Abstract
Germline mutations in the PTEN gene, which cause Cowden syndrome (CS), are known to be one of the genetic factors for primary thyroid and breast cancers, however, PTEN mutations are found in only a small subset of research participants with non-syndrome breast and thyroid cancers. In this study, we aimed to identify germline variants that may be related to genetic risk of primary thyroid and breast cancers. Genomic DNAs extracted from peripheral blood of 14 PTEN-wild-type female research participants with primary thyroid and breast cancers were analyzed by whole-exome sequencing. Gene-based case control association analysis using the information of 406 Europeans obtained from the 1000 Genomes Project database identified 34 genes possibly associated with the phenotype with P<1.0×10−3. Among them, rare variants in the PARP4 gene were detected at significant high frequency (odds ratio = 5.2, P = 1.0×10−5). The variants, G496V and T1170I, were found in 6 of the 14 study participants (43%) while their frequencies were only 0.5% in controls. Functional analysis using HCC1143 cell line showed that knockdown of PARP4 with siRNA significantly enhanced the cell proliferation, compared with the cells transfected with siControl (P = 0.02). Kaplan-Meier analysis using GEO, EGA and TCGA datasets showed poor progression-free survival (P = 0.006, Hazard ratio 0.71) and overall survival (P < 0.0001, Hazard ratio 0.79) in a PARP4 low-expression group, suggesting that PARP4 may function as a tumor suppression. In conclusion, we identified PARP4 as a possible susceptibility gene of primary thyroid and breast cancer.
Keywords: Thyroid, Breast, Cancer, PARP4, Germline Mutation
Introduction
A cancer of a different site and histologic type that develops in a person who already has a cancer diagnosis is considered a second primary cancer. Certain primary cancers are associated with a high risk of developing a second primary cancer. Specifically, individuals with a thyroid cancer primary have a high incidence of second primary cancers even after adjusting for surveillance bias. One study of a cohort of 39,002 research participants suggested that when compared to the general population thyroid cancer survivors have a 30% increased risk of a breast cancer second primary (Sandeep, et al. 2006). The incidence of second primary cancers is also high even in participants with thyroid micro-carcinoma (Hsu, et al. 2014; Kim, et al. 2013). Because of the high incidence of thyroid cancer in women (Davies and Welch 2014; Siegel, et al. 2015), it is important to understand why there is an increased risk of second primary breast cancers (Aschebrook-Kilfoy, et al. 2013).
One possible cause for the increased rate of breast second primary cancers is a common genetic pathway. Cowden syndrome (CS) is an autosomal dominant disorder characterized by developing multiple primary cancers in various sites including thyroid and breast (Eng 1993). PTEN is one of the causative genes of CS or Cowden-like syndrome (CLS) (Liaw, et al. 1997), and germline mutations are found in 25–85% of CS patients and <5% of CLS patients (Marsh, et al. 1998; Tan, et al. 2011). Germline mutations in other genes, such as succinate dehydrogenase genes (SDHx) (8%), PIK3CA (8.8%), AKT1 (2.2%) and SEC23B (4%) are also known as CS/CLS susceptibility genes (Ngeow, et al. 2011; Ni, et al. 2008; Orloff, et al. 2013). CS/CLS is one of the plausible causes of primary thyroid and breast cancers; and the standardized incidence rate ratio of second primary cancers in research participants with thyroid cancer is increased to 5.83 per 10000 person-years (95% CI, 3.01 to 10.18) by PTEN mutations (Ngeow, et al. 2014). However, PTEN germline mutations are responsible for only a small portion of participants who present with non-syndromic primary thyroid and breast cancers (Ngeow et al. 2014; Pal, et al. 2001). We hypothesize that germline mutations in genes other than PTEN, SDHx, PIK3CA, and AKT may explain the high rate of primary thyroid and breast cancer. In this study, we sought to identify possible germline mutations associated with thyroid and breast cancers using whole-exome sequencing.
Materials & Methods
Samples
The study cohort consisted of 14 women known to have both breast and thyroid cancer primary malignancies. They were identified from a cohort of patients followed by the Cleveland Clinic Cancer Genetics Clinic. These 14 research participants were selected under informed consent due to characteristics making them high risk for a possible genetic association. Particularly, they all developed both primary cancers prior to 50 years of age, none had radiation exposure to the head and neck or chest prior to developing the cancers and there was a high frequency of familial breast and thyroid cancers in the cohort. In addition all participants had already been ruled out for a PTEN germline mutation (Ngeow et al. 2014). Genomic DNA was extracted from peripheral blood samples, and the quality and quantity of each DNA sample were examined by Qubit 2.0 Fluorometer (Life Technologies, Carlsbad, CA) and 2200 Tape Station (Agilent Technologies, Santa Clara, CA). This study is approved by the Institutional Review Board at the University of Chicago (No.8962) and Cleveland Clinic (No.8458).
Whole-exome sequencing
DNA libraries for whole-exome sequencing were prepared from 200–1000 ng of genomic DNA using SureSelet XT Human All Exon V5 (Agilent Technologies). The prepared whole-exome libraries were quantified on the 2200 Tape Station (Agilent Technologies), and then sequenced by 150-bp paired-end reads on NextSeq 500 Desktop Sequencer (Illumina, San Diego, CA). All procedures were performed according to the manufacturers’ protocol.
Read mapping
After the exclusion of low-quality reads (base quality of < 20 for more than 80% of bases) using FASTX toolkit, sequence reads were mapped to the human reference genome GRCh37/hg19 using Burrows-Wheeler Aligner (BWA) (v0.7.10) (Li 2014). After read pairs with a mapping quality of < 30 and with mismatches more than 5% of lead length were excluded, BAM files were generated using SAMTools (v1.1), and possible PCR duplicated reads were removed using Picard v1.91 (http://broadinstitute.github.io/picard/). As a population control, BAM files of 406 European (including CEU, GBR, IBS and TSI, except for FIN) subjects, which were indicated to be healthy volunteers, were obtained from the 1000 Genomes Project database (www.1000genomes.org)
Variant calling
BCFtools (v1.1) was used to call variants (SNVs and indels) with the following criteria of (i) sequence depth of ≥ 10, (ii) variant depth of ≥ 4 and (iii) base quality of ≥ 15 (Li, et al. 2009). Variants in 14 cases and 406 controls were merged into a single vcf file, and further filtering was performed to identify high-confident variants, requiring that each variant (i) have a call rate of ≥ 0.8, (ii) found ≤ 1% in controls, and (iii) found ≤ 1% in the American, African and Asian populations in 1000 Genomes Project.
Variant annotation
Variants were annotated using the hg19 database in SNP effect prediction tools (SnpEff) (Cingolani, et al. 2012). The primary SnpEff genomic effects include splice-site acceptor, splice-site donor, indel frameshift, indel non-frameshift, non-sense, non-synonymous and synonymous variants. For variants that have multiple different annotations, the highest impact effect was selected. The predicted impact of amino acid substitutions was annotated using 5 algorithms of LRT score, MutationTaster, PolyPhen-2 HumDiv, PolyPhen-2 HumVar and SIFT. Variants predicted as “deleterious” in LRT, “disease causing automatic” and “disease causing” in Mutation Taster, “probably damaging” and “possibly damaging” in PolyPhen-2 HumDiv and PolyPhen-2 HumVar, and “damaging” in SIFT were considered as “deleterious”.
Sanger sequencing
Target regions were amplified by polymerase chain reaction (PCR) from genomic DNA. The amplified products were sequenced using BigDye Terminator v3.1 Cycle Sequencing Kit and 3500 Series Genetic Analyzer (Life Technologies) according to the manufacturer’s protocols. Variants not validated by Sanger sequencing were excluded from the following analysis.
Association analysis
Gene-based association analysis was performed using a burden test with 100,000-time permutation in PLINK/SEQ v0.10. In case of multiple results due to alternative transcripts, we selected the result with the smaller P-value. P-values of < 0.05 were considered as statistically significant. To adjust multiple testing by the strict Bonferroni correction, we used significance level of P < 3.65×10−6 (0.05/13,705) in the gene-based association analysis.
Quantitative Real-time PCR
Total RNAs from 18 cell lines previously extracted (Park, et al. 2010) were reverse-transcribed to cDNA using SuperScript III (LifeTechnologies). Real-time PCR using TaqMan gene expression assay for PARP4 (Hs00173105__m1) and GAPDH (Hs02758991_g1) was conducted on ViiA7 instrument (Life Technologies). mRNA levels of PARP4 were normalized to GAPDH expression.
siRNA transfection and cell proliferation assay
HCC 1143 cell line authenticated by STR analysis was cultured under the recommendations of their respective depositors. siRNA oligonucleotides for targeting PARP4 transcripts were purchased from Sigma-Aldrich. siNegative control (Cosmo Bio, Tokyo, Japan) was used as a control siRNA. siRNAs were transfected into cells with Lipofectamine RNAi max (Life Technologies). After 48-h incubation, cell viability assay was performed with the Cell Counting Kit-8 (Dojindo, Kumamoto, Japan).
Statistical analysis
Expression analysis and cell viability experiment were repeated three times at triplicates, and evaluated using the student t-test. The prognostic value of PARP4 was analyzed by Kaplan-Meier Plotter (http://kmplot.com/), which includes gene expression data of 4,142 breast cancer cases from GEO, EGA and TCGA using Affymetrix HG-U133A, HG-U133 Plus 2.0 and HG-U133A 2.0 (Gyorffy, et al. 2010; Gyorffy, et al. 2013).
Results
The median age at diagnosis for thyroid and breast cancer was 38 (range; 23–49) and 46 (range; 36–49) years old, respectively (Table 1). Five participants were diagnosed with both thyroid and breast cancer at the same age. Six cases were diagnosed to have thyroid cancer before breast cancer diagnosis. Thirteen thyroid cancers were either follicular or papillary subtype, and 11 breast cancers were a ductal subtype (Table 1). Seven (50%) and 5 (36%) participants had a family history of thyroid or breast cancer within 3 generations of the proband, respectively, although no additional family members had primary thyroid and breast cancers.
Table 1.
Demographics of the 14 participants with thyroid and breast double primary cancers
| ID | Race | Cancer type | Age of diagnosis | Family history of thyroid and breast cancer | Histology | Metastasis |
|---|---|---|---|---|---|---|
| #1 | White | Thyroid | 36 | Papillary | Unknown | |
|
| ||||||
| Breast | 36 | Medullary | Unknown | |||
|
| ||||||
| #2 | White | Thyroid | 38 | Papillary | Unknown | |
|
| ||||||
| Breast | 48 | Ductal | Unknown | |||
|
| ||||||
| #3 | Hispanic | Thyroid | 40 | Follicular | Unknown | |
|
| ||||||
| Breast | 40 | Mother(63yr), Aunt (maternal; 60’s) | Unknown | Unknown | ||
|
| ||||||
| #4 | American Indian | Thyroid | 25 | Follicular | No | |
|
| ||||||
| Breast | 43 | Aunt (paternal; 50’s), Cousin (maternal; 60’s), Cousin (maternal, 40’s) | Ductal | No | ||
|
| ||||||
| #5 | White | Thyroid | 47 | Follicular/Papillary | No | |
|
| ||||||
| Breast | 47 | Ductal | No | |||
|
| ||||||
| #6 | White | Thyroid | 25 | Daughter (30yr), Sister (45yr) | Follicular/Papillary | No |
|
| ||||||
| Breast | 46 | Ductal | No | |||
|
| ||||||
| #7 | White | Thyroid | 46 | Papillary | No | |
|
| ||||||
| Breast | 46 | Ductal | Lymph node | |||
|
| ||||||
| #8 | White | Thyroid | 37 | Uncle (maternal; 51yr) | Papillary | Unknown |
|
| ||||||
| Breast | 38 | Aunt (maternal; 49yr) | Mucinous | Unknown | ||
|
| ||||||
| #9 | Unknown | Thyroid | 48 | Papillary | Lymph node | |
|
| ||||||
| Breast | 46 | Aunt (paternal; 56yr) | Ductal | No | ||
|
| ||||||
| #10 | White | Thyroid | 35 | Sister (19yr), Sister (38yr) | Papillary | Lymph node |
|
| ||||||
| Breast | 41 | Grandmother (paternal 84yr/maternal; 45yr), Aunt (paternal 54yr/maternal; 65yr), | Ductal | No | ||
|
| ||||||
| #11 | Black | Thyroid | N/A | Aunt (maternal; 50’s), Aunt (maternal; 50’s) | Hurthle cell | Unknown |
|
| ||||||
| Breast | N/A | Ductal | No | |||
|
| ||||||
| #12 | White | Thyroid | 23 | Uncle (maternal; 72yr), Cousin (paternal; 28yr) | Papillary | No |
|
| ||||||
| Breast | 49 | Aunt (maternal; 65yr) | Ductal | No | ||
|
| ||||||
| #13 | American Indian | Thyroid | 49 | Aunt (maternal; 53yr) | Papillary | No |
|
| ||||||
| Breast | 49 | Ductal | No | |||
|
| ||||||
| #14 | American Indian | Thyroid | 48 | Uncle (maternal; 40’s) | Follicular/Papillary | Unknown |
|
| ||||||
| Breast | 44 | Ductal | No | |||
N/A: Not available yr: years old
In exome sequencing of the 14 cases, we obtained a total sequencing output with an average read depth of 138× per base. We also analyzed 406 European white controls obtained from the 1000 Genomes Project database using the same algorithm to minimize the calling discordance due to different algorithms. We identified a total of 105,064 and 345,793 variants in our cases and the 1000 Genomes controls, respectively (Table 2). Of these variants, 4.8% and 18.8% were rare (defined as having a minor allele frequency (MAF) of < 1% in the controls in both cases and controls. After excluding common SNPs with MAF of ≥ 1% in the controls, 5,073 variants in 2,909 genes in cases and 65,170 variants in 14,232 genes in controls were identified within the coding regions (Table 2). In the 14 participants we inspected rare variants known to be responsible for hereditary cancer syndromes such as SDHx and KLLN (for Cowden syndrome), and BRCA1/2 (for hereditary breast and ovarian cancer syndrome) (Supplementary Figure 1), however, no participant had more than one of these rare variants identified. In addition, no statistically-significant enrichment of variants was observed in the recurrently somatically-mutated genes (known to occur in more than 10% of cancers) that have been previously reported in epithelial thyroid cancer (such as BRAF, RET, TERT) and breast cancers (such as PIK3CA, TP53, CDH1) (Supplementary Figure 2).
Table 2.
Summary of number of cases, variants and genes with rare variants
| Subjects | Variants | Rare variants | Genes with rare variants | Genes with variants predicted as deleterious | |
|---|---|---|---|---|---|
| Cases | 14 | 105,064 | 5,073 | 2,909 | 2,316 |
| Controlsa | 406 | 345,793 | 65,170 | 14,232 | 13,554 |
Controls are 406 European individuals in the 1000 Genomes Project database
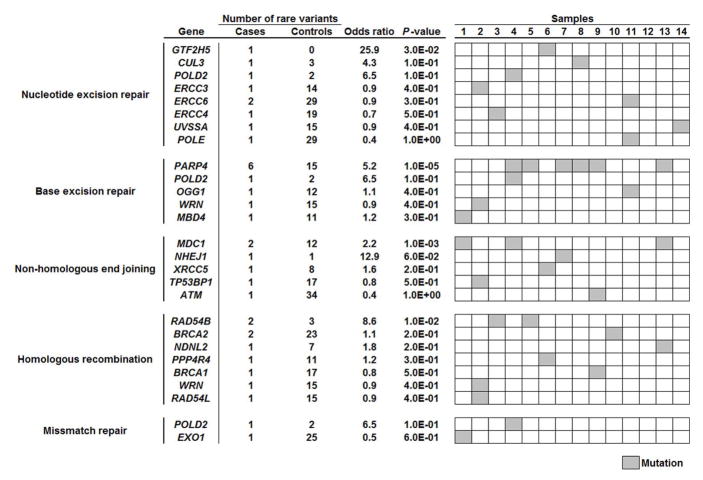
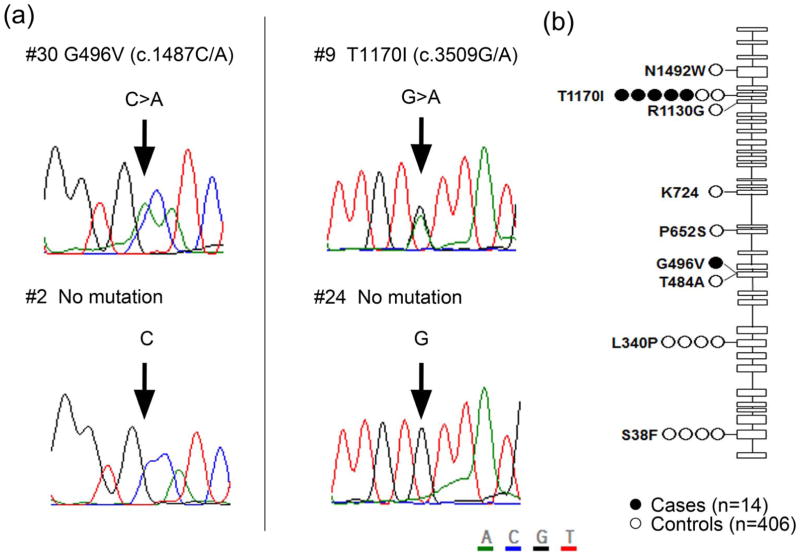
Using five different prediction tools, variants predicted as deleterious substitutions in at least one algorithm were identified in 2,316 and 13,554 genes in cases and controls, respectively (Table 2). Gene-based association analysis of these deleterious variants identified 34 genes, in which rare variants were enriched with a P value of < 1.0×10−3. However, none of these genes showed a genome-wide significant level of association when we considered the Bonferroni correction for multiple testing (P < 3.65×10−6) (Supplementary Figure 3). Since several previous reports indicated that the mutations in the genes involved in DNA repair pathways are genetic risk factors for several types of familial cancer, we focused on the DNA repair pathway, and found that 13 of the 14 participants had rare variants in at least one DNA repair-related gene (Figure 1). Among them, PARP4 showed the most significant association with the risk for primary thyroid and breast cancers (odds ratio = 5.2, P = 1.0×10−5; Figure 1). In PARP4, 2 different variants, G496V and T1170I, were found in 6 of the 14 (43%) participants, and both of these were validated by Sanger sequencing (Figure 2a). The majority of these were T1170I substitutions found in 5 of the 14 (36%) cases, including 3 White, 1 American Indian, 1 unknown race case. In contrast, this variant was found in only 2 of the 406 (0.5%) control individuals. Interestingly, among the 17 mutations found in 960 breast cancer cases in TCGA database, 7 cases (41%) showed the two-hit mutation; a somatic mutation and loss of one PARP4 allele. Furthermore, one of these 17 somatic mutations, p.T1170I, is identical to one of germline mutations we detected (Supplementary Table 2). G496V substitution was uniquely found in the remaining one American Indian case, and not found in any of the 406 controls (Figure 2b).
Figure 1.
Association of the burden of rare variants in DNA repair related genes with risk for primary thyroid and breast cancer
Figure 2.
Mutations in PARP4 gene in 14 participants with synchronous thyroid and breast cancers and 406 control individuals. (a) Validation of 2 variants, G496V and T1170I, by Sanger sequencing. (b) Location of each mutation in PARP4. Closed and open circles represent the location of PARP4 mutations found in cases and controls, respectively.
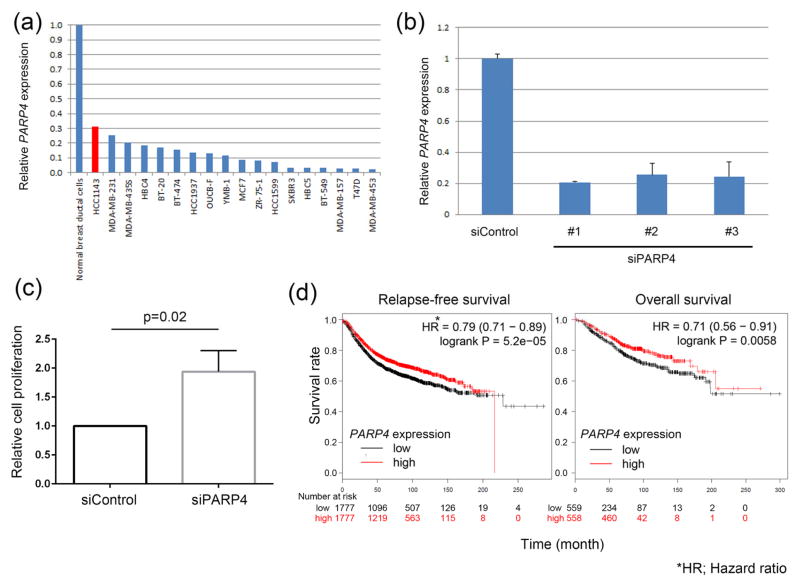
Next, we examined whether PARP4 shows tumor-suppressive function (Figure 3). mRNA expression of PARP4 in 18 breast cancer cell lines was examined by qPCR. Since HCC1143 cells showed the highest expression of PARP4 among the 18 cell lines tested, we used HCC1143 cells for the further experiments. When PARP4 expression was knocked down by siRNA, the proliferation of HCC1143 cells was significantly enhanced three-fold compared to the cells transfected with siControl (P = 0.02, Figure 3c), suggesting that PARP4 works as a tumor suppressor.
Figure 3.
Functional analysis of PARP4. (a) PARP4 mRNA expression in 18 breast cancer cell lines. (b) Knockdown of PARP4 expression by PARP4-specific siRNAs in HCC1143 breast cancer cells. (c) Effects of PARP4 knockdown on the proliferation of HCC1143 cells. (d) Kaplan-Meier analysis of progression-free survival and overall survival in 4,142 patients with breast cancer patients (from GEO, EGA and TCGA datasets) according to the expression of PARP4. High and low expressions were defined as above and below median, respectively.
Discussion
In this study, we comprehensively analyzed exome variants in 14 participants with primary thyroid and breast cancer, and identified PARP4 as a possible susceptibility gene candidate for primary thyroid and breast cancer. PARP4, also known as VPARP, is a family member of the poly (ADP-ribose) polymerase (PARP) (Kickhoefer, et al. 1999). The PARP protein superfamily has 17 members and controls a wide array of cellular processes such as DNA repair, transcriptional regulation and RNA interference (Gibson and Kraus 2012). So far, the function of a few PARPs including PARP1, PARP2 and PARP5 have been well characterized (Gibson and Kraus 2012). PARP1 and PARP4 contain BRCA1 carboxy-terminal domain (BRCT) repeats, which are thought to bind phosphorylated DNA damage-sensing proteins recruiting PARPs to sites of DNA damage (Manke, et al. 2003). Although little is known about the biological function of PARP4, it is suspected to be involved in the DNA repair pathway due to its BRCT domain. Many hereditary diseases responsible for synchronous cancers such as Lynch syndrome (MLH1, MSH2, MSH6 and PMS2 are responsible genes) and hereditary breast and ovarian cancer (BRCA1 and BRCA2) are known to be caused by germline mutations in genes involved in the DNA repair pathway (Lynch, et al. 2009). Moreover, several DNA repair genes are reported to be mutated to a significant degree in both aggressive papillary thyroid carcinoma and breast cancers (Cancer Genome Atlas 2012; Cancer Genome Atlas Research 2014). As shown in Figure 3, we suggest that PARP4 might have a tumor-suppressor function. In addition, Kaplan-Meier analysis using gene expression data from 4,142 cases of breast cancer which was available on Kaplan-Meier Plotter (http://kmplot.com/) showed worse progression-free survival (P = 0.006, Hazard ratio 0.71) and overall survival (P < 0.0001, Hazard ratio 0.79) in the PARP4 low-expression group. These lines of evidence support the possibility that PARP4 plays a critical role in thyroid and breast tumorigenesis.
Radiation therapy is known to increase the risk of second primary cancers (Tucker, et al. 1991). Generally, a radiation-induced second primary cancer arising at the radiation-associated site typically arises 10 years after the initial radiation exposure. (Grantzau and Overgaard 2015). However, since both thyroid and breast cancers were diagnosed prior to any therapeutic radiation exposure, including RAI, our cohort does not reflect a link between radiation-induced secondary primary cancer risk and PARP4 germline mutations.
In conclusion, through the whole-exome sequencing approach, we have implicated PARP4 germline mutations as possible susceptibility factors for the risk of synchronous thyroid and breast cancers. It is obvious that our study has a limitation in its sample size, thus further analysis is warranted with a larger number of samples to verify the results obtained in this study. Although we show evidence of the tumor suppressor function of PARP4 in breast cancer cells, further analysis is warranted to clarify the development of primary thyroid and breast cancer. Regardless, the high rate of the rare variant of PARP4 found in our study when compared to the very low rate in the controls is compelling evidence of a possible new genetic syndrome that is responsible for both primary thyroid and breast cancer.
Supplementary Material
Supplementary figure 1. Rare variants in hereditary cancer associated genes
(a) Rare variants in hereditary cancer associate genes found in 14 samples from double primary thyroid and breast cancers (b) Functional prediction of each variant found in our cases
Supplementary figure 2. Rare variants in recurrently mutated genes
Supplementary figure 3. Top 34 genes with P-value less than 1.0 × 10−3
Acknowledgments
Funding
Author RHG was supported by Award Number K12CA139160 from the National Cancer Institute. Research samples were accrued, in part, under the Doris Duke Distinguished Clinical Scientist Award and P01124570 from the National Cancer Institute (both to CE). The content is solely the responsibility of the authors and does not necessarily represent the official view of the National Cancer Institute or the National Institute of Health. CE is the Sondra J and Stephen R Hardis Endowed Chair of Caner Genomic Medicine at the Cleveland Clinic, and an ACS Clinical Research Professor.
Super-computing resources were provided by the Human Genome Center, the Institute of Medical Science, the University of Tokyo (http://sc.hgc.jp/shirokane.html). Additional thanks to Victoria Raymond for assistance with medical records review.
Footnotes
Declaration of interest
The authors have nothing to disclose
References
- Aschebrook-Kilfoy B, Schechter RB, Shih YC, Kaplan EL, Chiu BC, Angelos P, Grogan RH. The clinical and economic burden of a sustained increase in thyroid cancer incidence. Cancer Epidemiol Biomarkers Prev. 2013;22:1252–1259. doi: 10.1158/1055-9965.EPI-13-0242. [DOI] [PubMed] [Google Scholar]
- Cancer Genome Atlas N. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer Genome Atlas Research N. Integrated genomic characterization of papillary thyroid carcinoma. Cell. 2014;159:676–690. doi: 10.1016/j.cell.2014.09.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cingolani P, Platts A, Wang le L, Coon M, Nguyen T, Wang L, Land SJ, Lu X, Ruden DM. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin) 2012;6:80–92. doi: 10.4161/fly.19695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies L, Welch HG. Current thyroid cancer trends in the United States. JAMA Otolaryngol Head Neck Surg. 2014;140:317–322. doi: 10.1001/jamaoto.2014.1. [DOI] [PubMed] [Google Scholar]
- Eng C. PTEN Hamartoma Tumor Syndrome (PHTS) In: Pagon RA, Adam MP, Ardinger HH, Wallace SE, Amemiya A, Bean LJH, Bird TD, Fong CT, Mefford HC, Smith RJH, et al., editors. GeneReviews(R) Seattle (WA): 1993. [Google Scholar]
- Gibson BA, Kraus WL. New insights into the molecular and cellular functions of poly(ADP-ribose) and PARPs. Nat Rev Mol Cell Biol. 2012;13:411–424. doi: 10.1038/nrm3376. [DOI] [PubMed] [Google Scholar]
- Grantzau T, Overgaard J. Risk of second non-breast cancer after radiotherapy for breast cancer: a systematic review and meta-analysis of 762,468 patients. Radiother Oncol. 2015;114:56–65. doi: 10.1016/j.radonc.2014.10.004. [DOI] [PubMed] [Google Scholar]
- Gyorffy B, Lanczky A, Eklund AC, Denkert C, Budczies J, Li Q, Szallasi Z. An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1,809 patients. Breast Cancer Res Treat. 2010;123:725–731. doi: 10.1007/s10549-009-0674-9. [DOI] [PubMed] [Google Scholar]
- Gyorffy B, Surowiak P, Budczies J, Lanczky A. Online survival analysis software to assess the prognostic value of biomarkers using transcriptomic data in non-small-cell lung cancer. PLoS One. 2013;8:e82241. doi: 10.1371/journal.pone.0082241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu CH, Huang CL, Hsu YH, Iqbal U, Nguyen PA, Jian WS. Co-occurrence of second primary malignancy in patients with thyroid cancer. QJM. 2014;107:643–648. doi: 10.1093/qjmed/hcu051. [DOI] [PubMed] [Google Scholar]
- Kickhoefer VA, Siva AC, Kedersha NL, Inman EM, Ruland C, Streuli M, Rome LH. The 193-kD vault protein, VPARP, is a novel poly(ADP-ribose) polymerase. J Cell Biol. 1999;146:917–928. doi: 10.1083/jcb.146.5.917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim C, Bi X, Pan D, Chen Y, Carling T, Ma S, Udelsman R, Zhang Y. The risk of second cancers after diagnosis of primary thyroid cancer is elevated in thyroid microcarcinomas. Thyroid. 2013;23:575–582. doi: 10.1089/thy.2011.0406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. Toward better understanding of artifacts in variant calling from high-coverage samples. Bioinformatics. 2014;30:2843–2851. doi: 10.1093/bioinformatics/btu356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R Genome Project Data Processing S. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liaw D, Marsh DJ, Li J, Dahia PL, Wang SI, Zheng Z, Bose S, Call KM, Tsou HC, Peacocke M, et al. Germline mutations of the PTEN gene in Cowden disease, an inherited breast and thyroid cancer syndrome. Nat Genet. 1997;16:64–67. doi: 10.1038/ng0597-64. [DOI] [PubMed] [Google Scholar]
- Lynch HT, Casey MJ, Snyder CL, Bewtra C, Lynch JF, Butts M, Godwin AK. Hereditary ovarian carcinoma: heterogeneity, molecular genetics, pathology, and management. Mol Oncol. 2009;3:97–137. doi: 10.1016/j.molonc.2009.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manke IA, Lowery DM, Nguyen A, Yaffe MB. BRCT repeats as phosphopeptide-binding modules involved in protein targeting. Science. 2003;302:636–639. doi: 10.1126/science.1088877. [DOI] [PubMed] [Google Scholar]
- Marsh DJ, Coulon V, Lunetta KL, Rocca-Serra P, Dahia PL, Zheng Z, Liaw D, Caron S, Duboue B, Lin AY, et al. Mutation spectrum and genotype-phenotype analyses in Cowden disease and Bannayan-Zonana syndrome, two hamartoma syndromes with germline PTEN mutation. Hum Mol Genet. 1998;7:507–515. doi: 10.1093/hmg/7.3.507. [DOI] [PubMed] [Google Scholar]
- Ngeow J, Mester J, Rybicki LA, Ni Y, Milas M, Eng C. Incidence and clinical characteristics of thyroid cancer in prospective series of individuals with Cowden and Cowden-like syndrome characterized by germline PTEN, SDH, or KLLN alterations. J Clin Endocrinol Metab. 2011;96:E2063–2071. doi: 10.1210/jc.2011-1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ngeow J, Stanuch K, Mester JL, Barnholtz-Sloan JS, Eng C. Second malignant neoplasms in patients with Cowden syndrome with underlying germline PTEN mutations. J Clin Oncol. 2014;32:1818–1824. doi: 10.1200/JCO.2013.53.6656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni Y, Zbuk KM, Sadler T, Patocs A, Lobo G, Edelman E, Platzer P, Orloff MS, Waite KA, Eng C. Germline mutations and variants in the succinate dehydrogenase genes in Cowden and Cowden-like syndromes. Am J Hum Genet. 2008;83:261–268. doi: 10.1016/j.ajhg.2008.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orloff MS, He X, Peterson C, Chen F, Chen JL, Mester JL, Eng C. Germline PIK3CA and AKT1 mutations in Cowden and Cowden-like syndromes. Am J Hum Genet. 2013;92:76–80. doi: 10.1016/j.ajhg.2012.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal T, Hamel N, Vesprini D, Sanders K, Mitchell M, Quercia N, Ng Cheong N, Murray A, Foulkes W, Narod SA. Double primary cancers of the breast and thyroid in women: molecular analysis and genetic implications. Fam Cancer. 2001;1:17–24. doi: 10.1023/a:1011541424424. [DOI] [PubMed] [Google Scholar]
- Park JH, Nishidate T, Kijima K, Ohashi T, Takegawa K, Fujikane T, Hirata K, Nakamura Y, Katagiri T. Critical roles of mucin 1 glycosylation by transactivated polypeptide N-acetylgalactosaminyltransferase 6 in mammary carcinogenesis. Cancer Res. 2010;70:2759–2769. doi: 10.1158/0008-5472.CAN-09-3911. [DOI] [PubMed] [Google Scholar]
- Sandeep TC, Strachan MW, Reynolds RM, Brewster DH, Scelo G, Pukkala E, Hemminki K, Anderson A, Tracey E, Friis S, et al. Second primary cancers in thyroid cancer patients: a multinational record linkage study. J Clin Endocrinol Metab. 2006;91:1819–1825. doi: 10.1210/jc.2005-2009. [DOI] [PubMed] [Google Scholar]
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA Cancer J Clin. 2015;65:5–29. doi: 10.3322/caac.21254. [DOI] [PubMed] [Google Scholar]
- Tan MH, Mester J, Peterson C, Yang Y, Chen JL, Rybicki LA, Milas K, Pederson H, Remzi B, Orloff MS, et al. A clinical scoring system for selection of patients for PTEN mutation testing is proposed on the basis of a prospective study of 3042 probands. Am J Hum Genet. 2011;88:42–56. doi: 10.1016/j.ajhg.2010.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker MA, Jones PH, Boice JD, Jr, Robison LL, Stone BJ, Stovall M, Jenkin RD, Lubin JH, Baum ES, Siegel SE, et al. Therapeutic radiation at a young age is linked to secondary thyroid cancer. The Late Effects Study Group. Cancer Res. 1991;51:2885–2888. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary figure 1. Rare variants in hereditary cancer associated genes
(a) Rare variants in hereditary cancer associate genes found in 14 samples from double primary thyroid and breast cancers (b) Functional prediction of each variant found in our cases
Supplementary figure 2. Rare variants in recurrently mutated genes
Supplementary figure 3. Top 34 genes with P-value less than 1.0 × 10−3