Abstract
To model how interactions among enteropathogens and gut microbial community members contribute to undernutrition, we colonized gnotobiotic mice fed representative Bangladeshi diets with sequenced bacterial strains cultured from the fecal microbiota of two 24-month-old Bangladeshi children: one healthy, the other underweight. The undernourished donor’s bacterial collection contained an enterotoxigenic Bacteroides fragilis strain (ETBF), whereas the healthy donor’s bacterial collection contained two nontoxigenic strains of B. fragilis (NTBF). Analyses of mice harboring either the unmanipulated culture collections or systematically manipulated versions revealed that ETBF was causally related to weight loss in the context of its native community, but not when introduced into the healthy donor’s community. This phenotype was transmissible from the dams their offspring and was associated with derangements in host energy metabolism manifest by impaired tricarboxylic acid cycle activity and decreased acyl-CoA utilization. NTBF reduced ETBF’s expression of its enterotoxin and mitigated the effects of ETBF on the transcriptomes of other healthy donor community members. These results illustrate how intraspecific (ETBF-NTBF) and interspecific interactions impact the effects of harboring B. fragilis..
INTRODUCTION
Undernutrition is the leading cause of childhood mortality worldwide; it is not due to food insecurity alone, but rather reflects a complex and incompletely understood set of interactions involving a variety of factors that operate within and across generations (1–4). Among these factors is the gut microbiota. A recent culture-independent study of fecal microbiota samples collected monthly from members of a Bangladeshi birth cohort with healthy growth phenotypes identified age-associated changes in the representation of bacterial species during the first two years of postnatal life. These age-discriminatory species together define a developmental program for the microbiota that is shared across biologically unrelated Bangladeshi infants and children (5). Children with moderate or severe undernutrition exhibit disruptions in this program resulting in microbial communities that appear younger than those of chronologically age-matched healthy individuals (5, 6). Clinical studies have provided evidence that microbiota immaturity in children with severe acute undernutrition is not repaired when they are fed current therapeutic foods (5). Transplantation of fecal microbiota from children with healthy growth phenotypes, or immature microbiota from chronologically-aged matched undernourished children, into germ-free mice fed diets similar to those consumed by the human donors has provided preclinical evidence for a causal role of the gut community in disease pathogenesis. Young mouse recipients of undernourished donor microbiota exhibit growth faltering, manifest in part by a reduced rate of lean body mass gain compared to recipients of healthy donor microbiota. These differences are not associated with differences in food consumption (6). Adult mouse recipients of undernourished donor microbiota exhibit a weight loss (wasting) phenotype (7, 8).
A large enteropathogen burden is associated with childhood undernutrition (9, 10). However, the effects of microbiota configuration on enteropathogen invasion and burden, or how community context influences the expressed properties of organisms classified as enteropathogens in children at risk for or already showing undernutrition, remain poorly understood. Identifying gut community configurations that are resistant to invasion or that accommodate organisms currently classified as enteropathogens without supporting their ability to produce deleterious host effects has potential diagnostic and therapeutic implications.
Bacteroides fragilis provides a model for examining how community context, including intraspecific interactions involving different strains of a given species as well as interspecific interactions involving different species, impacts the properties of an enteropathogen and its effects on the host. Enterotoxigenic B. fragilis (ETBF) strains contain one of three alleles of B. fragilis toxin (bft-1, bft-2, and bft-3) in their 6 kb pathogenicity island (BfPAI). bft encodes a zinc-dependent metalloprotease, fragilysin, that perturbs gut barrier function through cleavage of E-cadherin at epithelial cell adherence junctions (11). ETBF strains cause diarrhea in children residing in Bangladesh (12, 13). Nontoxigenic B. fragilis strains (NTBF) lack bft. Thus, B. fragilis can be viewed as a potentially pathogenic symbiont (pathobiont) based on the presence or absence of this gene, or more broadly, based on its community context (i.e., its compositional and functional configuration).
Here, we describe a gnotobiotic mouse model for analyzing how intraspecific and interspecific interactions determine the effects of ETBF. We started with fecal samples collected from Bangladeshi children who were participants in a previously completed birth-cohort study (14). Using anthropometric scores to define healthy growth versus stunting and wasting (15), and a PCR-based assay that targeted bft, we selected two chronologically age-matched individuals: one markedly stunted, underweight, and ETBF-positive; and the other with a healthy growth phenotype who was ETBF-negative but NTBF-positive. We transplanted intact uncultured fecal microbiota samples, collected from these two children at 24 months of age, into adult germ-free mice. These gnotobiotic mice were fed cooked diets containing ingredients embodying those consumed by the population from which the microbiota donors were selected. Finding that the severely stunted and underweight but not the healthy donor’s microbiota transmitted a weight loss phenotype to recipient animals and subsequently to their offspring, we performed follow-up transplant studies using clonally arrayed collections of sequenced anaerobic bacterial strains cultured from the donors’ fecal samples. These culture collections allowed us to dissect microbe-microbe and host-microbe interactions by testing the effects of several types of manipulations: (i) removing the ETBF strain from the stunted donor’s cultured community, (ii) introducing the ETBF strain into the healthy donor’s culture collection together with or in lieu of its own NTBF strains, and (iii) introducing the NTBF strain into the stunted donor’s culture collection together with or in lieu of its own ETBF strain. Microbiota community structure and gene expression, microbial and host metabolism, and host weight and immune phenotypes were characterized as a function of these alterations.
RESULTS
Uncultured fecal gut microbiota from an underweight donor confers weight loss on gnotobiotic mice
We used anthropometric data collected from members of a birth cohort study (14) of 100 children living in Mirpur thana in Dhaka, Bangladesh, to define whether they were healthy or undernourished (Table S1). Those with height-for-age z-scores (HAZ) greater than or equal to -2 were classified as ‘healthy’ while those with scores less than or equal to -3 were deemed severely stunted. At 18 months, 30 and 25 children satisfied these criteria for healthy versus severely stunted, respectively, while at 24 months, 27 and 20 received these designations; the remaining children were classified as moderately stunted (HAZ between -2 and -3). A PCR-based screen for ETBF targeting all three fragilysin gene subtypes (14) was performed using DNA isolated from fecal samples that had been collected from these children at 18 months and 24 months of age. The results revealed that ETBF was variably present across individuals and within a given individual over time, with a total of 25% of 18-month old and 14% of 24-month old children having a positive test (Table S1). In this small cohort, ETBF carriage was not significantly correlated with indices of linear or ponderal growth [HAZ, weight-for-age z-score (WAZ), and weight-for-height z-score (WHZ) measured at 12 and 24 months of age (p=0.8 and p=0.4; p=0.7 and p=0.2; p=0.5 and p=0.2, respectively; two-tailed Student’s t-test)]. We combined anthropometric and PCR data to select fecal samples collected at 24 months from two children: (i) a healthy individual (Child ID 7114 in Table S1) with an HAZ score of −0.71, a WAZ score of −1.49, and a WHZ score of −1.62 who was ETBF-negative at the two time points tested, and (ii) a severely stunted and moderately underweight individual (Child ID 7004) with a HAZ score of −3.02, a WAZ score of −2.51, and a WHZ score of −1.34 who was ETBF-positive at both time points. Of the 35 individuals with a positive ETBF test at either time point, only this stunted/underweight child was positive at both 18 and 24 months of age. Fecal samples obtained from members of this singleton birth cohort were screened for parasites using microscopic methods (5); neither of the two donors tested positive (see Materials and Methods for details).
To define the effects of diet and these two donors’ gut microbiota on host biology, we generated three representative versions (embodiments) of the diets consumed by the population represented by the donors. To do so, we determined the relative daily caloric contributions of various selected ingredient types, based on a study by Arsenault and coworkers (16). Selection of specific food items as representative of each ingredient type was based on consumption incidence surveys tabulated by Islam et al. (17) and incorporated into a database consisting of 54 food ingredients. We filtered this database to remove items consumed by <20% of households and categorized each of the remaining 39 items (see Materials and Methods for additional details). From the resulting diet ingredient matrix, we randomly sampled (without replacement) one item each from cereals, pulse vegetables, roots/tubers, leafy vegetables, fruits, and fish, plus three non-leafy vegetables, to populate three separate diet lists. Using the USDA National Nutrient Database for Standard References (18), we determined the caloric information for each ingredient and subsequently calculated proportions required to match the pre-determined contributions of each ingredient type. Food items were cooked in a manner intended to simulate Bangladeshi practices, and the resulting three embodiments of a Bangladeshi diet were sterilized by irradiation. This approach allowed us to generate several representative Bangladeshi diets that were not dominated by the idiosyncrasies of a single individual’s diet or by our own biases. The composition and results of nutritional analysis of the three diet embodiments are described in Table S2A,B. The nutritional requirements of mice and children are compared in Table S2C.
The results of a 12-year survey of demographic variations in the nutritional status of 16,278 Bangladeshi children found no significant sex differences in WHZ, WAZ or HAZ scores (19). Therefore, in these and subsequent experiments, we eliminated gender as an experimental variable and only studied male mice. We gavaged separate groups of 8–9 week old germ-free C57BL/6 mice with the intact uncultured fecal microbiota samples obtained from the healthy or stunted/underweight Bangladeshi donors (2 independent experiments; n=4 singly-caged mice/donor microbiota/experiment; see Figure S1A for study design). Fecal microbiota transplantation occurred 2 days after mice had been switched from an irradiated, nutritionally complete, low-fat/high plant polysaccharide (LF/HPP) mouse chow that they had received since weaning to the first of the three embodiments of the Bangladeshi diet. Animals were subsequently fed, ad libitum, embodiment 1 for one week, followed by embodiment 2 for one week, and finally embodiment 3 for one week, with frequent sampling of their fecal microbiota during the course of each diet. Sequencing PCR amplicons generated from variable region 4 (V4) of bacterial 16S rRNA genes present in the donor fecal sample and in fecal samples collected over time from recipient gnotobiotic mice (Table S3) provided an in vivo assay of colonization efficiency for each human donor sample. 16S rRNA sequencing reads were grouped into operational taxonomic units (OTUs) based on a threshold of ≥97% nucleotide sequence identity (97% ID). The results revealed that at the conclusion of the experiment 65.8±2.5% (mean ± s.e.m.) of OTUs in the stunted/underweight donor’s fecal microbiota sample and 68.4±8.8% (mean ± s.e.m.) of the OTUs in the healthy donor’s microbiota were detectable in recipient mice (i.e., each OTU had a relative abundance of ≥0.1% in ≥1% of fecal samples obtained from the mice).
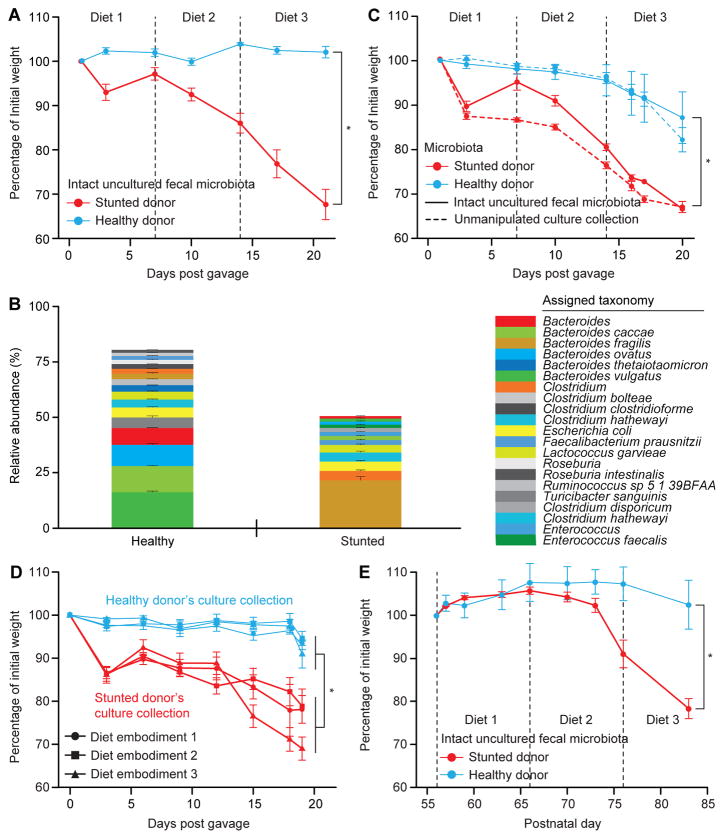
While gnotobiotic animals colonized with the healthy donor’s intact uncultured fecal microbiota maintained weight, recipients of the severely stunted/underweight donor’s intact uncultured fecal microbiota exhibited progressive and significant weight loss (p<0.005, paired two-tailed Student’s t-test, comparing final versus initial weights between the two treatment groups; Fig. 1A). In contrast to mice colonized with the healthy donor’s microbiota, those that received the stunted donor’s microbiota exhibited statistically significant weight loss by 10 days post gavage (dpg), during consumption of diet embodiment 2. Weight loss in this group worsened progressively, reaching 31±6% (mean ± s.e.m.) of original starting weight by 21 days post gavage (dpg) (p<0.001, two-tailed Student’s t-test, comparison of final weights; Fig. 1A; in a linear mixed effects model, both dpg as well as the interaction between microbiota and dpg were significant factors impacting weight throughout the experiment (p<1x10−7 for each). Food consumption was not different between the two treatment groups as their weight phenotypes diverged. The relative abundance of B. fragilis, defined by V4-16S rRNA analysis of fecal samples obtained at the time of sacrifice, was significantly greater in mice colonized with the stunted/underweight donor’s microbiota than in mice colonized with the healthy donor’s microbiota (p=1.9x10−6, Student’s two-tailed t-test; Fig. 1B).
Fig. 1. Intact uncultured human fecal microbiota and derived culture collections from healthy and undernourished Bangladeshi children transmit discordant weight phenotypes to gnotobiotic mice.
(A) 8–9 week old germ-free male C57BL/6J mice (n=8/treatment group) gavaged with intact uncultured fecal microbiota from Bangladeshi donors were fed a sequence of three embodiments of a representative Bangladeshi diet consumed by members of the donor population. See Fig. S1A for experimental design. Mean weights (± s.e.m.) as a function of days post gavage (dpg) are shown as percentages of weights immediately prior to fecal microbiota transplantation. (B) Efficiency of capture of bacterial OTUs present in the donor’s intact uncultured fecal samples in gnotobiotic mice. Mean relative abundances (± s.e.m.) of 97%ID OTUs representing ≥1% of the total fecal microbial communities in recipient animals. Results are based on V4-16S rRNA datasets and summarized at the species level (or genus when species could not be determined). OTUs present at lower abundances are not shown and account for the proportion not represented in each stacked barplot. (C) Transplantation of culture collections (dashed lines) generated from the fecal microbiota of the healthy or stunted/underweight donors recapitulated the discordant weight phenotype seen with the corresponding intact uncultured microbiota (solid lines). (n=6 mice/treatment group, mean weights ± s.e.m. plotted). *, p<0.05 (paired two-tailed Student’s t-test and linear mixed effects model as above). (D) The weight-loss phenotype observed in recipients of the stunted/underweight donor’s culture collection is not significantly different between the three Bangladeshi diet embodiments tested (p>0.05; two-tailed Student’s t-test). Mean weights (± s.e.m.) are plotted as a function of dpg (n=6 mice/culture collection/diet embodiment). Significant weight differences were seen between mice colonized with the healthy donor’s compared to the stunted/underweight donor’s culture collection in the context of all three embodiments of the Bangladeshi diet. *, p<0.05 (paired two-tailed Student’s t-test and linear mixed effects model as above). (E) Intergenerational transmission of discordant weight phenotypes. See Fig. S1C for experimental design. Mean weights (± s.e.m.) of offspring of female gnotobiotic mice colonized with the indicated donors’ microbiota (n=3–4 mice/treatment group) are plotted as a function of age. Animals were switched from a nutrient sufficient low fat, high plant polysaccharide mouse chow to embodiments of the Bangladeshi diets beginning on postnatal day 56. *, p<0.05 (tested by both paired two-tailed Student’s t-test comparing weights at sacrifice, and linear mixed effects model assessing interaction of weight, dpg, and microbiota through the experiment). The efficiency of intergenerational transmission of 97% ID OTUs was 96±1.8% and 88±2.3% (mean ± s.e.m.) for the healthy and stunted/underweight donor’s microbiota, respectively (defined at the time of euthanasia).
Bacterial culture collections from donor fecal microbiota transmit contrasting weight phenotypes
We next cultured bacterial strains from the healthy and stunted/underweight donors’ fecal samples (20, 21). Each collection of cultured strains was clonally arrayed in multi-well plates so that each well contained a monoculture of a given bacterial isolate (20). Each culture collection consisted of organisms that had co-existed in the donor’s gut and thus were the products of the donor’s history of environmental exposures to various microbial reservoirs (including those of family members and various enteropathogens endemic to the Mirpur thana), as well as the selective pressures and evolutionary events placed on and operating within their microbiota (e.g., immune, antibiotic, dietary, horizontal gene transfer). Individual isolates in the clonally arrayed culture collection were grouped into ‘strains’ if they shared an overall level of nucleotide sequence identity of >96% across their assembled draft genomes (21). Based on this criterion and the results of sequencing amplicons generated from the isolates’ 16S rRNA genes, we determined that the healthy and stunted donors’ culture collections contained 53 and 37 strains, respectively. Only one strain was shared between the two culture collections: Bifidobacterium breve hVEW9 [see Table S4 for a list of all isolates in the culture collection derived from the stunted/underweight child and ref. (21) for details of the healthy donor’s culture collection]. The two B. fragilis strains present in the healthy donor’s culture collection (hVEW46 and hVEW47) lacked a BfPAI and were therefore classified as NTBF. The stunted donor’s collection contained a single B. fragilis strain (mVEW4) with a bft-3 allele. ETBF strains of this type are globally distributed but most common in Southeast Asia (22). (See Table S5 for a comparison of the functions encoded by genes in the genomes of these ETBF and NTBF strains and the reference B. fragilis type strain ATCC 25285.)
To ascertain whether the contrasting weight phenotypes conferred by the two intact uncultured fecal microbiota samples could be transmitted by the strains captured in their derivative culture collections, we colonized 8-week-old adult germ-free C57BL/6 mice with all members of either of these two culture collections (n=6 singly-caged mice/collection; all mice receiving a given culture collection were maintained in a single gnotobiotic isolator). As a reference control for this experiment, and to compare results between this and the previous experiment, we colonized mice with the corresponding intact uncultured fecal microbiota samples, housing these mice in separate isolators from those used for the culture collection transplants. All mice were fed the three embodiments of the Bangladeshi diet (1 week/diet) in the same order described for the previous experiment. As with the intact uncultured microbiota, the corresponding culture collections transmitted discordant weight phenotypes to recipient animals (p<0.002, two-tailed Student’s t-test, comparison of final weights; Fig. 1C). Moreover, the weight phenotypes (change in body weight over time as a percentage of initial weight before gavage) observed with each intact uncultured fecal microbiota and the corresponding derivative culture collection were not significantly different (p>0.05 for both microbiota donors, two-tailed Student’s t-test; Fig. 1C). The difference in weight phenotypes first became statistically significant between the two groups of mice midway through consumption of diet embodiment 2, continued to increase with diet embodiment 3 (Fig. 1C), and again were not attributable to differences in food consumption.
Effect of diet
To test whether the weight loss phenotype was sensitive or robust to diet embodiment-type, the two clonally-arrayed bacterial culture collections were gavaged into separate groups of 8-week-old adult male germ-free C57BL/6 mice who were monotonously fed Bangladeshi diet embodiment 1, 2, or 3 for three weeks (n=6 singly caged recipient mice/culture collection/diet embodiment; Fig. S1B). The discordant weight phenotype observed previously was preserved irrespective of the Bangladeshi diet embodiment consumed (p<0.01, two-tailed Student’s t-test, comparison of final weights of mice regardless of diet embodiment consumed; n=18 mice/culture collection; Fig. 1D). Moreover, no significant differences in weights were noted between groups of mice colonized with the same culture collection but fed different diet embodiments (p>0.05 for embodiments 1 versus 2, 1 versus 3, and 2 versus 3 for mice receiving either culture collection, two-tailed Student’s t-test, comparison of final weights; Fig. 1D).
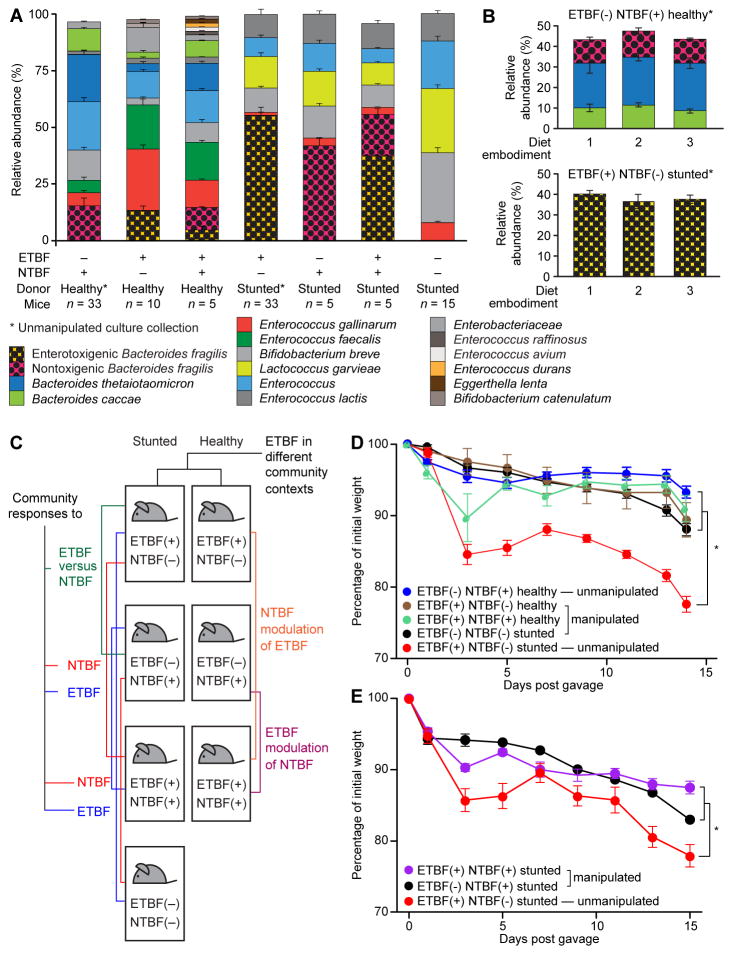
Transmission of strains was assessed by short read shotgun sequencing of DNA isolated from fecal samples collected at the end of the experiment. This method, known as COmmunity PROfiling by Sequencing (COPRO-Seq) (21), maps reads onto the draft genome assemblies of community members. At the depth of sequencing employed [354,352±23,216 (mean ± s.e.m.) 50 nucleotide (nt) unidirectional reads/fecal DNA sample], we could reliably detect strains whose relative abundance is ≥0.1%. COPRO-Seq demonstrated that transplantation of the culture collections was efficient and reproducible, with 98.1±0.6% and 94.5±1.6% (mean ± s.e.m.) of strains in the collections derived from the healthy and stunted donors, respectively, appearing in recipient animals. The relative abundance of ETBF in the fecal microbiota of mice containing the stunted/underweight donor’s culture collection was significantly greater than the cumulative relative abundance of the two NTBF strains in recipients of the healthy culture collection irrespective of the diet embodiment consumed (75.5±4.1% versus 17.0±4.4%, p=2.8x10−9, two-tailed Student’s t-test; Fig. 2A). The relative abundances of the ETBF strain in recipients of the stunted/underweight donor’s culture collection, the two NTBF strains in the healthy donor’s collection, and all other Bacteroides species did not differ significantly between diet embodiments (p>0.2 for all Bacteroides, one-way ANOVA; Fig. 2B).
Fig. 2. Enterotoxigenic B. fragilis (ETBF) is necessary but not sufficient to produce weight loss in recipient gnotobiotic mice.
(A) Gut microbial community composition, defined by COPRO-Seq, in mice colonized with either of the two unmanipulated culture collections or the derived manipulated versions. Mean values for relative abundances ± s.e.m. are plotted using aggregate data generated from fecal samples collected from mice colonized with a given community. Taxa present at abundances lower than 1% are not represented in the stacked barplots. (B) The proportional representation of Bacteroides taxa in unmanipulated culture collections installed in gnotobiotic mice do not differ significantly as a function of the diet embodiments animals were fed. Mean values ± s.e.m. for data generated from feces are shown (n=5–6/group; one-way ANOVA). (C) Schematic illustrating the different groups of gnotobiotic mice generated by manipulating the presence/absence of ETBF and NTBF within the stunted/underweight or healthy donors’ culture collections, and the questions addressed by the indicated comparisons. (D) Removal of ETBF prevents weight loss in mice colonized with the stunted/underweight donor’s culture collection. In contrast, addition of ETBF with the simultaneous removal of NTBF does not significantly impact weight in mice colonized with the culture collection derived from the healthy child (n=5–6 mice/treatment group). Mean values ± s.e.m. are plotted. *, p<0.05 (paired two-tailed Student’s t-test and linear mixed effects model as above). (E) Addition of NTBF to the stunted/underweight donor’s culture collection ameliorates ETBF-associated weight loss in gnotobiotic mice fed embodiment 2 of a representative Bangladeshi diet. (n=6 mice/treatment group). Mean values ± s.e.m. are plotted. *, p<0.05 (paired two-tailed Student’s t-test and linear mixed effects model as above).
Intergenerational transmission of weight phenotypes
To assess whether this weight loss phenotype was transmissible across generations of mice, two C57BL/6 males from the transplant experiment, one containing the stunted/underweight donor’s culture collection and the other the healthy donor’s collection, were switched to and subsequently maintained on an irradiated nutritionally enhanced mouse breeder chow from 21 to 48 dpg, at which time they were each co-housed with two germ-free 6-week old females that had received breeder chow since weaning. Seven days after co-housing, each male mouse was withdrawn from each mating trio, and the females were subsequently maintained on breeder chow throughout their pregnancy and as their pups completed the suckling period (Fig. S1C). Male pups (n=3–4/litter) were then weaned onto an irradiated, nutritionally sufficient, low fat/high plant polysaccharide (LF/HPP) chow, until they were 9-weeks old, at which time they were switched to the Bangladeshi diets (10 days/diet; same order of sequential presentation of the embodiments as before). Mice born to mothers colonized with either of these arrayed culture collections experienced identical weight gain profiles while consuming the LF/HPP diet (p=0.9, two-tailed Student’s t-test; Table S6). However, once they were transitioned to the sequence of three Bangladeshi diet embodiments (consumed from postnatal days 56 to 86), mice born to mothers harboring a stunted/underweight donor’s microbial community exhibited significantly greater weight loss (p=0.03, two-tailed Student’s t-test comparing weights at sacrifice). The total relative abundance of the two NTBF strains in fecal samples obtained from recipients of the healthy donor’s culture collection was 4.2±0.7% at the conclusion of the LF/HPP diet period and 4.6±0.9% at the conclusion of the Bangladeshi diet embodiment sequence, while the relative abundance of ETBF at these two time points was 34.3±4.2% and 50.0±0.7%, respectively, in mice colonized with the stunted/underweight donor’s culture collection.
An independent intergenerational transfer experiment was performed, in this case using the donors’ intact uncultured fecal microbiota. The efficiency of ETBF and NTBF transmission from mothers to pups was 100%. As with the culture collections, there was diet-dependent transmission of the discordant weight loss phenotype (Fig. 1E; compare with Fig. 1A).
Microbial community context impacts the effects of ETBF on community members and host
To establish whether ETBF is necessary and sufficient to cause dramatic weight loss in multiple community contexts, we performed a series of manipulations that involved removing the ETBF strain from the stunted/underweight donor’s culture collection and adding it to the healthy donor’s culture collection, with or without subtraction of its two NTBF strains (Fig. 2C). These manipulations allowed us to characterize (i) the role of community context in determining ETBF pathogenicity, (ii) community/host responses to ETBF, (iii) the ability of NTBF to modulate ETBF effects, and (iv) the effects of ETBF on NTBF. Recipient C57BL/6 male mice in each of the different treatment groups were 8–9 weeks old at the time of colonization; all were placed on diet embodiment 2 for two days prior to gavage and subsequently maintained on this diet for 14 days until they were euthanized (n=5 singly-caged animals/treatment group maintained separate gnotobiotic isolators). Fecal samples were collected at the time points described in Fig. S1D.
Weight phenotypes
Removal of the ETBF strain from the stunted/underweight donor’s culture collection prevented the transmissible weight loss phenotype (Fig. 2D; p=5.9x10−8, two-tailed Student’s t-test comparing weights at sacrifice). However, addition of the ETBF strain to the healthy donor’s culture collection did not produce significant weight loss, regardless of whether the NTBF strains were present or absent (p=0.3 and p=0.2, respectively, two-tailed Student’s t-test comparing weights at sacrifice; Fig. 2D). Based on these findings, we concluded that whether ETBF is necessary and sufficient for producing weight loss is dependent upon microbial community context.
COPRO-Seq analysis of the fecal microbiota of recipients of the unmanipulated ETBF(−) NTBF(+) healthy donor’s culture collection revealed that it contained the two NTBF strains [total relative abundance 14.5±3.0% (mean ± s.e.m.), with B. fragilis hVEW46 and B. fragilis hVEW47 comprising 1.1% and 13.5%, respectively], two other Bacteroides (B. thetaiotaomicron and B. caccae), plus Bifidobacterium breve and Enterococcus. The relative abundance of B. fragilis was not significantly different between mice harboring the transplanted unmanipulated healthy donor’s culture collection and its two manipulated ETBF(+) NTBF(−) and ETBF(+) NTBF(+) versions (p>0.5, two-tailed Student’s t-test) (Fig. 2A). (The term ‘unmanipulated’ indicates that all bacterial isolates that comprise a culture collection were pooled prior to transplantation, while ‘manipulated’ refers to the inclusion and/or exclusion of B. fragilis strains as part of the gavaged consortium.) The fecal microbiota of recipients of the unmanipulated stunted donor’s culture collection was dominated by ETBF (relative abundance 62.3±4.0%). Removal of ETBF led to significant increases in the relative abundances of Bifidobacterium breve another Bifidobacterium strain, Enterococcus lactis, and Enterococcus gallinarum (p<0.02, two-tailed Student’s t-test; Fig. 2A).
To determine whether NTBF alone is sufficient to protect mice from ETBF’s cachectic effects, we colonized three groups of C57BL/6 male gnotobiotic mice, each with a different version of the stunted donor’s culture collection: the unmanipulated culture collection containing ETBF alone or one of two manipulated versions, one with NTBF alone and the other with both ETBF and NTBF strains. Mice were placed on diet embodiment 2 for two days prior to gavage and maintained on this diet for 2 weeks until euthanized (n=6 animals/treatment group; all singly-caged; one treatment group/gnotobiotic isolator; Fig. S1D). We observed a significant difference in weight phenotypes between mice colonized with the unmanipulated undernourished donor’s ETBF(+) NTBF(−) culture collection compared to the manipulated ETBF(−) NTBF(+) version (p=0.01, one-tailed Student’s t-test; Fig. 2E. Addition of NTBF [yielding the ETBF(+) NTBF(+) community] dramatically ameliorated the weight loss phenotype (p=0.0004 for weights at sacrifice compared to mice with the unmanipulated community, one-tailed Student’s t-test; Fig. 2E). Follow-up COPRO-Seq analysis revealed that the relative abundances of ETBF at the conclusion of the experiment were 38.9±3.9% and 39.0±3.5% when animals were colonized with and without NTBF, respectively. Thus, NTBF does not appear to mediate its effects by reducing the fractional representation of ETBF in the community. However, ETBF appears to reduce the relative abundance of NTBF, which constituted 41.8±3.2% of the total community when ETBF was absent but only 19.2±2.6% when ETBF was present (p=0.04; one-tailed Student’s t-test).
The effects of intraspecific interactions on microbial gene expression
We performed microbial RNA-Seq of cecal contents harvested at sacrifice in order to characterize the meta-transcriptomes of members of the unmanipulated and manipulated versions of the healthy and stunted communities. Our goal was to assess (i) the effects of intraspecific competition (NTBF on ETBF and vice versa) in the healthy and stunted community contexts, (ii) the effects of the cultured stunted/underweight versus healthy donor community on ETBF, and (iii) the effects of co-colonization with ETBF on other bacterial members (including other Bacteroides). ETBF genes with significant differential expression attributable to the presence or absence of NTBF, in both healthy and stunted community contexts, are listed in Table S8B,F. Conversely, NTBF genes with significant differential expression attributable to the presence or absence of ETBF, in both healthy and stunted community contexts, are highlighted in Table S8C,E.
Fragipain is a cysteine protease that activates fragilysin by removing its auto-inhibitory prodomain. In mouse models of colitis, host proteases can also serve this function, but fragipain is required for sepsis to occur (23, 24). In the presence of NTBF, ETBF expression of fragilysin (bft-3) in the cecal meta-transcriptome of mice harboring the manipulated ETBF(+) NTBF(+) healthy donor’s community was significantly decreased compared to the manipulated version of the community where ETBF but not NTBF was present (39-fold, based on normalized transcript counts, p=0.002, one-tailed Student’s t-test). Fragipain expression was also significantly reduced (14.2-fold, p=0.0005, one-tailed Student’s t-test) (Table S8B). In the context of the stunted community, the reduction in bft-3 expression associated with introducing NTBF was considerably more modest (5.9-fold; p=0.09, one-tailed Student’s t-test) while fragipain expression was not significantly different between the two treatment groups (p>0.5; one-tailed Student’s t-test; Table S8).
When we abrogated fragilysin (bft-3) expression through insertional mutagenesis (Fig. S2), the mutant Δbft-3 strain grew robustly in vitro. However, when germ-free mice were gavaged with a manipulated version of the stunted donor’s culture collection containing this isogenic strain with a disrupted bft-3 locus substituted for the wild-type ETBF strain, we observed no detectable colonization of the mutant (n=5 mice fed diet embodiment 2 for 14 days with fecal samples collected); the number of COPRO-Seq reads mapping to the bft knockout strain was no greater than background, and a PCR assay that used B. fragilis-specific bft primers was negative. These results led us to conclude that this locus functions as an important colonization factor for this particular ETBF strain in this community context. These experiments did not allow us to directly address the hypothesis that attenuation of bft-3 expression produced by inclusion of NTBF in the stunted community contributed to the observed mitigation of weight loss.
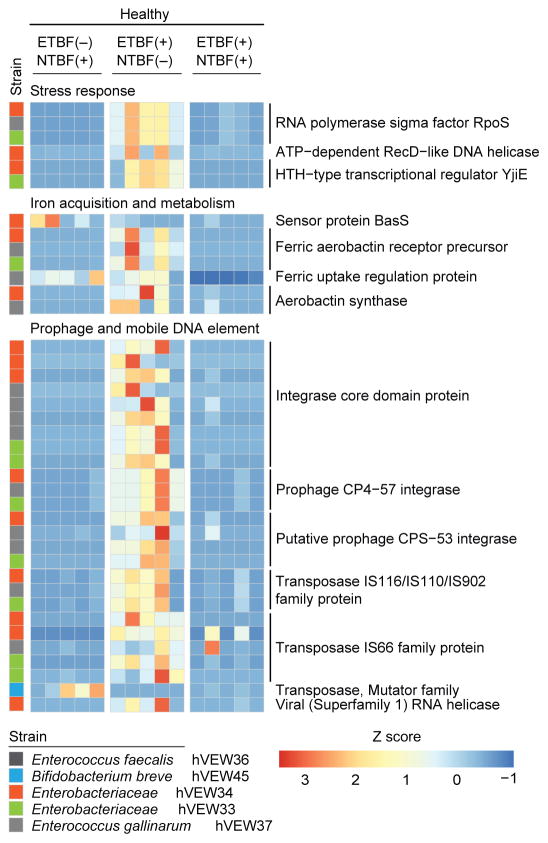
Looking beyond the effects of intraspecific interactions on btf-3 expression, we compared the cecal meta-transcriptomes of gnotobiotic mice colonized with the unmanipulated NTBF(+) ETBF(−) healthy donor’s culture collection versus mice harboring the two manipulated versions where ETBF was added, with or without removal of the two NTBF strains. The results revealed that ETBF in the absence of NTBF produced significant alterations in the expression of a number of transcripts related to various features of stress responses in several community members (Enterococcus faecalis, Enterococcus gallinarum, Bifidobacterium breve and two members of Enterobacteriaceae; differentially expressed genes identified using the Robinson and Smyth exact negative binomial test (25), with Bonferroni correction for multiple hypotheses) (Fig. 3). Both rpoS, a key general stress response sigma factor that positively controls expression of genes involved in transport of carbon sources and iron acquisition, and recD, which is involved in DNA repair, exhibited significant increases in their expression in the setting of ETBF without NTBF (p<0.05). Several genes involved in the acquisition and metabolism of iron were either upregulated in the presence of ETBF (e.g., ferric aerobactin receptor, ferric uptake regulation protein, aerobactin synthase) or repressed (e.g., Enterobacteriaceae homolog of the E. coli BasSR system component BasS, which is normally induced under high iron conditions) (26). ETBF’s effect on expression of these latter genes was mitigated when NTBF was present (Fig. 3), highlighting the importance of iron in intraspecific as well as interspecific interactions in the healthy donor’s consortium of transplanted cultured bacterial strains. In contrast, the presence or absence of ETBF or NTBF did not evoke significant changes in the expression of these or other genes involved in iron metabolism in the context of the stunted/underweight donor’s community. Numerous genes related to prophage and mobile DNA element biology were also expressed at significantly higher levels by healthy community members when ETBF was present in the absence of NTBF (p<0.05; Fig. 3). Prophage activation occurs in response to stress; some studies have postulated that phage induction can ‘shuffle’ community structure to favor an increased proportion of pathobionts (27).
Fig. 3. The effects of intraspecific NTBF-ETBF interactions on the community meta-transcriptome.
Adult mice were colonized with the indicated unmanipulated and manipulated versions of the healthy donor’s culture collection. All treatment groups were monotonously fed diet embodiment 2. Cecal contents collected at the time of sacrifice 14 days after initial colonization and gene expression in the community were analyzed by microbial RNA-Seq. Each column represents data from an individual mouse, while each row represents the levels of a given transcript, normalized across that row. Addition of ETBF to and removal of NTBF from the healthy donor’s culture collection produced an increase in expression of the indicated genes in strains whose identity is denoted by the color code on the left, compared to their expression in the (i) unmanipulated ETBF(−) NTBF(−) version or (ii) the manipulated version where the NTBF strains were retained when ETBF was added. UniProt-based annotations are shown on the right.
Studies in gnotobiotic mice have shown that signaling by members of the human gut microbiota involving the quorum sensing molecule, autoinducer-2 (AI-2), can alter virulence factor expression in enteropathogens (28) and have linked AI-2 signaling to modulation of the levels of Bacteroidetes in the gut (29). LuxQ is involved in the detection of AI-2. In the context of the healthy community, expression of three of the four luxQ homologs in the ETBF genome were decreased when NTBF was present [log2(fold-change) of −2.8, −4.4, and −9.5, p<0.005, exact negative binomial test; Table S8B]. Comparing the mice colonized with the unmanipulated ETBF(−) NTBF(+) and manipulated ETBF(+) NTBF(+) versions of the healthy donor’s culture collection revealed differential regulation of five other luxQ transcripts encoded by Bacteroides members (three in B. thetaiotaomicron hVEW3, and two in B. caccae hVEW51; Table S8D). In the context of the stunted donor’s community, the presence of ETBF had no significant effects on Lux gene expression in NTBF or any other community members, nor did the presence of NTBF have any effect on Lux expression in ETBF (Table S8E,F). Together, these results illustrate the importance of community context in determining the transcriptional effects of intraspecific (and interspecific) interactions involving ETBF.
Metabolism
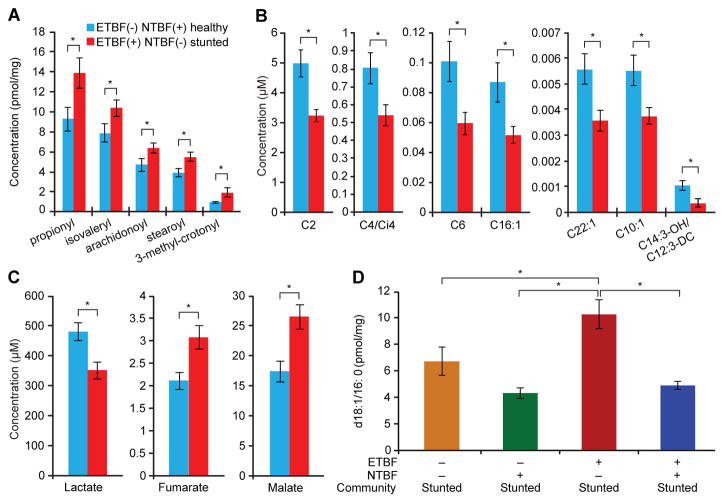
The metabolic effects of manipulating the representation of ETBF and NTBF in the healthy and stunted donor’s communities were studied by targeted mass spectrometry of tissue samples obtained from mice in the fed state (Table S7). Quantifying amino acids, organic acids, acylcarnitines, and acyl-CoAs in livers obtained from animals colonized with either of the two unmanipulated culture collections disclosed that compared to mice harboring the healthy donor’s ETBF(−) NTBF(+) culture collection, those colonized with the stunted donor’s ETBF(+) NTBF(−) culture collection had higher concentrations of propionyl-CoA and isovaleryl-CoA (byproducts of oxidation of branched-chain and other amino acids; p<0.05, FDR-adjusted Student’s two-tailed t-test; Fig. 4A), and lower concentrations of acetyl-CoA (p=0.07) and its cognate metabolite acetyl carnitine (i.e., C2 acylcarnitine; p=0.001; Fig. 4B). Mirroring these trends, cecal contents harvested at the time of euthanasia from mice harboring the stunted donor’s unmanipulated culture collection contained higher concentrations of branched-chain amino acids (p=0.066 for isoleucine/leucine, p=0.1 for valine) and lower concentrations of acetyl-carnitine (p=0.067). However, these trends were not observed in skeletal muscle.
Fig. 4. Host metabolic abnormalities associated with the stunted donor’s community.
Mice were colonized with the indicated culture collections and fed diet embodiment 2. Animals were sacrificed 21 days after colonization and metabolites in their livers were quantified by mass spectrometry. n=5–6 mice/treatment group. Mean values ± s.e.m. are plotted. Altered concentrations of (A) acyl-CoA, (B) acylcarnitine and (C) organic acids in the livers of mice colonized with the unmanipulated ETBF(+) NTBF(−) stunted donor’s culture collection versus animals harboring the unmanipulated ETBF(−) NTBF(+) healthy donor’s culture collection provide evidence of metabolic dysfunction involving the TCA cycle and mitochondrial fatty acid oxidation in the former group. (D) ETBF in the setting of the unmanipulated stunted donor’s community is associated with greater hepatic concentrations of the d18:1/16:0 ceramide known to inhibit mitochondrial oxidative metabolism compared to manipulated versions of the culture collection where ETBF has been removed with or without replacement by NTBF or when NTBF and ETBF are both present. Key for all panels: *, p<0.05 (two-tailed Student’s t-test).
Acetyl-CoA and acetyl-carnitine are generated by glucose, fatty acid, or amino acid metabolism, with a primary contribution from glucose metabolism in the fed state. A deficit in these metabolite pools suggests impaired flux of glucose to acetyl-CoA in mice with the stunted compared to healthy donor’s unmanipulated culture collection. Interestingly, the decline in acetyl-CoA and acetyl-carnitine levels occurred in concert with significant increases in hepatic levels of the late TCA cycle intermediates malate and fumarate (p<0.005 compared to mice with the healthy donor’s culture collection; Fig. 4C), as well as a trend toward increased succinate (p=0.1). These patterns were again mirrored in the cecum but not in skeletal muscle. Early-stage TCA cycle intermediates citrate and α-ketoglutarate were not different across these groups. Conversely, and consistent with impaired glucose availability or metabolism, lactate concentrations were significantly lower in the livers of mice colonized with the stunted donor’s culture collection (p=0.002; Fig. 4C), with an associated decrease in cecal lactate concentrations (p=0.0005; Table S7B). Hepatic pyruvate also showed a down trend (p=0.16) (Table S7B).
Combining the acyl-CoA and organic acid data, this metabolic profile is consistent with a scenario in which mice colonized with the unmanipulated ETBF-containing stunted donor’s culture collection have decreased capacity for metabolism of glucose to acetyl CoA. It appears that these animals attempt to compensate by an increase in amino acid metabolism that is sufficient to produce enough acetyl CoA to maintain normal citrate and α-ketoglutarate levels. However, use of these fuels also creates anaplerotic substrates that lead to production of distal TCA cycle intermediates, e.g., via propionyl CoA conversion to succinate, and aspartate conversion to oxaloacetate, fumarate, and malate.
Comparing mice colonized with the unmanipulated stunted donor’s culture collection versus the manipulated ETBF(−) NTBF(−) version of the stunted donor’s culture collection revealed that removal of ETBF was not sufficient to reverse the metabolic abnormalities described above. ETBF removal did produce a significant elevation in hepatic levels of d18:1/16:0 ceramide, a known inhibitor of mitochondrial electron transport complex IV (p=0.02; Fig. 4D) and consistent with decreased oxidative metabolism. Ceramides are involved in key intracellular stress response pathways, including those associated with immuno-inflammatory responses. This effect was strain-specific: NTBF alone did not produce the same result in mice colonized with the manipulated ETBF(−) NTBF(+) version of the stunted donor’s culture collection. Moreover, the ETBF-associated elevation in d18:1/16:0 was prevented by the presence of NTBF [p=0.007, FDR-adjusted Student’s two-tailed t-test in a comparison of mice colonized with the unmanipulated stunted community versus the manipulated ETBF(+) NTBF(+) community].
Gut barrier/immune function
One view of the effects of B. fragilis enterotoxin is that it perturbs gut barrier function, increasing permeability via cleavage of E-cadherin (11), triggering the release of pro-inflammatory cytokines including TNF-α (30) and augmenting β-catenin signaling (31). Treatment of chow-fed, conventionally-raised mice with antibiotics followed by introduction of wild-type NTBF, NTBF with an engineered active bft allele, wild-type ETBF with an active bft allele, or ETBF with a Δbft1 allele revealed that under these conditions bft was necessary to produce colitis (32). Small-scale human studies have demonstrated a greater representation of ETBF in individuals with active inflammatory bowel disease (33) and colorectal cancer (34). Strains harboring bft-3 have been reported to generate less biologically active toxin than strains possessing bft-1 or bft-2 alleles (22).
Analyses of hematoxylin and eosin-stained sections of small intestine and colon obtained from mice consuming the representative Bangladeshi diet and colonized with either of the two unmanipulated culture collections or two manipulated versions [ETBF(−) NTBF(−) stunted or ETBF(+) NTBF(+) healthy] revealed no hallmarks of inflammation such as an overabundance of neutrophils in the gut mucosa, crypt hyper-proliferation or loss, or loss/damage of villi in the small intestine (n=5 mice examined/treatment group).
We performed a follow-up FACS analysis of immune cell populations in the colon, mesenteric lymph nodes, and spleen of four groups of mice: those colonized with the stunted donor’s culture collection with or without ETBF, and those harboring the unmanipulated healthy donor’s culture collection or its manipulated ETBF(+) NTBF(−) version. Consistent with a previous study of specified pathogen-free mice treated with antibiotics to boost colonization with an ETBF isolate (34), colonic Th17 cell populations (TCR-β+CD4+CD8a−IL-17A+) were significantly increased in animals colonized with manipulated ETBF(+) NTBF(−) healthy and unmanipulated ETBF(+) NTBF(−) stunted culture collections compared to the unmanipulated ETBF(−) NTBF(+) healthy and manipulated ETBF(−) NTBF(−) stunted culture collections (p<0.001 and p<0.02, respectively, Student’s t-test; Fig. S3A,B). The presence of ETBF was not associated with significant differences in the size of the anti-inflammatory FoxP3+ regulatory T-cell population in mesenteric lymph nodes in either the stunted/underweight or healthy community contexts (p>0.05, ANOVA corrected for multiple comparisons; Fig. S3C).
B. fragilis polysaccharide A (PSA) is the product of one of the organism’s capsular polysaccharide synthesis (CPS) loci. A number of studies have shown that PSA functions as an immunomodulatory factor mitigating inflammatory responses in the gut via its effects on reducing IL-17 and inducing FoxP3+ regulatory T-cells (35, 36). RNA-Seq disclosed that the expression of genes in this CPS locus (37) was not significantly different in the ETBF and NTBF strains as a function of community context (unmanipulated versions of the healthy or stunted donor consortia) or engineered intraspecific interactions (Table S8B).
We expanded our analysis to examine the effects of intraspecific interactions on serum cytokine profiles in three groups of mice: those harboring the unmanipulated stunted donor’s culture collection, and those colonized with the manipulated ETBF(+) NTBF(+) and EBTF(−) NTBF(+) versions. ETBF(+) NTBF(−) mice had significantly higher levels of the pro-inflammatory cytokines IL-17A, TNF-α, and IFN-γ compared to ETBF(−) NTBF(+) mice (p=0.02, 0.03, and 0.03, respectively; Student’s one-tailed t-test) (Fig. S3D). However, these effects were not reversed with NTBF co-colonization (p>0.05 for all comparisons) despite NTBF’s protective effect with respect to weight loss, leading us to postulate that the weight loss phenotype was a reflection, at least in part, of the combined effects of disturbances in central metabolism and subtle perturbations in gut barrier function. Moreover, other members of the undernourished donor’s transplanted culture collection, besides ETBF, were contributors to these abnormalities.
DISCUSSION
The question of what determines the effects of a large enteropathogen burden in children at risk for undernutrition, or with already overt disease, is rooted in the definition of ‘pathogen’ – both conceptually and operationally. Koch’s postulates invoke the requirement for an isolated candidate pathogen to produce a disease when introduced into a host species, without necessarily considering the microbial ecology of the body habitat that the organism invades and establishes itself. The approach described in this study, involving generation of sequenced, clonally arrayed culture collections from the fecal microbiota of healthy and stunted/underweight Bangladeshi children and subsequent introduction of these consortia, with or without addition or subtraction of ETBF and NTBF strains, into germ-free mice fed embodiments of the diets consumed by the microbiota donors, allowed us to characterize how B. fragilis functions as a pathobiont in a microbial community context-dependent manner.
A topic of interest to those studying the pathogenesis of undernutrition is the relative effects of enteropathogen burden in non-diarrheal fecal samples and the presence or absence of environmental enteric dysfunction, an enigmatic disorder associated with populations where sanitation is poor and enteropathogen burden is high (38). While ETBF itself was not correlated with stunting in our small study cohort of 100 children, additional studies in larger populations coupled with quantitative PCR assays for other enterpathogens are needed to ascertain the contributions of ETBF to disease. Results from our gnotobiotic mouse studies emphasize that microbiota community context also needs to be considered when understanding the potential effects of this and other enteropathogens (39). The approach described in this report should help shed light on this issue. Constructing communities from culture collections capture a donor’s history of microbial exposures in their places of residency, as well as the evolutionary events that have shaped the genetic features of the gut strains they harbor. Culture collections generated from healthy and undernourished donors from this or other populations can be introduced into young rapidly growing animals (either through direct gavage or maternal transmission), or into adult animals, to better understand how intraspecific and interspecific interactions with one or more enteropathogens impact host biology in specified and/or systematically manipulated dietary and microbial community settings. A limitation of the present study is that the microbiota dissection was limited to two donors from a small cohort of 100 children. An obvious next step is to address the generalizability of our findings; this can be accomplished in a variety of ways, including analyses of the effects of introducing different ETBF and NTBF strains recovered from other Bangladeshi donors into the existing culture collections, as well as culture collections generated from individuals representing other populations.
An intriguing difference transmitted by the healthy and undernourished donor’s culture collections involves the host metabolic phenotype. Based on our findings, we propose that the limited rate of acetyl-CoA production in mice receiving the stunted donor’s culture collection cannot keep pace with the increased influx of substrates that replenish TCA cycle intermediates (anaplerosis), explaining the increase in distal TCA cycle substrates. At a more global level, increased oxidation of amino acids to maintain a minimal acetyl-CoA pool may divert amino acids away from protein synthesis, contributing to a wasting phenotype. Furthermore, concentrations of amino acids were modestly but consistently lower in the sera of mice colonized with the culture collection from the stunted compared to the healthy donor. This is consistent with use of amino acids for anaplerosis, thereby contributing to lower amino acid supplies for host growth and metabolism.
Some bacterial members of the gut microbiota contain the complete enzymatic apparatus for executing the TCA reaction sequence. The finding that there is marked inhibition of the distal TCA cycle in harvested cecal contents and liver but not skeletal muscle leads to the following testable hypotheses: (i) interactions between the undernourished donor’s microbiota and dietary components yield products that are present at high enough levels to inhibit the TCA cycle in community members; (ii) these products are transported from the gut by the portal circulation in sufficient quantities to inhibit the TCA cycle in hepatocytes; (iii) either first pass metabolism of these products by the liver or other processes limit their levels in the systemic circulation so that their effects on the TCA cycle are not observed in muscle; (iv) TCA cycle inhibition negatively impacts the ability to harvest energy from an already impoverished diet.
In our previous study of Malawian twins discordant for severe acute malnutrition, 1H NMR analyses of urine obtained from adult gnotobiotic mice colonized with intact uncultured microbiota indicated that TCA cycle inhibition was a feature of animals harboring the undernourished co-twin’s microbiota and consuming a macro- and micronutrient deficient prototypic Malawian diet. These animals but not their counterparts that had been colonized with the healthy co-twin’s microbiota exhibited a pronounced diet-dependent weight loss phenotype that did not occur in the context of a macro- and micronutrient sufficient diet (7). Together, these findings suggest that metabolic studies of serum, urine, and feces obtained from children prior to, during, and after treatment for their undernutrition should include quantitative assessment of analytes linked to TCA cycle activity.
MATERIALS AND METHODS
Human study design
Samples were obtained from an already completed observational birth cohort study (Field Studies of Human Immunity to Amebiasis in Bangladesh; 14). The study was conducted using protocols for obtaining informed consent, clinical samples, and clinical metadata that were approved by institutional review boards from the International Centre for Diarrhoeal Disease Research, Bangladesh (icddr,b)(study ID number 2007-041), the University of Virginia, Charlottesville (study ID 7563), and Washington University in St. Louis (study ID 201111065). Fecal specimens used in this study were covered by a Materials Transfer Agreement between icddr,b and Washington University in St. Louis. Fecal samples from 100 of the 147 children enrolled in this study were used for the analysis described in the current report; this number was not based on a prior power calculation derived from knowledge of ETBF carriage rate but rather represented samples from individuals who had been surveyed during the second year of postnatal life.
bft PCR assay
DNA isolated from fecal samples collected at 18 and 24 months of age from members of the birth cohort was used for a bft PCR assay employing primer pairs 5′-GAACCTAAAACGGTATATGT-3′ (GBF-201) and 5′-GTTGTAGACATCCCACTGGC-3′ (GBF-210) (40), OneTaq 2x master mix (New England Biolabs) and the following thermocycling conditions: after initial melting at 95°C for 30 s, 30 cycles of 95°C for 30 s, 53°C for 30 s, and 68°C for 30 s, with a final annealing time of 7 min at 68°C.
Testing fecal samples for parasites
A clinical microscopy-based screen for Entamoeba histolytica, Entamoeba dispar, Blastocystis hominis, Trichomonas hominis, Blastocystis hominis, Coccidian-like bodies, Giardia lamblia, Ascaris lumbricoides, Trichuris Tricuria, Ancylostoma duodenale/Necator americanus, Hymenolepsis nana, Endolimax nana, Iodamoeba butschlii and Chilomastix mesnili was also performed on these fecal samples as reported in ref. 5.
Preparation of human fecal samples for transplantation to germ-free mice
Aliquots (1g) of previously frozen fecal samples were resuspended under anaerobic conditions (77%N2, 20%CO,, 3%H2) in 15 mL of gut microbiota medium (GMM) (20) and homogenized at a setting of ‘high’ in a sterilized blender (Waring, Torrington, CT). Homogenates were clarified by passage through 100 μm pore-diameter nylon filters (BD Falcon, Franklin Lakes, NJ). Five mL of sterile 2 mm-diameter glass beads were added and remaining cell clumps were disrupted by vortexing (four on/off cycles, each 30 s). A final filtration through a 40 μm pore-diameter nylon filter (BD Falcon) was performed before storage in GMM containing glycerol (final concentration 15% v/v) at −80°C in 2 mL amber glass vials with a crimp-top butyl septum (Wheaton Scientific, Millville, NJ).
Preparation of Bangladeshi diet embodiments
Diet embodiments with ingredient and nutritional content described in Table S2 were prepared in 15 kg batches. Tilapia fish filets (Whole Foods Markets, St. Louis, MO) were simmered for 30 min in a 20 L stainless steel pot over a Corning hotplate (temperature setting 4). Fruits and vegetables were combined and simmered in a separate pot for 45 min on a Corning hotplate (temperature set at ‘4’). Parboiled rice (Delta Star, Stuttgart, AR) was cooked in a rice cooker (KRUPS, Medford, MA). Lentils were simmered for 90 min (temperature set at ‘2’). All ingredients for a given diet embodiment were combined in a vertical cutter-mixer (Robot Coupe Model R23, Jackson, MS) and pureed for 5 min. Aliquots were cooled for 12 hours in large plastic containers at 4°C. Aliquots (500 g) were then vacuum-sealed in 8″x10″ plastic bags (U-line, Pleasant Prairie, WI). The vacuum-sealed food paste was placed in a second 8″x10″ plastic bag (as an added barrier against incidental contamination), and the contents of the packages were sterilized by irradiation (20–50 kGy) within 24 h of food production (Steris Co, Chicago, IL). Sterility was verified by culturing samples of the irradiated diet in BHI medium for 5 days at 37°C under anaerobic and aerobic conditions (21). In addition, B. subtilis spore strips that had been included along with the food during the irradiation were cultured under the same conditions. Final nutritional profiles of the diets were obtained by submitting 100 g samples of each embodiment to Nestle Purina Analytical Labs (St. Louis, MO).
Clonally arrayed bacterial culture collections
Collections of cultured anaerobic bacterial strains were generated from frozen fecal samples according to previously published methods (20, 21). Cultures were arrayed in 384-well plates in Coy chambers under strict anaerobic conditions (77%N2, 20%CO, 3%H2) using a Precision XS liquid handling robot (BioTek, Winooski, VT). Bacteria isolates occupying each well were first grouped into 100%ID OTUs based on the results of V4-16S rRNA amplicon sequencing. Most OTUs were observed more than once across an arrayed library. Next, 3–4 isolates representing each OTU were picked robotically from the 384-well arrays and struck out individually onto 8-well agar plates containing GMM (20). DNA from isolates was subjected to whole genome shotgun sequencing using an Illumina HiSeq 2000 instrument (101 nt paired-end reads) or a MiSeq machine (150 nt paired-end reads). Sequences were assembled using MIRA (41), version 4.02 (parameters: “-NW:cnfs=warn, -NW:cmrnl=no, –GE:not=4”; template_size = “150 500 autorefine”). Coverage of isolate genomes from the severely stunted/underweight donor’s culture collection was 38.6±5.0-fold (mean±SEM) with an N50 contig length of 28.5±3.5kb (mean±SEM) (Table S3B). The corresponding values for members of the healthy donor’s collection are described in ref. 21. Genes were annotated using Prokka v1.10 (42). Predicted genes in each isolate’s genome assemblies were mapped to KEGG pathways and assigned KEGG Ortholog (KO) groups by querying the KEGG reference database (release 72.1) (BLAST 2.2.29+, blastp E-value ≤10−10, single best hit defined by E-value and bit score). Putative virulence factors were identified using the Virulence Factor Database [(43); BLAST hits with E-value < 10−17 bearing UniProt database annotations].
Full-length 16S rRNA gene amplicons were generated from isolates using primers 8F and 1391R. Isolates sharing ≥99% nucleotide sequence identity in their 16S rRNA genes and ≥96% sequence identity throughout their genomes as determined by NUCmer (44) were defined as a unique strain. Full-length 16S rRNA sequences were used to define taxonomy (Ribosomal Database Project (RDP) version 2.4 classifier (45)). Clonally arrayed, sequenced culture collections were stored in GMM/15% glycerol at 80° C in 96-well plates (TPP Tissue Culture Test Plates, Switzerland).
Generation of a bft-3:pGERM mutant in the ETBF strain cultured from the stunted donor’s microbiota
bft gene disruption was accomplished using the Bacteroides suicide vector pGERM. We introduced a 380-bp fragment of the 5′ end of bft-3 into BamH1-digested pGERM using the Gibson Assembly Cloning Kit (NEB). Primers to create the PCR product for the Gibson reaction were created using the NEBuilder Assembly Tool (v1.7.2). Assembled constructs were introduced by electroporation into kanamycin-resistant, electrocompetent E. coli DH5-α RK231 (46). Transformants were plated on LB-agar containing IPTG/X-gal (Sigma-Aldrich, St. Louis, MO) and ampicillin (100 μg/mL). Individual colonies were picked and grown in LB medium plus ampicillin; successful transformation was verified by PCR. The pGERM-transformed E. coli DH5-α RK231 and the ETBF strain (isolate mB11 of strain Bacteroides fragilis mVEW4) were conjugated by filter-mating on BHI-blood agar plates incubated under anaerobic conditions at 37° C for 24 h. Bacterial cells from the resulting lawn were collected by scraping and plated on BHI agar containing erythromycin/gentamycin (25 μg/mL and 100 μg/mL, respectively). Plates were incubated under anaerobic conditions at 37° C for 48 h. Picked colonies were sequenced to confirm successful pGERM insertion into bft-3 and positive colonies were stored in TYG containing 15% (v/v) glycerol.
The mutant and wild-type strains were cultured at 37°C under anaerobic conditions in TYG liquid medium to mid-log phase. Cells were then harvested and RNA was extracted using Trizol reagent (Life Technologies, Carlsbad, CA). To confirm that bft expression had been disrupted, cDNA was generated from the extracted RNA (see RNA-Seq section below) and used as a template for a PCR-based assay that used ReddyMix PCR Mastermix (Thermo Fisher Scientific, Waltham, MA), a forward primer that spans the vector insertion site (5′-ATGAAGAATGTAAAGTTACTTTTAATGCTAGGAACCG-3′), a reverse primer 3′ to the crossover site (5′-CTCCACTTTGTACTTTATACTACTGAATATGCTTG-3′) and the following cycling conditions: 94°C for 10 min followed by 30 cycles of 94°C for 30 s, 56°C for 30 s, 72°C for 60 s, with a final extension of 72°C for 5 min.
Assembling consortia of cultured bacterial strains for gavage
Archived 96-well culture collection plates were thawed in an anaerobic chamber and aliquots of each well inoculated into 600 μL of GMM in 1 mL 96-deep-well culture plates (Thermo Fisher Scientific, Rochester, NY). Cultures were grown anaerobically to stationary phase at 37°C, with growth measured at OD600. Equal amounts of selected isolates (100 μL) were pooled using the Precision XS liquid handling robot. For manipulations involving addition of NTBF, both strains of the nontoxigenic strain were used. For manipulations involving subtraction of the single ETBF strain or both NTBF strains, the strains were simply excluded from the pooling step. Final pools were mixed 1:1 with sterile PBS plus glycerol (to achieve a final concentration of 15%) and aliquots (1 mL) placed in 2 mL butyl septum-stoppered amber glass vials (Wheaton, Melville NJ) for storage or for gavage into germ-free animals.
Gnotobiotic mouse experiments
All mouse experiments were performed using protocols approved by Washington University Animal Studies Committee. Prior to the initiation of experiments, germ-free adult male C57BL/6 mice were maintained in plastic flexible film gnotobiotic isolators under a strict 12-hour light cycle and fed an autoclaved low-fat, high-plant polysaccharide (LF/HPP) chow ad libitum (B&K Universal, East Yorkshire, U.K; diet 7378000). Different treatment groups (i.e., mice gavaged with different microbial communities) were maintained in separate gnotobiotic isolators. All animals studied were included in subsequent analyses. Male mice were age- and weight- matched prior to gavage of human donor microbiota. Investigators were not blinded to either diet or microbiota treatments.
Feeding Bangladeshi diet embodiments was initiated two days prior to colonization with either uncultured fecal microbiota samples or pools generated from arrayed bacterial culture collections. Diets were extruded as a paste from their plastic bags into food trays. Fresh 30 g aliquots of the paste were provided to each singly caged animal/day. For experiments where diet embodiments were changed, autoclaved Aspen hardwood lab bedding (NEPCO, Warrensburg, NY) was replaced at the start of each embodiment transition to minimize carryover of ingredients from the preceding embodiment exposure.
For the intergenerational transmission experiments, male pups (n=3–4 for each intact donor microbiota; n=5 for each culture collection), representing the combined litter from trio matings were weaned onto LF/HPP chow and transitioned to the Bangladeshi diet embodiments at 8 weeks of age (embodiments 1→2→3; 10 d/diet phase).
Characterizing microbial community composition and gene expression
Multiplex sequencing of 16S rRNA PCR amplicons —
Genomic DNA was extracted from mouse fecal pellets using a phenol-chloroform and beat-beating protocol (45). Barcoded primers 515F and 806R were used to generate PCR amplicons covering the V4 region of bacterial 16S rRNA genes present in the samples. Multiplex sequencing of pooled amplicons with sample-specific barcodes was performed using an Illumina MiSeq instrument (250 bp paired-end reads). Reads were trimmed in silico to 200 bases to retain the highest-quality base calls, assembled with FLASH v1.2.11, and de-multiplexed in QIIME v1.8.0 (21). The 16S rRNA sequence datasets were first analyzed using open-reference OTU picking (97%ID OTUs) (uclust-ref against the Greengenes reference database). 98.7% of the 26,419,497 sequences that passed QIIME’s default quality filters were successfully clustered with a reference sequence of ≥97%ID. For all subsequent analyses, OTU tables were rarefied to 4000 reads per sample and filtered to retain only those OTUs whose relative abundances in fecal microbiota were ≥0.1% in ≥1% of all samples sequenced. Taxonomies were assigned using RDP classifier 2.4 trained on the manually curated Greengenes database ‘Isolated named strains 16S’.
Community profiling by sequencing (COPRO-Seq) of fecal samples collected from colonized with unmanipulated and manipulated culture collections
75 nt unidirectional reads were generated from fecal DNA samples using an Illumina MiSeq instrument and trimmed to 50 nt to preserve the highest quality reads before mapping onto the genomes of culture community members. The analytic pipeline for COPRO-Seq is described in ref. 21 and employs software available at https://github.com/nmcnulty/COPRO-Seq.
Microbial RNA-Seq
Procedures for performing microbial RNA-Seq are detailed in previous publications (21). Aliquots of cecal contents (~50 mg) that had been collected at sacrifice (14 dpg) and stored at −80° C, were suspended in 500 μL of extraction buffer (200 mM NaCl, 20 mM EDTA), 210 μL of 20% SDS, 500 μL of phenol:chloroform:isoamyl alcohol (pH 4.5, 125:24:1, Ambion/Life Technologies, Carlsbad, CA), and 150 μL of acid-washed glass beads (212–300 μm diameter, Sigma-Aldrich, St. Louis, MO). Cells were lysed by mechanical disruption using a bead beater (maximum setting; 5 min at room temperature; BioSpec Products), followed by phenol:chloroform:isoamyl alcohol extraction and isopropanol precipitation on ice. After treatment with RNase-free TURBO-DNase (Ambion/Life Technologies, Carlsbad, CA), MEGAClear columns (Life Technologies) were employed to remove 5S rRNA and tRNAs. A second DNase treatment (Baseline-Zero DNase; Epicentre/Illumina, San Diego, CA) was performed before a second MEGAClear purification followed by rRNA depletion (Ribo-Zero rRNA removal kits; Epicentre/Illumina, San Diego, CA). cDNA was synthesized (SuperScript II; Invitrogen, Carlsbad, CA), and second strand synthesis was performed with RNaseH, E. coli DNA polymerase and E. coli DNA ligase (all from New England Biolabs). cDNA libraries were prepared by shearing samples using a BioRuptor Pico sonicator (Diagenode, Denville, NJ), then size-selecting 150–200 bp fragments for blunting, A-tailing, and ligating sample-specific barcoded sequencing adapters, and finally PCR enrichment. cDNA libraries were pooled for multiplex sequencing using an Illumina NextSeq instrument (13.4 ±1.1 million unidirectional 75 nt reads/cecal RNA sample), with the exception of the sampleIDs ETBF.expt.112–116 (Table S3D), which were sequenced on the MiSeq platform (6.6±0.7 million unidirectional 75 nt reads/ sample). Identification of differentially expressed genes was performed in R (version 3.2.3) using the Robinson and Smyth exact negative binomial test with Bonferroni correction (edgeR package, version 3.10.2) as described previously (21).
Mass spectrometry
Tissues previously stored at −80°C were weighed while frozen and immediately homogenized in a solution of 50% aqueous acetonitrile/0.3% formic acid (50mg tissue/ml solution; procedure done at 4°C using a IKA T25 Ultra-Turrax high speed homogenizer at maximum setting for 30–45 s). Amino acids, acylcarnitines, organic acids, acyl-CoAs, and ceramides were analyzed using stable isotope dilution techniques. Amino acids and acylcarnitine measurements were made by flow injection tandem mass spectrometry using sample preparation methods described previously (47, 48). Data were acquired using a Waters Acquity UPLC system (Waters, Milford, MA) equipped with a TQ (triple quadrupole) detector and a data system controlled by the MassLynx 4.1 operating system (Waters). Organic acids were quantified (49) using a Trace Ultra GC coupled to ISQ MS operating under Xcalibur 2.2 (Thermo Fisher Scientific, Austin, TX). Acyl-CoAs were extracted and purified as described previously (50), and analyzed by flow injection analysis using positive electrospray ionization on a Xevo TQ-S, triple quadrupole mass spectrometer (Waters). Heptadecanoyl CoA was employed as an internal standard. Ceramides were extracted (51) and analyzed by flow injection tandem mass spectrometry using a Xevo TQS spectrometer (Waters).
Histologic analysis
Intestines were harvested and placed in 10% neutral-buffered formalin for 3 h at room temperature, washed in 70% ethanol, prepared as ‘Swiss-rolls’, paraffin-embedded, and 5 μm-thick sections were cut. Hematoxylin and eosin-stained sections were evaluated for alterations in crypt drop out (evaluated by crypt density), crypt depth, villus height, epithelial proliferation (measured by M-phase cells/crypt, n=100 crypts/specimen evaluated)) and neutrophils/crypt unit (n=100 crypt units evaluated per specimen). All slides were blinded prior to quantification.
Florescence-activated cell sorting
Cell isolation
Cells from the spleen, mesenteric lymph node and lamina propria of the colon were isolated as described previously (52) with two minor modifications; (i) Hank’s Balanced Salt Solution (HBSS) was used in place of Dulbecco’s phosphate buffered saline, and (ii) collagenase type VIII (Sigma Aldrich) was used at a concentration of 0.1mg/ml for isolation of colonic lamina propria cells.
Cell re-stimulation
Intracellular cytokine staining was performed on cell suspensions from the spleen, mesenteric lymph nodes and colonic lamina propria following re-stimulation in round bottom 96 well plates in complete RPMI containing β-mercaptoethanol (0.05 mM; Sigma-Aldrich) supplemented with 50 ng/ml phorbol-myrisotol-acetate (Sigma-Aldrich), 750 ng/ml ionomycin (Sigma-Aldrich) and Brefeldin A (eBioscience, San Diego, CA; diluted according to manufacturers recommendations). Cells were re-stimulated at 37°C for 3 h in a humidified CO2 tissue culture incubator. Cells in which FoxP3 was assessed were not re-stimulated.
Flow cytometry
Cell surface and intracellular staining was performed as described previously (52) with the following two modifications; (i) cells were washed free of excess protein from re-stimulation medium, or from the HBSS/0.1% BSA (w/v) buffer used for assaying FoxP3, using HBSS without BSA, and (ii) the incubation containing anti-CD16/CD32 [Fc block (2.4G2); BD Pharmingen, San Jose, CA] also included Live/Dead Fixable Aqua Dead Cell Stain (Life Technologies, Carlsbad, CA) to allow identification and removal of dead cells during the analysis that followed fixation and permeabilization. Cells were stained with the following antibodies; CD4 eFluor 450 (GK1.5; eBioscience), CD4 allophycocyanin (RM-4.5; BD Pharmingen) TCR-β Alexa Fluor 488 (H57–597; Biolegend), CD8a allophycocyanin (53–6.7; Biolegend), FoxP3 eFluor 450 (FJK-16s; eBioscience) and IL-17A phycoerythrin (TC11-18H10.1; Biolegend). Appropriate isotype control antibodies were purchased from the same vendors that supplied antibodies targeting molecules of interest. Cells were analyzed with an Aria III flow cytometer (BD Biosciences) and data were analyzed with FlowJo software (TreeStar; version 7.6.1).
Measurements of serum cytokine levels
Blood was collected via retro-orbital phlebotomy at the time of euthanasia, and derived serum samples were analyzed, alongside standards, using the Bio-Plex Pro™ Mouse Cytokine 23-plex Assay (Bio-Rad, Hercules, CA) per the manufacturer’s instructions. Technical duplicates were performed for each sample.
Statistical analyses
Routine statistical analyses were performed in R (version 3.1.2). P < 0.05 was considered to be statistically significant. Specific statistical tests are noted in the figure legends and throughout the text. Bonferroni correction for multiple hypotheses was applied to identify genes that exhibited significant differences in their expression between treatment groups.
Supplementary Material
Fig. S1. Experimental designs.
Fig. S2. Generation of a Δbft-3 strain of ETBF.
Fig. S3. Assessing the effects of ETBF on gut barrier/immune function.
Table S1. Summary of anthropometric scores and results of PCR assays for bft in the fecal microbiota of members of a birth cohort of 100 Bangladeshi children.
Table S2. Composition and nutritional content of Bangladeshi diet embodiments, and nutritional requirements of mice and children.
Table S3. V4-16S rRNA, COPRO-Seq, microbial RNA-Seq, and cultured bacterial strain genome sequencing datasets.
Table S4. Bacterial culture collection generated from the fecal microbiota of a severely stunted Bangladeshi child.
Table S5. Genome features of isolates in the clonally arrayed bacterial culture collection generated from the fecal microbiota of a severely stunted 2-year-old Bangladeshi child.
Table S6. Weights of all individual mice as a function of diet, microbiota, and days post gavage.
Table S7. Mass spectrometry-based quantitation of metabolites in host tissues, serum, and cecum obtained from mice in the different treatment groups.
Table S8. Effects of intraspecific and interspecific interactions on ETBF, NTBF, and other Bacteroides gene expression profiles in healthy or stunted donor community contexts.
Overline: Microbiome.
Single Sentence Summary
Addition, subtraction, and replacement of enterotoxigenic and non-toxigenic strains of Bacteroides fragilis in defined gut microbial communities from undernourished and healthy Bangladeshi children, reveals that the ability of an enterotoxigenic strain to produce cachexia in gnotoboitic mice is regulated by other community members.
Editor’s Summary
A big unanswered question is what determines the effects of enteropathogen burden in children who are undernourished or who are at risk for undernutrition. In a new study, Wagner, Dey and their colleagues introduce collections of sequenced gut bacterial strains cultured from healthy or underweight Bangladeshi children into germ-free mice fed diets resembling those consumed by the children.. The gut bacterial strains were transplanted with or without non-toxigenic or enterotoxigenic Bacteroides fragilis strains. Addition of enterotoxigenic Bacteroides fragilis induced cachexia in the transplanted mice, and altered gene expression and metabolic activity of the transplanted bacterial strains. These effects were mitigated by co-colonization with non-toxigenic Bacteroides fragilis, illustrating the influence of intra- and interspecies interactions in determining the impact of an enteropathogen on its host.
Acknowledgments
We thank David O’Donnell, Maria Karlsson, Sabrina Wagoner, and Justin Serugo for assistance with gnotobiotic mouse husbandry, Marty Meier, Su Deng, and Jessica Hoisington-Lopez, for superb technical assistance and Mark Charbonneau and Michael Barratt for providing valuable insights during the course of this study. The birth cohort study was conducted using protocols for obtaining informed consent, clinical samples, and clinical metadata that were approved by institutional review boards from the icddr,b (study ID number 2007-041), the University of Virginia, Charlottesville (study ID 7563), and Washington University in St. Louis (study ID 201111065). Fecal specimens characterized in this report were covered by a Materials Transfer Agreement between icddr,b and Washington University in St. Louis.
Funding: N.D. is the recipient of a Young Investigator Grant for Probiotics Research from the Global Probiotics Council. P.P.A is the recipient of a Sir Henry Wellcome Postdoctoral Fellowship from the Wellcome Trust (096100). This study was supported by the Bill & Melinda Gates Foundation and the NIH (DK30292).
Footnotes
Author Contributions: V.E.W., N.D., and J.I.G. designed the experiments. R.H., T.A. and B.P. directed the clinical study design, enrollment, plus clinical data and sample collection for the Bangladeshi birth cohort. V.E.W. and N.D., performed gnotobiotic mouse experiments and generated 16S rRNA, COPRO-Seq, microbial RNA-Seq, and bacterial isolate genome sequencing datasets. V.E.W. and J.G. produced culture collections from the fecal microbiota of the healthy and stunted Bangladeshi children. J.C., O.I. and L.B. conducted mass spectrometry-based studies of biospecimens obtained from gnotobiotic mice. P.A. performed fluorescence activated cell sorting of immune cell populations. N.S. and N.D. measured serum cytokine levels. N.D., V.E.W., A.H., T.S.S., J.C., P.A., C.N., and J.I.G. analyzed the data. N.D., V.E.W., and J.I.G. wrote the paper.
Competing interests: J.I.G. is co-founder of Matatu, Inc., a company characterizing the role of diet-by-microbiota interactions in animal health. The other authors declare that they have no competing interests.
Data and materials availability: 16S rRNA, COPRO-Seq, and microbial RNA-Seq datasets, plus whole genome shotgun sequencing datasets from cultured bacterial strains are available through the European Nucleotide Archive (ENA Study Accession Number PRJEB9703).
References
- 1.Christian P, Mullany LC, Hurley KM, Katz J, Black RE. Nutrition and maternal, neonatal, and child health. Semin Perinatol. 2015;39:361–372. doi: 10.1053/j.semperi.2015.06.009. [DOI] [PubMed] [Google Scholar]
- 2.Black RE, Victora CG, Walker SP, Bhutta ZA, Christian P, de Onis M, Ezzati M, Grantham-McGregor S, Katz J, Martorell R, Uauy R Maternal and Child Nutrition Study Group. Maternal and child undernutrition and overweight in low-income and middle-income countries. Lancet. 2013;382:427–451. doi: 10.1016/S0140-6736(13)60937-X. [DOI] [PubMed] [Google Scholar]
- 3.Martorell R, Zongrone A. Intergenerational influences on child growth and undernutrition. Paediatr Perinat Epidemiol. 2012;26(Suppl 1):302–314. doi: 10.1111/j.1365-3016.2012.01298.x. [DOI] [PubMed] [Google Scholar]
- 4.Victora CG, Adair L, Fall C, Hallal PC, Martorell R, Richter L, Sachdev HS Maternal and Child Undernutrition Study Group. Maternal and child undernutrition: consequences for adult health and human capital. Lancet. 2008;371:340–357. doi: 10.1016/S0140-6736(07)61692-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Subramanian S, Huq S, Yatsunenko T, Haque R, Mahfuz M, Alam MA, Benezra A, DeStefano J, Meier MF, Muegge BD, Barratt MJ, VanArendonk LG, Zhang Q, Province MA, Petri WA, Jr, Ahmed T, Gordon JI. Persistent gut microbiota immaturity in malnourished Bangladeshi children. Nature. 2014;510:417–421. doi: 10.1038/nature13421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blanton LV, Charbonneau MR, Salih T, Barratt MJ, Venkatesh S, Ilkaveya O, Subramanian S, Manary MJ, Trehan I, Jorgensen JM, Fan Y, Henrissat B, Leyn SA, Rodionov DA, Osterman AL, Maleta KM, Newgard CB, Ashorn P, Dewey KG, Gordon JI. Gut bacteria that prevent growth impairments transmitted by microbiota from malnourished children. Science. 2016;351:aad3311. doi: 10.1126/science.aad3311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Smith MI, Yatsunenko T, Manary MJ, Trehan I, Mkakosya R, Cheng J, Kau AL, Rich SS, Concannon P, Mychaleckyj JC, Liu J, Houpt E, Li JV, Holmes E, Nicholson J, Knights D, Ursell LK, Knight R, Gordon JI. Gut Microbiomes of Malawian Twin Pairs Discordant for Kwashiorkor. Science. 2013;339:548–554. doi: 10.1126/science.1229000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kau AL, Planer JD, Liu J, Rao S, Yatsunenko T, Trehan I, Manary MJ, Liu TC, Stappenbeck TS, Maleta KM, Ashorn P, Dewey KG, Houpt ER, Hsieh CS, Gordon JI. Functional characterization of IgA-targeted bacterial taxa from undernourished Malawian children that produce diet-dependent enteropathy. Sci Transl Med. 2015;7:276ra24. doi: 10.1126/scitranslmed.aaa4877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guerrant RL, Oriá RB, Moore SR, Oriá MOB, Lima AAM. Malnutrition as an enteric infectious disease with long-term effects on child development. Nutr Rev. 2008;66:487–505. doi: 10.1111/j.1753-4887.2008.00082.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee G, Paredes Olortegui M, Peñataro Yori P, Black RE, Caulfield L, Banda Chavez C, Hall E, Pan WK, Meza R, Kosek M. Effects of Shigella-Campylobacter- and ETEC-associated diarrhea on childhood growth. Pediatr Infect Dis J. 2014;33:1004–1009. doi: 10.1097/INF.0000000000000351. [DOI] [PubMed] [Google Scholar]
- 11.Sears CL, Geis AL, Housseau F. Bacteroides fragilis subverts mucosal biology: from symbiont to colon carcinogenesis. J Clin Invest. 2014;124:4166–4172. doi: 10.1172/JCI72334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pathela P, Hasan KZ, Roy E, Alam K, Huq F, Siddique AK, Sack RB. Enterotoxigenic Bacteroides fragilis-associated diarrhea in children 0–2 years of age in rural Bangladesh. J Infect Dis. 2005;191:1245–1252. doi: 10.1086/428947. [DOI] [PubMed] [Google Scholar]
- 13.Sears CL, Islam S, Saha A, Arjumand M, Alam NH, Faruque ASG, Salam MA, Shin J, Hecht D, Weintraub A, Sack RB, Qadri F. Association of enterotoxigenic Bacteroides fragilis infection with inflammatory diarrhea. Clin Infect Dis. 2008;47:797–803. doi: 10.1086/591130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mondal D, Minak J, Alam M, Liu Y, Dai J, Korpe P, Liu L, Haque R, Petri WA. Contribution of enteric infection, altered intestinal barrier function and maternal malnutrition to infant malnutrition in Bangladesh. Clin Infect Dis. 2012;54:185–192. doi: 10.1093/cid/cir807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.World Health Organization. WHO Child Growth Standards: Growth velocity based on weight, length and head circumference: Methods and development. WHO; 2009. (available at http://www.who.int/childgrowth/standards/velocity/technical_report/en/) [Google Scholar]
- 16.Arsenault JE, Yakes EA, Hossain MB, Islam MM, Ahmed T, Hotz C, Lewis B, Rahman AS, Jamil KM, Brown KH. The current high prevalence of dietary zinc inadequacy among children and women in rural Bangladesh could be substantially ameliorated by zinc biofortification of rice. J Nutr. 2010;140:1683–1690. doi: 10.3945/jn.110.123059. [DOI] [PubMed] [Google Scholar]
- 17.Islam SN, Khan MNI, Akhtaruzzaman M. A Food Composition Database for Bangladesh with Special reference to Selected Ethnic Foods. INFS-NFPCSP-FAO; Dhaka, Bangladesh: 2010. [Google Scholar]
- 18.US Department of Agriculture, Agricultural Research Service, Nutrient Data Laboratory. USDA National Nutrient Database for Standard Reference, Release. 2015:28. (available at http://www.ars.usda.gov/nea/bhnrc/ndl)
- 19.Mohsena M, Goto R, Mascie-Taylor CGN. Socioeconomic and demographic variation in nutritional status of under-five Bangladeshi children and trend over the twelve-year period 1996–2007. J Biosoc Sci. 2016:1–17. doi: 10.1017/S0021932016000328. [DOI] [PubMed] [Google Scholar]
- 20.Goodman AL, Kallstrom G, Faith JJ, Reyes A, Moore A, Dantas G, Gordon JI. Extensive personal human gut microbiota culture collections characterized and manipulated in gnotobiotic mice. Proc Natl Acad Sci U S A. 2011;108:6252–6257. doi: 10.1073/pnas.1102938108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dey N, Wagner VE, Blanton LV, Cheng J, Fontana L, Haque R, Ahmed T, Gordon JI. Regulators of Gut Motility Revealed by a Gnotobiotic Model of Diet-Microbiome Interactions Related to Travel. Cell. 2015;163:95–107. doi: 10.1016/j.cell.2015.08.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Scotto d’Abusco AS, Del Grosso M, Censini S, Covacci A, Pantosti A. The alleles of the bft gene are distributed differently among enterotoxigenic Bacteroides fragilis strains from human sources and can be present in double copies. J Clin Microbiol. 2000;38:607–612. doi: 10.1128/jcm.38.2.607-612.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Choi VM, Herrou J, Hecht AL, Teoh WP, Turner JR, Crosson S, Bubeck Wardenburg J. Activation of Bacteroides fragilis toxin by a novel bacterial protease contributes to anaerobic sepsis in mice. Nat Med. 2016;22:563–567. doi: 10.1038/nm.4077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Herrou J, Choi VM, Bubeck Wardenburg J, Crosson S. Activation Mechanism of the Bacteroides fragilis Cysteine Peptidase, Fragipain. Biochemistry (Mosc) 2016 doi: 10.1021/acs.biochem.6b00546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Robinson MD, Smyth GK. Small-sample estimation of negative binomial dispersion with applications to SAGE data. Biostatistics. 2008;9:321–332. doi: 10.1093/biostatistics/kxm030. [DOI] [PubMed] [Google Scholar]
- 26.Hagiwara D, Yamashino T, Mizuno T. A Genome-wide view of the Escherichia coli BasS-BasR two-component system implicated in iron-responses. Biosci Biotechnol Biochem. 2004;68:1758–1767. doi: 10.1271/bbb.68.1758. [DOI] [PubMed] [Google Scholar]
- 27.Selva L, Viana D, Regev-Yochay G, Trzcinski K, Corpa JM, Lasa I, Novick RP, Penadés JR. Killing niche competitors by remote-control bacteriophage induction. Proc Natl Acad Sci U S A. 2009;106:1234–1238. doi: 10.1073/pnas.0809600106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hsiao A, Ahmed AMS, Subramanian S, Griffin NW, Drewry LL, Petri WA, Haque R, Ahmed T, Gordon JI. Members of the human gut microbiota involved in recovery from Vibrio cholerae infection. Nature. 2014;515:423–426. doi: 10.1038/nature13738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thompson JA, Oliveira RA, Djukovic A, Ubeda C, Xavier KB. Manipulation of the quorum sensing signal AI-2 affects the antibiotic-treated gut microbiota. Cell Rep. 2015;10:1861–1871. doi: 10.1016/j.celrep.2015.02.049. [DOI] [PubMed] [Google Scholar]
- 30.Kim JM, Cho SJ, Oh YK, Jung HY, Kim YJ, Kim N. Nuclear factor-kappa B activation pathway in intestinal epithelial cells is a major regulator of chemokine gene expression and neutrophil migration induced by Bacteroides fragilis enterotoxin. Clin Exp Immunol. 2002;130:59–66. doi: 10.1046/j.1365-2249.2002.01921.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wu S, Morin PJ, Maouyo D, Sears CL. Bacteroides fragilis enterotoxin induces c-Myc expression and cellular proliferation. Gastroenterology. 2003;124:392–400. doi: 10.1053/gast.2003.50047. [DOI] [PubMed] [Google Scholar]
- 32.Rhee K-J, Wu S, Wu X, Huso DL, Karim B, Franco AA, Rabizadeh S, Golub JE, Mathews LE, Shin J, Sartor RB, Golenbock D, Hamad AR, Gan CM, Housseau F, Sears CL. Induction of persistent colitis by a human commensal, enterotoxigenic Bacteroides fragilis in wild-type C57BL/6 mice. Infect Immun. 2009;77:1708–1718. doi: 10.1128/IAI.00814-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Prindiville TP, Sheikh RA, Cohen SH, Tang YJ, Cantrell MC, Silva J. Bacteroides fragilis enterotoxin gene sequences in patients with inflammatory bowel disease. Emerg Infect Dis. 2000;6:171–174. doi: 10.3201/eid0602.000210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu S, Rhee KJ, Albesiano E, Rabizadeh S, Wu X, Yen HR, Huso DL, Brancati FL, Wick E, McAllister F, Housseau F, Pardoll DM, Sears CL. A human colonic commensal promotes colon tumorigenesis via activation of T helper type 17 T cell responses. Nat Med. 2009;15:1016–1022. doi: 10.1038/nm.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mazmanian SK, Round JL, Kasper DL. A microbial symbiosis factor prevents intestinal inflammatory disease. Nature. 2008;453:620–625. doi: 10.1038/nature07008. [DOI] [PubMed] [Google Scholar]
- 36.Round JL, Mazmanian SK. Inducible Foxp3+ regulatory T-cell development by a commensal bacterium of the intestinal microbiota. Proc Natl Acad Sci U S A. 2010;107:12204–12209. doi: 10.1073/pnas.0909122107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Coyne MJ, Tzianabos AO, Mallory BC, Carey VJ, Kasper DL, Comstock LE. Polysaccharide Biosynthesis Locus Required for Virulence of Bacteroides fragilis. Infect Immun. 2001;69:4342–4350. doi: 10.1128/IAI.69.7.4342-4350.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Blanton LV, Barratt MJ, Charbonneau MR, Ahmed T, Gordon JI. Childhood undernutrition, the gut microbiota and microbiota-directed therapeutics. Science. 2016;352:1533. doi: 10.1126/science.aad9359. [DOI] [PubMed] [Google Scholar]
- 39.Bäumler AJ, Sperandio V. Interactions between the microbiota and pathogenic bacteria in the gut. Nature. 2016;535:85–93. doi: 10.1038/nature18849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kato N, Liu C, Kato H, Watanabe K, Nakamura H, Iwai N, Ueno K. Prevalence of enterotoxigenic Bacteroides fragilis in children with diarrhea in Japan. J Clin Microbiol. 1999;37:801–803. doi: 10.1128/jcm.37.3.801-803.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chevreux B, Wetter T, Suhai S. Computer Science and Biology: Proceedings of the German Conference on Bioinformatics (GCB); 1999; pp. 45–56. [Google Scholar]
- 42.Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinformatics. 2014;30:2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 43.Chen L, Xiong Z, Sun L, Yang J, Jin Q. VFDB 2012 update: toward the genetic diversity and molecular evolution of bacterial virulence factors. Nucleic Acids Res. 2012;40:D641–645. doi: 10.1093/nar/gkr989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kurtz S, Phillippy A, Delcher AL, Smoot M, Shumway M, Antonescu C, Salzberg SL. Versatile and open software for comparing large genomes. Genome Biol. 2004;5:R12. doi: 10.1186/gb-2004-5-2-r12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ridaura VK, Faith JJ, Rey FE, Cheng J, Duncan AE, Kau AL, Griffin NW, Lombard V, Henrissat B, Bain JR, Muehlbauer MJ, Ilkayeva O, Semenkovich CF, Funai K, Hayashi DK, Lyle BJ, Martini MC, Ursell LK, Clemente JC, Treuren WV, Walters WA, Knight R, Newgard CB, Heath AC, Gordon JI. Gut Microbiota from Twins Discordant for Obesity Modulate Metabolism in Mice. Science. 2013;341:1241214. doi: 10.1126/science.1241214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shoemaker NB, Getty C, Gardner JF, Salyers AA. Tn4351 transposes in Bacteroides spp. and mediates the integration of plasmid R751 into the Bacteroides chromosome. J Bacteriol. 1986;165:929–936. doi: 10.1128/jb.165.3.929-936.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.An J, Muoio DM, Shiota M, Fujimoto Y, Cline GW, Shulman GI, Koves TR, Stevens R, Millington D, Newgard CB. Hepatic expression of malonyl-CoA decarboxylase reverses muscle liver and whole-animal insulin resistance. Nat Med. 2004;10:268–274. doi: 10.1038/nm995. [DOI] [PubMed] [Google Scholar]
- 48.Ferrara CT, Wang P, Neto EC, Stevens RD, Bain JR, Wenner BR, Ilkayeva OR, Keller MP, Blasiole DA, Kendziorski C, Yandell BS, Newgard CB, Attie AD. Genetic networks of liver metabolism revealed by integration of metabolic and transcriptional profiling. PLoS Genet. 2008;4:e1000034. doi: 10.1371/journal.pgen.1000034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jensen MV, Joseph JW, Ilkayeva O, Burgess S, Lu D, Ronnebaum SM, Odegaard M, Becker TC, Sherry AD, Newgard CB. Compensatory responses to pyruvate carboxylase suppression in islet beta-cells. Preservation of glucose-stimulated insulin secretion. J Biol Chem. 2006;281:22342–22351. doi: 10.1074/jbc.M604350200. [DOI] [PubMed] [Google Scholar]
- 50.Monetti M, Levin MC, Watt MJ, Sajan MP, Marmor S, Hubbard BK, Stevens RD, Bain JR, Newgard CB, Farese RV, Hevener AL, Farese RV. Dissociation of hepatic steatosis and insulin resistance in mice overexpressing DGAT in the liver. Cell Metab. 2007;6:69–78. doi: 10.1016/j.cmet.2007.05.005. [DOI] [PubMed] [Google Scholar]
- 51.Merrill AH, Sullards MC, Allegood JC, Kelly S, Wang E. Sphingolipidomics: high-throughput, structure-specific and quantitative analysis of sphingolipids by liquid chromatography tandem mass spectrometry. Methods. 2005;36:207–224. doi: 10.1016/j.ymeth.2005.01.009. [DOI] [PubMed] [Google Scholar]
- 52.Faith JJ, Ahern PP, Ridaura VK, Cheng J, Gordon JI. Identifying gut microbe-host phenotype relationships using combinatorial communities in gnotobiotic mice. Sci Transl Med. 2014;6:220ra11. doi: 10.1126/scitranslmed.3008051. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Experimental designs.
Fig. S2. Generation of a Δbft-3 strain of ETBF.
Fig. S3. Assessing the effects of ETBF on gut barrier/immune function.
Table S1. Summary of anthropometric scores and results of PCR assays for bft in the fecal microbiota of members of a birth cohort of 100 Bangladeshi children.
Table S2. Composition and nutritional content of Bangladeshi diet embodiments, and nutritional requirements of mice and children.
Table S3. V4-16S rRNA, COPRO-Seq, microbial RNA-Seq, and cultured bacterial strain genome sequencing datasets.
Table S4. Bacterial culture collection generated from the fecal microbiota of a severely stunted Bangladeshi child.
Table S5. Genome features of isolates in the clonally arrayed bacterial culture collection generated from the fecal microbiota of a severely stunted 2-year-old Bangladeshi child.
Table S6. Weights of all individual mice as a function of diet, microbiota, and days post gavage.
Table S7. Mass spectrometry-based quantitation of metabolites in host tissues, serum, and cecum obtained from mice in the different treatment groups.
Table S8. Effects of intraspecific and interspecific interactions on ETBF, NTBF, and other Bacteroides gene expression profiles in healthy or stunted donor community contexts.