Abstract
Cushing’s syndrome is caused by excessive adrenocorticotropic hormone (ACTH) secretion derived from pituitary corticotroph tumors (Cushing disease) or from non-pituitary tumors (ectopic Cushing’s syndrome). Hypercortisolemic features of ectopic Cushing’s syndrome are severe, and no definitive treatment for paraneoplastic ACTH excess is available. We aimed to identify subcellular therapeutic targets by elucidating transcriptional regulation of the human ACTH precursor POMC (proopiomelanocortin) and ACTH production in non-pituitary tumor cells as well as in cell lines derived from patients with ectopic Cushing’s syndrome. We show that ectopic hPOMC transcription proceeds independently of pituitary-specific Tpit/Pitx1, and demonstrate a novel E2F1-mediated transcriptional mechanism regulating hPOMC. We identify an E2F1 cluster binding to the proximal hPOMC promoter region (−42 to +68), with DNA binding activity determined by phosphorylation at Ser-337. hPOMC mRNA expression in cancer cells was upregulated (up to 40-fold) by co-expression of E2F1 and its heterodimer partner DP1. Direct and indirect inhibitors of E2F1 activity suppressed hPOMC gene expression and ACTH by modifying E2F1 DNA binding activity in ectopic Cushing’s cell lines and primary tumor cells, and also suppressed paraneoplastic ACTH and cortisol levels in xenografted mice. E2F1-mediated hPOMC transcription is a potential target for suppressing ACTH production in ectopic Cushing’s syndrome.
Keywords: ectopic Cushing’s syndrome, proopiomelanocortin (POMC), E2F1, Ser-337 E2F1
Introduction
ACTH-dependent Cushing’s syndrome and consequent hypercortisolemia leads to critical metabolic and cardiovascular complications (Chrousos 2009; Nieman, et al. 2015; Raff and Carroll 2015). High ACTH levels induce adrenal cortisol secretion, and may derive from pituitary corticotroph adenomas, i.e., Cushing disease, or, less commonly, from non-pituitary tumors, i.e., ectopic or paraneoplastic production (Brown 1928). Ectopic ACTH-secreting tumors can arise from a variety of sites, such as breast, lung (Ilias, et al. 2005), colon, pancreas, thyroid, ovary, and thymus. Features of hypercortisolemia are usually florid and rapidly progressive, with concomitant severe hypokalemia and higher morbidity and mortality than observed with pituitary-derived Cushing disease. The source of excess ACTH production may be occult, and pose a challenge to identify, localize, and treat. As many such tumors are already metastatic at the time of diagnosis, surgical resection is not usually an option, and failure to control ACTH levels contributes to high morbidity and mortality. Current medical therapies, which are frequently associated with significant side effects and are often poorly tolerated, address peripheral effects of excess ACTH by blocking adrenal steroidogenesis (Newell-Price 2014) (Preda, et al. 2012) or by antagonizing cortisol actions (Fleseriu, et al. 2012; Fleseriu, et al. 2015), but do not directly target the ACTH source (Colao, et al. 2012).
Subcellular mechanisms for ectopic Cushing’s syndrome are poorly defined. ACTH produced by post-translational cleavage from the proopiomelanocortin (POMC) precursor is abundantly expressed in pituitary corticotroph cells and in the hypothalamus (Stevens and White 2010). The rodent pituitary Pomc promoter (rPomc), mostly elucidated in AtT20 mouse corticotroph adenoma cells, is regulated by Tpit/Pitx1 (Lamolet, et al. 2001; Liu, et al. 2001), NeuroD1 (Poulin, et al. 1997), nuclear receptors (Maira, et al. 1999), TR4 (Du, et al. 2013), STAT3 (Bousquet, et al. 2000), and cytokines and hormones such as LIF (Bousquet, et al. 1999) and CRH (Karalis, et al. 2004). However, regulation of the human POMC (hPOMC) gene is not well understood. It is also not known whether paraneoplastic hPOMC in ectopic Cushing’s tumors is regulated by the same factors identified in pituitary cells, nor are pathways controlling POMC/ACTH expression in non-pituitary Cushing’s tumor cells well defined (Newell-Price 2003; Picon, et al. 1995; Ray, et al. 1996). Moreover, investigation of hPOMC gene regulation has been hampered due to lack of a functional human pituitary cell line.
With a view to identifying a molecular target for controlling ACTH production in patients with ectopic Cushing’s syndrome, we analyzed hPOMC expression using primary tumor cell cultures and human cell lines derived from non-pituitary ACTH-secreting tumors. We show here that ectopic hPOMC regulation differs from pituitary regulation, and report an E2F1-mediated mechanism for hPOMC transcription. We also show that E2F1 antagonists markedly suppress hPOMC expression and paraneoplastic ACTH secretion in human derived tumor primary cultures. Our results indicate that E2F1-mediated hPOMC signaling is a potential target for suppressing ACTH production in ectopic Cushing’s syndrome.
Materials and Methods
Cell culture
After obtaining informed consent, ectopic/pituitary ACTH-secreting surgical specimens were processed using Tissue Dissociate Kits (Miltenyi Biotec), cultured in low glucose DMEM 10% FBS for 48 hours, cell viability monitored by trypan blue staining, and culture medium collected for RIA. DMS79 cells (human small cell lung carcinoma), and COLO320 cells (human carcinoid-like colorectal carcinoma) were cultured in RPMI 1640 with L-glutamine and 10% FBS. Human samples are described in Supplement 6.
Radioimmunoassay (RIA)
Human ACTH and corticosterone levels were analyzed by RIA (MP Biomedicals).
DMS79 xenografts
DMS79 cells (1.5×106 cells) were injected subcutaneously to 24 five-week-old male Nu/J mice (Jackson Laboratory). 7 days later, mice were treated by oral gavage with R-roscovitine (200 mg/kg, n=12) or vehicle (0.5% methylcellose, 0.5% tween80/PBS; 100 µl, n=12) for 17 days. Animals were then sacrificed, tumors were measured and weighed, and blood was collected for ACTH and corticosterone RIA. Pre- and post-treatment ACTH and corticosterone levels are depicted in Supplement 3.
RT-PCR
POMC, PTH, and GH mRNA levels were analyzed by RT-PCR. Total RNA was isolated using RNeasy mini kit (Qiagen), and cDNA prepared using a QuantiTect reverse transcription kit with random oligo primers (Applied Biosystems). Quantitative RT-PCR was performed by Taqman Gene Expression Assays (hPOMC probe; Hs01596743_m1, hGAPDH probe; Hs02758991_g1, hPTH probe; Hs00757710_g1, hGH probe; Hs00792200_g1, Taqman Gene Expression Master mix; 439016, Applied Biosystems). POMC RNA levels were normalized by GAPDH. Each figure depicting RT-PCR results is compiled from at least three independent experiments, and each sample was analyzed in triplicate or quadruplicate.
Immunoblotting
Immunoblotting was performed using anti-ACTH (ab20358; Abcam), anti-E2F1 (sc-251; Santa Cruz Biotechnology), anti-phospho-Serine 364-E2F1 (ab5391; Abcam), anti-phospho-Serine337-E2F1 (ab135549; Abcam), LaminA/C (sc-20681; Santa Cruz Biotechnology), and GAPDH (sc-25778; Santa Cruz Biotechnology). Ab binding was detected using Mini PROTEAN electrophoresis protocol (BioRad).
Small interfering RNA (siRNA) transfection
E2F1 siRNA (107660; Thermo Fisher) transfections were performed using 4D-Nucleofector (Lonza).
Flow cytometry
After drug treatment, cell viability was analyzed by flow cytometer (CyAn) using FITC Annexin V detection kit with 7-AAD (BioLegend).
Mapping of hPOMC RNA start sites
The human POMC transcription start site was determined by 5’ RACE (Frohman, et al. 1988) in DMS79 and COLO320 cells, with minor modifications (5). Briefly, cDNA was prepared using an oligo primer (CAGTCAGCTCCCTCTTGAACT CCA) binding to POMC RNA. The G-tail was added at the 5’-end of the resulting cDNA, and 5’-ends were PCR amplified using a poly(C) primer and an antisense POMC primer (ACTCCAGGGGGAAGGCCTCGGCCGA). To concentrate the hPOMC cDNA fragments, a second PCR was performed using the antisense POMC primer (TTGCCCTCGCGCGGGCCC GGCT) and the poly(C) promer. Resulting PCR products were cloned and 5’-ends determined by DNA sequencing. To avoid mis-mapping due to PCR artifacts, three independent PCR products were analyzed.
Construction of promoters and luciferase assays
The hPOMC promoter (−1200 to +83) was PCR amplified from genomic DNA (Jurkat cells) and cloned. Wild type (Wt) and 5’ deletion mutants were amplified using 5’-primers binding to −1140, −428, −303, −102, −42, (containing Sal I site at the 5’-end) and 3’-primer binding to +68 (containing Hind III sites at the 3’-end). Resulting amplified fragments were digested with Sal I and Hind III and cloned into Xho I and Hind III sites in pGL3 basic vector. Point mutations in the −428/+68 sequence (Fig. 1C) were introduced by PCR assembly, and mutated fragments integrated into Xho I and Hind III sites in pGL3 basic vector. DNA sequences of all inserted fragments were determined to remove defective fragments generated by PCR errors. For minimal promoters, −42 to +68 probes were divided into seven small fragments (Fig. 2B). Sense and antisense oligonucleotides encoding these sequences were synthesized, duplicated, and ligated upstream of the minimal promoter in pGL3 as described (Ogawa, et al. 2014). Luciferase reporter plasmids containing five direct repeat inserts were selected by DNA sequencing (Fig. 2B). DNA sequences of inserted fragments were determined to remove defective fragments generated by PCR errors.
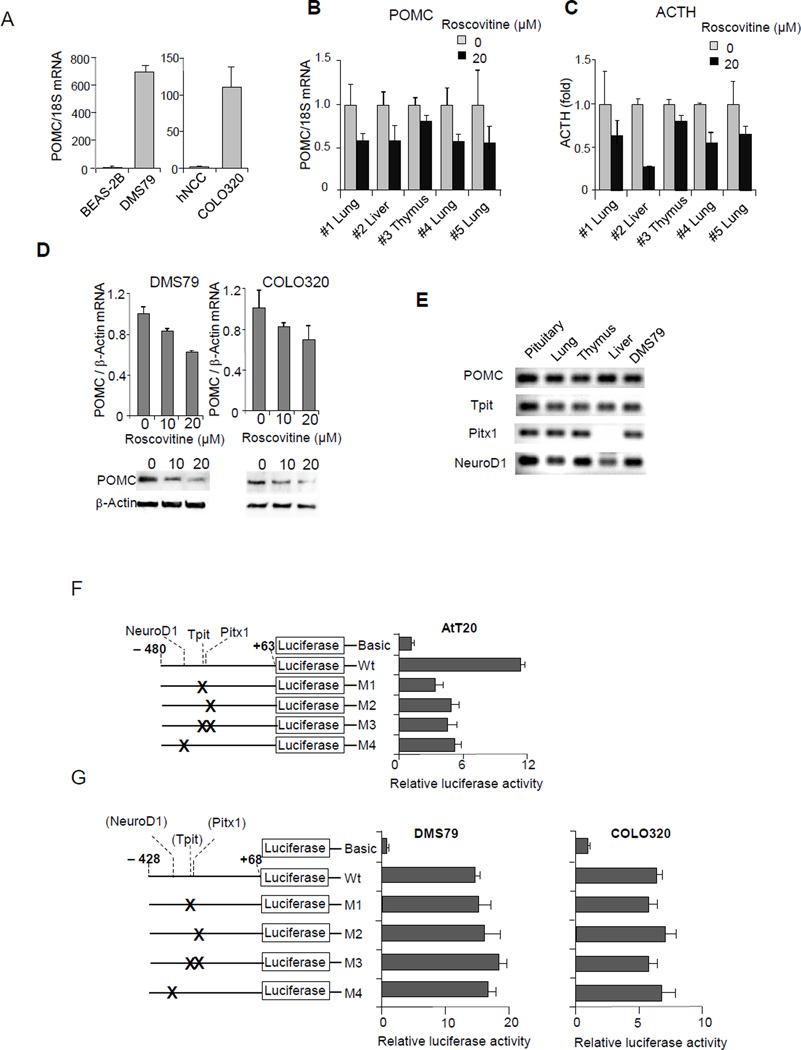
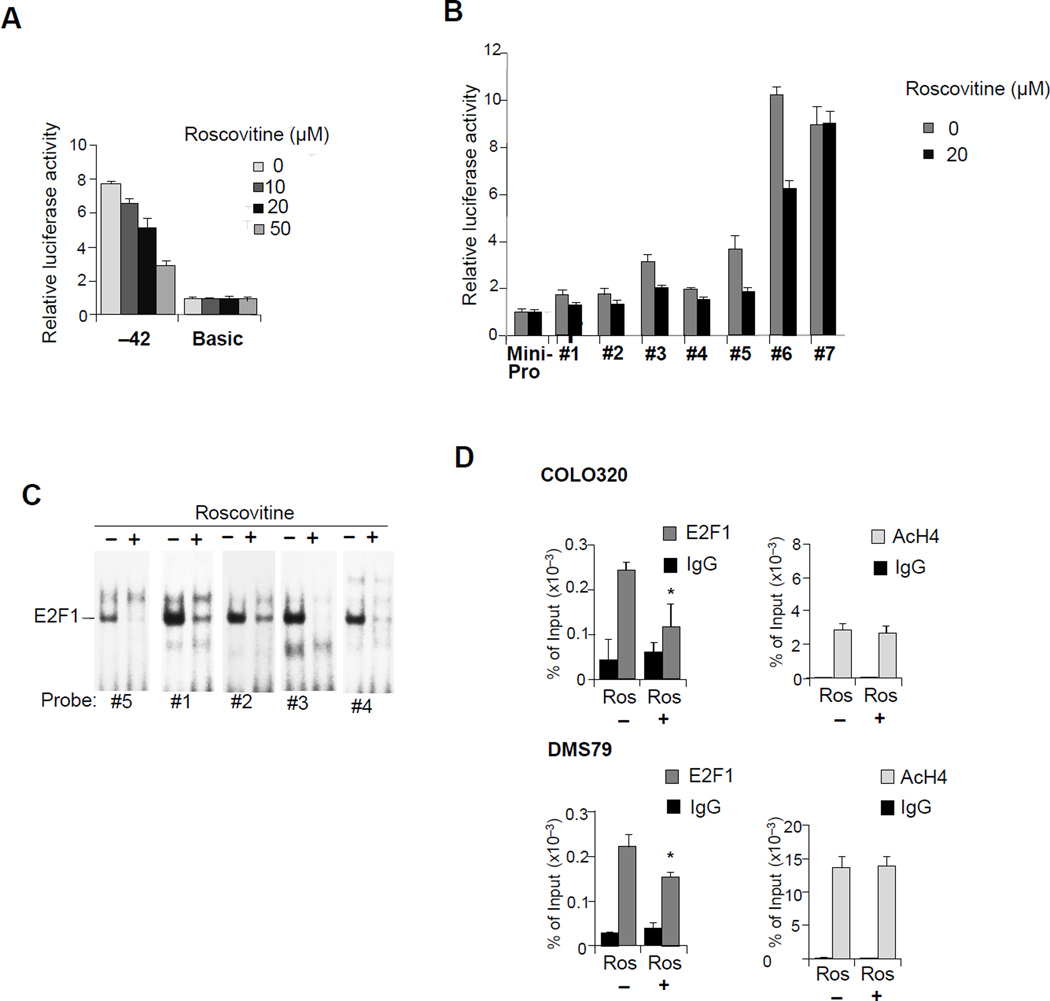
Figure 1. hPOMC promoter activity in ectopic Cushing’s cell lines.
(A) POMC expression in lung (DMS79) and colon (COLO320) cancer cell lines were analyzed by RT-PCR and compared with those in normal lung (BEAS-2B) and normal colon (hNCC) cells. Expression levels were normalized with 18S rRNA. (B-C) Primary cultures of cells from five human ectopic ACTH producing tumors (#1, #4, #5 lung; #2 liver; #3 thymus) treated with or without R-roscovitine for 48 hours. POMC mRNA were measured by RT-PCR (B) and medium ACTH concentrations in primary cultures were measured by RIA (normalized for viable cell numbers; n=5 tumors, mean ± SE) (C). (D) POMC mRNA levels were analyzed by RT-PCR using RNA derived from DMS79 and COLO320 cells treated with or without indicated doses of R-roscovitine. POMC protein levels in derived whole cell extracts were also analyzed by immunoblotting with anti-POMC. (E) POMC, Tpit, Pitx1, and NeuroD1 expression in pituitary Cushing’s tumor, ectopic Cushing’s tumors (lung, thymus, and liver), and DMS79 cells were analyzed by RT-PCR. (F) Structure of point mutated luciferase reporter plasmids of the rPomc promoter (−480/+63) (left). Luciferase assays performed using point mutations in AtT20 cells (right) were compared with those generated using negative control plasmid basic (no promoter). (G) Structure of point mutated luciferase reporter plasmids of the hPOMC promoter (−428/+68) (left). Luciferase assays performed using point mutants in DMS79 cells and COLO320 cells (right) were compared with those generated using negative control plasmid basic (no promoter). Luciferase (F, G) and RT-PCR (A, D) results are representative of four independent experiments, RIA was performed in duplicate tubes; values are mean ± SE (B, C), and immunoblotting results are representative of three independent experiments (D).
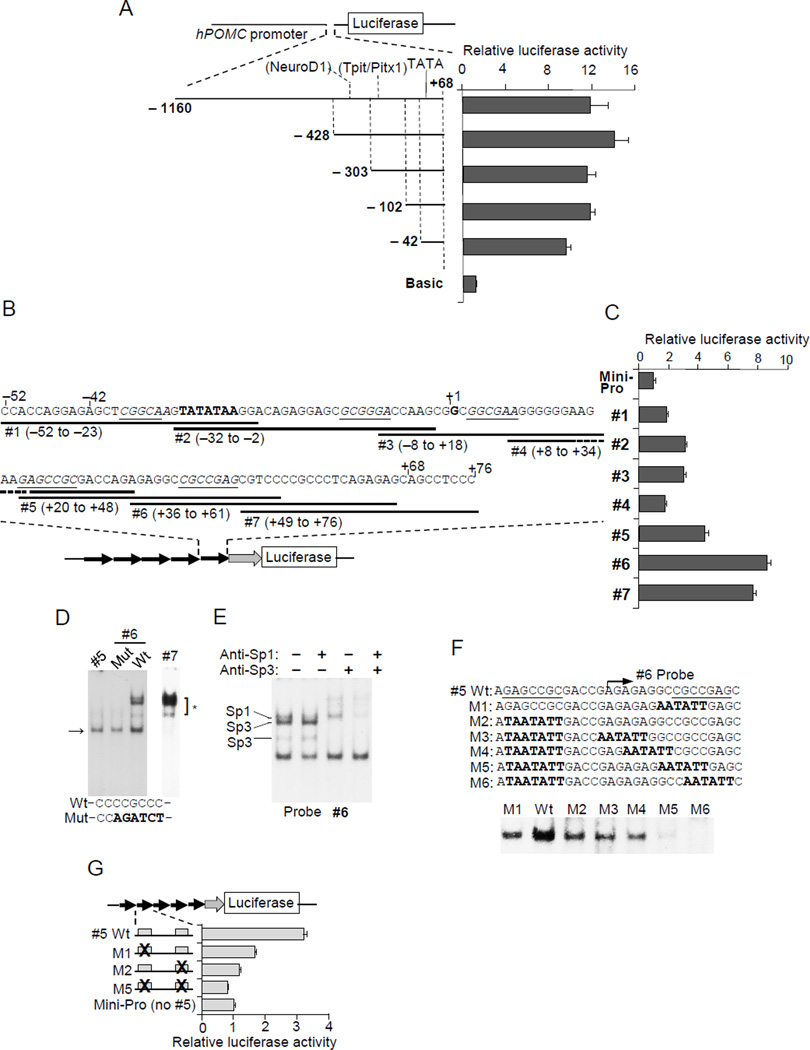
Figure 2. Nuclear factor binding to the proximal hPOMC promoter (−42 to +68).
(A) Structures of luciferase reporter plasmids using human POMC promoter 5’-deletion mutants. Positions of 5’-ends are indicated (all deletion mutants contain the same 3’-end, +68). Luciferase assays using 5’ deletion mutants in DMS79 cells (right), and activities compared with negative control plasmid basic (no promoter). (B) hPOMC DNA sequence (−52/+76). The transcription start site is defined as position +1. Minimal promoter plasmids contain five direct repeat copies of the indicated fragments (#1 to #7) and oligonucleotide orientation is indicated by an arrow (direct repeat). (C) Luciferase assays in DMS79 cells using minimal promoter plasmids shown in B, compared with mini-Pro (no inserted fragment). (D) EMSA performed using probes #5, #6, mutant probe #6, and #7 in DMS79 cells. The mutated sequence in probe #6 is shown in bold. The three upper bands are indicated by * (the top thick band consists of two bands) and the common band from probes #5 and #6 is indicated by the arrow. (E) Super-shift EMSA was performed using probe #6 with nuclear extracts from DMS79 cells, and anti-Sp1, and anti-Sp3. Positions of Sp1 and two different Sp3 sizes are shown. (F) EMSA was performed with Wt and M1-6 probes with nuclear extracts from DMS79 cells. Mutant sequences are in bold; bands are shown under the probe sequences. (G) Luciferase assays were performed in DMS79 cells using the indicated reporter plasmids. Inserted fragments (five direct repeat copies) are shown, and mutated sequences (shown in F) are indicated by ×. Luciferase activities were compared with those without inserted fragment. EMSA results are representative of three independent experiments (D, E) and luciferase results are representative of four independent experiments, each depicted as mean ± SE of triplicate samples (A, C, G).
Luciferase reporter assays were performed in 2.5 × 105 DMS79 cells with 0.7 µg luciferase reporter plasmids, and 50 ng pRL-CMV used as an internal control plasmid. Cells were transfected using lipofectamine 2000 (Invitrogen) and cultured in 0.5 ml medium in 24-well plates. Forty-eight hours post-transfection, cells were harvested and luciferase activity analyzed by Dual-Luciferase Reporter Assay System (Promega). Luciferase assays were repeated more than three times and luciferase activities normalized by internal renilla activity. For co-transfection experiments, 0.5 µg luciferase reporter plasmids were co-transfected with 1.5 µg E2F1, E2F3, and/or DP1 expression plasmids constructed using pMF vector carrying the EF-1α promoter. Total DNA (2.0 µg) was adjusted with the empty vector.
EMSA
Nuclear extracts were prepared as described (Tone, et al. 2002). 5 µg nuclear extracts were incubated in 14 µl pre-incubation buffer [5 mM Tris (pH 8.0) and 14% glycerol] for 20 min; 0.5 µg poly (dI-dC), 1.2µl of 5 x EMSA standard buffer (Tris-HCl 4mM, 12 mM HEPES (pH 7.9), 60 mM KCl, 0.2 mM EDTA, 2mM DTT and 12% glycerol), and 32P-labeled probes were added for a further 15 minutes. For competition assays, 100-fold excess unlabeled competitor primer was added to EMSA reaction mixtures. For supershift assays, nuclear extracts in EMSA reaction buffer were incubated for 15 min with anti-E2F1 (sc-251X; Santa Cruz Biotechnology), anti-E2F2 (sc-633X; Santa Cruz Biotechnology), anti-E2F3 (sc-878X; Santa Cruz Biotechnology), anti-E2F4 (sc-866X; Santa Cruz Biotechnology), anti-Sp1 (07-645; Millipore), or anti-Sp3 (sc-644X; Santa Cruz Biotechnology), and probes then added. DNA-protein complexes were analyzed by 4% DNA retardation gel. Probe and competitor sequences (sense strand) used were: # 1; CCACCAGGAGAGCTCGGCAAGTATATAAGG, # 2; GTATATAAGGACAG AGGAGCGCGGGACCAA, # 3; GACCAAGCGGCGGCGAAGGAGGGGAAG, # 4; AA GGAGGGGAAGAAGAGCCGCGACCGAG, #5; AAGAGCCGCGACCGAGAGAG GCCGCCGAGC, # 6; AGAGGCCGCCGAGCGTCCCCGCCCTCA, # 7; GTCCCC GCCCTCAGAGAGCAGCCTCCC.
EMSA was also performed using CIP-treated nuclear extract from DMS79 cells as described (Tone et al. 2002). Briefly, 5 µg nuclear extract was treated with 0.6 units CIP (New England BioLabs) in EMSA buffer at room temperature for 1 hour, and the reaction terminated by adding phosphatase inhibitor cocktail (Sigma-Aldrich). To prepare control nuclear extracts, phosphatase inhibitor cocktail was added before the addition of CIP and incubated for 1 hour.
Chromatin immunoprecipitation assay
ChIP assay was performed using DMS79 and COLO320 cells as described (Tone, et al. 2008). Cells were fixed in 1% formaldehyde, 4.5 mM HEPES pH8.0, 9 mM NaCl, 0.09 mM EDTA, and 0.045 mM EGTA for 10 min at room temperature, and sonicated (Bioruptor) in lysis buffer (1% SDS, 10 mM EDTA, and 50 mM Tris-HCl pH 8.0) with proteinase inhibitor (Sigma-Aldrich P8340). Pre-cleared lysates were incubated overnight at 4 °C with anti-E2F1 (sc-251X; Santa Cruz Biotechnology); anti-acetyl histone H4 (06-866; Millipore); and normal rabbit IgG (sc-2027; Santa Cruz Biotechnology) polyclonal. DNA fragments were isolated from immunoprecipitated chromatin, and analyzed by RT-PCR analysis with SYBR Green PCR Master Mix (Applied Biosystems). PCR primers used were: hPOMC 5’-CCCCTCCTACCCTTGCTGTA, 5’-GGGCACAGCTCCCATAATCA, 5’-CAAAGAGGGCTTCAGAGAACA, 5’-CACTTCAGCATCCACAACATTC, 5’-GAGAGCTCGGCAAGTATAAAG, 5’-TCGGCGCAGAAAGTTTG.
DNA pull-down assay
DNA pull-down assay was performed as described (Ogawa et al. 2014). A 74-bp DNA fragment containing the E2F1 binding site (-24/+50) (Fig. 2B) was amplified by PCR using biotinylated primers. 30 µl nuclear extract (100 µg) was added to 180 µl of 5 mM Tris (pH 8.0)-14% glycerol buffer and pre-incubated on ice. 15 µl of poly(dI-dC)(dI-dC) (7.5 µg) and 18 µl of 5 x binding buffer (300 mM KCl, 60 mM HEPES [pH 7.9], 20 mM Tris-HCl [pH 8], 0.5 mM EDTA, 25% glycerol and 5mM DTT) and 6 µl of probe (1 µg) were incubated at room temperature for 15 min. DNA–protein complexes were collected with 10 µl Dynabeads M-280 Streptavidin magnetic beads (Life Technologies) and analyzed by immunoblotting.
Statistics
Differences between vehicle and R-roscovitine groups were analyzed using ANOVA followed by nonparametric t test (Mann-Whitney) or Student t test. Probability of P < 0.05 was considered significant. In bronchial carcinoid tumors, the correlation of POMC and E2F1 expression were assessed by Spearman’s correlation.
Study approval
Protocols were approved by the Cedars-Sinai Institutional Review Board (protocol #30753). Male and female patients over 18 years of age with a clinical diagnosis of ectopic or pituitary ACTH-producing tumors and lung carcinoid tumors were screened and informed consent obtained. Human samples (age, gender, diagnosis) are described in Supplement 6. Animal protocol (protocol #2603) was approved by the Cedars-Sinai Institutional Animal Care and Use Committee.
Results
hPOMC promoter activity in non-pituitary cells
We used transfection-based luciferase assays to analyze hPOMC promoter activity and investigate transcriptional regulation in non-pituitary human cells. As primary tumor cells are not suitable for these experiments, we employed human small-cell lung cancer DMS79 cells widely used in studies of ectopic Cushing’s syndrome (Pettengill, et al. 1980) (White, et al. 1989) (Newell-Price, et al. 2006) (Picon, et al. 1999). We also found that ACTH-positive human colon carcinoid-like cancer cells COLO320 (Quinn, et al. 1979) express POMC mRNA. Therefore, we analyzed expression of hPOMC mRNA in these two cell lines by RT-PCR. Compared with normal lung BEAS-2B cells (Lechner, et al. 1983) and normal colon hNCC cells (Chesnokova, et al. 2013), DMS79 and COLO320 cells expressed hPOMC mRNA at high and intermediate levels, respectively (Fig. 1A).
Previously, we showed that POMC gene expression in pituitary cells is suppressed by the cyclin-dependent kinase/cyclin E inhibitor R-roscovitine (Liu, et al. 2015; Liu, et al. 2011) We therefore analyzed hPOMC gene expression in human tumor cells derived from five patients with ectopic Cushing’s syndrome and in DMS79 and COLO320 cells. Similar to what we found in pituitary cells, hPOMC mRNA expression was suppressed with R-roscovitine treatment (Fig. 1B-1D), suggesting that hPOMC gene expression is, at least in part, regulated by mechanisms similar to those in human ectopic primary cells. We therefore employed these cell lines for subsequent hPOMC gene analysis.
We next considered whether the transcription factors Tpit, Pitx1, and NeuroD1 regulate in POMC expression in pituitary corticotrophs are similarly involved in ectopic Cushing’s syndrome. We found mRNA expression of all three transcription factors in patient-derived ectopic Cushing’s tumors arising from the lung, thymus, and liver as well as in DMS79 cells (Fig. 1E), although Pitx1 mRNA was undetectable in the liver tumor cells. We therefore analyzed whether these transcription factors regulate hPOMC promoter activity in non-pituitary cells, as they do in pituitary cells.
To map the hPOMC promoter region, we determined the major transcription start site in non-pituitary tumor cells by 5’-RACE (rapid amplifications of cDNA ends) (Frohman et al. 1988). This start site, which we defined as position +1, is 4-bp downstream from the site previously defined by primer extension (Takahashi, et al. 1983) (Fig. 2A). A TATA-like sequence was identified 32-bp upstream of the start site, similar to the previously identified rat promoter (Drouin, et al. 1985). We then analyzed hPOMC promoter activity in non-pituitary tumor cells by performing luciferase reporter assays with point mutations within the Tpit/Pitx1 and NeuroD1 sites (Fig. 1F). These sequences identified in the rPomc promoter are conserved in the hPOMC promoter (Supplement 1). When similar point mutations were introduced in the rPomc promoter in AtT20 pituitary cells, promoter activity was attenuated (Fig. 1F), but mutated Tpit/Pitx1 and NeuroD1 sites did not reduce hPOMC promoter activity in ectopic cell lines (Fig. 1G). Next we performed hPOMC promoter analysis using a series of 5’-deletion mutants. Previously, the hPOMC promoter was analyzed up to −428 bp (Picon et al. 1995); we now extended the analysis to −1160/+68 (Fig. 2A). Similar promoter activity was observed with all deletion mutants. Seventy percent of maximum promoter activity was maintained in the shortest −42/+68 promoter, suggesting this is a key proximal regulatory region.
Taken together, these results suggest that hPOMC promoter activity is not similarly controlled by the same pituitary-specific factors that regulate promoter activity in Cushing’s disease. Rather, it is mainly controlled by a proximal regulatory region located between −42/+68 bp in non-pituitary cells.
As this short proximal promoter comprises only a 42-bp sequence upstream of the transcription start site, including the TATA (−31 to −25 bp), we could not generate further 5’-deletion mutants. To identify the binding of transcription factors, we therefore constructed a luciferase reporter system with a minimal promoter containing the TATA sequence and transcription start site (Tone et al. 2008) (Fig. 2B). This method was used previously to identify other transcription factors (Ogawa et al. 2014; Tone, et al. 2000; Tone et al. 2008). The putative regulatory region [−52 (−42 plus additional 10-bp) to +76 (+68 plus additional 8-bp)] was systematically divided into seven fragments with partially overlapping sequences (Fig. 2B). These short (24-bp to 30-bp) DNA fragments (#1 to #7) were integrated upstream of the minimal promoter, and five sequence copies added as direct repeats (Fig. 2B). When the resulting luciferase reporter plasmids were transfected into DMS79 cells (Fig. 2C), moderate-to-strong luciferase activity was detected with fragments #5 to #7, and mild increments were detected with fragments #1 to #4. We then performed electrophoretic mobility shift assays (EMSA) with fragments #5 to #7 (Fig. 2D). Several bands were detected; the lower bands were consistently observed with #5 and #6 fragments, and the three upper bands (the top thick band consists of two bands) were detected in #6 and #7 fragments. Overlapping regions within #6 and #7 include a CCCCGCCC sequence similar to the Sp1 consensus, and mobility patterns resemble those identified for Sp1/Sp3 binding (Tone et al. 2002). Mutating this sequence resulted in dissipation of the top three bands (Fig. 2D). Sp1/Sp3 binding was also confirmed by super-shift EMSA using anti-Sp1 and anti-Sp3 (Fig. 2E). The lower single band detected with #5 and #6 regions (Fig. 2D, 2E, arrow) was also seen with the #6 Sp1 mutant probe. We therefore reasoned that a common factor, different from SP1/Sp3, binds this overlapping #5 and #6 sequence region (AGAGAGGCCGCCGAG).
hPOMC expression is regulated by E2F1-mediated transcriptional mechanisms
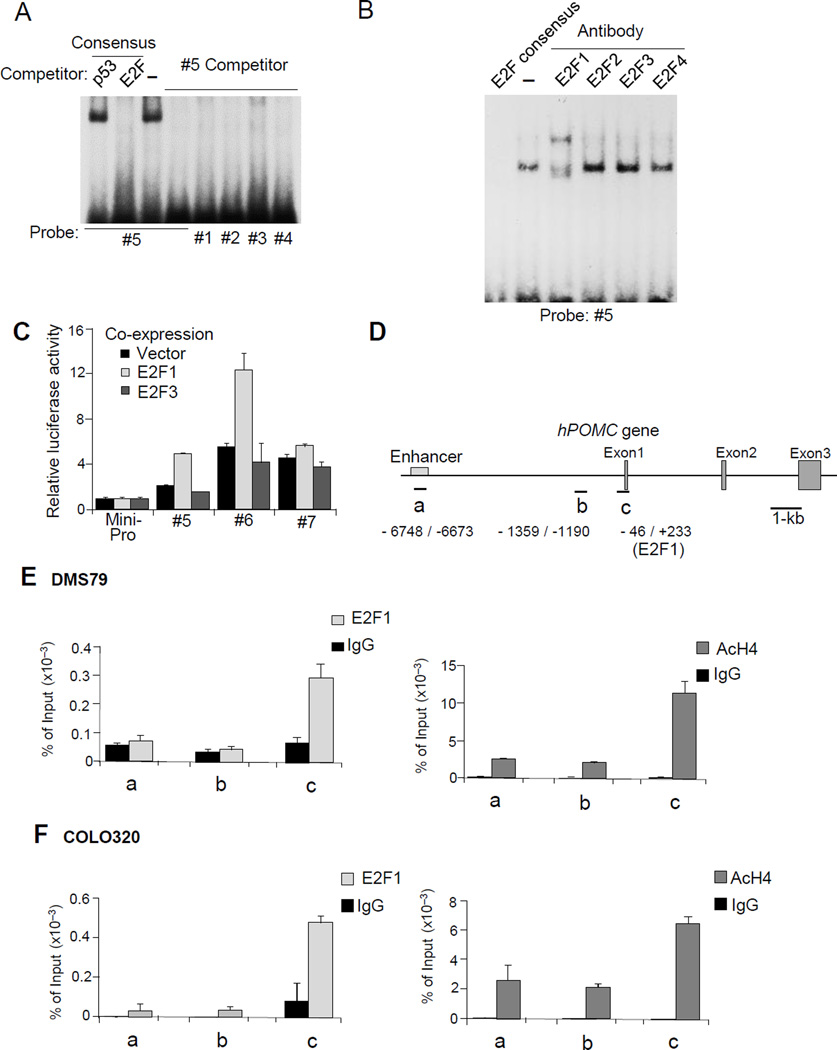
We introduced a mutation (GCCGCC to AATATT) in the middle of the overlapping #5 and #6 sequence, and, using EMSA, showed partially attenuated binding (about 50%) (Fig 2F), suggesting a nuclear factor binding to GCCGCC, and also indicating the likely presence of another binding site for this probe. Indeed, we observed a similar GCCGC sequence near the 5’-end of the probe (Fig. 2F), with binding reduced to half by mutating GAGCCGC to TAATATT. Using EMSA with additional mutant probes (Fig. 2F), we observed that binding was abolished with M5 and M6 probes, suggesting the presence of two core binding motifs: CGCCGAG in the 3’- site and GAGCCGC (mirror image to the 3’-site core sequence) in the 5’ region of the probe (Fig. 2F). Functions of these two core-binding sequences were further confirmed by luciferase reporter assays, which showed that mutating these sequences led to reduced promoter activity of probe #5 (Fig. 2G).
Although no potential transcription factor binding to the identified sequences (GAGCCGC and CGCCGAG) was suggested using databases, we noted that these sequences were similar to the 3’-half of the E2F transcription family consensus motif, i.e., AGTGCCGC (Tao, et al. 1997). Using EMSA competition assays with a labeled #5 probe and an unlabeled E2F consensus sequence as competitor, transcription factor binding was abolished by both the #5 probe as well as by E2F consensus competitors, but not by unrelated p53 consensus competitors (Fig. 3A left), suggesting that E2F member(s) bind the #5 probe. Using super-shift EMSA assays with antibodies to E2Fs (E2F1 to E2F4), we detected a super-shifted band with anti-E2F1 (Fig. 3B), but not with other antibodies. E2F1-mediated hPOMC promoter activity was also confirmed by luciferase reporter assay using the #5, #6, and #7 promoter plasmids with co-transfection of either E2F1 or E2F3 expression plasmid. Both promoters #5 and #6 were upregulated by co-expression of E2F1 but not by E2F3, while promoter #7 (as a negative control with no E2F1 binding site) was not regulated by either E2F1 or E2F3 co-expression (Fig. 3C).
Figure 3. E2F1 binding to the hPOMC promoter.
(A) Results of EMSA performed with indicated probes and consensus competitors. No competitor is indicated by −. (B) EMSA and supershift-EMSA were performed with nuclear extracts prepared from DMS79 cells using probe #5, E2F consensus competitor, and indicated antibodies to E2F molecules. (C) Results of luciferase assays performed using the indicated reporter plasmids constructed with hPOMC promoter fragments (#5, #6, and #7) and the minimal promoter shown in Fig. 2B. Reporter plasmids were co-transfected with E2F1 or E2F3 expression plasmids or negative control empty vector (Vector). (D-F) ChIP assays were performed with anti-E2F1 (E2F1), anti-acetyl histone H4 (AcH4), or negative control (IgG) using primer sets shown in D (a-c). The analyzed ChIP regions are from DMS79 (E) and COLO320 cells (F). EMSA and ChIP results are each representative of triplicate independent experiments (A, B, E, F) and luciferase results are representative of four independent experiments, each depicted as mean ± SE of triplicate samples (C).
E2F1 binding to the proximal promoter region was further analyzed (Fig. 2B). Similar sequences of the 3’-half of the E2F consensus (CGGCAA in #1, GCGGGA in #2, GGCGAA in #3, and GAGCCGC in #4) were identified in the −42/+68 hPOMC region, and minimal promoter activities were also moderately upregulated in these fragments (Fig. 2C). We therefore performed EMSA competition assays using the labeled probe #5 with un-labeled competitor (#1 to #4) (Fig. 3A right), and observed that E2F1 binding to probe #5 was blocked by #1 to #4 competitors, suggesting that the E2F1 cluster binds the −42/+68 region.
E2F1 binding to the proximal hPOMC in vivo was confirmed by ChIP assay using DMS79 and COLO320 cells (Fig. 3D-F), and histone H4 was highly acetylated in this region, but not in upstream control regions (−1.2-kb or − 6.7-kb) identified as an rPomc Tpit palindrome enhancer (Langlais, et al. 2011) (Fig. 3E, 3F).
E2F1 determines hPOMC expression in non-pituitary ACTH-producing tumor cells
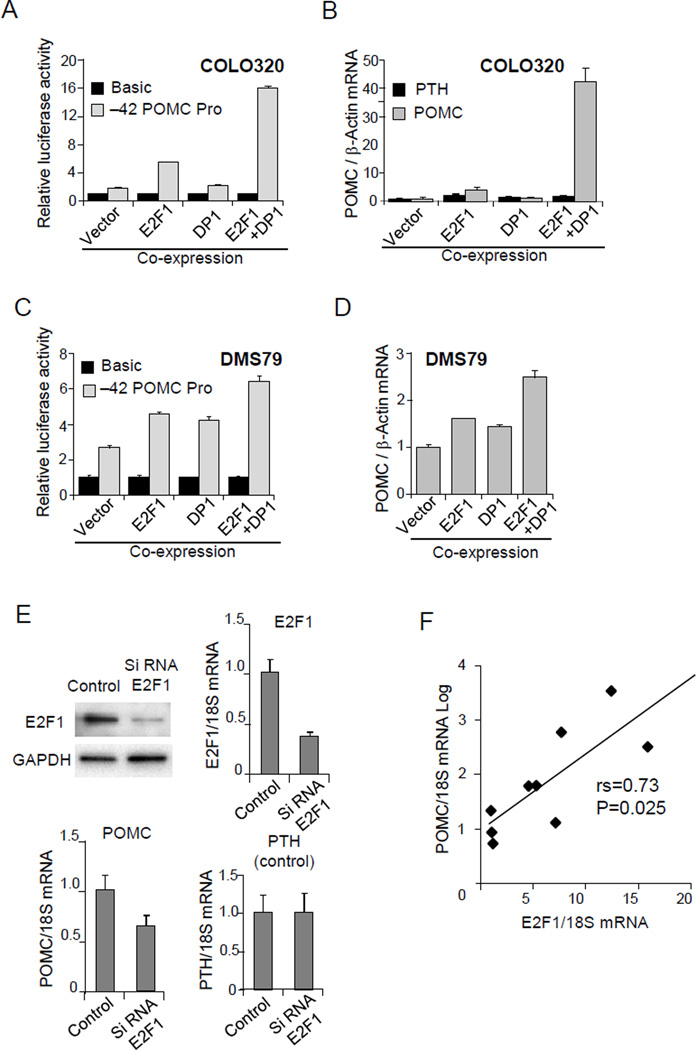
To define the functional role of E2F1, we performed luciferase assays and RT-PCR after co-expressing E2F1 and its heterodimer DP1 (Girling, et al. 1993). hPOMC promoter activity (−42/+68) and POMC mRNA levels in COLO320 cells were markedly upregulated by co-expression of E2F1 and DP1, but parathyroid hormone, which served as a control gene, was not upregulated (Fig. 4A, 4B). In DMS79 cells, hPOMC promoter activity and mRNA expression were also upregulated by E2F1 and DP1 co-expression (Fig. 4C, 4D). Gene specific POMC knock down was performed by using Si-RNA E2F1, while the control gene (PTH) was unchanged, further suggesting a specific contribution of E2F1 to POMC gene regulation (Fig. 4E).
Figure 4. E2F1-mediated hPOMC transcription in ectopic Cushing’s tumor cells.
(A) Results of luciferase assays performed in COLO320 cells using the −42 hPOMC promoter (−42/+68) reporter plasmid (Fig. 2A) or negative control basic (no promoter). E2F1 and/or DP1 expression plasmids were co-transfected, and total DNA abundance adjusted with empty vector. (B) POMC and PTH mRNA levels were analyzed by RT-PCR using RNA derived from COLO320 cells transfected with E2F1 and/or DP1 expression plasmids. (C) Luciferase assays were performed using the −42 hPOMC promoter in DMS79 cells as described in A. (D) POMC mRNA levels were analyzed in DMS79 cells as described in B. (E) Immunoblotting of protein extracts derived from DMS79 cells transfected with control siRNA (control) or E2F1 siRNA. E2F1, POMC, and PTH expression measured by RT-PCR of cDNA derived from DMS79 cells transfected with control siRNA (control) or E2F1 siRNA. Expression levels were normalized with 18S rRNA. (F) POMC and E2F1 mRNA levels were analyzed by RT-PCR using human lung carcinoid tumors. Expression levels were normalized with 18S rRNA. Immunoblotting results are representative of three independent experiments (E) and RT-PCR and luciferase results are representative of three independent experiments (A-F), each depicted as mean ± SE of triplicate samples (A-E).
We next confirmed the role of E2F1 in regulation of hPOMC gene expression using 12 human bronchial carcinoid tumor samples, known to exhibit a propensity for ectopic ACTH production (Aniszewski, et al. 2001; Isidori, et al. 2006). hPOMC expression was detected in 9 of 12 tumors and, importantly, E2F1 and POMC expression levels correlated strongly (P = 0.025) (Fig. 4F).
Taken together, the results suggest that hPOMC expression levels are determined by E2F1, which mediates transcription through the −42/+68 proximal promoter.
E2F1 binding to hPOMC proximal promoter decreased by R-roscovitine treatment
Given the role of E2F1 in regulating hPOMC expression, we considered whether E2F1 could serve as a target in ectopic Cushing’s tumors. As shown in Fig. 1B-D, we used R-roscovitine, known to suppress mPomc in pituitary corticotrophs (Liu et al. 2015), to elucidate the mechanism. Cell viability assays were used to determine doses for in vitro experiments (Supplement 2). We confirmed suppression of paraneoplastic hPOMC mRNA and ACTH production in surgically excised ectopic ACTH-producing samples derived from lung, liver, and thymus tumors (n=5) treated with R-roscovitine (Fig. 1B, 1C). Dose-dependent hPOMC expression was seen in DMS79 and COLO320 cells treated with R-roscovitine for 48 hours (Fig. 1D). We also confirmed in vivo efficacy of this drug in DMS79 cell xenografted mice, showing decreased plasma ACTH and corticosterone levels (Supplement 3).
Next, using minimal promoters and the proximal −42/+68 hPOMC promoter, we showed that R-roscovitine dose-dependently suppressed −42/+68 hPOMC promoter activity (Fig. 5A). Marked inhibition of promoter activity by R-roscovitine was observed, especially with fragments #5 and #6 (E2F1 sites), while promoter activity was unaltered with fragment # 7 (Sp1/Sp3 site) (Fig. 5B), confirming that R-roscovitine acts on E2F1 binding sites, not via SP1/SP3. To elucidate a mechanism for the observed inhibition, we performed EMSA using the same minimal promoter fragments with or without R-roscovitine treatment. E2F1 binding to probe #1 to #5 (E2F1 cluster) (Fig. 3A) was dissipated by R-roscovitine (Fig. 5C). Reduction of in vivo E2F1 binding to this region was also observed using ChIP assays (Fig. 5D) in DMS79 and COLO320 cells, while highly acetylated histone H4 was unaltered by R-roscovitine, suggesting that E2F1 determines hPOMC transcriptional levels without affecting chromatin remodeling. Taken together, these results provide evidence that E2F1 is an appropriate molecular target for suppressing hPOMC.
Figure 5. hPOMC is suppressed by R-roscovitine treatment through E2F1 DNA binding.
(A) Luciferase assays were performed using −42 hPOMC promoter in DMS79 cells treated with (+) or without (−) R-roscovitine at the indicated concentrations, and luciferase activities compared to negative control plasmid basic (no promoter). (B) Luciferase assays using the minimal promoter (Fig. 2B) were performed in DMS79 cells treated with (+) or without (−) R-roscovitine at the indicated concentrations. (C) Results of EMSA performed using indicated probes (#1 through #5, shown in Fig. 2A) with nuclear extracts derived from DMS79 cells treated with (+) or without (−) R-roscovitine (50 µM). (D) ChIP assays were performed with anti-E2F1 (E2F1), anti-acetyl histone H4 (AcH4), or negative control (IgG) using the primers ™46/+233 (Fig. 3D). Analyzed ChIP regions from R-roscovitine (50 µM) treated (Ros+) or non-treated (Ros−) DMS79 cells and COLO320 cells are shown. Luciferase results are representative of four independent experiments (A,B), EMSA results are representative of three independent experiments (C), and ChIP assays results are representative of three independent experiments (D).
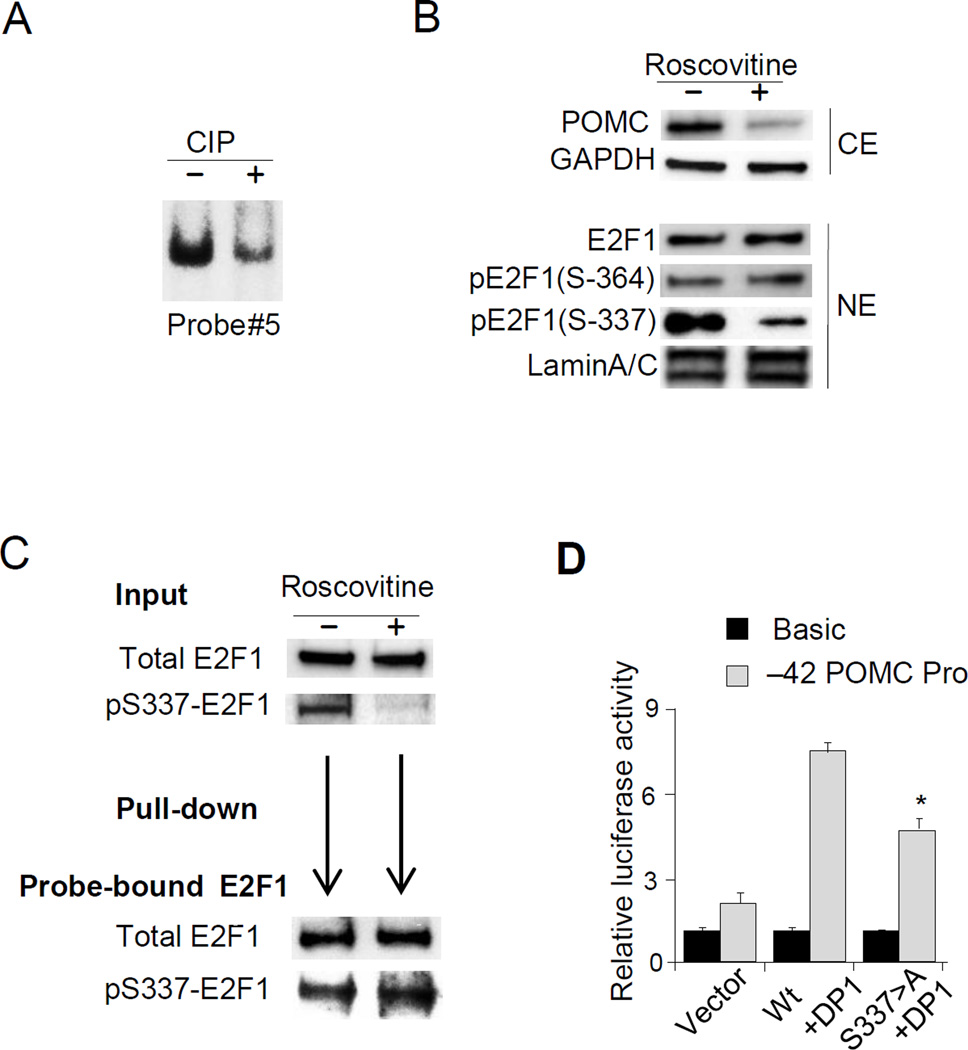
hPOMC transcription is regulated by E2F1 Ser-337 phosphorylation
Since E2F1 DNA binding was modulated by R-roscovitine, we assessed whether E2F1 phosphorylation status affects E2F1 DNA binding. EMSA with nuclear extracts dephosphorylated by CIP (Tone et al. 2002) reduced E2F1 DNA binding to the hPOMC promoter after dephosphorylation (Fig. 6A). To test the site-specificity of E2F1 dephosphorylation, we performed a series of immunoblotting experiments in DMS79 cells. Total nuclear E2F1 and phosphorylated Ser-364 E2F1 (pSer364-E2F1) were not altered by R-roscovitine treatment. However, R-roscovitine reduced nuclear phosphorylated Ser-337 E2F1 (pSer337-E2F1) (Fig. 6B). We further assessed the role of phosphorylated Ser337-E2F1 to DNA binding activity by pull down assay using excess nuclear extract to saturate the hPOMC probe (−24/+50) (Fig. 6C). Although pSer337-E2F1 input was markedly reduced by R-roscovitine, DNA-bound pSer337-E2F1 was still clearly detected (Fig. 6C, Supplement 4), suggesting highly selective binding of pSer337-E2F1 to the proximal hPOMC region. Point mutated E2F1 (Serine 337>Alanine) also decreased hPOMC activity by luciferase assays (Fig. 6D).
Figure 6. E2F1 Phosphorylation is required for E2F1 binding to the hPOMC promoter.
(A) Results of EMSA performed using probe #5 and nuclear extracts treated with CIP (+) or control nuclear extracts (−) incubated with CIP and CIP inhibitor (see Methods). The E2F1 complex band is shown. (B) Immunoblotting using indicated antibodies (anti-POMC, anti-GAPDH, anti-E2F1, anti-pE2F1-Ser364, anti-pE2F1-Ser337, and anti-Lamin A/C) performed using whole cell (CE) or nuclear (NE) extracts derived from DMS79 cells treated with (+) or without (−) R-roscovitine (50µM). (C) Results of immunoblotting using anti-total E2F1 and anti-pSer337-E2F1 performed with nuclear extracts (input) derived from DMS79 cells treated with (+) or without (−) R-roscovitine (50 µM). Nuclear extracts from R-roscovitine treated cells were used for DNA pull-down assay, and proteins precipitated with biotinylated probe containing E2F1 binding sites (−24/+50) (shown in Fig. 2B) were analyzed by anti-total E2F1 and anti-pSer337-E2F1. (D) Results of luciferase assays performed in COLO320 cells using the −42 hPOMC promoter (−42/+68) reporter plasmid or negative control. Wild type E2F1 or S337>A E2F1 and DP1 expression plasmids were co-transfected, and total DNA abundance adjusted with empty vector. EMSA, immunoblotting, pull-down assay, and luciferase assay results are representative of three independent experiments (A-D).
Suppression of POMC expression by targeting E2F binding inhibitor HLM006474
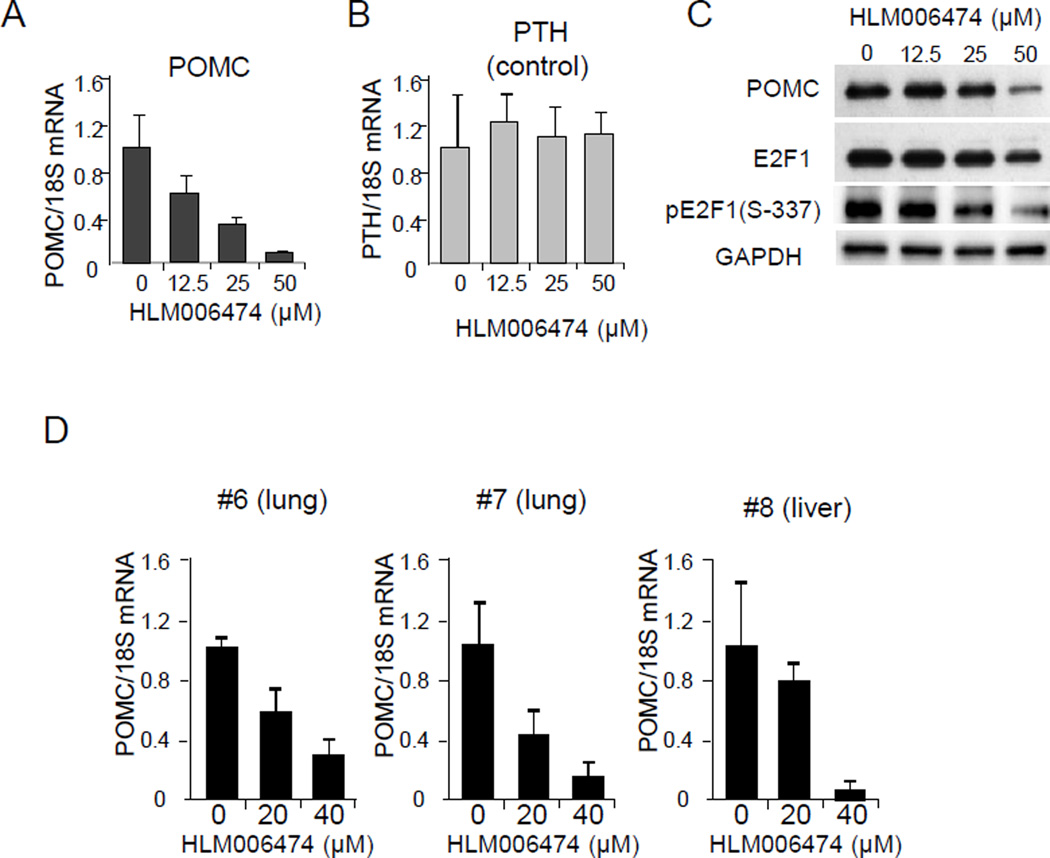
As HLM006474, a direct inhibitor of the E2F family, interrupts DNA binding in melanoma cells (Ma, et al. 2008), and also decreases cell proliferation in DMS79 cells (Kurtyka, et al. 2014), we investigated the effects of this small molecule on hPOMC. After testing cell viability to determine the appropriate dose for in vitro experiments (Supplement 2), similar to our results with R-roscovitine, we found that HLM006474 strongly suppressed POMC mRNA levels in a dose-dependent manner in DMS79 cells analyzed by RT-PCR (Fig. 7A). mRNA expression of PTH, used as a negative control, was unaltered (Fig. 7B), suggesting that E2F1-mediated antagonists elicit gene-specific hPOMC actions. POMC reduction was also confirmed by immunoblotting, and total E2F1 and pSer337-E2F1 were also both dose-dependently reduced (Fig. 7C). Finally, HLM006474 also suppressed hPOMC mRNA expression in three ACTH-secreting tumor primary cultures derived from liver and lung (Fig. 7D), while expression of PTH remained unaltered (Supplement 5).
Figure 7. E2F1 inhibitors HLM006474 suppress hPOMC expression.
(A-C) DMS79 cells were treated with HLM006474 at the indicated concentrations and hPOMC (A) and hPTH (B) mRNA levels analyzed by RT-PCR. Expression levels are normalized with 18S rRNA. Immunoblotting using indicated antibodies (anti-POMC, anti-E2F1, anti-pE2F1-Ser337, and anti-GAPDH) was performed using protein extracts derived from DMS79 cells treated with HLM006474 at indicated concentrations (C). (D) Primary cultures derived from non-pituitary ACTH secreting tumors (#1-3) treated with HLM006474. hPOMC and hPTH mRNA levels were analyzed by RT-PCR and normalized with 18S rRNA. RT-PCR results from DMS79 cells are representative of four independent experiments and are depicted as mean ± SE of triplicate samples (A, B). For patient samples, RT-PCR and immunoblotting were performed in triplicate.
Discussion
Ectopic Cushing’s syndrome is an as yet incurable disease. We elucidated mechanisms underlying hPOMC transcription and ACTH production in non-pituitary tumor cells.
Unlike the rat pituitary Pomc promoter, paraneoplastic hPOMC expression does not seem to be regulated by major pituitary specific transcription factors (Lamolet et al. 2001; Lamonerie, et al. 1996; Liu et al. 2001; Poulin et al. 1997). Rather, we show that hPOMC expression in non-pituitary tumors is regulated by E2F1 and Sp1/Sp3 binding to the proximal hPOMC promoter region (−42 to +68). Since these binding sites are closely located, these two transcription factors may mutually interact (Lin, et al. 1996). As hPOMC expression is regulated by E2F1 expression levels and activity, E2F1-mediated POMC transcription is a potential target for ectopic Cushing’s tumors.
In earlier reports, an E2F1 binding site at position −376/−417 in the POMC promoter had been identified by luciferase assays, showing a 1.2-fold to 1.5-fold decrease in activity with deletion of the site (Picon et al. 1999; Picon et al. 1995). We found a similar mild reduction of luciferase activity by deletion of the promoter from −303 to −428 (Fig. 2A). However, we also detected much stronger activity (70% of all activity) at our newly identified proximal E2F1 cluster binding site (Fig. 2A), which was not analyzed in previous studies. We therefore focused on that proximal promoter site as being a major site affecting hPOMC expression in ectopic Cushing’s tumors, rather than the weaker upstream E2F family binding site.
We previously found that R-roscovitine targets pituitary specific Tpit/Pitx1-mediated rat Pomc transcription and suppresses Tpit mRNA expression in human pituitary corticotroph tumors (Liu et al. 2015). Pituitary-specific rat Pomc gene expression is also regulated by Tpit/Pitx1 and NeuroD1 (Poulin et al. 1997), binding at −316/−297 and − 377/−370 (Poulin et al. 1997). Yet, despite these sequences being conserved in the human POMC promoter (located at −375/−369 and −287/−281), our current data indicate that hPOMC transcription is not altered by deletions nor by point mutations of these potential sites in two ectopic tumor cell lines. Therefore, hPOMC promoter activity in ectopic Cushing’s tumors seems not to be similarly regulated as pituitary corticotrophs. Alternatively, hPOMC promoter activity in human pituitary corticotrophs may differ from rodent pituitary expression. Indeed, clustered E2F1 binding sequences in the human promoter region are only partially conserved in the rodent Pomc promoter, even though E2F1 binds rodent Pomc in AtT20 cells (Liu et al. 2015). Epigenetic modifications may also contribute to the species differences and tissue specific regulation of POMC. For example, CpG sites are more abundant in the hPOMC compared to the rPomc promoter, and differences in tissue specific methylation status were also reported for hPOMC (Newell-Price, et al. 2001). However, further elucidation of tissue specificity (i.e., pituitary vs. non-pituitary) and species specificity (i.e., human vs. rodent) of POMC regulation will be challenging to distinguish because of the lack of functional human pituitary corticotroph cell lines.
We show the functional phenotypes of E2F1 on hPOMC using both over-expression and knock-down studies in DMS79 and in COLO320 cells, and also show a strong positive correlation between E2F1 and POMC expression in human bronchial carcinoid tumors, confirming a role for E2F1 on POMC expression in non-pituitary cells and supporting our in vitro results in human tumors using primary cell cultures. These results, together with our ChIP results showing that E2F1 controls hPOMC transcriptional levels without altering AcH4 levels (chromatin remodeling), further support the rationale for E2F1 as a potential molecular target.
Our observation of ACTH suppression by R-roscovitine in both patient tumors and in mouse models elucidates a novel mechanism that E2F1 DNA binding and POMC expression are regulated by pSer337-E2F1. E2F1 Ser-337 phosphorylation may also directly modify E2F1 structure, including the DNA binding domain, or it may prevent Rb/E2F1 binding (Fagan, et al. 1994), and therefore increase E2F1 DNA binding (Robertson, et al. 2000). As DMS79 and COLO320 cells also express several Rb family members (e.g., p130 and p107) (Helin, et al. 1997), these may also be associated with E2F1 Ser-337 phosphorylation status in ectopic Cushing’s tumors.
Controlling E2F1 activity may be a promising therapeutic strategy for ectopic Cushing’s syndrome, because expression levels of POMC seem to be determined by E2F1 activity in POMC positive tumor cells. However, E2F1 is a ubiquitously expressed transcription factor, and inhibition of E2F1 binding to the −42/+68 promoter region did not affect acetylation of histone H4 in this region (Fig. 5D), suggesting E2F1 does not regulate open chromatin status in the proximal promoter region. Induction of POMC may be cell type dependent and regulated by other transcription factors, including STAT3 (Bousquet et al. 2000), through chromatin remodeling and epigenetic modifications such as DNA methylation (Newell-Price et al. 2001). Thus, although we cannot exclude involvement of other transcription factors, E2F1 activity seems to be a common mechanism controlling POMC expression, and E2F1 over-activity may be integral to induction of ectopic Cushing’s syndrome. Indeed, E2F1 is highly expressed in small-cell lung cancer (Eymin, et al. 2001), which may explain relatively high occurrence of ectopic Cushing’s syndrome associated with these tumors, and may also underlie suppression of cell proliferation seen with the E2F inhibitor HLM006474 in small-cell lung cancer(Kurtyka et al. 2014).
POMC expression strongly correlates with E2F1 in a variety of POMC expressing cells. The direct E2F inhibitor HLM006474 robustly suppressed POMC/ACTH levels in DMS79 cells, and also reduced total E2F1 and pSer337-E2F1, suggesting that both E2F1 expression and phosphorylation status are important to hPOMC gene regulation. We recognize that our study employed a limited number of human samples to support mechanistic findings derived from POMC-expressing cell lines. Ectopic Cushing’s syndrome is rare and up to 50% are occult (Ilias et al. 2005), therefore often treated based on signs and symptoms rather than definitive localized pathological diagnosis, making tissue samples scarce. It is also likely that such tumors are under-diagnosed, as screening for ACTH oversecretion is typically not performed until fulminant signs of disease develop, further limiting availability of tissue samples. Confirmation for the role of E2F1 in regulating POMC expression and ACTH secretion in paraneoplastic tumors may best be demonstrated clinically by targeting kinases involved in E2F1 regulation. Our results represent a unique mechanism for E2F1 action on paraneoplastic hPOMC/ACTH hormonal production. Elucidation of subcellular mechanisms for ectopic Cushing’s syndrome and our finding of E2F1 DNA binding as a potential target for excess hPOMC expression provide new insights into this challenging disorder.
Supplementary Material
Acknowledgments
We thank Drs. Odelia Cooper, Run Yu, and Harmik Soukiasian for patient samples, and Norika Chiba, Masato Tsuda, Anat Ben-Shlomo, and Shira Berman for technical advice and assistance. Financial support was provided by NIH grants DK103198 and DK007770 to SM and by the Doris Factor Molecular Endocrinology Laboratory.
The funding sources had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Disclosures
The authors have declared that no conflict of interest exists.
Author contributions
TA, MT, NL and SM designed the research studies; TA, YT, DC, and RH conducted experiments; TA, NL, MT, and SM analyzed the data, and TA, NL, MT, and SM wrote the manuscript.
References
- Aniszewski JP, Young WF, Jr, Thompson GB, Grant CS, van Heerden JA. Cushing syndrome due to ectopic adrenocorticotropic hormone secretion. World J Surg. 2001;25:934–940. doi: 10.1007/s00268-001-0032-5. [DOI] [PubMed] [Google Scholar]
- Bousquet C, Susini C, Melmed S. Inhibitory roles for SHP-1 and SOCS-3 following pituitary proopiomelanocortin induction by leukemia inhibitory factor. J Clin Invest. 1999;104:1277–1285. doi: 10.1172/JCI7924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bousquet C, Zatelli MC, Melmed S. Direct regulation of pituitary proopiomelanocortin by STAT3 provides a novel mechanism for immuno-neuroendocrine interfacing. J Clin Invest. 2000;106:1417–1425. doi: 10.1172/JCI11182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown WH. A case of pluriglandular syndrome: “ Diabetes of bearded women. “. The Lancet. 1928;212:1022–1023. [Google Scholar]
- Chesnokova V, Zhou C, Ben-Shlomo A, Zonis S, Tani Y, Ren SG, Melmed S. Growth hormone is a cellular senescence target in pituitary and nonpituitary cells. Proc Natl Acad Sci U S A. 2013;110:E3331–E3339. doi: 10.1073/pnas.1310589110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chrousos GP. Stress and disorders of the stress system. Nat Rev Endocrinol. 2009;5:374–381. doi: 10.1038/nrendo.2009.106. [DOI] [PubMed] [Google Scholar]
- Colao A, Petersenn S, Newell-Price J, Findling JW, Gu F, Maldonado M, Schoenherr U, Mills D, Salgado LR, Biller BM. A 12-month phase 3 study of pasireotide in Cushing’s disease. N Engl J Med. 2012;366:914–924. doi: 10.1056/NEJMoa1105743. [DOI] [PubMed] [Google Scholar]
- Drouin J, Chamberland M, Charron J, Jeannotte L, Nemer M. Structure of the rat pro-opiomelanocortin (POMC) gene. FEBS Lett. 1985;193:54–58. doi: 10.1016/0014-5793(85)80078-8. [DOI] [PubMed] [Google Scholar]
- Du L, Bergsneider M, Mirsadraei L, Young SH, Jonker JW, Downes M, Yong WH, Evans RM, Heaney AP. Evidence for orphan nuclear receptor TR4 in the etiology of Cushing disease. Proc Natl Acad Sci U S A. 2013;110:8555–8560. doi: 10.1073/pnas.1306182110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eymin B, Gazzeri S, Brambilla C, Brambilla E. Distinct pattern of E2F1 expression in human lung tumours: E2F1 is upregulated in small cell lung carcinoma. Oncogene. 2001;20:1678–1687. doi: 10.1038/sj.onc.1204242. [DOI] [PubMed] [Google Scholar]
- Fagan R, Flint KJ, Jones N. Phosphorylation of E2F-1 modulates its interaction with the retinoblastoma gene product and the adenoviral E4 19 kDa protein. Cell. 1994;78:799–811. doi: 10.1016/s0092-8674(94)90522-3. [DOI] [PubMed] [Google Scholar]
- Fleseriu M, Biller BM, Findling JW, Molitch ME, Schteingart DE, Gross C. Mifepristone, a glucocorticoid receptor antagonist, produces clinical and metabolic benefits in patients with Cushing’s syndrome. J Clin Endocrinol Metab. 2012;97:2039–2049. doi: 10.1210/jc.2011-3350. [DOI] [PubMed] [Google Scholar]
- Fleseriu M, Pivonello R, Young J, Hamrahian AH, Molitch ME, Shimizu C, Tanaka T, Shimatsu A, White T, Hilliard A, et al. Osilodrostat, a potent oral 11beta-hydroxylase inhibitor: 22-week, prospective, Phase II study in Cushing’s disease. Pituitary. 2015 doi: 10.1007/s11102-015-0692-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frohman MA, Dush MK, Martin GR. Rapid production of full-length cDNAs from rare transcripts: amplification using a single gene-specific oligonucleotide primer. Proc Natl Acad Sci U S A. 1988;85:8998–9002. doi: 10.1073/pnas.85.23.8998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girling R, Partridge JF, Bandara LR, Burden N, Totty NF, Hsuan JJ, La Thangue NB. A new component of the transcription factor DRTF1/E2F. Nature. 1993;362:83–87. doi: 10.1038/362083a0. [DOI] [PubMed] [Google Scholar]
- Helin K, Holm K, Niebuhr A, Eiberg H, Tommerup N, Hougaard S, Poulsen HS, Spang-Thomsen M, Norgaard P. Loss of the retinoblastoma protein-related p130 protein in small cell lung carcinoma. Proc Natl Acad Sci U S A. 1997;94:6933–6938. doi: 10.1073/pnas.94.13.6933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilias I, Torpy DJ, Pacak K, Mullen N, Wesley RA, Nieman LK. Cushing’s syndrome due to ectopic corticotropin secretion: twenty years’ experience at the National Institutes of Health. J Clin Endocrinol Metab. 2005;90:4955–4962. doi: 10.1210/jc.2004-2527. [DOI] [PubMed] [Google Scholar]
- Isidori AM, Kaltsas GA, Pozza C, Frajese V, Newell-Price J, Reznek RH, Jenkins PJ, Monson JP, Grossman AB, Besser GM. The ectopic adrenocorticotropin syndrome: clinical features, diagnosis, management, and long-term follow-up. J Clin Endocrinol Metab. 2006;91:371–377. doi: 10.1210/jc.2005-1542. [DOI] [PubMed] [Google Scholar]
- Karalis KP, Venihaki M, Zhao J, van Vlerken LE, Chandras C. NF-kappaB participates in the corticotropin-releasing, hormone-induced regulation of the pituitary proopiomelanocortin gene. J Biol Chem. 2004;279:10837–10840. doi: 10.1074/jbc.M313063200. [DOI] [PubMed] [Google Scholar]
- Kurtyka CA, Chen L, Cress WD. E2F inhibition synergizes with paclitaxel in lung cancer cell lines. PLoS One. 2014;9:e96357. doi: 10.1371/journal.pone.0096357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamolet B, Pulichino AM, Lamonerie T, Gauthier Y, Brue T, Enjalbert A, Drouin J. A pituitary cell-restricted T box factor, Tpit, activates POMC transcription in cooperation with Pitx homeoproteins. Cell. 2001;104:849–859. doi: 10.1016/s0092-8674(01)00282-3. [DOI] [PubMed] [Google Scholar]
- Lamonerie T, Tremblay JJ, Lanctot C, Therrien M, Gauthier Y, Drouin J. Ptx1, a bicoid-related homeo box transcription factor involved in transcription of the pro-opiomelanocortin gene. Genes Dev. 1996;10:1284–1295. doi: 10.1101/gad.10.10.1284. [DOI] [PubMed] [Google Scholar]
- Langlais D, Couture C, Sylvain-Drolet G, Drouin J. A pituitary-specific enhancer of the POMC gene with preferential activity in corticotrope cells. Mol Endocrinol. 2011;25:348–359. doi: 10.1210/me.2010-0422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechner JF, McClendon IA, LaVeck MA, Shamsuddin AM, Harris CC. Differential control by platelet factors of squamous differentiation in normal and malignant human bronchial epithelial cells. Cancer Res. 1983;43:5915–5921. [PubMed] [Google Scholar]
- Lin SY, Black AR, Kostic D, Pajovic S, Hoover CN, Azizkhan JC. Cell cycle-regulated association of E2F1 and Sp1 is related to their functional interaction. Mol Cell Biol. 1996;16:1668–1675. doi: 10.1128/mcb.16.4.1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Lin C, Gleiberman A, Ohgi KA, Herman T, Huang HP, Tsai MJ, Rosenfeld MG. Tbx19, a tissue-selective regulator of POMC gene expression. Proc Natl Acad Sci U S A. 2001;98:8674–8679. doi: 10.1073/pnas.141234898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu NA, Araki T, Cuevas-Ramos D, Hong J, Ben-Shlomo A, Tone Y, Tone M, Melmed S. Cyclin E - mediated human proopiomelanocortin regulation as a therapeutic target for Cushing disease. J Clin Endocrinol Metab. 2015 doi: 10.1210/jc.2015-1606. jc20151606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu NA, Jiang H, Ben-Shlomo A, Wawrowsky K, Fan XM, Lin S, Melmed S. Targeting zebrafish and murine pituitary corticotroph tumors with a cyclin-dependent kinase (CDK) inhibitor. Proc Natl Acad Sci U S A. 2011;108:8414–8419. doi: 10.1073/pnas.1018091108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y, Kurtyka CA, Boyapalle S, Sung SS, Lawrence H, Guida W, Cress WD. A small-molecule E2F inhibitor blocks growth in a melanoma culture model. Cancer Res. 2008;68:6292–6299. doi: 10.1158/0008-5472.CAN-08-0121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maira M, Martens C, Philips A, Drouin J. Heterodimerization between members of the Nur subfamily of orphan nuclear receptors as a novel mechanism for gene activation. Mol Cell Biol. 1999;19:7549–7557. doi: 10.1128/mcb.19.11.7549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newell-Price J. Proopiomelanocortin gene expression and DNA methylation: implications for Cushing’s syndrome and beyond. J Endocrinol. 2003;177:365–372. doi: 10.1677/joe.0.1770365. [DOI] [PubMed] [Google Scholar]
- Newell-Price J. Ketoconazole as an adrenal steroidogenesis inhibitor: effectiveness and risks in the treatment of Cushing’s disease. J Clin Endocrinol Metab. 2014;99:1586–1588. doi: 10.1210/jc.2014-1622. [DOI] [PubMed] [Google Scholar]
- Newell-Price J, Bertagna X, Grossman AB, Nieman LK. Cushing’s syndrome. Lancet. 2006;367:1605–1617. doi: 10.1016/S0140-6736(06)68699-6. [DOI] [PubMed] [Google Scholar]
- Newell-Price J, King P, Clark AJ. The CpG island promoter of the human proopiomelanocortin gene is methylated in nonexpressing normal tissue and tumors and represses expression. Mol Endocrinol. 2001;15:338–348. doi: 10.1210/mend.15.2.0599. [DOI] [PubMed] [Google Scholar]
- Nieman LK, Biller BM, Findling JW, Murad MH, Newell-Price J, Savage MO, Tabarin A. Treatment of Cushing’s Syndrome: An Endocrine Society Clinical Practice Guideline. J Clin Endocrinol Metab. 2015;100:2807–2831. doi: 10.1210/jc.2015-1818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa C, Tone Y, Tsuda M, Peter C, Waldmann H, Tone M. TGF-beta-mediated Foxp3 gene expression is cooperatively regulated by Stat5, Creb, and AP-1 through CNS2. J Immunol. 2014;192:475–483. doi: 10.4049/jimmunol.1301892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettengill OS, Sorenson GD, Wurster-Hill DH, Curphey TJ, Noll WW, Cate CC, Maurer LH. Isolation and growth characteristics of continuous cell lines from small-cell carcinoma of the lung. Cancer. 1980;45:906–918. doi: 10.1002/1097-0142(19800301)45:5<906::aid-cncr2820450513>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- Picon A, Bertagna X, de Keyzer Y. Analysis of the human proopiomelanocortin gene promoter in a small cell lung carcinoma cell line reveals an unusual role for E2F transcription factors. Oncogene. 1999;18:2627–2633. doi: 10.1038/sj.onc.1202635. [DOI] [PubMed] [Google Scholar]
- Picon A, Leblond-Francillard M, Raffin-Sanson ML, Lenne F, Bertagna X, de Keyzer Y. Functional analysis of the human pro-opiomelanocortin promoter in the small cell lung carcinoma cell line DMS-79. J Mol Endocrinol. 1995;15:187–194. doi: 10.1677/jme.0.0150187. [DOI] [PubMed] [Google Scholar]
- Poulin G, Turgeon B, Drouin J. NeuroD1/beta2 contributes to cell-specific transcription of the proopiomelanocortin gene. Mol Cell Biol. 1997;17:6673–6682. doi: 10.1128/mcb.17.11.6673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preda VA, Sen J, Karavitaki N, Grossman AB. Etomidate in the management of hypercortisolaemia in Cushing’s syndrome: a review. Eur J Endocrinol. 2012;167:137–143. doi: 10.1530/EJE-12-0274. [DOI] [PubMed] [Google Scholar]
- Quinn LA, Moore GE, Morgan RT, Woods LK. Cell lines from human colon carcinoma with unusual cell products, double minutes, and homogeneously staining regions. Cancer Res. 1979;39:4914–4924. [PubMed] [Google Scholar]
- Raff H, Carroll T. Cushing’s syndrome: from physiological principles to diagnosis and clinical care. J Physiol. 2015;593:493–506. doi: 10.1113/jphysiol.2014.282871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray DW, Davis JR, White A, Clark AJ. Glucocorticoid receptor structure and function in glucocorticoid-resistant small cell lung carcinoma cells. Cancer Res. 1996;56:3276–3280. [PubMed] [Google Scholar]
- Robertson KD, Ait-Si-Ali S, Yokochi T, Wade PA, Jones PL, Wolffe AP. DNMT1 forms a complex with Rb, E2F1 and HDAC1 and represses transcription from E2F-responsive promoters. Nat Genet. 2000;25:338–342. doi: 10.1038/77124. [DOI] [PubMed] [Google Scholar]
- Stevens A, White A. ACTH: cellular peptide hormone synthesis and secretory pathways. Results Probl Cell Differ. 2010;50:63–84. doi: 10.1007/400_2009_30. [DOI] [PubMed] [Google Scholar]
- Takahashi H, Hakamata Y, Watanabe Y, Kikuno R, Miyata T, Numa S. Complete nucleotide sequence of the human corticotropin-beta-lipotropin precursor gene. Nucleic Acids Res. 1983;11:6847–6858. doi: 10.1093/nar/11.19.6847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao Y, Kassatly RF, Cress WD, Horowitz JM. Subunit composition determines E2F DNA-binding site specificity. Mol Cell Biol. 1997;17:6994–7007. doi: 10.1128/mcb.17.12.6994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tone M, Powell MJ, Tone Y, Thompson SA, Waldmann H. IL-10 gene expression is controlled by the transcription factors Sp1 and Sp3. J Immunol. 2000;165:286–291. doi: 10.4049/jimmunol.165.1.286. [DOI] [PubMed] [Google Scholar]
- Tone M, Tone Y, Babik JM, Lin CY, Waldmann H. The role of Sp1 and NF-kappa B in regulating CD40 gene expression. J Biol Chem. 2002;277:8890–8897. doi: 10.1074/jbc.M109889200. [DOI] [PubMed] [Google Scholar]
- Tone Y, Furuuchi K, Kojima Y, Tykocinski ML, Greene MI, Tone M. Smad3 and NFAT cooperate to induce Foxp3 expression through its enhancer. Nat Immunol. 2008;9:194–202. doi: 10.1038/ni1549. [DOI] [PubMed] [Google Scholar]
- White A, Stewart MF, Farrell WE, Crosby SR, Lavender PM, Twentyman PR, Rees LH, Clark AJ. Pro-opiomelanocortin gene expression and peptide secretion in human small-cell lung cancer cell lines. J Mol Endocrinol. 1989;3:65–70. doi: 10.1677/jme.0.0030065. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.