Abstract
Alzheimer’s disease (AD) is an irreversible neurodegenerative disease and affects a large population in the world. Cognitive scores at multiple time points can be reliably used to evaluate the progression of the disease clinically. In recent studies, machine learning techniques have shown promising results on the prediction of AD clinical scores. However, there are multiple limitations in the current models such as linearity assumption and missing data exclusion. Here, we present a nonlinear supervised sparse regression–based random forest (RF) framework to predict a variety of longitudinal AD clinical scores. Furthermore, we propose a soft-split technique to assign probabilistic paths to a test sample in RF for more accurate predictions. In order to benefit from the longitudinal scores in the study, unlike the previous studies that often removed the subjects with missing scores, we first estimate those missing scores with our proposed soft-split sparse regression–based RF and then utilize those estimated longitudinal scores at all the previous time points to predict the scores at the next time point. The experiment results demonstrate that our proposed method is superior to the traditional RF and outperforms other state-of-art regression models. Our method can also be extended to be a general regression framework to predict other disease scores.
Keywords: Alzheimer’s disease, Clinical scores, Longitudinal study, Random forest, Sparse representation, Soft-split
1. Introduction
Alzheimer’s disease (AD), the most common cause of dementia, is an irreversible and progressive neurodegenerative disease that destroys memory and other important mental functions, resulting in the loss of intellectual and social skills. Mild memory problems are usually the first warning signs of AD. As the disease progresses, dementia symptoms gradually worsen. A convenient strategy for a physician is to conduct neuropsychological tests that can be used to identify behavioral and mental abnormalities associated with the disease. A variety of clinical assessment criteria or scores have been developed for that purpose, such as Mini-Mental State Examination (MMSE) (Folstein et al., 1975), Clinical Dementia Rating–Global and Sum of Boxes (CDR-GLOB and CDR-SOB) (Morris, 1993), and Alzheimer’s Disease Assessment Scale–Cognitive Subscale (ADAS-cog) (Rosen et al., 1984). Many studies have shown the reliable correlation between these clinical scores and the prognosis of AD (Doraiswamy et al., 1996; O’Bryant et al., 2008). They can thus be considered as the quantifiable measurements of the disease progression. Since cognitive problems resulting from neurodegeneration usually begin many years before the clinical onset of AD, it is desirable to accurately predict the disease progression that can be quantitatively represented by the corresponding clinical scores, so that the appropriate treatment plans can be initiated or adjusted.
The progression of AD can also be monitored with medical imaging such as magnetic resonance imaging (MRI). Alterations of image features in gray matter (GM) such as hippocampus atrophy (Morra et al., 2009) and in white matter (WM) such as fornix degeneration (Jin et al., 2015) and network connectivity breakdown (Li et al., 2013; Wang et al., 2016b) have been linked to AD or used to classify AD (Thung et al., 2013, 2014, 2015a, b; Zhan et al., 2014, 2015; Zhu et al., 2014, 2015). In recent years, machine learning techniques have been applied to predict AD-related future clinical scores by imaging-related features, such as average regional GM density and tissue volume of MRI (Duchesne et al., 2009; Fan et al., 2010; Stonnington et al., 2010; Wang et al., 2010; Zhang et al., 2012a), average regional intensity of positron emission tomography (Zhang et al., 2012a), average regional cortical thickness, average regional WM volume, and total cortical surface area (Zhou et al., 2013).
However, there are a few limitations about the current prediction methods. First, those studies assumed linear relationship between the applied features and the clinical scores. Therefore, their proposed regression models were linear, such as generalized linear model (Duchesne et al., 2009), linear support vector machine (SVM) (Fan et al., 2010; Zhang et al., 2012a), relevance vector machine (Stonnington et al., 2010; Wang et al., 2010), or Lasso regression (Zhou et al., 2013). Unfortunately, this critical assumption usually does not hold in the reality. Second, since AD is a slowly progressive disease, it may take many years or even decades for the disease to be developed from its earliest stage to the latest. During that process, multiple MRI scans and scores are usually acquired to monitor the patient’s mental health. Therefore, the longitudinal scores from other time points provide valuable information to predict a future score more accurately. However, many studies used either only the baseline image features to predict baseline scores (Stonnington et al., 2010) or the baseline image features and scores to predict the future scores (Duchesne et al., 2009; Fan et al., 2010; Wang et al., 2010). Although Zhou et al., (2013) included the longitudinal image and score information in their framework, they enforced a temporal smoothness prior to discourage the big discrepancy between the scores of 2 neighboring time points, which may not be the case for some individuals. Finally, the longitudinal scores are often missing at some time points for some patients due to different reasons. A simple strategy is to remove those subjects with missing data (Zhang et al., 2012a). However, this may reduce the number of samples greatly and thus the prediction power of the model. It is highly preferred that the partial set of scores from those subjects with missing data can still be utilized in the algorithm.
To overcome the first aforementioned limitation of model complexity, here, we propose to use a nonlinear learning model–random forest (RF)–to describe the complex relationship between the applied features and the future clinical scores. RF (Breiman, 2001) is a kind of ensemble learning models that has been widely used in brain tissue segmentation (Zhang et al., 2014, 2016). It can generate a nonlinear decision boundary which is more suitable for describing complex patterns such as the relationship between brain image features and the AD clinical scores.
However, the axis-aligned split function in the traditional RF generates a stair-like splitting boundary that causes poor generalization. An oblique split function can generate a more complex boundary with better generalization. Nevertheless, the search of its parameters is difficult in the high dimensional space. Therefore, we propose our sparse regression–based RF and improve the traditional RF model by incorporating it with the Lasso regression. More specifically, first of all, the Lasso regression (Tibshirani, 1996) was used before RF to act as a supervised filter to select the informative features, instead of exhaustive search in the traditional RF. Then, the principal component analysis (PCA) is applied to find the best oblique split function for RF. Another improvement we introduce in our framework is to use the soft-split strategy, instead of hard-split in the traditional RF. Compared to hard split, the soft-split method considers the ambiguity for those hard-to-split samples near the splitting hyperplane at each split node. In soft split, each test sample is allowed to go through both left and right nodes with certain probabilities, which depends on the distance of this sample to the learned splitting decision boundary. Then, the target (e.g., clinical scores) of the sample can be estimated as the weighted average of the statistics in all leaf nodes this sample visits.
For the second and the third limitations, we propose a 2-stage prediction procedure that first estimates the missing scores at different time points by our proposed soft-split sparse regression–based RF, so that a complete score table becomes available to all the subjects at all time points. Then, we make use of the baseline features and also the longitudinal scores at all the previous time points to predict the scores at the next time point.
2. Methods
2.1. Subjects and image acquisition
The data set in this study was obtained from the Alzheimer’s Disease Neuroimaging Initiative (ADNI) databases (http://adni.loni.usc.edu). A total of 805 subjects were included in this study. Multiple sources of imaging data or biological samples were acquired from the participants, such as MRI and positron emission tomography scans and cerebrospinal fluid samples. The cognitive tests were conducted and the corresponding clinical scores were recorded. A series of longitudinal data including those imaging scans and clinical scores were obtained when the subjects were enrolled in the program for the first time (the baseline) and every 12 months at the follow-up visits. Due to different reasons such as subject dropouts, the number of subjects was gradually reduced at the follow-up time points. Here, we selected the baseline MRI T1-weighted (T1w) images and the 4 types of clinical scores (MMSE, CDR-SOB, CDR-GLOB, and ADAS-cog) at the baseline and the sequential 4 follow-up time points with a year apart from each other. The complete subject demography and the clinical score statistics are listed in Table 1.
Table 1.
The demography and the clinical score information of the subjects in this study
| Month | Groups | Number of subjects | Age | MMSE | CDR-SOB | CDR-GLOB | ADAS-cog |
|---|---|---|---|---|---|---|---|
| 0 | NC | 226 (M/F: 122/104) | 75.4 ± 7.0 | 29.1 ± 1.0 | 0.0 ± 0.1 | 0.0 ± 0.0 | 6.2 ± 2.9 |
| MCI | 393 (M/F: 231/162) | 75.3 ± 6.9 | 27.0 ± 1.8 | 1.0 ± 0.8 | 0.5 ± 0.0 | 11.6 ± 4.4 | |
| AD | 186 (M/F: 117/69) | 75.4 ± 6.5 | 23.3 ± 2.0 | 3.3 ± 1.4 | 0.7 ± 0.3 | 18.6 ± 6.3 | |
| 12 | NC | 201 (M/F: 106/95) | 75.0 ± 7.1 | 29.2 ± 1.1 | 0.1 ± 0.2 | 0.0 ± 0.1 | 5.5 ± 2.8 |
| MCI | 351 (M/F: 214/137) | 75.2 ± 6.9 | 26.3 ± 3.0 | 1.6 ± 1.3 | 0.5 ± 0.2 | 12.6 ± 6.2 | |
| AD | 149 (M/F: 90/59) | 75.5 ± 6.3 | 21.3 ± 4.3 | 4.5 ± 2.3 | 1.0 ± 0.5 | 22.3 ± 8.8 | |
| 24 | NC | 192 (M/F: 100/92) | 75.0 ± 7.2 | 29.1 ± 1.1 | 0.1 ± 0.4 | 0.1 ± 0.2 | 5.8 ± 3.0 |
| MCI | 289 (M/F: 179/110) | 75.3 ± 7.0 | 25.4 ± 4.0 | 2.2 ± 1.9 | 0.6 ± 0.3 | 14.1 ± 7.5 | |
| AD | 122 (M/F: 73/49) | 75.5 ± 6.1 | 19.3 ± 5.6 | 6.0 ± 3.0 | 1.3 ± 0.6 | 27.4 ± 11.3 | |
| 36 | NC | 172 (M/F: 93/79) | 74.8 ± 7.4 | 29.0 ± 1.3 | 0.1 ± 0.5 | 0.1 ± 0.2 | 5.3 ± 2.9 |
| MCI | 221 (M/F: 133/88) | 75.6 ± 7.1 | 24.9 ± 4.6 | 2.6 ± 2.3 | 0.7 ± 0.5 | 15.0 ± 8.8 | |
| AD | 9 (M/F: 8/1) | 73.8 ± 4.2 | 17.4 ± 6.6 | 9.0 ± 3.0 | 1.9 ± 0.6 | 30.0 ± 12.7 | |
| 48 | NC | 51 (M/F: 29/22) | 75.0 ± 7.4 | 29.2 ± 1.1 | 0.1 ± 0.3 | 0.1 ± 0.2 | 6.4 ± 2.8 |
| MCI | 50 (M/F: 33/17) | 74.9 ± 6.9 | 24.6 ± 4.3 | 2.9 ± 2.3 | 0.7 ± 0.4 | 14.8 ± 7.3 | |
| AD | 2 (M/F: 1/1) | 75.0 ± 0.0 | 21.0 ± 5.7 | 6.0 ± 4.2 | 1.5 ± 0.7 | 20.0 ± 8.0 |
Age, MMSE, CDR-SOB, CDR-GLOB, and ADAS-cog are listed as mean ± standard deviation.
Key: AD, Alzheimer’s disease; ADAS-cog, Alzheimer’s disease assessment scale–cognitive subscale; CDR-GLOB, clinical dementia rating–global; CDR-SOB, clinical dementia rating–sum of boxes; F, female; M, male; MCI, mild cognitive impairment; MMSE, Mini-Mental State Examination; NC, normal control.
All subjects were scanned with a standardized MRI protocol developed for ADNI. High-resolution structural T1w MRI scans were acquired at 58 sites using 1.5 Tesla MRI scanners. The T1w images were acquired with a sagittal 3D magnetization-prepared rapid gradient-echo (MPRAGE) sequence. Acquisition parameters were as follows: repetition time of 2400 ms, echo time of 3 ms, inversion time of 1000 ms, flip angle of 8°, 24-cm field of view, and a 192 × 192 × 166 acquisition matrix with the voxel size of 1.25 × 1.25 × 1.2 mm3.
2.2. Image preprocessing
Each subject’s T1w images were first corrected for intensity inhomogeneity using the N3 method, and the skull of the brain was also removed. Next, the tissue of the brain was segmented into GM, WM, and cerebrospinal fluid with FAST (automated segmentation tool) in FSL, an open source software package for neuroimaging data analysis (http://fsl.fmrib.ox.ac.uk/fsl/fslwiki/). Then the segmented T1w images were registered to a publicly available atlas Automatic Anatomical Labeling (AAL; Tzourio-Mazoyer et al., 2002) with Hierarchical Attribute Matching Mechanism for Elastic Registration (HAMMER; Shen and Davatzikos, 2002; Shen et al., 1999; Xue et al., 2006). The AAL atlas is a widely used high-resolution T1w GM parcellation with 90 cerebral regions of interest (ROIs) based on anatomical definitions on a single adult subject. The 90 AAL ROIs were assigned to each subject after registration through the deformation fields. The GM regions were selected because GM is affected by AD the most and has been widely used as features in the AD-related studies (Zhang and Shen, 2012b). The volumes of all the 90 ROIs were computed for each subject. In order to filter out the bias caused by the variability of individual brain sizes, each ROI volume was normalized by the total intracranial volume. Finally, the normalized volumes of those ROIs were utilized as features in our prediction algorithm.
2.3. Random forest
An RF consists of a set of decision trees where each tree consists of split nodes and leaf nodes. The split nodes evaluate each arriving sample and depending on the features of the sample, pass it to the left or right child. Each leaf node stores the statistic of the sample that arrives. The training stage of RF is to construct such multiple decision trees. Let Γ denote the entire feature set (the attributes such as ROI volumes, clinical scores, age, sex, etc.). If T trees are generated, then, in the ith tree, a subset of training data Di⊂D is randomly sampled with a replacement from the entire data set D∈ℝ|Γ|×n, where n is the number of subjects. Then, starting from the root node (the top split node), each node in the tree is trained recursively. Taking the jth node as an example, a split function is generated to split the data Di,j at the jth node into the left and right child split nodes with the samples and . To learn the split function, a subset of features Γi,j⊂Γ is selected by random sampling, which further enhances the randomness and diversity among different decision trees in the forest. There are 2 types of split functions, that is, the axis-aligned function and the oblique function. With the traditional axis-aligned split function, only 1 feature and the corresponding threshold are learned as the splitting parameters for each split function, and it can be expressed as:
| (1) |
where x∈ℝ|Γij|×1 is a feature vector; w∈ℝ|Γij|×1 is an indicator vector; is the L0 constraint, which limits to only 1 feature that can be selected; t is the threshold; and H (a) is the Heaviside function (Abramowitz and Stegun, 1965) denoted as:
| (2) |
Therefore, and .
Similarly, the oblique split function can be formulated without the L0 constraint:
| (3) |
To find the optimal w and t, the purity of the split samples is often used as a criterion. It can be defined as (Criminisi et al., 2011):
| (4) |
where V (D) is the variance defined as:
| (5) |
where (x, y) means a pair of feature and target vectors of a sample. Finally, the objective of each split function is to maximize the purity of the samples after the split:
| (6) |
In this way, each node will continue splitting until it reaches certain stopping criteria such as the minimum number of samples or the maximum of tree depth, and then the statistical information (e.g., mean and variance) of the inseparable target group in the sub-data set for each tree will be stored in the leaf nodes for future prediction.
The testing process of RF is very similar to the training process. First, a new test sample is fed into the root node of each tree. By using the split function learned in the training stage, this new sample is then classified into either the left or the right child node until it reaches a leaf node. For each decision tree, the prediction result for the new sample is presented by the statistic of the training samples stored in the leaf node and the final result of RF is obtained by averaging the prediction results across all the decision trees.
2.4. Sparse regression in random forest
In RF, each split function breaks down the remaining samples at the node into 2 subsets. According to Equation 6, given a data set, the best split function can be learned by searching a pair of optimal splitting parameters w and t that can maximize the purity. Here, w can be considered as the normal of the splitting hyperplane and the threshold t as the intercept of the hyperplane along the normal direction.
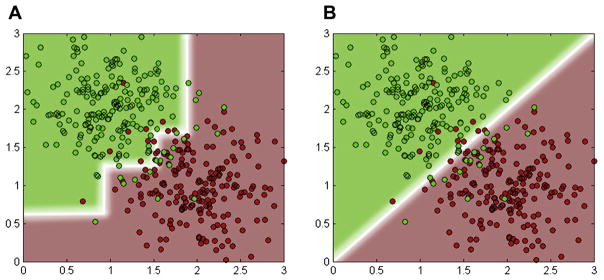
The axis-aligned split function uses only 1 feature at a time and separates the feature space of the training samples by a hyperplane that is aligned to the feature axes (Fig. 1A). The axis-aligned split function is efficient to compute, but it generates an unsmooth blocky decision boundary in the feature space. Sometimes, it might be different from the real situation and tends to result in bad generalization (Hastie et al., 2009). In contrast, the oblique split function uses multiple features and learns a hyperplane oblique to the axes (Fig. 1B). It outperforms the axis-aligned function by generating a smoother and more complex boundary (Breiman, 2001). The RF algorithms with oblique split function show better generalization capability and are not easily susceptible to over-fitting, compared to the traditional axis-aligned split function (Menze et al., 2011; Tan and Dowe, 2006). However, considering the fact that the conventional RF uses exhaustive search to find the optimal splitting parameters, the search space grows exponentially with the dimension of w. This makes the optimization of the oblique split function computationally demanding in practice.
Fig. 1.
The decision boundaries generated by (A) an axis-aligned split function and (B) an oblique split function.
To solve this drawback in the oblique split function, we propose to use the Lasso regression (Tibshirani, 1996) as a supervised filter to select the informative features, instead of exhaustive search, when learning the oblique split function. Meanwhile, the Lasso regression also maps the original features into the target-like features, which are expected to be more discriminative, thus better suited for splitting the samples. Afterwards, an oblique hyperplane can be easily found by PCA in the mapped feature space.
More specifically, in each node, we first randomly resample a feature subset Γ’ from the entire feature set Γ (Γ’⊂Γ). Then the new feature vector x∈ℝ|Γ’|×1 is regarded as the input and the d-dimentional target vector y∈ℝd×1 (e.g., the d types of clinical scores) as the regression target for the Lasso regression. The optimal selected feature set can be obtained by solving the following objective function:
| (7) |
where B∈ℝ|Γ’|×d is the sparse coefficient matrix; the L1 norm of B; λ is a regularization parameter that controls the sparsity of B.
By solving Equation 7, the mapped target-like feature ỹ∈ℝd×1 can be obtained by multiplying the original feature vector with the sparse coefficients matrix:
| (8) |
In the mapped feature space, based on the definition of oblique split function or splitting hyperplane, the original one in Equation 3 becomes:
| (9) |
where u is the maximum purity direction in the space spanned by ỹ. Then, the search of the splitting hyperplane that separates the sample set with the maximization of purity is equivalent to clustering the mapped samples into 2 clusters. According to Ding and He (2004), the normal of the hyperplane obtained by k means clustering when k = 2 is equal to the first principal component of PCA. Therefore, we first apply PCA to the set of y˜s and find the first principal component p∈ℝd×1 that is considered as the maximum purity direction, that is, u = p. Now plugging Equation 8 into Equation 9, the splitting function becomes:
| (10) |
By comparing Equation 3 with Equation 10, we can readily see that:
| (11) |
After solving w analytically, the only remaining unknown parameter t, that is, the intercept of the hyperplane, can be determined by exhaustive search. That is, we randomly sample a set of hyperplanes orthogonal to w and pick the one that leads to the maximum purity as defined in Equation 4. Since t is a scalar, the computation is much less demanding. After obtaining both parameters {w,t}, they are saved in the split node for prediction at the testing stage.
2.5. Soft split in random forest
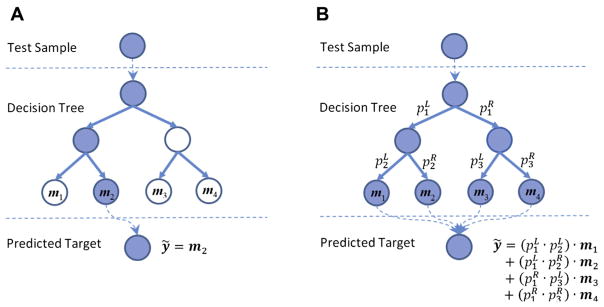
In the conventional RF, when a test sample comes to a split node, it will be pushed toward either the left or the right child node, based on the split function. By recursively passing this sample from the root node toward the bottom of the tree, it will finally reach a leaf node, where a particular group of the training samples lie. The statistic of those samples (e.g., the mean of the regression targets such as clinical scores) is then retrieved as the prediction result of this tree. Fig. 2A illustrates such a process.
Fig. 2.
A testing procedure of (A) the hard-split decision tree and (B) the soft-split decision tree. The paths connecting those blue nodes are the paths that the test sample goes through during the testing procedure. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
The above split criterion is called hard split. It works well when the samples can be well split by the split function with a big margin. However, sometimes it is not the case in practice. There can be many “hard-to-split” samples close to the splitting hyperplane, and they will make a clear separation or split difficult. The conventional hard split tends to ignore this fact and may misclassify samples to a wrong side, thus leading to inaccurate prediction.
To solve this problem, we introduce a novel concept of soft split. When a test sample comes to a split node, instead of classifying it into only 1 child node, we implement the probabilistic split and assign this sample to both child nodes with certain probabilities. Consequently, unlike the conventional RF in which each test sample is predicted by only 1 leaf node, our soft-split strategy predicts the regression target of a test sample by a linear fusion of the targets in multiple leaf nodes it visits with nonzero probabilities.
Mathematically, for each split node, the probabilities of a test sample x assigned to the left (pL) and the right (pR) child nodes are defined as:
| (12) |
| (13) |
where r = wTx−t is the signed distance of this sample to the splitting hyperplane (Eq. 3); r’∈[−1,1] is the normalized signed distance by the minimum distance (rmin) and the maximum distance (rmax) of the training samples at this node to the hyperplane; s is the parameter that controls the slope of the sigmoid function; and ε is an infinitesimal number to prevent the division by 0. In Equation 12, the further this sample is from the hyperplane, the more extreme the probabilities are. For those hard-to-split samples, as they are often located close to the splitting hyperplane, they tend to have a similar probability to be assigned to the left or the right child node. By combining Equation 10 and Equation 12, the sparse oblique split function with soft split can be formulated as follows:
| (14) |
Therefore, and , where c is a small number (e.g., c = 0.1) controlling the selection between soft split and hard split. Specifically, if either pL or pR is smaller than c, that is, f (x|w; t) ≥ 1 − c or f (x|w; t) < c, the soft split will be replaced with hard split for improving the computing efficiency.
Finally, at each leaf node, its weight is computed as the multiplication of all probabilities along the path the sample goes through starting from the root node. The prediction of the test target by this tree is calculated as the weighted average of the mean statistics contained in the leaf nodes with nonzero probabilities. Fig. 2B illustrates our proposed soft-split RF.
In the end, our proposed regression model is named soft-split sparse regression–based RF by incorporating both techniques in Section 2.4 and Section 2.5 into the traditional RF described in Section 2.3.
2.6. Longitudinal score prediction
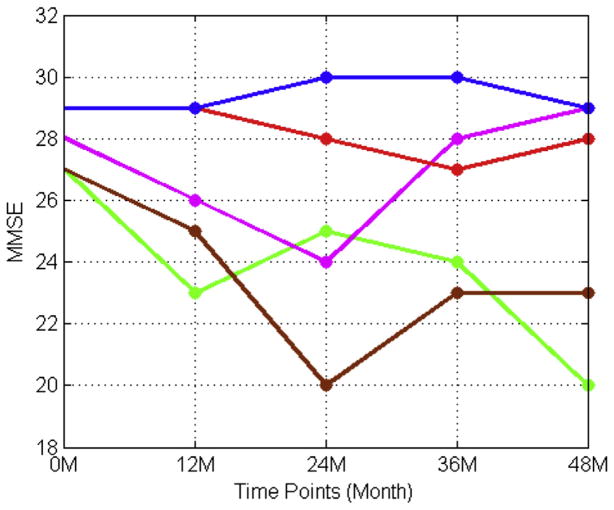
Since the progression of AD is a slow process, patients are usually evaluated on their cognitive wellness at multiple time points throughout the prognosis of the disease. Utilizing the information from multiple previous time points may provide a more accurate result of predicting the scores at the next time point than only using the baseline scores. In general, it is reasonable to assume that those scores from different time points are not independent. Zhou et al. (2013) assumed that the scores between 2 successive time points should be close. Therefore, they introduced a temporal smoothness prior into their regression model to discourage large deviations between predictions at neighboring time points. However, in clinical practice, this assumption may not always hold. Fig. 3 shows how the real MMSE scores of several subjects from our data set were changed over the years. Steady periods and sharp declines intertwined with occasional reverse improvements. This indicates that longitudinal clinical scores may have more complex relations than a simple linear correlation.
Fig. 3.
The change patterns of several patients’ MMSE scores over the 5 time points including the baseline examination. Abbreviation: MMSE, Mini-Mental State Examination.
RF is a more complex model that can handle nonlinearity in the data better than the Lasso regression used by Zhou et al. (2013). In order to more accurately predict the scores at a time point, we adopted the scores from all previous time points before this target time point as the context features to train our regression model. However, it is common to encounter missing clinical scores for some subjects in such a large longitudinal study. A common strategy is to keep only those subjects who have scores at every time point and remove the others; however, the participants with missing clinical scores accounted for a large percentage of the ADNI data set. Simply removing these subjects would significantly reduce the size of the training samples and consequently undermine the power of our learned regression model.
An alternative way is to estimate the missing scores for each subject before training the regression model. The most direct way is to perform the linear interpretation. Nevertheless, the performance would suffer on those subjects whose scores do not change linearly. Here, we propose a 2-stage longitudinal score prediction scheme. First, we train our regression model using only the baseline features to predict the future clinical scores at each follow-up time point using our proposed algorithm. Since the baseline data are complete, we are able to predict the missing clinical scores of any subject. Then, we conduct a second-round training process in which instead of using only the baseline features as inputs, we also include either the ground-truth scores if existing or the predicted scores from the first training stage at each future time point into the training features to build our regression model. Similarly, in the prediction or testing stage, during the first prediction process, we only use the baseline features of a testing subject to predict his or her clinical scores at all the previous time points before the target time point, if any of them is missing, with the regression model obtained from the first-round training. Then, the predicted missing scores along with the baseline features and the known longitudinal scores are fed into the regression model obtained from the second-round training to predict the score at the target time point for that subject.
3. Results
3.1. Experiment setup
We applied our soft-split sparse regression–based RF to predict 4 types of clinical scores (MMSE, CDR-SOB, CDR-GLOB, and ADAS-cog) at the future time points from the baseline time point with the data set downloaded from ADNI (Table 1). The features we used were the normalized volumes of the 90 ROIs transformed from the AAL atlas and the baseline scores. The prediction performance was measured by a 10-fold cross validation. The RF parameters were selected as follows: the number of trees = 10, the number of candidates for the search of the optimal t = 10, the number of the randomly sampled feature variables at each node = 60, the lower bound of the sample size at each node = 3, the number of the randomly drawn samples in each tree = 720, and the upper bound of tree depth = 20. The weights λ of the regularization term in Equation 7 and the slopes s of the sigmoid function in Equation 12 were automatically determined by a 2-fold cross validation on the training set.
The mean absolute error (MAE) and the Pearson’s correlation coefficient (R) between the predicted scores and the ground truth were used to evaluate the prediction performance, respectively. At the single point, MAE and R are defined as:
| (15) |
| (16) |
where yis are the ground-truth clinical scores, ŷis are the corresponding predicted scores, ŷ and are the mean values of yis and ŷis, respectively, and n is the number of subjects.
The overall performance at multiple time points can be evaluated with the total MAE (i.e., the MAE across all the subjects at multiple time points) and the weighted R (wR) that can be defined as:
| (17) |
where Ri is the Pearson’s correlation coefficient defined in Equation 16 at the ith time point, ni is the number of subjects at the ith time point, and t is the number of the time points.
3.2. Prediction with baseline scores
First, we predicted the clinical scores of the subjects in our data set at the 5 future time points (the 12th, 24th, 36th, and 48th month) with only the baseline clinical scores and the MRI ROI volume features. To demonstrate the performance of our proposed sparse regression–based RF (oblique RF with the L1 norm constraint [oRF-L1]) and soft-split sparse regression–based RF methods (oblique RF with L1 and soft split [oRF-L1-soft]), we compared their performance with several other RF regression models, that is, the conventional axis-aligned RF, the oblique RF with the L2 norm constraint (oRF-L2), the oblique RF with L2 and soft split (oRF-L2-soft).
Table 2 shows the MAEs of the 5 methods on the 4 types of clinical scores per each time point, respectively. Table 3 shows the corresponding Rs, respectively. In terms of the MAE, the proposed oRF-L1 and oRF-L1-soft achieved the best performance at most of the time points for each type of clinical scores. For example, for MMSE, oRF-L1-soft had a relative prediction accuracy improvement of 6%, 4%, 11%, and 15% at the 12th, 24th, 36th, and 48th month time points, respectively, compared to the traditional RF. In addition, oRF-L1-soft achieved the highest R at all the time points for all types of clinical scores. For example, the least correlation increase was 1% at the 24th month for CDR-GLOB, and the most increase was 14% at the 48th month for CDR-SOB, compared to the traditional RF.
Table 2.
The MAEs of the proposed methods (oRF-L1 and oRF-L1-soft) and other RF models (RF, oRF-L2, and oRF-L2-soft)
| Month | Method | MAE (mean ± standard deviation)
|
|||
|---|---|---|---|---|---|
| MMSE | CDR-SOB | CDR-GLOB | ADAS-cog | ||
| 12 | RF | 1.778 ± 0.198 | 0.724 ± 0.088 | 0.142 ± 0.023 | 3.426 ± 0.372 |
| oRF-L2 | 1.764 ± 0.235 | 0.743 ± 0.135 | 0.154 ± 0.030 | 3.440 ± 0.337 | |
| oRF-L2-soft | 1.722 ± 0.218 | 0.737 ± 0.134 | 0.158 ± 0.028 | 3.444 ± 0.357 | |
| oRF-L1 | 1.744 ± 0.163 | 0.692 ± 0.053 | 0.137 ± 0.022a | 3.422 ± 0.231 | |
| oRF-L1-soft | 1.675 ± 0.176a | 0.685 ± 0.068 | 0.146 ± 0.021 | 3.275 ± 0.249a | |
| 24 | RF | 2.118 ± 0.270 | 0.999 ± 0.159 | 0.222 ± 0.032 | 4.298 ± 0.589 |
| oRF-L2 | 2.117 ± 0.371 | 1.031 ± 0.143 | 0.229 ± 0.031 | 4.194 ± 0.701 | |
| oRF-L2-soft | 2.093 ± 0.367 | 1.020 ± 0.138 | 0.236 ± 0.034 | 4.098 ± 0.686 | |
| oRF-L1 | 2.101 ± 0.314 | 1.014 ± 0.127 | 0.226 ± 0.024 | 4.227 ± 0.555 | |
| oRF-L1-soft | 2.043 ± 0.294a | 0.979 ± 0.122a | 0.229 ± 0.027 | 4.070 ± 0.483a | |
| 36 | RF | 2.028 ± 0.346 | 0.975 ± 0.150 | 0.223 ± 0.030 | 3.965 ± 0.985 |
| oRF-L2 | 2.009 ± 0.328 | 0.912 ± 0.129 | 0.223 ± 0.028 | 3.887 ± 0.756 | |
| oRF-L2-soft | 1.950 ± 0.325 | 0.894 ± 0.124 | 0.224 ± 0.026 | 3.753 ± 0.651 | |
| oRF-L1 | 1.838 ± 0.249 | 0.893 ± 0.150 | 0.215 ± 0.033a | 3.770 ± 0.909 | |
| oRF-L1-soft | 1.808 ± 0.247a | 0.883 ± 0.113a | 0.220 ± 0.023 | 3.653 ± 0.778a | |
| 48 | RF | 1.939 ± 0.593 | 1.160 ± 0.470 | 0.250 ± 0.104 | 3.653 ± 0.866 |
| oRF-L2 | 1.871 ± 0.580 | 0.998 ± 0.497 | 0.243 ± 0.106 | 3.785 ± 0.824 | |
| oRF-L2-soft | 1.830 ± 0.568 | 1.006 ± 0.511 | 0.244 ± 0.107 | 3.729 ± 0.821 | |
| oRF-L1 | 1.693 ± 0.512 | 0.977 ± 0.472 | 0.236 ± 0.095a | 3.635 ± 0.833 | |
| oRF-L1-soft | 1.656 ± 0.469a | 0.978 ± 0.475 | 0.240 ± 0.092 | 3.565 ± 0.847a | |
The best results are shown in bold.
Key: ADAS-cog, Alzheimer’s Disease Assessment Scale–Cognitive Subscale; CDR-GLOB, Clinical Dementia Rating–Global; CDR-SOB, Clinical Dementia Rating–Sum of Boxes; MAE, mean absolute error; MMSE, Mini-Mental State Examination; oRF-L1, oblique RF with the L1 norm constraint; oRF-L1-soft, oblique RF with L1 and soft split; oRF-L2, oblique RF with the L2 norm constraint; oRF-L2-soft, oblique RF with L2 and soft split; RF, random forest.
Represents that the result in bold is statistically significantly better than other comparison methods (p < 0.05).
Table 3.
The Pearson’s correlation coefficients (R) of the proposed methods (oRF-L1 and oRF-L1-soft) and other RF models (RF, oRF-L2, and oRF-L2-soft)
| Month | Method | R
|
|||
|---|---|---|---|---|---|
| MMSE | CDR-SOB | CDR-GLOB | ADAS-cog | ||
| 12 | RF | 0.807 | 0.840 | 0.813 | 0.840 |
| oRF-L2 | 0.800 | 0.829 | 0.798 | 0.828 | |
| oRF-L2-soft | 0.816 | 0.836 | 0.805 | 0.833 | |
| oRF-L1 | 0.813 | 0.849 | 0.823 | 0.842 | |
| oRF-L1-soft | 0.826 | 0.855 | 0.830 | 0.852 | |
| 24 | RF | 0.809 | 0.835 | 0.804 | 0.828 |
| oRF-L2 | 0.810 | 0.833 | 0.802 | 0.827 | |
| oRF-L2-soft | 0.82 | 0.845 | 0.812 | 0.839 | |
| oRF-L1 | 0.809 | 0.841 | 0.804 | 0.828 | |
| oRF-L1-soft | 0.822 | 0.852 | 0.816 | 0.841 | |
| 36 | RF | 0.765 | 0.785 | 0.778 | 0.789 |
| oRF-L2 | 0.775 | 0.825 | 0.788 | 0.800 | |
| oRF-L2-soft | 0.792 | 0.842 | 0.806 | 0.819 | |
| oRF-L1 | 0.821 | 0.831 | 0.802 | 0.824 | |
| oRF-L1-soft | 0.832 | 0.843 | 0.813 | 0.836 | |
| 48 | RF | 0.742 | 0.653 | 0.679 | 0.731 |
| oRF-L2 | 0.731 | 0.717 | 0.689 | 0.697 | |
| oRF-L2-soft | 0.747 | 0.722 | 0.693 | 0.708 | |
| oRF-L1 | 0.800 | 0.738 | 0.718 | 0.745 | |
| oRF-L1-soft | 0.804 | 0.746 | 0.725 | 0.752 | |
The best results are shown in bold. The p-values associated with each R is less than 10 −5 (statistically significant)
Key: ADAS-cog, Alzheimer’s Disease Assessment Scale–Cognitive Subscale; CDR-GLOB, clinical dementia rating–global; CDR-SOB, clinical dementia rating–sum of boxes; MMSE, Mini-Mental State Examination; oRF-L1, oblique RF with the L1 norm constraint; oRF-L1-soft, oblique RF with L1 and soft split; oRF-L2, oblique RF with the L2 norm constraint; oRF-L2-soft, oblique RF with L2 and soft-split; RF, random forest.
We also compared the performance of oRF-L1 and oRF-L1-soft with that of other state-of-art regression models, such as Lasso regression, ridge regression, and SVM. The MAEs and the wRs across all the 4 time points for all these methods per each type of clinical scores are listed in Table 4 and Table 5, respectively. oRF-L1 outperformed other regression models, that is, Lasso regression, ridge regression, and SVM. And, with the inclusion of the soft-split technique, oRF-L1-soft further improved the performance. For instance, for MMSE, the MAE of oRF-L1 was decreased by 12%, 18%, and 5%, compared to that of Lasso regression, ridge regression, and SVM, respectively, while the wR increased by 5%, 3%, and 3%, respectively. oRF-L1-soft further decreased the MAE by 3% and increased the wR by 1% from those of oRF-L1.
Table 4.
The total MAEs of the proposed methods (oRF-L1-soft) and other regression methods (Lasso regression, ridge regression, and SVM) across all the 4 future time points
| Method | MAE (mean ± standard deviation)
|
|||
|---|---|---|---|---|
| MMSE | CDR-SOB | CDR-GLOB | ADAS-cog | |
| LASSO | 2.088 ± 0.347 | 1.002 ± 0.180 | 0.215 ± 0.044 | 3.794 ± 0.614 |
| Ridge | 2.029 ± 0.353 | 0.994 ± 0.180 | 0.215 ± 0.043 | 3.731 ± 0.585 |
| SVM | 1.945 ± 0.380 | 0.970 ± 0.223 | 0.205 ± 0.052 | 3.818 ± 0.622 |
| oRF-L1 | 1.844 ± 0.310 | 0.894 ± 0.200 | 0.203 ± 0.043 | 3.764 ± 0.632 |
| oRF-L1-soft | 1.796 ± 0.297a | 0.881 ± 0.195a | 0.209 ± 0.041 | 3.641 ± 0.589a |
The best results are shown in bold.
Key: ADAS-cog, Alzheimer’s Disease Assessment Scale–Cognitive Subscale; CDR-GLOB, clinical dementia rating–global; CDR-SOB, clinical dementia rating–sum of boxes; MAE, mean absolute error; MMSE, Mini-Mental State Examination; oRF-L1, oblique RF with the L1 norm constraint; oRF-L1-soft, oblique RF with L1 and soft split; SVM, support vector machine.
Represents that the result in bold is statistically significantly better than other comparison methods (p < 0.05).
Table 5.
The wRs of the proposed methods (oRF-L1-soft) and other regression methods (Lasso regression, ridge regression, and SVM) across all the 4 future time points
| Method | wR
|
|||
|---|---|---|---|---|
| MMSE | CDR-SOB | CDR-GLOB | ADAS-cog | |
| Lasso | 0.775 | 0.793 | 0.777 | 0.802 |
| Ridge | 0.789 | 0.799 | 0.781 | 0.809 |
| SVM | 0.786 | 0.788 | 0.775 | 0.793 |
| oRF-L1 | 0.811 | 0.815 | 0.787 | 0.810 |
| oRF-L1-soft | 0.821 | 0.824 | 0.796 | 0.820 |
The best results are shown in bold.
Key: ADAS-cog, Alzheimer’s disease assessment scale–cognitive subscale; CDR-GLOB, clinical dementia rating–global; CDR-SOB, clinical dementia rating–sum of boxes; MMSE, Mini-Mental State Examination; oRF-L1, oblique RF with the L1 norm constraint; oRF-L1-soft, oblique RF with L1 and soft split; SVM, support vector machine; wRs, weighted Pearson’s correlation coefficients.
At last, we compared the prediction accuracy of our method with that of other works. For example, Duchesne et al. (2009) used the baseline MRI, age, gender, and years of education as features to predict the MMSE scores at the 12th month (75 normal control subjects-NC, 49 mild cognitive impairment patients-MCI, and 75 AD patients) and only achieved a correlation of 0.31. The R of MMSE at the 12th month with our proposed oRF-L1-soft was 0.83, which was a relative gain of 168%.
3.3. Most discriminative regions
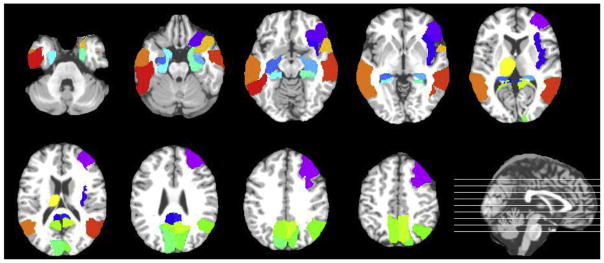
The Lasso regression can effectively select the most discriminative image features relevant to score prediction. Here, we counted the MRI features (the normalized ROI volumes) that were selected via the Lasso regression described in Section 2.4 at each root node of a tree with oRF-L1-soft over all the 10 trees throughout the 10-fold cross validation. We repeated this procedure on the predictions of all 4 future time points and obtained the total counts for each MRI feature. The top 20 most selected regions are shown in color in Fig. 4. Those regions included the right middle frontal lobe, the right inferior orbitofrontal cortex, the right insula, the bilateral posterior cingulate cortices, the bilateral hippocampi, the bilateral parahippocampal gyri, the bilateral cunei, the right superior parietal lobe, the bilateral precunei, the left thalamus, the right superior temporal pole, the bilateral middle temporal lobes, and the left inferior temporal lobe. Most of these regions are well known to show the abnormality in AD.
Fig. 4.
The top 20 most discriminative regions from the AAL atlas are shown in color in the axial slices. The position of each slice is marked by a line on the sagittal image at the bottom right corner. The lines from the bottom to the top correspond to the axial slices from the upper-left corner to the bottom-right corner, respectively. Abbreviation: AAL, automatic anatomical labeling. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
3.4. Validation with scanning sites
Although cross validation is a reliable technique to fairly evaluate the performance of an algorithm, in order to address the issue of possible overfitting, we designed an alternative scenario with the current data set to further validate our framework. The current ADNI data set was acquired from 58 different sites across North America. Images were acquired with different scanners at different sites, so the data from each site can be considered as an independent data set. We grouped the subjects from 5 randomly selected sites as our test data set (18 NC, 33 MCI, and 11 AD).We used the remaining subjects tobuild our RF model and then tested on the test data set. The results of MAE and R are shown in Table 6 and Table 7, respectively. These results were in line with those using the entire data set, which demonstrated the robustness and reliability of our proposed methods.
Table 6.
The MAEs of the proposed methods (oRF-L1 and oRF-L1-soft) on the data set from the randomly selected 5 acquisition sites
| Month | Method | MAE
|
|||
|---|---|---|---|---|---|
| MMSE | CDR-SOB | CDR-GLOB | ADAS-cog | ||
| 12 | oRF-L1 | 1.293 | 0.707 | 0.149 | 3.090 |
| oRF-L1-soft | 1.285 | 0.694 | 0.132 | 3.000 | |
| 24 | oRF-L1 | 1.760 | 0.915 | 0.224 | 3.421 |
| oRF-L1-soft | 1.739 | 0.866 | 0.222 | 3.186 | |
| 36 | oRF-L1 | 1.759 | 0.824 | 0.219 | 3.069 |
| oRF-L1-soft | 1.701 | 0.780 | 0.231 | 2.879 | |
| 48 | oRF-L1 | 0.533 | 0.250 | 0.120 | 1.202 |
| oRF-L1-soft | 0.481 | 0.249 | 0.131 | 0.862 | |
The best results are shown in bold.
Key: ADAS-cog, Alzheimer’s disease assessment scale–cognitive subscale; CDR-GLOB, clinical dementia rating–global; CDR-SOB, clinical dementia rating–sum of boxes; MAE, mean absolute error, MMSE, Mini-Mental State Examination; oRF-L1, oblique RF with the L1 norm constraint; oRF-L1-soft, oblique RF with L1 and soft split.
Table 7.
The Rs of the proposed methods (oRF-L1 and oRF-L1-soft) on the data set from the randomly selected 5 acquisition sites
| Month | Method | R
|
|||
|---|---|---|---|---|---|
| MMSE | CDR-SOB | CDR-GLOB | ADAS-cog | ||
| 12 | oRF-L1 | 0.847 | 0.815 | 0.810 | 0.877 |
| oRF-L1-soft | 0.852 | 0.836 | 0.807 | 0.887 | |
| 24 | oRF-L1 | 0.873 | 0.849 | 0.822 | 0.898 |
| oRF-L1-soft | 0.873 | 0.879 | 0.859 | 0.918 | |
| 36 | oRF-L1 | 0.888 | 0.930 | 0.866 | 0.926 |
| oRF-L1-soft | 0.894 | 0.935 | 0.877 | 0.930 | |
| 48 | oRF-L1 | n/a | n/a | n/a | n/a |
| oRF-L1-soft | n/a | n/a | n/a | n/a | |
The best results are shown in bold. Since there are only 3 subjects at the 48th month, the Rs are not applicable.
Key: ADAS-cog, Alzheimer’s disease assessment scale–cognitive subscale; CDR-GLOB, clinical dementia rating–global; CDR-SOB, clinical dementia rating–sum of boxes; MMSE, Mini-Mental State Examination; oRF-L1, oblique RF with the L1 norm constraint; oRF-L1-soft, oblique RF with L1 and soft split; Rs, Pearson’s correlation coefficients.
3.5. Prediction with longitudinal scores
We applied the proposed strategy in Section 2.6 to utilize the longitudinal score information at all previous time points to predict the scores at the next time point. For example, we first predicted the missing scores at the 12th month with the baseline features (the scores and the MRI ROI volumes) and then combined both the baseline features and the complete scores at the 12th month to predict the scores at the 24th month. We refer this strategy as oRF-L1-soft-long in the rest of the article. To evaluate the performance of oRF-L1-soft-long, we compared it with oRF-L1-soft (by using only the baseline features) and oRF-L1-soft-interp (by linearly interpolating the missing longitudinal scores at the previous time points). The total MAEs across all the 4 time points with the 3 methods are listed in Table 8. The comparison of the wRs with the 3 methods is shown in Table 9.
Table 8.
The total MAEs of oRF-L1-soft (only baseline features), oRF-L1-soft-interp (linear interpolation), and oRF-L1-soft-long (our proposed algorithm with longitudinal data) across all the 4 future time points
| Method | MAE (mean ± standard deviation)
|
|||
|---|---|---|---|---|
| MMSE | CDR-SOB | CDR-GLOB | ADAS-cog | |
| oRF-L1-soft | 1.796 ± 0.297 | 0.881 ± 0.195 | 0.209 ± 0.041 | 3.641 ± 0.589 |
| oRF-L1-soft-interp | 1.633 ± 0.263 | 0.793 ± 0.182 | 0.220 ± 0.042 | 3.201 ± 0.496 |
| oRF-L1-soft-long | 1.548 ± 0.214a | 0.703 ± 0.160a | 0.187 ± 0.035a | 2.913 ± 0.404a |
The best results are shown in bold.
Key: ADAS-cog, Alzheimer’s disease assessment scale–cognitive subscale; CDR-GLOB, clinical dementia rating–global; CDR-SOB, clinical dementia rating–sum of boxes; MAE, mean absolute error; MMSE, Mini-Mental State Examination; oRF-L1-soft, oblique RF with L1 and soft split.
Represents that the result in bold is statistically significantly better than other comparison methods (p < 0.05).
Table 9.
The wRs of oRF-L1-soft (only baseline features), oRF-L1-soft-interp (linear interpolation), and oRF-L1-soft-long (our proposed algorithm with longitudinal data) across all the 4 future time points
| Method | wR
|
|||
|---|---|---|---|---|
| MMSE | CDR-SOB | CDR-GLOB | ADAS-cog | |
| oRF-L1-soft | 0.821 | 0.824 | 0.796 | 0.613 |
| oRF-L1-soft-interp | 0.867 | 0.882 | 0.835 | 0.709 |
| oRF-L1-soft-long | 0.877 | 0.897 | 0.857 | 0.715 |
The best results are shown in bold.
Key: ADAS-cog, Alzheimer’s disease assessment scale–cognitive subscale; CDR-GLOB, clinical dementia rating–global; CDR-SOB, clinical dementia rating–sum of boxes; MMSE, Mini-Mental State Examination; oRF-L1-soft, oblique RF with L1 and soft split; wRs, weighted Pearson’s correlation coefficients.
oRF-L1-soft-long showed the best performance in both of the measures for all the 4 types of clinical scores. With the inclusion of the longitudinal scores, the MAE was reduced 14% for MMSE, 21% for CDR-SOB, 10% for CDR-GLOB, and 20% for ADAS-cog, compared to oRF-L1-soft with only using the baseline features. The same trend was observed in the wR. The wRs of oRF-L1-soft-long increased by 7%–9% versus that of oRF-L1-soft.
Zhou et al. (2013) also utilized the longitudinal clinical scores to predict the clinical scores at the future time points along with the baseline MRI features (such as clinical scores, cortical thickness, cortical volumes, WM ROI volumes, and the total cortical surface area) using the similar number of subjects from the ADNI database. Their proposed methods were named Temporal Group Lasso (TGL) and convex Fused Sparse Group Lasso (cFSGL). In order to have a fair comparison between our method and their methods (TGL and cFSGL), we adapted their strategy at the training and testing stages with oRF-L1-soft-long and re-evaluated the performance of our method with their proposed measures. They used root mean square error (rMSE) instead of MAE to evaluate the error of prediction. It is defined as:
| (18) |
where ys are the ground-truth clinical scores at a single time point, ŷs are the corresponding predicted scores, and n is the number of subjects.
Table 10 lists the performance comparison between their methods and ours with the overall wR and the rMSEs at the 4 time points, respectively. Except that at the 24th month, our method achieved the comparable performance as theirs in terms of rMSE, our method showed its superiority to theirs in rMSE at any other time points (15% reduced at the 12th month, 12% at the 36th month, and 18% at the 48th month, compared to that of cFSGL, respectively) and the overall wR (4% better than cFSGL).
Table 10.
The performance comparison of our proposed method (oRF-L1-soft-long) and TGL and cFSGL used in Zhou et al. (2013) on the prediction of MMSE at the 12th, 24th, 36th and 48th month, in terms of rMSE and the overall wR
| Method | wR | rMSE
|
|||
|---|---|---|---|---|---|
| M12 | M24 | M36 | M48 | ||
| TGL | 0.755 | 2.923 | 3.363 | 3.768 | 3.631 |
| cFSGL | 0.796 | 2.762 | 3.000 | 3.265 | 2.871 |
| oRF-L1-soft-long | 0.825 | 2.352 | 3.069 | 2.871 | 2.362 |
The best results are shown in bold.
Key: cFSGL, convex Fused Sparse Group Lasso; MMSE, Mini-Mental State Examination; rMSE, root mean square error; TGL, Temporal Group Lasso; wR, weighted Pearson’s correlation coefficient.
4. Discussion
In this study, we propose to predict 4 types of clinical scores: MMSE, CDR-SOB, CDR-GLOB, and ADAS-cog at the future time points with the baseline scores and the MRI-derived features. In previous studies, the relationship has been reported between those cognitive clinical scores and MRI-based imaging features such as regional volume change (Duchesne et al., 2009), GM density (Apostolova et al., 2006), local shape variations of ventricles (Ferrarini et al., 2008), etc. Therefore, it is reasonable to assume that MRI image features can be used to predict clinical scores. However, the linear relationship between them may not hold, resulting in the suboptimal performance of the regression models with linear characteristics (Duchesne et al., 2009; Fan et al., 2010; Stonnington et al., 2010; Wang et al., 2010; Zhang et al., 2012a; Zhou et al., 2013). On the other hand, RF has many advantages over linear regression. First, it does not expect any linear features or even features that interact linearly. Second, due to how it is constructed (using bagging or boosting), it can handle very well the high dimensional space as well as the large number of training examples. This has been demonstrated in the results listed in Tables 4 and 5 when comparing our results with those by different linear regression models.
Our proposed sparse regression–based RF first transforms the original features (the scores and the MRI features) to the target-like features, and then the traditional RF is performed on those target-like features for score prediction. At each split node, the Lasso regression is performed to select the best target-like features based on the samples at that node. At its child nodes, the new Lasso regressions are performed based on the subset of samples the child nodes get after the split. It forms a set of hierarchical sparse regressions to ensure the best feature selection for sample separation at each node. As the nodes go down from the root to the leafs, the formed target-like features from sparse regressions change from the general representation from a large group of subjects to the specific representation from a few individual subjects. With such a technique, our method oRF-L1 shows better performance than the traditional RF or other regression models such as Lasso regression, ridge regression, and SVM.
In principle, the performance of RF improves along the increase of the number of trees, the tree depth, and the number of features or training samples at each node. However, the increase of accuracy is at the expense of computational cost. The set of RF parameters in our experiment was chosen under the consideration of balancing this trade-off. Our results will not be sensitive to small fluctuations of the chosen parameters. It is worth noting that the performance with this set of parameters has been widely evaluated in our prior studies (Huynh et al., 2016; Wang et al., 2015).
The top 20 most discriminative regions used in score prediction are consistent with the abnormal regions in AD reported from previous studies. The atrophy of hippocampus, parahippocampal gyrus, precuneus, cuneus, temporal lobe, and parietal lobe are well known in the pathological pathway of AD (He et al., 2007; Karas et al., 2007; Scahill et al., 2002; Visser et al., 2002). The volume of the cingulate gyrus has been found to be significantly reduced, especially in the posterior regions in AD (Jones et al., 2006). Studies also showed that the atrophy of the frontal lobe occurred later in the disease (Scahill et al., 2002). The dementia of AD often includes behavioral dyscontrol and visceral dysfunction, which could be related to the pathologic changes with the insula (Bonthius et al., 2005). Recently, researchers revealed that, besides the cortical regions, some of the subcortical regions were affected by AD as well, such as the degeneration of the thalamus (de Jong et al., 2008).
Soft split is another technique we propose in our algorithm. A similar concept was proposed in previous literature. Tu (2005) presented a probabilistic boosting-tree framework in which the probability was computed at each node by integrating the probabilities gathered from its child nodes so that a strong classifier can be built at that node as a combination of weak classifiers at leaf nodes. The technique was successfully applied to classification and object recognition. Here, we apply this soft-split concept to a regression task that has not been done before. Meanwhile, by using RF, the depth of the tree is much deeper than that of probabilistic boosting tree (usually only 3–4 layers), resulting in more accurate information gathered from more finely divided classes of samples. oRF-L1-soft further improves the prediction performance by integrating both sparse regression and soft split into the original RF framework.
With utilizing the longitudinal information from all the previous time points, the accuracy of prediction at the future time points is greatly improved. The largest obstacle for using the longitudinal information is the missing scores. The previous studies either discarded all the subjects with missing scores (Zhang et al., 2012a) or removed the missing scores at the specific time points (Zhou et al., 2013). We took a different strategy by filling out the missing scores with our regression method and then using the complete score sequences from all the previous time points to predict the scores at the next time point. With the addition of the longitudinal scores, oRF-L1-soft-long outperforms oRF-L1-soft (with only baseline features), as well as both TGL and cFSGL proposed in Zhou et al. (2013) that also took advantage of the longitudinal information.
A practical concern in a clinical setting is how an algorithm performs on the previously unseen data. In the study by Bron et al. (2015), the algorithms of AD classification were tested on a multicenter data set consisting of 354 T1w MRI scans, and the AD DREAM challenge was reported in the study by Allen et al. (2016), where participants estimated MMSE scores with shape measures (volume, thickness, area, etc.) derived from MR images. These algorithms were trained on the ADNI data set and then tested on the AddNeuroMed data set (Lovestone et al., 2009). As some of our future work, we will use those additional independent data sets to further validate the robustness of our current framework. Furthermore, it is also desired to develop a more sophisticated algorithm to fully take advantage of longitudinal information, including not only clinical scores but also image features at different time points (Chincarini et al., 2016). Recently, genetic information has been increasingly explored to predict changes in cognitive examination performance (Allen et al., 2016). Therefore, the potential gene candidates closely associated to AD can also be incorporated into our algorithm to form a multimodality framework for better accuracy (Peng et al., 2016; Wang et al., 2016a).
5. Conclusion
We presented an RF regression framework with sparse regression and soft split for longitudinal AD clinical score prediction. There are 3 contributions in our framework. First, the oblique split function in RF can generate better separation at each node than the traditional axis-aligned function, but the exhaustive search of its parameters causes heavy computational load and often becomes unrealistic in reality. Here, we proposed a supervised sparse regression model with PCA to locate the slope of the splitting hy-perplane analytically. Second, we adopted a probabilistic splitting strategy called soft split during the testing stage to increase the robustness of prediction. At each split node, the test sample can be assigned to both left and right child nodes with a probability and the predicted score was the linear combination of multiple leaf nodes, instead of only 1 node in the traditional RF. Our algorithm outperformed the traditional RF and other popular regression results such as Lasso regression, ridge regression, and SVM with the integration of these improvements. Finally, we utilized not only the baseline features but also the longitudinal scores at all the previous time points to predict the scores at the next future time point. In order to deal with the missing scores, we used the predicted values with our proposed method as the replacements. Our results showed higher accuracy, compared to those with only baseline features and the algorithms proposed in other studies. Furthermore, the proposed framework can be extended to a general regression tool for other regression tasks, not only limited to AD clinical score prediction.
Acknowledgments
This work was supported by the National Institute of Health grants (EB006733, EB008374, EB009634, AG041721, AG049371, AG042599). The authors thank the Alzheimer’s Disease Neuro-imaging Initiative (ADNI) for providing the data to this research.
Footnotes
Disclosure statement
The authors have no conflicts of interest to disclose.
References
- Abramowitz M, Stegun IA. Handbook of Mathematical Functions: With Formulas, Graphs, and Mathematical Tables. Dover Publications; New York: 1965. [Google Scholar]
- Allen GI, Amoroso N, Anghel C, Balagurusamy V, Bare CJ, Beaton D, Bellotti R, Bennett DA, Boehme KL, Boutros PC, Caberlotto L, Caloian C, Campbell F, Chaibub Neto E, Chang YC, Chen B, Chen CY, Chien TY, Clark T, Das S, Davatzikos C, Deng J, Dillenberger D, Dobson RJ, Dong Q, Doshi J, Duma D, Errico R, Erus G, Everett E, Fardo DW, Friend SH, Fröhlich H, Gan J, St George-Hyslop P, Ghosh SS, Glaab E, Green RC, Guan Y, Hong MY, Huang C, Hwang J, Ibrahim J, Inglese P, Iyappan A, Jiang Q, Katsumata Y, Kauwe JS, Klein A, Kong D, Krause R, Lalonde E, Lauria M, Lee E, Lin X, Liu Z, Livingstone J, Logsdon BA, Lovestone S, Ma TW, Malhotra A, Mangravite LM, Maxwell TJ, Merrill E, Nagorski J, Namasivayam A, Narayan M, Naz M, Newhouse SJ, Norman TC, Nurtdinov RN, Oyang YJ, Pawitan Y, Peng S, Peters MA, Piccolo SR, Praveen P, Priami C, Sabelnykova VY, Senger P, Shen X, Simmons A, Sotiras A, Stolovitzky G, Tangaro S, Tateo A, Tung YA, Tustison NJ, Varol E, Vradenburg G, Weiner MW, Xiao G, Xie L, Xie Y, Xu J, Yang H, Zhan X, Zhou Y, Zhu F, Zhu H, Zhu S Alzheimer’s Disease Neuroimaging Initiative. Crowdsourced estimation of cognitive decline and resilience in Alzheimer’s disease. Alzheimers Dement. 2016;12:645–653. doi: 10.1016/j.jalz.2016.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apostolova LG, Lu PH, Rogers S, Dutton RA, Hayashi KM, Toga AW, Cummings JL, Thompson PM. 3D mapping of mini-mental state examination performance in clinical and preclinical Alzheimer disease. Alzheimer Dis Assoc Disord. 2006;20:224–231. doi: 10.1097/01.wad.0000213857.89613.10. [DOI] [PubMed] [Google Scholar]
- Bonthius DJ, Solodkin A, Van Hoesen GW. Pathology of the insular cortex in Alzheimer disease depends on cortical architecture. J Neuropathol Exp Neurol. 2005;64:910–922. doi: 10.1097/01.jnen.0000182983.87106.d1. [DOI] [PubMed] [Google Scholar]
- Breiman L. Random forests. Mach Learn. 2001;45:5–32. [Google Scholar]
- Bron EE, Smits M, van der Flier WM, Vrenken H, Barkhof F, Scheltens P, Papma JM, Steketee RM, Méndez Orellana C, Meijboom R, Pinto M, Meireles JR, Garrett C, Bastos-Leite AJ, Abdulkadir A, Ronneberger O, Amoroso N, Bellotti R, Cárdenas-Peña D, Álvarez-Meza AM, Dolph CV, Iftekharuddin KM, Eskildsen SF, Coupé P, Fonov VS, Franke K, Gaser C, Ledig C, Guerrero R, Tong T, Gray KR, Moradi E, Tohka J, Routier A, Durrleman S, Sarica A, Di Fatta G, Sensi F, Chincarini A, Smith GM, Stoyanov ZV, Sørensen L, Nielsen M, Tangaro S, Inglese P, Wachinger C, Reuter M, van Swieten JC, Niessen WJ, Klein S Alzheimer’s Disease Neuroimaging Initiative. Standardized evaluation of algorithms for computer-aided diagnosis of dementia based on structural MRI: the CAD dementia challenge. Neuroimage. 2015;111:562–579. doi: 10.1016/j.neuroimage.2015.01.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chincarini A, Sensi F, Rei L, Gemme G, Squarcia S, Longo R, Brun F, Tangaro S, Bellotti R, Amoroso N, Bocchetta M, Redolfi A, Bosco P, Boccardi M, Frisoni GB, Nobili F Alzheimer’s Disease Neuroimaging Initiative. Integrating longitudinal information in hippocampal volume measurements for the early detection of Alzheimer’s disease. Neuroimage. 2016;125:834–847. doi: 10.1016/j.neuroimage.2015.10.065. [DOI] [PubMed] [Google Scholar]
- Criminisi A, Shotton J, Konukoglu E. Decision forests for classification, regression, density estimation, manifold learning and semi-supervised learning. Microsoft Tech Rep TR-2011-114. 2011:146. [Google Scholar]
- de Jong LW, van der Hiele K, Veer IM, Houwing JJ, Westendorp RG, Bollen EL, de Bruin PW, Middelkoop HA, van Buchem MA, van der Grond J. Strongly reduced volumes of putamen and thalamus in Alzheimer’s disease: an MRI study. Brain. 2008;131:3277–3285. doi: 10.1093/brain/awn278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding C, He X. K-means clustering via principal component analysis. Proceedings of International Conference on Machine Learning. 2004:29–37. [Google Scholar]
- Doraiswamy PM, Bieber F, Kaiser L, Krishnan KR, Reuning-Scherer J, Gulanski B. The Alzheimer’s Disease Assessment Scale: patterns and predictors of baseline cognitive performance in multicenter Alzheimer’s disease trials. Neurology. 1996;48:1511–1517. doi: 10.1212/wnl.48.6.1511. [DOI] [PubMed] [Google Scholar]
- Duchesne S, Caroli A, Geroldi C, Collins DL, Frisoni GB. Relating one-year cognitive change in mild cognitive impairment to baseline MRI features. Neuroimage. 2009;47:1363–1370. doi: 10.1016/j.neuroimage.2009.04.023. [DOI] [PubMed] [Google Scholar]
- Fan Y, Kaufer D, Shen D. Joint estimation of multiple clinical variables of neurological diseases from imaging patterns. Proceedings of IEEE International Symposium Biomedical Imaging. 2010:852–855. [Google Scholar]
- Ferrarini L, Palm WM, Olofsen H, van der Landen R, Jan Blauw G, Westendorp RG, Bollen EL, Middelkoop HA, Reiber JH, van Buchem MA, Admiraal-Behloul F. MMSE scores correlate with local ventricular enlargement in the spectrum from cognitively normal to Alzheimer disease. Neuroimage. 2008;39:1832–1838. doi: 10.1016/j.neuroimage.2007.11.003. [DOI] [PubMed] [Google Scholar]
- Folstein MF, Folstein SE, McHugh PR. “Mini-mental state”: a practical method for grading the cognitive state of patients for the clinician. J Psychiatr Res. 1975;12:189–198. doi: 10.1016/0022-3956(75)90026-6. [DOI] [PubMed] [Google Scholar]
- Hastie T, Tibshirani R, Friedman J. The Elements of Statistical Learning: Data Mining, Inference, and Prediction. Springer-Verlag; New York: 2009. [Google Scholar]
- He Y, Wang L, Zang Y, Tian L, Zhang X, Li K, Jiang T. Regional coherence changes in the early stages of Alzheimer’s disease: a combined structural and resting-state functional MRI study. Neuroimage. 2007;35:488–500. doi: 10.1016/j.neuroimage.2006.11.042. [DOI] [PubMed] [Google Scholar]
- Huynh T, Gao Y, Kang J, Wang L, Zhang P, Lian J, Shen D Alzheimer’s Disease Neuroimaging Initiative. Estimating CT image from MRI data using structured random forest and auto-context model. IEEE Trans Med Imaging. 2016;35:174–183. doi: 10.1109/TMI.2015.2461533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Y, Shi Y, Zhan L, Thompson PM. Automated Multi-Atlas Labeling of the Fornix and its Integrity in Alzheimer’s Disease. Proceedings of IEEE International Symposium Biomedical Imaging. 2015:140–143. doi: 10.1109/ISBI.2015.7163835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones BF, Barnes J, Uylings HB, Fox NC, Frost C, Witter MP, Scheltens P. Differential regional atrophy of the cingulate gyrus in Alzheimer disease: a volumetric MRI study. Cereb Cortex. 2006;16:1701–1708. doi: 10.1093/cercor/bhj105. [DOI] [PubMed] [Google Scholar]
- Karas G, Scheltens P, Rombouts S, van Schijndel R, Klein M, Jones B, van der Flier W, Vrenken H, Barkhof F. Precuneus atrophy in early-onset Alzheimer’s disease: a morphometric structural MRI study. Neuroradiology. 2007;49:967–976. doi: 10.1007/s00234-007-0269-2. [DOI] [PubMed] [Google Scholar]
- Li J, Jin Y, Shi Y, Dinov ID, Wang DJ, Toga AW, Thompson PM. Voxelwise spectral diffusional connectivity and its application to Alzheimer’s disease and intelligence prediction. Med Image Comput Comput Assist Interv. 2013;16:655–662. doi: 10.1007/978-3-642-40811-3_82. LNCS 8149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovestone S, Francis P, Kloszewska I, Mecocci P, Simmons A, Soininen H, Spenger C, Tsolaki M, Vellas B, Wahlund LO, Ward M, AddNeuroMed Consortium. AddNeuroMed–the European collaboration for the discovery of novel biomarkers for Alzheimer’s disease. Ann N Y Acad Sci. 2009;1180:36–46. doi: 10.1111/j.1749-6632.2009.05064.x. [DOI] [PubMed] [Google Scholar]
- Menze BH, Kelm BM, Splitthoff DN, Koethe U, Hamprecht FA. On Oblique Random Forests. Machine Learning and Knowledge Discovery in Databases LNCS. 2011;6912:453–469. [Google Scholar]
- Morra JH, Tu Z, Apostolova LG, Green AE, Avedissian C, Madsen SK, Parikshak N, Hua X, Toga AW, Jack CR, Jr, Schuff N, Weiner MW, Thompson PM Alzheimer’s Disease Neuroimaging Initiative. Automated 3D mapping of hippocampal atrophy and its clinical correlates in 400 subjects with Alzheimer’s disease, mild cognitive impairment, and elderly controls. Hum Brain Mapp. 2009;30:2766–2788. doi: 10.1002/hbm.20708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris JC. Clinical dementia rating (CDR): current version and score rules. Neurology. 1993;43:2412–2414. doi: 10.1212/wnl.43.11.2412-a. [DOI] [PubMed] [Google Scholar]
- O’Bryant SE, Waring SC, Cullum CM, Hall J, Lacritz L, Massman PJ, Lupo PJ, Reisch JS, Doody R Texas Alzheimer’s Research Consortium. Staging dementia using Clinical Dementia Rating Scale Sum of Boxes scores: a Texas Alzheimer’s research consortium study. Arch Neurol. 2008;65:1091–1095. doi: 10.1001/archneur.65.8.1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng J, An L, Zhu X, Jin Y, Shen D. Structured sparse kernel learning for imaging genetics based Alzheimer’s disease diagnosis. Med Image Comput Comput Assist Interv. 2016 doi: 10.1007/978-3-319-46723-8_9. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosen WG, Mohs RC, Davis KL. A new rating scale for Alzheimer’s disease. Am J Psychiatry. 1984;141:1356–1364. doi: 10.1176/ajp.141.11.1356. [DOI] [PubMed] [Google Scholar]
- Scahill RI, Schott JM, Stevens JM, Rossor MN, Fox NC. Mapping the evolution of regional atrophy in Alzheimer’s disease: unbiased analysis of fluid-registered serial MRI. Proc Natl Acad Sci U S A. 2002;99:4703–4707. doi: 10.1073/pnas.052587399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen D, Davatzikos C. HAMMER: hierarchical attribute matching mechanism for elastic registration. IEEE Trans Med Imaging. 2002;21:1421–1439. doi: 10.1109/TMI.2002.803111. [DOI] [PubMed] [Google Scholar]
- Shen D, Wong WH, Ip HHS. Affine-invariant image retrieval by correspondence matching of shapes. Image Vis Comput. 1999;17:489–499. [Google Scholar]
- Stonnington CM, Chu C, Klöppel S, Jack CR, Jr, Ashburner J, Frachowiak RS Alzheimer Disease Neuroimaging Initiative. Predicting clinical scores from magnetic resonance scans in Alzheimer’s disease. Neuroimage. 2010;51:1405–1413. doi: 10.1016/j.neuroimage.2010.03.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan PJ, Dowe DL. Decision forests with oblique decision trees. Proceedings of Mexican International Conference on Artificial Intelligence, LCNS. 2006;4293:593–603. [Google Scholar]
- Tibshirani R. Regression shrinkage and selection via the Lasso. J R Stat Soc Ser B. 1996;58:267–288. [Google Scholar]
- Thung KH, Wee CY, Yap PT, Shen D Alzheimer’s Disease Neuroimaging Initiative. Identification of Alzheimer’s disease using incomplete multimodal dataset via matrix shrinkage and completion. Mach Learn Med Imaging LNCS. 2013;8184:163–170. [Google Scholar]
- Thung KH, Wee CY, Yap PT, Shen D Alzheimer’s Disease Neuroimaging Initiative. Neurodegenerative disease diagnosis using incomplete multimodality data via matrix shrinkage and completion. Neuroimage. 2014;91:386–400. doi: 10.1016/j.neuroimage.2014.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thung KH, Wee CY, Yap PT, Shen D. Identification of progressive mild cognitive impairment patients using incomplete longitudinal MRI scans. Brain Struct Funct. 2015a doi: 10.1007/s00429-015-1140-6. http://dx.doi.org/10.1007/s00429-015-1140-6. [DOI] [PMC free article] [PubMed]
- Thung KH, Yap PT, Adeli ME, Shen D. Joint diagnosis and conversion time prediction of progressive mild cognitive impairment (pMCI) using low-rank subspace clustering and matrix completion. Med Image Comput Comput Assist Interv. 2015b;9351:527–534. doi: 10.1007/978-3-319-24574-4_63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu Z. Probabilistic boosting-tree: learning discriminative models for classification, recognition, and clustering. Proceedings of IEEE International Conference on computer vision 2005;2:1589–1596. [Google Scholar]
- Tzourio-Mazoyer N, Landeau B, Papathanassiou D, Crivello F, Etard O, Delcroix N, Mazoyer B, Joliot M. Automated anatomical labeling of activations in SPM using a macroscopic anatomical parcellation of the MNI MRI single-subject brain. Neuroimage. 2002;15:273–289. doi: 10.1006/nimg.2001.0978. [DOI] [PubMed] [Google Scholar]
- Visser PJ, Verhey FR, Hofman PA, Scheltens P, Jolles JJ. Medial temporal lobe atrophy predicts Alzheimer’s disease in patients with minor cognitive impairment. Neurol Neurosurg Psychiatry. 2002;72:491–497. doi: 10.1136/jnnp.72.4.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Fan Y, Bhatt P, Davatzikos C. High-dimensional pattern regression using machine learning: from medical images to continuous clinical variables. Neuroimage. 2010;50:1519–1535. doi: 10.1016/j.neuroimage.2009.12.092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Gao Y, Shi F, Li G, Gilmore JH, Lin W, Shen D. LINKS: Learning-based multi-source IntegratioN frameworK for Segmentation of infant brain images. Neuroimage. 2015;108:160–172. doi: 10.1016/j.neuroimage.2014.12.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, Shi F, Jin Y, Jiang W, Shen D, Xiao S. Abnormal changes of brain cortical anatomy and the association with plasma MicroRNA107 level in amnestic mild cognitive impairment. Front Aging Neurosci. 2016a;8:112. doi: 10.3389/fnagi.2016.00112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, Shi F, Jin Y, Yap PT, Wee CY, Zhang J, Yang C, Li X, Xiao S, Shen D. Multilevel deficiency of white matter connectivity networks in Alzheimer’s disease: a diffusion MRI study with DTI and HARDI models. Neural Plast. 2016b;2016:2947136. doi: 10.1155/2016/2947136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue Z, Shen D, Karacali B, Stern J, Rottenberg D, Davatzikos C. Simulating deformations of MR brain images for validation of atlas-based segmentation and registration algorithms. Neuroimage. 2006;33:855–866. doi: 10.1016/j.neuroimage.2006.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhan L, Nie Z, Ye J, Wang Y, Jin Y, Jahanshad N, Prasad G, de Zubicaray GI, McMahon KL, Martin NG, Wright MJ, Thompson PM. Multiple stages classification of Alzheimer’s disease based on structural brain networks using generalized low rank approximations (GLRAM) Computational Diffusion MRI, Mathematics Visualization. 2014:35–44. [Google Scholar]
- Zhan L, Zhou J, Wang Y, Jin Y, Jahanshad N, Prasad G, Nir TM, Leonardo CD, Ye J, Thompson PM Alzheimer’s Disease Neuroimaging Initiative. Comparison of nine tractography algorithms for detecting abnormal structural brain networks in Alzheimer’s disease. Front Aging Neurosci. 2015;7:48. doi: 10.3389/fnagi.2015.00048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D, Shen D Alzheimer’s Disease Neuroimaging Initiative. Predicting future clinical changes of MCI patients using longitudinal and multimodal biomarkers. PLoS One. 2012a;7:e33182. doi: 10.1371/journal.pone.0033182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D, Shen D. Multi-modal multi-task learning for joint prediction of multiple regression and classification variables in Alzheimer’s disease. Neuroimage. 2012b;59:895–907. doi: 10.1016/j.neuroimage.2011.09.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Wang Q, Gao Y, Wu G, Shen D. Automatic labeling of MR brain images by hierarchical learning of atlas forests. Med Phys. 2016;43:1175–1186. doi: 10.1118/1.4941011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Wang Q, Gao Y, Wu G, Shen D. Learning of atlas forest hierarchy for automatic labeling of MR brain images. Mach Learn Med Imaging LNCS. 2014;8679:323–330. [Google Scholar]
- Zhou J, Liu J, Narayan VA, Ye J. Alzheimer’s Disease Neuroimaging Initiative. Modeling disease progression via multi-task learning. Neuroimage. 2013;78:233–248. doi: 10.1016/j.neuroimage.2013.03.073. [DOI] [PubMed] [Google Scholar]
- Zhu X, Suk HI, Shen D. A novel matrix-similarity based loss function for joint regression and classification in AD diagnosis. Neuroimage. 2014;100:91–105. doi: 10.1016/j.neuroimage.2014.05.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X, Suk HI, Zhu Y, Thung KH, Wu G, Shen D. Multi-view classification for identification of Alzheimer’s disease. Mach Learn Med Imaging LNCS. 2015;9352:255–262. doi: 10.1007/978-3-319-24888-2_31. [DOI] [PMC free article] [PubMed] [Google Scholar]