary Fig. S1, Supplemental digital content 1, http://links.lww.com/FPC/B103). However, among the nonresponders (n=14), concordance was only moderate (Tb=0.86, P=0.005; κ=0.39, P=0.006), although all nonresponders were prescribed the CNSDose predicted dose or a higher dose by week 10 (Supplementary Fig. S2, Supplemental digital content 2, http://links.lww.com/FPC/B104). Performance estimates (i.e. sensitivity, specificity, and accuracy) were not calculated within the nonremitted responder and nonresponder samples because of concerns of the reliability of such estimates, given the extremely small sample sizes 30.
Discussion
Our results tentatively suggest that the CNSDose tool may have clinical utility in guiding desvenlafaxine dosing in a subset of individuals with moderate to severe depressive symptoms. We found that clinically driven (unguided by CNSDose) dosing of desvenlafaxine needed, on average, 8 weeks to find the dose required for remission. Importantly, the CNSDose predicted dose had high concordance with the actual dose required for remission, suggesting that the use of CNSDose at the commencement of desvenlafaxine treatment has the potential to shorten the time to remission, particularly among patients requiring a high dose (≥150 mg). To our knowledge, no other genetically based desvenlafaxine dosing tools have been reported in the literature. However, genetic-based dosing tools for drugs other than antidepressants such as warfarin have reported comparable concordance between actual and predicted dose (Pearson’s r=0.54–0.67) 31.
Our results, in part, also support findings from a double-blinded, randomized clinical trial that showed that the CNSDose tool improved MDD outcomes among individuals prescribed a variety of first-generation and second-generation antidepressant pharmacotherapy, although few received desvenlafaxine 22. As noted above, desvenlafaxine is not subject to phase I CYP450 metabolism 9 and the evidence supporting ABCB1 and ABCC1 as regulators of desvenlafaxine concentrations in the brain is modest. Thus, two of the genes (CYP2D6 and CYP2C19) included in the CNSDose tool are not used to predict desvenlafaxine dosing and the relevance of ABCB1 and ABCC1 is uncertain because of conflicting results in the literature. Therefore, the underlying mechanism(s) by which CNSDose confers its predictive value would presumably involve the phase II hepatic UGT1A1 gene. However, our results suggest that UGT1A1 on its own has limited ability to predict the actual dose needed to achieve remission, suggesting that the predictive value of CNSDose requires a combinatorial approach. This notion is supported by a previous work by Assurex Health (Mason, Ohio, USA), developers of the GeneSight test, that showed that a combinatorial pharmacogenetic approach had superior predictive value compared with a single-gene approach 32, albeit single genes/variants not tested to date may prove to have stronger predictive value for particular drugs in particular settings.
Interestingly, UGT1A1, ABCB1, and ABCC1 are under-represented in the antidepressant pharmacogenetic literature 1 and are typically not included in commercially available pharmacogenetic gene panels. In fact, of the 22 commercially available pharmacogenetic tools relevant to psychiatry, UGT1A1 is included on two (CNSDose and PGxOne; Admera Health, South Plainfield, New Jersey, USA), ABCB1 on three (CNSDose; PGxPredict, Transgenomic, Omaha, Nebraska, USA; and HMNC Brain Health, Munich, Germany), and ABCC1 on one (CNSDose) pharmacogenetic gene panel 7. Arguably, the exclusion of these genes in previous antidepressant pharmacogenetic studies may, in part, influence the mixed findings in the literature to date. Further, commercial pharmacogenetic gene panels including these genes may be more clinical applicable, particularly for clinicians who prescribe desvenlafaxine.
The current study does have some notable limitations. The exclusion of patients with current or previous exposure to antidepressants, a history of childhood trauma and comorbidities, particularly personality disorders with dysthymia and adjustment disorder with depressed mood, may limit the application of these findings to larger real-world clinical settings – settings where comorbidity is very common. This is supported by a response and remission rate that was considerably higher than that observed in most antidepressant trials. In addition to these exclusion criteria, the high response and remission rate could, in part, be attributed to the use of doses up to 150 mg above the recommended effective dose (i.e. 50 mg) 33. In addition, dose adjustments were based on clinical judgment rather than specific criteria, which may hamper the reproducibility of our findings. Our findings are also limited to Caucasians of a relatively older and more chronic population than may be seen in other settings. Furthermore, our trial used an open-label design and as such study participants were not blinded to the dosage adjustments, which may have influenced the symptom rating. Thus, generalization of our findings should be performed with caution. It should also be noted that only a small selection of the known polymorphisms in ABCB1, ABCC1, and UGT1A1 were assessed. It is likely that other polymorphisms in these genes as well as other unexamined genes are relevant to desvenlafaxine pharmacokinetics. In fact, several ABCB1 polymorphisms have been linked to antidepressant efficacy 19 and four other UGTs (UGT1A3, UGT2B4, UGT2B15, and UGT2B17) have been implicated in the metabolism of desvenlafaxine, with genetic variation in UGT1A3 and UGT2B17 linked to the mRNA expression of these genes 34. In fact, one in every 10 of our participants did not respond to desvenlafaxine despite being prescribed the CNSDose predicted dose or higher dose. This may suggest that genetic variation in the above-mentioned genes may improve dose prediction or could indicate that nonresponse was a result of nongenetic factors such as adherence or tolerability. Unfortunately, measurements of treatment adherence and tolerability as well as desvenlafaxine blood levels were not available. Although typical adverse effects reported included diaphoresis, constipation, light-headedness, and agitation, no severe adverse reactions occurred. Furthermore, the CNSDose tool, unlike other currently available tools 7, does not include genes associated with the pharmacodynamics of antidepressants, which raises the question of whether the CNSDose dosing support tool represents a significant improvement over other currently available tools. Addressing this issue was beyond the scope of the current study, but future head-to-head trials with other tools are warranted. Importantly, personal (e.g. age, sex) and environmental factors (e.g. abuse history) were not included in the CNSDose dosing tool. Given the known role that personal and environmental factors play in antidepressant response, inclusion of such factors may further improve the performance of the tool 35.
Conclusion
Our results serve as initial evidence for the clinical validity of CNSDose for the dosing of desvenlafaxine and, pending replication, suggest potential clinical utility. However, future pharmacogenetic-based dosing support tool development and evaluation that address the limitations of this study are warranted and are necessary before universal adoption into clinical practice.
Supplementary Material
Supplemental digital content is available for this article. Direct URL citations appear in the printed text and are provided in the HTML and PDF versions of this article on the journal's website (www.pharmacogeneticsandgenomics.com).
Acknowledgements
This study was supported in part by a 2012 Pfizer Australia NSR grant (A.S.). C.B. was supported by a University of Melbourne, Ronald Phillip Griffith Fellowship. M.B. is supported by an NHMRC Senior Principal Research Fellowship (1059660).
Authors’ contribution: A.S. designed the study, K.B. performed genotyping, and C.B. analyzed the data and wrote the first draft. D.M., C.N., K.B., M.B., and A.S. revised the first draft. All authors approved the final draft of the manuscript.
Conflicts of interest
A.S. owns shares in the ABC Life Pty Ltd and Baycrest Technology Pty Ltd (developer of the CNSDose tool), and is on the speakers bureau for Servier, Australia. For the remaining authors there are no conflicts of interest.
References
- 1.Singh AB, Bousman CA, Ng C, Berk M. Antidepressant pharmacogenetics. Curr Opin Psychiatry 2014; 27:43–51. [DOI] [PubMed] [Google Scholar]
- 2.Kirchheiner J, Nickchen K, Bauer M, Wong ML, Licinio J, Roots I, Brockmöller J. Pharmacogenetics of antidepressants and antipsychotics: the contribution of allelic variations to the phenotype of drug response. Mol Psychiatry 2004; 9:442–473. [DOI] [PubMed] [Google Scholar]
- 3.Muller DJ, Kekin I, Kao AC, Brandl EJ. Towards the implementation of CYP2D6 and CYP2C19 genotypes in clinical practice: update and report from a pharmacogenetic service clinic. Int Rev Psychiatry 2013; 25:554–571. [DOI] [PubMed] [Google Scholar]
- 4.Hicks JK, Bishop JR, Sangkuhl K, Müller DJ, Ji Y, Leckband SG, et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) Guideline for CYP2D6 and CYP2C19 genotypes and dosing of selective serotonin reuptake inhibitors. Clin Pharmacol Ther 2015; 98:127–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Caudle KE, Klein TE, Hoffman JM, Muller DJ, Whirl-Carrillo M, Gong L, et al. Incorporation of pharmacogenomics into routine clinical practice: the Clinical Pharmacogenetics Implementation Consortium (CPIC) guideline development process. Curr Drug Metab 2014; 15:209–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hicks JK, Swen JJ, Thorn CF, Sangkuhl K, Kharasch ED, Ellingrod VL, et al. Clinical Pharmacogenetics Implementation Consortium guideline for CYP2D6 and CYP2C19 genotypes and dosing of tricyclic antidepressants. Clin Pharmacol Ther 2013; 93:402–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bousman CA, Hopwood M. Commercial pharmacogenetic-based decision-support tools in psychiatry. Lancet Psychiatry 2016; 3:585–590. [DOI] [PubMed] [Google Scholar]
- 8.Fromm MF. Importance of P-glycoprotein at blood-tissue barriers. Trends Pharmacol Sci 2004; 25:423–429. [DOI] [PubMed] [Google Scholar]
- 9.Preskorn S, Patroneva A, Silman H, Jiang Q, Isler JA, Burczynski ME, et al. Comparison of the pharmacokinetics of venlafaxine extended release and desvenlafaxine in extensive and poor cytochrome P450 2D6 metabolizers. J Clin Psychopharmacol 2009; 29:39–43. [DOI] [PubMed] [Google Scholar]
- 10.Baird-Bellaire S, Behrle JA, Parker VD, Patat A, Paul J, Nichols AI. An open-label, single-dose, parallel-group study of the effects of chronic hepatic impairment on the safety and pharmacokinetics of desvenlafaxine. Clin Ther 2013; 35:782–794. [DOI] [PubMed] [Google Scholar]
- 11.Shin HJ, Kim JY, Cheong HS, Na HS, Shin HD, Chung MW. Functional study of haplotypes in UGT1A1 promoter to find a novel genetic variant leading to reduced gene expression. Ther Drug Monit 2015; 37:369–374. [DOI] [PubMed] [Google Scholar]
- 12.Karlsson L, Schmitt U, Josefsson M, Carlsson B, Ahlner J, Bengtsson F, et al. Blood–brain barrier penetration of the enantiomers of venlafaxine and its metabolites in mice lacking P-glycoprotein. Eur Neuropsychopharmacol 2010; 20:632–640. [DOI] [PubMed] [Google Scholar]
- 13.Kato M, Fukuda T, Serretti A, Wakeno M, Okugawa G, Ikenaga Y, et al. ABCB1 (MDR1) gene polymorphisms are associated with the clinical response to paroxetine in patients with major depressive disorder. Prog Neuropsychopharmacol Biol Psychiatry 2008; 32:398–404. [DOI] [PubMed] [Google Scholar]
- 14.Uhr M, Tontsch A, Namendorf C, Ripke S, Lucae S, Ising M, et al. Polymorphisms in the drug transporter gene ABCB1 predict antidepressant treatment response in depression. Neuron 2008; 57:203–209. [DOI] [PubMed] [Google Scholar]
- 15.Lin KM, Chiu YF, Tsai IJ, Chen CH, Shen WW, Liu SC, et al. ABCB1 gene polymorphisms are associated with the severity of major depressive disorder and its response to escitalopram treatment. Pharmacogenet Genomics 2011; 21:163–170. [DOI] [PubMed] [Google Scholar]
- 16.Singh AB, Bousman CA, Ng CH, Byron K, Berk M. ABCB1 polymorphism predicts escitalopram dose needed for remission in major depression. Transl Psychiatry 2012; 2:e198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee SH, Lee MS, Lee JH, Kim SW, Kang RH, Choi MJ, et al. MRP1 polymorphisms associated with citalopram response in patients with major depression. J Clin Psychopharmacol 2010; 30:116–125. [DOI] [PubMed] [Google Scholar]
- 18.Breitenstein B, Scheuer S, Bruckl TM, Meyer J, Ising M, Uhr M, Holsboer F. Association of ABCB1 gene variants, plasma antidepressant concentration, and treatment response: results from a randomized clinical study. J Psychiatr Res 2015; 73:86–95. [DOI] [PubMed] [Google Scholar]
- 19.Breitenstein B, Bruckl TM, Ising B, Müller-Myhsok M, Holsboer F, Czamara D. ABCB1 gene variants and antidepressant treatment outcome: a meta-analysis. Am J Med Genet B Neuropsychiatr Genet 2015; 168B:274–283. [DOI] [PubMed] [Google Scholar]
- 20.Jelen AM, Salagacka A, Zebrowska MK, Mirowski M, Talarowska M, Gałecki P, Balcerczak EI. The influence of C3435T polymorphism of the ABCB1 gene on genetic susceptibility to depression and treatment response in Polish population – preliminary report. Int J Med Sci 2015; 12:974–979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chang HH, Chou CH, Yang YK, Lee IH, Chen PO. Association between ABCB1 polymorphisms and antidepressant treatment response in Taiwanese major depressive patients. Clin Psychopharmacol Neurosci 2015; 13:250–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Singh AB. Improved antidepressant remission in major depression via a pharmacokinetic pathway polygene pharmacogenetic report. Clin Psychopharmacol Neurosci 2015; 13:150–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bushra R, Aslam N, Khan AY. Food-drug interactions. Oman Med J 2011; 26:77–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tsai HH, Lin HW, Simon Pickard A, Tsai HY, Mahady GB. Evaluation of documented drug interactions and contraindications associated with herbs and dietary supplements: a systematic literature review. Int J Clin Pract 2012; 66:1056–1078. [DOI] [PubMed] [Google Scholar]
- 25.Akamine Y, Yasui-Furukori N, Ieiri I, Uno T. Psychotropic drug-drug interactions involving P-glycoprotein. CNS Drugs 2012; 26:959–973. [DOI] [PubMed] [Google Scholar]
- 26.Tod M, Nkoud-Mongo C, Gueyffier F. Impact of genetic polymorphism on drug–drug interactions mediated by cytochromes: a general approach. AAPS J 2013; 15:1242–1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Weiss J, Kerpen CJ, Lindenmaier H, Dormann SM, Haefeli WE. Interaction of antiepileptic drugs with human P-glycoprotein in vitro. J Pharmacol Exp Ther 2003; 307:262–267. [DOI] [PubMed] [Google Scholar]
- 28.MIMS Australia. 2016. Desvenlafaxine. Available at: http://www.mims.com.au. [Accessed 2 August 2016].
- 29.Riedel M, Moller HJ, Obermeier M, Schennach-Wolff R, Bauer M, Adi M, et al. Response and remission criteria in major depression – a validation of current practice. J Psychiatr Res 2010; 44:1063–1068. [DOI] [PubMed] [Google Scholar]
- 30.Hajian-Tilaki K. Sample size estimation in diagnostic test studies of biomedical informatics. J Biomed Inform 2014; 48:193–204. [DOI] [PubMed] [Google Scholar]
- 31.Langley MR, Booker JK, Evans JP, McLeod HL, Weck KE. Validation of clinical testing for warfarin sensitivity: comparison of CYP2C9-VKORC1 genotyping assays and warfarin-dosing algorithms. J Mol Diagn 2009; 11:216–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Altar CA, Carhart JM, Allen JD, Hall-Flavin D, Dechairo BM, Winner JG. Clinical validity: combinatorial pharmacogenomics predicts antidepressant responses and healthcare utilizations better than single gene phenotypes. Pharmacogenomics J 2015; 15:443–451. [DOI] [PubMed] [Google Scholar]
- 33.DeMartinis NA, Yeung PP, Entsuah R, Manley AL. A double-blind, placebo-controlled study of the efficacy and safety of desvenlafaxine succinate in the treatment of major depressive disorder. J Clin Psychiatry 2007; 68:677–688. [DOI] [PubMed] [Google Scholar]
- 34.Liu W, Ramirez J, Gamazon ER, Mirkov S, Chen P, Wu K, et al. Genetic factors affecting gene transcription and catalytic activity of UDP-glucuronosyltransferases in human liver. Hum Mol Genet 2014; 23:5558–5569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dodd S, Berk M. Predictors of antidepressant response: a selective review. Int J Psychiatry Clin Pract 2004; 8:91–100. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental digital content is available for this article. Direct URL citations appear in the printed text and are provided in the HTML and PDF versions of this article on the journal's website (www.pharmacogeneticsandgenomics.com).


