i. Summary
Metastasis is the major cause of breast cancer deaths. To spread from the primary tumor sites to distant tissues, solid tumor cells need to degrade the surrounding extracellular matrix (ECM). The protrusive membrane structures named invadopodia have been shown to play a critical role in the degradation of the ECM and invasion of invasive cancer cells. In this chapter, we describe a detailed protocol to examine invadopodia in human breast cancer cells.
Keywords: Invadopodia, breast cancer, invasion, metastasis, extracellular matrix
1 Introduction
Metastasis is a process in which cancer cells spread from their primary sites to colonize at distant organs (1,2). Patients with metastatic tumor usually have poor prognosis. About 90 percent of breast cancer deaths are due to metastases (1,3). During metastasis, solid tumor cells invade the surrounding extracellular matrix (ECM), intravasate into the circulatory system through the endothelium, then extravasate into a distant tissue, where they eventually establish secondary tumors (1,2). One major way of invasion requires cancer cells to degrade ECM components through matrix-digesting proteases. In recent years, increasing evidences has shown that the subcellular structures known as invadopodia are critical for the breakdown of ECM in the multiple steps of metastasis, such as local invasion, intravasation and extravasation (4,5).
Invadopodia are protrusive membrane structures rich in actin cytoskeleton and metalloproteases in invasive cancer cells. The primary function of invadopodia is to degrade ECM through recruiting various matrix proteases to the contacting sites of the plasma membrane and ECM, thus allowing cancer cells to leave the primary sites during metastasis. Invadopodia have been shown to be critical for the invasion of many types of cancer cells, such as melanoma, breast cancer, colon cancer, lung cancer and prostate cancer (6–10). Disruption of the key players of invadopodia, such as Twist, cortactin, Tks5, and Tks4, blocked cancer metastasis in several cancer mouse models (11–14). Because of the specific presence of invadopodia in invasive cancer cells and their critical roles in tumor metastasis, inhibition of key components or regulators of invadopodia has become a very attractive and unique strategy to target tumor metastasis (4,5).
In this chapter, we describe a protocol for examining invadopodia formation in the invasive MDA-MB-231 human breast cancer cells using fluorescently labeled gelatin, one of the major components in the ECM (15). In combination with inhibitor treatment, overexpression and knockdown/knockout of the genes of interest, this protocol can be used to study the signaling pathways that regulate different steps of invadopodia formation.
2 Materials
The water used for all solutions in this protocol is from the purified deionized water with a sensitivity of 18 MΩ cm at 25°C. The storage condition varies for different solutions and reagents.
2.1 Reagents for gelatin labeling
Dylight 488 labeling kits (Thermo, Rockford, IL, 53024): The Dylight Fluor is activated N-hydroxysuccinimide (NHS) ester, which is the most commonly used reactive group for labeling protein. NHS esters react with primary amines; forming stable, covalent amide bonds and releasing the NHS groups. The kit contains: Dylight Alexa 488 labeled-NHS Ester, Borate Buffer (0.67M), Purification Resin, Spin Column and Microcentrifuge Collection Tubes (see Note 1).
Phosphate-Buffered Saline (PBS): Dissolve 8 g of NaCl, 0.2 g of KCl, 1.44 g of Na2HPO4 and 0.24 g of KH2PO4 in 800 mL distilled H2O. Adjust the volume to 1 L with additional distilled H2O. Adjust the pH to 7.4 with 1 M HCl. Store at room temperature.
Gelatin: Type A gelatin form porcine skin (Sigma, G-2500).
2.2 Reagents for coating coverslips with labeled gelatin
Parafilm “M” laboratory film (Bemis Flexible Packaging, Neenah, WI 54956).
12 mm diameter circular microscope coverslips (Fisher, Waltham, MA, USA).
Poly-L-lysine solution: Dilute the stock poly-L-lysine (Sigma, P8920) to 50 μg/mL in distilled H2O.
Glutaraldehyde solution: 0.5% glutaraldehyde is diluted from 25% glutaraldehyde stock using PBS. Store the diluted solution in refrigerator and use it in 1 month.
Gelatin mix solution: Mix fluorescent conjugated gelatin and 1 mg/mL unconjugated gelatin at a ratio of 1:25–1:2 (see Note 2). The optimal ratio of labeled to unlabeled gelatin is dependent on the signal intensity of labeled gelatin. We used 1:4 in this protocol (see Note 3).
Fluorescein-conjugated gelatin (Thermo Scientific, Rockford, IL, G13187): Fluorescent labeled gelatin from some companies may be also used for invadopodia assay.
Sodium borohydride (5 mg/mL): Dissolve 10 mg of sodium borohydride in 2 mlL PBS. The solution must be freshly made before use.
70% ethanol: Dilute 200 proof ethanol to 70% with distilled H2O.
2.3 Other reagents for cell culturing and the invadopodia assessment
Cell line: MDA-MB-231 human breast cancer cells (American Type Culture Collection, ATCC).
DMEM: Dulbecco’s Modified Eagle Medium (DMEM) supplemented with 10% fetal bovine serum (FBS) from Hyclone. Store at 4 °C.
Enzyme-free cell dissociation solution (Millipore, S-014-B).
4% paraformaldehyde (PFA) solution: Dissolve 0.4 g paraformaldehyde powder in 10 mL PBS. Heat the solution in 60 °C water bath, shake vigorously every few minutes to help dissolve. Store at 4 °C, use within 1 week.
0.1% Triton X-100: Dilute from 10 % Triton X-100 using PBS.
Alexa Fluor® 594 phalloidin (Life Technologies, A22287).
Mounting medium: Make 4% n-Propyl gallate (w/v) in 90% glycerol (v/v) and 10% PBS. Aliquot and store at −20 °C.
Clear nail polish (Electron Microscopy Sciences, 72180).
3 Methods
3.1 Label gelatin with Dylight 488
Prepare 2 mg/mL gelatin solution in PBS (put it in 37°C water bath to help dissolve).
Add 40 μL of Borate Buffer (0.67M) to 0.5 mL gelatin solution.
Take out the vial containing Dylight 488 NHS Ester from freezer, and let it warm to room temperature. Add the prepared gelatin solution to the vial and vortex gently or invert 10 times.
Briefly centrifuge the vial to collect the sample in the bottom of the tube.
Incubate the reaction mixture for 60 min at room temperature. Protect from light during incubation.
Place two spin columns in two microcentrifuge collection tubes.
Mix the purification resin to ensure uniform suspension and add 400 μL of the suspension into both spin columns. Centrifuge for 45 seconds at ~1000 × g to remove the storage solution. Discard the used collection tubes and place the columns in new collection tubes.
Add 250–270 μL of the labeling reaction mixture to each spin column and mix the sample with the resin by pipetting up and down or briefly vortexing.
Centrifuge columns for 45 seconds at ~ 1000 × g to collect the purified gelatin. Combine the samples from both columns (~ 0.5 mL total). Discard the used columns. The concentration of labeled gelatin is around 2 mg/mL.
Store the labeled gelatin at 4 °C for up to one month, protected from light. Alternatively, store labeled protein in single use aliquots at −20 °C. Avoid repeated freeze/thaw cycles.
3.2 Coat coverslips with fluorescently labeled gelatin
Place coverslips on a piece of parafilm with point-ended tweezers (see Note 4).
Pretreat coverslips with 50 μg/mL poly-L-lysine for 20 min. Wash coverslips twice with PBS, 3 min each time.
Crosslink coverslips with 0.5% glutaraldehyde for 15 min. Wash three times with PBS, 3 min each time (see Note 5).
Cut another parafilm and put it in a humid box with nontransparent cover. Drop 40μL of mixed gelatin solution on the parafilm (see Note 6). Invert the coverslips and place them face down onto the gelatin solution. After 20 min incubation, wash the coverslips with PBS once for 3 min.
Quench the autofluorescence with 5 mg/mL sodium borohydride for 3 min. Wash three times with PBS (see Note 7).
Sterilize the coverslips with 70% ethanol for 5 min (see Note 8).
Wash the coverslips once with PBS and once with complete medium.
Incubate the coverslips in complete growth medium for 1 hour before use.
3.3 Invadopodia formation assessment
One or two days before the experiment, plate MDA-MB-231 cells into a 6-well plate (4 × 105 cells/well)(see Note 9). Grow cells in 10% FBS DMEM in a 37°C 5% CO2 incubator (see Note 10).
On the day of experiment, treat cells with appropriate inhibitors for 0.5–1 hr.
Remove medium in the well and detach the cells with 500 μL enzyme-free cell dissociation solution. Let the plate stand for 5 min in 37°C 5% CO2 incubator (see Notes 11 and 12).
Check the cells under an inverted microscope. After the cells are completely detached from the bottom of well, collect the supernatant and spin at 500 × g for 5 min.
After centrifugation, remove the supernatant and re-suspend the cell pellet with 100 μL 10% FBS DMEM.
Count cells and seed 1 × 105 cells per well into a 12-well plate containing the coverslips coated with fluorescent labeled gelatin. Add appropriate inhibitors if needed.
Incubate cells in 37°C 5% CO2 for 3–4 hr (see Note 13).
Remove medium and fix cells with 4% PFA for 20 min at room temperature. Wash the coverslips with PBS for 5 min (see Note 14).
Permeabilize the cells with 0.1% Triton-X 100 for 10 min at room temperature.
Wash coverslips with PBS for 3 times, 5 min each time.
Incubate coverslips with 1:200 diluted Alexa 594 phalloidin in PBS for 1 hr at room temperature. Wash coverslips 3 times with PBS, 5 min each time.
After the final wash, carefully remove excess PBS by blotting the side of the coverslip with a kimwipe. Place coverslip face down onto a drop of anti-fading mounting medium on a microscope slide (see Note 15). Avoid any air bubbles during the process (see Note 16).
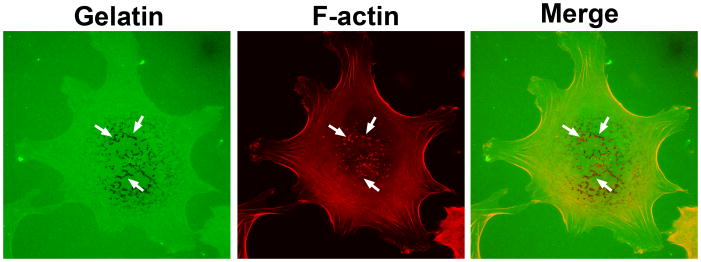
Aspirate excess mounting medium along the coverslip. Seal them by applying nail polish along the edge of coverslips. Observe the invadopodia formation under confocal laser scanning microscope with 100× objective (Fig. 1).
The formation of invadopodia will remove fluorescently labeled gelatin, leaving black spots on coverslips. Invadopodia formation can be quantified by the percentage of cells with invadopodia, and/or the number of invadopodia per cell. Alternatively, the total area of degraded gelatin may also be used to indicate the invasive activity.
Figure 1.
A representative picture of invadopodia. MDA-MB-231 human breast cancer cells were plated on Alexa 488 gelatin-coated coverslips for 3 hour. F-actin was stained with Alexa 594 phalloidin. The arrows show the degradation of Alexa 488 gelatin and F-actin staining, indicating the presence of active invadopodia. The images were captured by a 100X objective using a Nikon A1 laser confocal microscope.
Acknowledgments
This work was supported by a research grant RP130425 from the Cancer Prevention and Research Institute of Texas (CPRIT) and a research grant R01HL119478 from the National Heart, Lung, and Blood Institute of the National Institutes of Health to GD. The content is solely the responsibility of the authors and does not necessarily represent the official views of the CPRIT and National Institutes of Health.
Footnotes
NHS ester-activated fluorophores are moisture-sensitive. Keep them in a dark and dry environment.
Pre-warm the labeled gelatin and 1 mg/mL unlabeled gelatin in 37 °C water bath before mixing. Frozen gelatin must be thawed completely to prevent precipitation. Gelatin that was incompletely dissolved can cause uneven bright spots on coverslips.
The stiffness of ECM can directly affect the ability of breast cancer cells to form invadopodia. It is critical to use appropriate concentrations of gelatin in invadopodia experiments. The gelatin concentration (labeled and unlabeled) used for coating the slides in this protocol is about 1.2 mg/mL.
Coverslips with dirt can cause uneven spots with strong fluorescence. If necessary, clean the coverslips with acid alcohol (1 % HCl in 70% ethanol).
Glutaraldehyde is a divalent crosslinker. It is used here to irreversibly crosslink gelatin to the glutaraldehyde-pretreated poly-L-lysine coating. The gelatin itself is not crosslinked. It binds to activated aldehyde groups associated with the poly-L-lysine after the glutaraldehyde is washed away.
Gelatin-labeled with fluorescein or other fluorophores from commercial sources, such as Thermo Fisher can also be used.
Sodium borohydride is used to reduce the autofluorescence generated by reversible Schiff’s bases in the aldehyde-NH2 reaction. Prepare this solution on ice. Be cautious that sodium borohydride is highly caustic and prone to explode.
If the experiment lasts only several hours, this step is not necessary.
Invadopodia can be also easily examined in many other metastatic cancer cell lines, e.g., breast cancer cell line 4T1, melanoma cell line RPMI7951 and lung cancer cell line A549.
More invadopodia can be observed when the cells are healthy.
Cells may be also detached by trypsin-EDTA solution. However, enzyme-free cell dissociation solution causes minimal disruption of surface proteins; therefore greatly improves invadopodia formation.
The incubation time can be longer if the cells do not completely detach.
The time to form invadoposia varies in different cell types. An optimal plating time should be determined when a new cell line is used. For MDA-MB-231 cells, we generally analyze invadopodia plaques 3–4 hr after plating. Longer incubation can increase both number and size of the invadopodia plaques. However, cell migration and merging of individual invadopodium may occur after 5 hours, making the interpretation of result difficult.
The appropriate fixation method should be determined according to the experiment conditions, such as the antibodies used for staining. For invadopodia visualization using fluorescent microscopy, PFA is our preferred fixation reagent. Other fixation methods, such as cold methanol or glutaraldehyde, may be also used for visualizing specific antigens. However, cold methanol destroys the native quaternary structure of F-actin, and hence is not suitable for actin staining with phalloidin. In addition, cold methanol removes lipids and dehydrates the cells; therefore, it may also disrupt the fine cell structures.
It is important to choose a correct mounting medium for certain fluorophores. For example, many commercial anti-fade mounting media do not work well for far-red fluorophores. Propyl gallate is a very general anti-fade mounting medium that is suitable for most of the commonly used fluorophores.
It is very common that air bubbles are formed during slide mounting. To avoid air bubbles, place one side of the coverslip on the mounting solution first, then slowly lower the tweezers, do not release the coverslips from the tweezers until the mounting medium fully covers the coverslip.
References
- 1.Chaffer CL, Weinberg RA. A perspective on cancer cell metastasis. Science. 2011;331(6024):1559–1564. doi: 10.1126/science.1203543. [DOI] [PubMed] [Google Scholar]
- 2.Klein CA. Cancer. The metastasis cascade. Science. 2008;321(5897):1785–1787. doi: 10.1126/science.1164853. [DOI] [PubMed] [Google Scholar]
- 3.Nguyen DX, Massague J. Genetic determinants of cancer metastasis. Nat Rev Genet. 2007;8(5):341–352. doi: 10.1038/nrg2101. [DOI] [PubMed] [Google Scholar]
- 4.Murphy DA, Courtneidge SA. The ‘ins’ and ‘outs’ of podosomes and invadopodia: characteristics, formation and function. Nat Rev Mol Cell Biol. 2011;12(7):413–426. doi: 10.1038/nrm3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Paz H, Pathak N, Yang J. Invading one step at a time: the role of invadopodia in tumor metastasis. Oncogene. 2014;33(33):4193–4202. doi: 10.1038/onc.2013.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nakahara H, Howard L, Thompson EW, Sato H, Seiki M, Yeh Y, Chen WT. Transmembrane/cytoplasmic domain-mediated membrane type 1-matrix metalloprotease docking to invadopodia is required for cell invasion. Proc Natl Acad Sci U S A. 1997;94(15):7959–7964. doi: 10.1073/pnas.94.15.7959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kelly T, Yan Y, Osborne RL, Athota AB, Rozypal TL, Colclasure JC, Chu WS. Proteolysis of extracellular matrix by invadopodia facilitates human breast cancer cell invasion and is mediated by matrix metalloproteinases. Clin Exp Metastasis. 1998;16(6):501–512. doi: 10.1023/a:1006538200886. [DOI] [PubMed] [Google Scholar]
- 8.Vishnubhotla R, Sun S, Huq J, Bulic M, Ramesh A, Guzman G, Cho M, Glover SC. ROCK-II mediates colon cancer invasion via regulation of MMP-2 and MMP-13 at the site of invadopodia as revealed by multiphoton imaging. Lab Invest. 2007;87(11):1149–1158. doi: 10.1038/labinvest.3700674. [DOI] [PubMed] [Google Scholar]
- 9.Wang S, Li E, Gao Y, Wang Y, Guo Z, He J, Zhang J, Gao Z, Wang Q. Study on invadopodia formation for lung carcinoma invasion with a microfluidic 3D culture device. PLoS One. 2013;8(2):e56448. doi: 10.1371/journal.pone.0056448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Desai B, Ma T, Chellaiah MA. Invadopodia and matrix degradation, a new property of prostate cancer cells during migration and invasion. J Biol Chem. 2008;283(20):13856–13866. doi: 10.1074/jbc.M709401200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eckert MA, Lwin TM, Chang AT, Kim J, Danis E, Ohno-Machado L, Yang J. Twist1-induced invadopodia formation promotes tumor metastasis. Cancer Cell. 2011;19(3):372–386. doi: 10.1016/j.ccr.2011.01.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Seals DF, Azucena EF, Jr, Pass I, Tesfay L, Gordon R, Woodrow M, Resau JH, Courtneidge SA. The adaptor protein Tks5/Fish is required for podosome formation and function, and for the protease-driven invasion of cancer cells. Cancer Cell. 2005;7(2):155–165. doi: 10.1016/j.ccr.2005.01.006. [DOI] [PubMed] [Google Scholar]
- 13.Li CM, Chen G, Dayton TL, Kim-Kiselak C, Hoersch S, Whittaker CA, Bronson RT, Beer DG, Winslow MM, Jacks T. Differential Tks5 isoform expression contributes to metastatic invasion of lung adenocarcinoma. Genes Dev. 2013;27(14):1557–1567. doi: 10.1101/gad.222745.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Leong HS, Robertson AE, Stoletov K, Leith SJ, Chin CA, Chien AE, Hague MN, Ablack A, Carmine-Simmen K, McPherson VA, Postenka CO, Turley EA, Courtneidge SA, Chambers AF, Lewis JD. Invadopodia are required for cancer cell extravasation and are a therapeutic target for metastasis. Cell reports. 2014;8(5):1558–1570. doi: 10.1016/j.celrep.2014.07.050. [DOI] [PubMed] [Google Scholar]
- 15.McDonald JA, Kelley DG, Broekelmann TJ. Role of fibronectin in collagen deposition: Fab’ to the gelatin-binding domain of fibronectin inhibits both fibronectin and collagen organization in fibroblast extracellular matrix. J Cell Biol. 1982;92(2):485–492. doi: 10.1083/jcb.92.2.485. [DOI] [PMC free article] [PubMed] [Google Scholar]