Abstract
The transcription cycle of RNA polymerase II (Pol II) correlates with changes to the phosphorylation state of its large subunit C-terminal domain (CTD). We recently developed Native Elongation Transcript sequencing using mammalian cells (mNET-seq), which generates single-nucleotide–resolution genome-wide profiles of nascent RNA and co-transcriptional RNA processing that are associated with different CTD phosphorylation states. Here we provide a detailed protocol for mNET-seq. First, Pol II elongation complexes are isolated with specific phospho-CTD antibodies from chromatin solubilized by micrococcal nuclease digestion. Next, RNA derived from within the Pol II complex is size fractionated and Illumina sequenced. using mNET-seq, we have previously shown that Pol II pauses at both ends of protein-coding genes but with different CTD phosphorylation patterns, and we have also detected phosphorylation at serine 5 (Ser5-P) CTD-specific splicing intermediates and Pol II accumulation over co-transcriptionally spliced exons. With moderate biochemical and bioinformatic skills, mNET-seq can be completed in ~6 d, not including sequencing and data analysis.
Introduction
The RNA population (transcriptome) generated by Pol II transcription of mammalian genomes can be characterized by various next-generation sequencing technologies1,2. Very often total RNA-seq is used, which provides information predominantly about steady-state RNA, largely comprising mature isoforms. As a result, information on RNA processing events coupled to Pol II transcription—such as pre-mRNA splicing and pre-miRNA processing3–5—is lacking in total RNA-seq profiles. RNA processing is orchestrated by the extended CTD of the Pol II large subunit that comprises a heptad amino acid sequence (consensus YSPTSPS) repeated 52 times6. Although Pol II CTD is highly phosphorylated during transcription, the relationship between co-transcriptional RNA processing and CTD phosphorylation is poorly understood.
Several different techniques have been described that generate profiles of extremely unstable nascent RNA in eukaryotes (Table 1). In particular, global nuclear run-on sequencing (GRO-seq) and precision nuclear run-on sequencing (PRO-seq), using modified nucleotides, have mapped transcriptionally engaged Pol II occupancy and revealed Pol II pausing7,8. Similarly, the 3′ ends of nascent RNA isolated from insoluble chromatin have been sequenced at high resolution9,10. Finally, native elongating transcript sequencing (NET-seq), achieved by immunoprecipitation (IP) of flag-tagged Pol II, generates genome-wide nascent RNA maps in yeast11 and bacteria12. However, these techniques have not so far provided nascent transcription profiles linked to either Pol II CTD modifications or to RNA processing. To enable us to generate such profiles, we recently developed and published an advanced NET-seq method for mammalian cells, termed mammalian NET-seq13 (mNET-seq, see Fig. 1); this protocol is based on those published methods.
Table 1.
Comparison of non-cross-linking nascent RNA techniques.
| GRO-seq | PRO-seq | NET-seq | 3′NT | Human NET-seq | Mammalian NET-seq | |
|---|---|---|---|---|---|---|
| Nuclear run on | Yes | Yes | No | No | No | No |
| RNA fragmentation | Alkaline hydrolysis | Alkaline hydrolysis | Alkaline hydrolysis | Alkaline hydrolysis | Alkaline hydrolysis | MNase digestion |
| RNA selection | BrU antibody | Biotin-streptavidin | FLAG antibody | 5′ cap antibody | No selection | Endogenous Pol II antibody |
| Pol II modification analyses | No | No | No | No | No | Yes |
| RNA processing analyses | No | No | No | No | No | Yes (splicing and miRNA processing) |
| Resolution | 30–100 bases | 1 base | 1 base | 1 base | 1 base | 1 base |
| Tested species | Human20, mouse21, Drosophila22 and Caenorhabditis elegans23 | Human24 and Drosophila8 | Yeast11 and Bacteria12 | Drosophila9 | Human10 | Human13 |
mNET-seq is the only currently available method that allows analysis of both Pol II pausing and co-transcriptional RNA processing at single-nucleotide resolution in a CTD phosphorylation–specific manner.
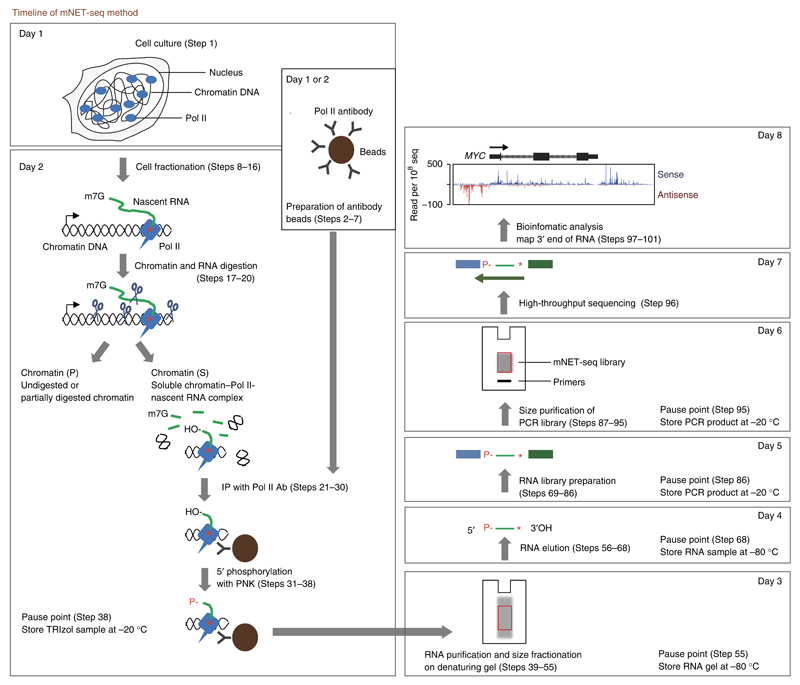
Figure 1.
Overview of the mNET-seq procedure. Adapted from ref. 13 with permission under the terms of the Creative Commons Attribution License. Copyright 2015, Nojima et al.
Development of mNET-Seq
Originally, NET-seq was established in yeast11 and used to isolate and sequence RNA in the Pol II active site by a Pol II IP approach. In essence, this technique involves cryogenic breakage of the yeast cell wall to allow the preparation of a crude whole-cell extract. Pol II complexes are immunoprecipitated from this extract using anti-FLAG antibodies against endogenous FLAG-tagged Rpb3. To extend the NET-seq approach to mammalian cells, we first isolate native chromatin using 1 M urea and 300 mM NaCl, as this fraction is enriched for nascent RNA and phosphorylated Pol II (ref. 14). After solubilization of chromatin by extensive micrococcal nuclease digestion, we then immunoprecipitated specific Pol II complexes from this chromatin fraction. Independently, Churchman and colleagues have also developed a so-called NET-seq method for human cells10. However, their method does not use IP of Pol II complexes, but instead it detects the 3′ end of chromatin-associated RNA, potentially including some degraded RNA. Consequently, our mNET-seq procedure is the only available method to profile Pol II CTD phosphorylation–specific nascent RNA genome-wide.
Consistent with previous nascent RNA-seq studies, mNET-seq detects both sense and antisense transcripts at transcription start sites (TSS), especially when using the 8WG16 Pol II antibody (specific for unphosphorylated CTD)13. Indeed, mNET-seq and GRO-seq give closely matching nascent RNA profiles(Supplementary Fig. 1). Interestingly, a splicing intermediate is readily detected by IP with Ser5-P CTD-specific Pol II antibody. This suggests that 5′ exons when cleaved by spliceosome are tethered to Pol II with Ser5-P–modified CTD13. Thus, mNET-seq provides comprehensive data not only on the precise Pol II occupancy of nascent RNA but also on RNA processing intermediates across the genome.
Overview of the procedure
An overview of the mNET-seq protocol is shown in Figure 1. Native chromatin fractions are isolated from purified nuclei using 1 M urea and 300 mM NaCl. Elongating Pol II complex is stably associated with this chromatin fraction, as judged by the abundance of unprocessed transcripts with retained introns and extended RNA 3′ ends uncleaved at polyA sites14–17. This chromatin fraction is then extensively digested with Micrococcal nuclease (MNase). This efficient nuclease digests all exposed DNA and RNA, effectively releasing the Pol II elongation complex from chromatin into a soluble fraction. Only RNA protected by protein complexes such as Pol II or the spliceosome will be resistant to MNase digestion. Pol II complex–associated nascent RNA fragments are purified from solubilized chromatin by Pol II IP using Pol II antibodies with different specificities. Consequently, Pol II modification–specific nascent RNA profiles are obtained. The 5′ hydroxyl (OH) generated by MNase digestion of RNA is converted to a 5′ phosphate by T4 polynucleotide kinase (PNK) treatment to allow 5′ linker ligation, whereas the 3′ linker is ligated to the 3′ OH end of the RNA present within the Pol II active site, thus enabling directional sequencing. Radioactive ATP (γ-32P ATP) is also used in the PNK reaction to facilitate size selection on denaturing polyacrylamide gels of 35- to 100-nt RNA before subsequent adapter ligation, PCR-based preparation of the cDNA sequencing library and multiplexed Illumina sequencing. The sequence readout can be bioinformatically aligned to the human genome sequence to effectively map all RNA 3′ ends. Note that co-transcriptional cleavage of 5′ splice sites (SS) additionally results in RNA 3′ OH ends that are protected by the spliceosome and are co-immunoprecipitated with Pol II, thus contributing to the mNET signal. It should also be emphasized that mNET-seq detects both protein-coding gene nascent transcripts and long noncoding RNA derived from intergenic regions.
Bioinformatic analysis allows the genomic alignment of nascent RNA profiles at single-nucleotide resolution. This effectively defines both the exact positions of elongating Pol II across the genome and the precise RNA cleavage positions of some co-transcriptional RNA processing factors. Remarkably, we can directly detect the initial 5′ SS cleavage step in pre-mRNA splicing and also observe active Drosha cleavage of pre-miRNA hairpin structures that are present in gene introns in a Pol II CTD phosphorylation-dependent manner13. Thus, the use of Pol II antibodies recognizing the full range of different CTD modifications will allow mNET-seq technology to uncover the highly complex CTD code of Pol II transcription.
Advantages of mNET-seq
mNET-seq is the only currently available method that allows examination of genome-wide nascent transcript profiles on the basis of Pol II CTD phosphorylation status and coupled RNA processing. The key features of this technique are as follows.
No cross-linking. No cross-linking is used, as the elongating Pol II complex is stable in chromatin14,16,17. Other techniques such as chromatin immunoprecipitation (ChIP) and in vivo cross-linking immunoprecipitation (CLIP) experiments require prior cross-linking (formaldehyde treatment for ChIP and UV cross-linking for CLIP) to examine DNA-protein and RNA-protein interactions. However, such cross-linking procedures may introduce artifactual interactions.
Chromatin digestion. MNase is a very efficient nuclease with little digestion bias on active chromatin regions, but it leaves the native Pol II complex intact. Furthermore, it is easy to inactivate by EGTA treatment. A short digestion time (90 s in our method) is used to minimize Pol II complex dissociation and consequent degradation of Pol II–protected RNA.
Pol II IP. Different Pol II–specific antibodies (Table 2) can be used in mNET-seq to explore Pol II modification–specific nascent RNA processing. For example, in our Ser5-P CTD-specific mNET-seq analysis, Pol II accumulation on co-transcriptionally spliced exons and a splicing intermediate tethered to the Pol II complex are detected. Furthermore, Ser2 and Ser5-P CTD-specific mNET-seq detects intermediates generated by co-transcriptional microprocessor-mediated cleavage13.
Table 2.
Appropriate RNA Pol II antibodies.
| Pol II antibody | Clonality | Supplier | Catalog number | Specificity | Beads to Conjugate |
|---|---|---|---|---|---|
| 8WG16 | Monoclonal | Abcam | Ab817 | Unphosphorylated CTD (low Ser2-P) | Dynabeads protein G |
| CMA601 | Monoclonal | MBL Life science | MABI0601 | Total CTD | Dynabeads M280 sheep anti-mouse IgG |
| CMA602 | Monoclonal | MBL Life science | MABI0602 | Ser2-P | Dynabeads M280 sheep anti-mouse IgG |
| CMA603 | Monoclonal | MBL Life science | MABI0603 | Ser5-P | Dynabeads M280 sheep anti-mouse IgG |
Tested Pol II antibodies are described. ELISA data and mNET-seq data are available13.
Limitations of mNET-seq
Some of the limitations of this technique are as follows:
In mNET-seq, 35- to 100-nt RNA fragments are selected and sequenced. Consequently, nascent RNAs shorter than 35 nt (i.e., those within 35 bp of the TSS) cannot be profiled. In theory, smaller fragments could also be selected, sequenced and mapped by extending the lower limit to 20 nt. However, this may result in the loss of unique sequence alignment18. In addition, it has been estimated that only nascent RNA longer than 15–17 nt extrudes from Pol II, to become accessible to 5′ capping enzymes19. This indicates that RNA shorter than 15–17 nt will not be accessible to 5′ phosphorylation by PNK. A possible modification of mNET-seq to enable investigation of promoter-proximal RNA includes removal of proteins after Pol II IP—e.g., by RNA extraction with TRIzol reagent or RNA phenol. Further, treatment of the purified RNA with tobacco acid pyrophosphatase before 5′ phosphorylation by PNK might enable detection of any 5′capped RNA present in this RNA fraction.
During cell fractionation, Pol II could potentially inefficiently elongate because of the presence of magnesium in the cell lysis buffer (HLB+N, HLB+NS and NUN2; see Reagent Setup). However, we consider this highly unlikely without a run-on reaction (as in GRO-seq8). Nevertheless, we recommend that the handling time should be minimized before chromatin digestion, and it should be consistent between samples. For this reason, we always use the same number of cells as starting material; fewer than 12 cell culture plates (150 mm) are recommended for each experiment. Alternatively, detergents (such as sarkosyl) and transcription inhibitors (such as actinomycin D) might be helpful additions to the cell lysis buffer to prevent any in vitro Pol II elongation.
The issue of RNA degradation is a potential concern, especially during Pol II IP. For the Pol II antibodies that we have used (Table 2), a 1-h incubation was identified as optimal for obtaining the maximum yield of Pol II complex under our IP conditions; this is a sufficiently short time period to avoid nonspecific IP products. However, some Pol II antibodies may be less efficient in Pol II IP, in which case longer incubations (>1 h) may be required. In this case, potent RNA inhibitors such as SUPERase-In (Thermo, cat. no. AM2696) should be added to the IP reaction buffer to prevent RNA degradation.
mNET-seq provides the genomic position of nascent RNA 3′ ends. Therefore, both elongating Pol II occupancy and RNA cleavage events, such as pre-mRNA splicing and pre-miRNA processing, can be determined. However, as Pol II–associated complexes, such as the spliceosome and microprocessor, can also give rise to RNA cleavage peaks, we were unable to distinguish whether individual mNET-seq peaks reflected Pol II accumulation or co-transcriptional RNA cleavage. It is possible that more stringent washing of the IP sample may allow separation of RNA processing–derived RNA 3′-ends from elongating Pol II. RNA 3′ ends in the Pol II active site could then be unambiguously determined in the mNET-seq profile.
mNET-seq shows Pol II distribution over a gene, with higher peaks at a specific location suggesting Pol II pausing. However, peaks might also arise from PCR bias that could dominate the true profile during library amplification. The use of a low number of PCR cycles (<15 cycles) to amplify the cDNA library is crucial to minimizing this effect. Primer bands should be detected on 6% (wt/vol) polyacrylamide gels when gel purification is performed as an indication that the PCR reaction is unsaturated. Addition of random nucleotides (barcodes) to the 3′ ends of RNA inserts before sequencing adapter ligation might help identify and remove PCR duplication problems10.
We use gentle MNase digestion conditions on the chromatin pellet to avoid overdigestion of Pol II–associated RNA. A possible limitation is that not all Pol II complexes are released under these conditions. However, a comparison between GRO-seq and mNET-seq suggests that mNET-seq detects virtually all engaged Pol II (Pearson’s correlation coefficient = 0.93, P < 2.2e−16, See Supplementary Fig. 1). We predict that undigested chromatin reflects mostly heterochromatic regions of the genome that are untranscribed, or that have low levels of transcription. However, it is possible that more potent nuclease treatment may be required to allow fuller recovery of Pol II complexes from chromatin.
Application of mNET-seq
We have so far used a variety of Pol II antibodies for mNET-seq (Table 2), which has allowed us to reveal genome-wide nascent transcription, as well as coupled RNA processing. In theory, a range of biological questions could be addressed by using other antibodies for mNET-seq. Antibodies specific to transcription factors and RNA processing factors associated with elongating Pol II could be used to determine where these factors are involved in Pol II elongation and pausing. In addition, Pol I and III antibodies would reveal nascent transcription maps for these separate polymerases. Nascent RNA profiles specifically associated with Pol II mutants (such as the kinetically slow Pol II and various Pol II CTD mutants) could be obtained by ectopically expressing tagged-Pol II mutants in cells, and performing IPs with antibodies specific to the tag.
Although we have analyzed nascent RNA in the elongating Pol II complex selected by specific Pol II antibodies, we have not analyzed other protein components of the complex. To understand which proteins are associated with particular co-transcriptional events using particular Pol II modifications, it will be important to examine all the proteins in the specific Pol II IP by mass spectrometry. It is also possible to compare mass spectrometry profiles using specific conditions, such as transcription elongation inhibition, to better understand which factor is important for the effect.
Experimental design
Chromatin and nascent RNA digestion
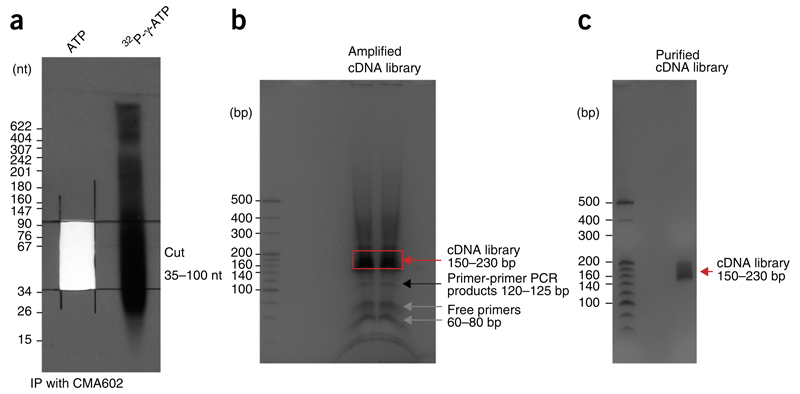
Before proceeding to a large-scale experiment, some of the mNET-seq cDNA library preparation stages should be carefully checked and optimized. First, digesting insoluble chromatin and nascent RNA is a crucial step. Even though MNase is a very efficient nuclease, its activity may sometimes vary. In particular, when using different cell lines, the optimal MNase concentration should be recalibrated by titrating MNase levels and by checking the efficiency of Pol II release from insoluble chromatin by western blotting. In addition, overly harsh MNase digestion could affect nascent RNA length. Determination of RNA length on denaturing gels after Pol II IP is recommended. To do this, radiolabel the mNET-seq RNA preparation using T4 PNK and γ-32P ATP, and check that the MNase digestion gives the expected smear pattern on a denaturing 8% (wt/vol) polyacrylamide gel (Fig. 2a).
Figure 2.
Examples of gel purification steps in the mNET-seq method. (a) RNA size purification of Pol II IP products from an 8% (wt/vol) urea gel (Step 55). The 35- to 100-nt-sized RNA should be cut from the unlabeled sample. At least one empty lane should separate the unlabeled and radiolabeled samples. (b) Gel analysis of amplified cDNA library in the mNET-seq method (Step 91). The library size should be 150–230 bp on the 6% (wt/vol) polyacrylamide gel (red arrow). (c) Gel analysis of purified cDNA library (Step 95). Make sure that there is no primer-dimer contamination.
Cell lines
It is important to test cell lines or tissue before mNET-seq, as their behavior in the protocol may vary. Before starting an experiment, cell fractionation and Pol II immunoprecipitated RNA recovery should be tested. HeLa13 and A549 (T.N., T.G., M.C.-F. and N.J.P., unpublished data) cells have been successfully used for mNET-seq by our group. However, the nucleus and chromatin pellet of A549 cell are much smaller than those of HeLa cells. Consequently, a lower MNase concentration (which depends on chromatin pellet size) is required for A549 cells.
Normally, 1.0 × 108 cells provides a good read coverage across the human genome in mNET-seq. Although the coverage level will be lower, 2.5 × 107 cells are sufficient for highly expressed genes. Transcription activity is different in each cell so that the minimum required cell number should be tested for each cell line.
Controls
The IP step is critical. IP efficiency and the specificity of the antibody should be checked by western blotting. An ideal IP efficiency is >50%. Immunoprecipitated RNA should be checked on a denaturing gel. Mock IPs (in which antibody is omitted from the IP reaction) or IPs using mouse IgG are useful negative controls; neither Pol II protein nor Pol II–associated RNA should be detected by western blotting or on denaturing gels, respectively13, for these controls. For mNET-seq, a mouse monoclonal antibody is preferable to a rabbit polyclonal antibody, as the latter may display significant lot variation.
Materials
Reagents
▲ CRITICAL Nuclease-free reagents are required. To avoid contamination and RNA degradation, do not share reagents and kits.
Cultured cells. We have used 2.5 × 107–1.0 × 108 HeLa or A549 cells successfully for cDNA library preparation ! CAUTION The cell lines used in your research should be regularly checked to ensure that they are authentic and not infected with mycoplasma.
DMEM with 4.5 mg/liter glucose (Sigma, cat. no. D6429)
FBS (Sigma, cat. no. F4135)
ATP, 10 mM (Cell Signaling, cat. no. 9804) ▲ CRITICAL ATP should be aliquotted (50 μl each) and stored at −20 °C for up to 1 year.
Boric acid (VWR Chemicals, cat. no. 20185.297)
Bromophenol blue (Sigma-Aldrich, cat. no. B0126)
Chloroform (Sigma-Aldrich, cat. no. C2432) ! CAUTION Chloroform is toxic and harmful. Wear gloves and eye glasses when you are using this compound. If the skin is exposed to chloroform, extensively wash it with running water.
Complete proteinase inhibitor cocktail, EDTA-free (Roche, cat. no. 11873580001)
Dulbecco’s PBS (Lonza, cat. no. BE17-516F)
d(+)-Saccharose, sucrose (VWR Chemicals, cat. no. 27480.294)
DTT, 0.1 M (accompanied with SuperScript III reverse transcriptase)
Dynabeads protein G (Life technologies, cat. no. 10004D)
Dynabeads M280 sheep Anti-mouse IgG (Life technologies, cat. no. 11202D)
EDTA (VWR Chemicals, cat. no. 20302.260)
EGTA (Sigma, cat. no. E3889)
Ethidium bromide (EtBr; Life technologies, cat. no. 15585-011) ! CAUTION EtBr is toxic and harmful. Wear gloves and eye glasses when you are using this compound. If skin is exposed to EtBr, extensively wash it with running water.
Ethanol (Sigma-Aldrich, cat. no. 32221)
Ficoll-400 (Sigma, cat. no. F-4375)
First-strand buffer, 5× (provided with SuperScript III reverse transcriptase)
Glycerol (Fisher Chemical, cat. no. G/0650/17)
GlycoBlue co-precipitant (Life technologies, cat. no. AM9515)
HEPES (Sigma-Aldrich, cat. no. H3375)
HCl, 36.5–38% (vol/vol; Sigma-Aldrich, cat.no. H1758) ! CAUTION HCl is toxic and harmful. Wear gloves and eye glasses when you are using this compound. If skin exposed to HCl, extensively wash it with running water. Handle the compound in hood space.
Isopropanol (Fisher Chemical, cat. no. P/7490/17)
Micrococcal nuclease (NEB, cat. no. M0247) ▲ CRITICAL Micrococcal nuclease should be aliquotted (50 μl each) and stored at −20 °C for up to 3 months.
MgCl2, 1 M (Sigma-Aldrich, cat. no. M1028)
NaCl (VWR Chemicals, cat. no. 27810.295)
NP-40, IGEPAL CA630 (Sigma, cat. no. I8896)
Nuclease-free water, not DEPC-Treated (Life technologies, cat. no. AM9937)
pBR322 MspI digest (NEB, cat. no. N3032S)
-
γ-32P ATP (Perkin Elmer, 6,000 Ci/mmol, cat. no. NEG502Z)
! CAUTION Consult local and institutional rules for detailed guidelines
▲ CRITICAL Always use fresh.
PhosSTOP, phosphatase inhibitor cocktail (Roche, cat. no. 04906837001)
Appropriate RNA polymerase II antibodies (Table 2)
RNaseZap (Ambion, cat. no. AM9782)
Sodium acetate (VWR Chemicals, cat. no. 27652.260)
SuperScript III reverse transcriptase (Life technologies, cat. no. 18080-044)
T4 polynucleotide kinase (PNK), 3′ phosphatase minus (NEB, cat. no. M0236S)
T4 RNA ligase, deletion mutant 2 (Epicentre, cat. no. LR2D1132K)
Tris, Trizmabase (Sigma-Aldrich, cat. no. T6066)
-
TRIzol reagent (Life Technologies, cat. no. 15596-026) ! CAUTION TRIzol is harmful. Wear gloves and eye glasses when you are using this compound.
If skin is exposed to TRIzol, extensively wash it with running water.
-
TruSeq small RNA library preparation kit (Illumina, cat. no. RS-200-0012)
▲ CRITICAL Prepare aliquots of RNA adapters and store them at −80 °C (up to 1 year). Always use fresh aliquots.
Tween 20 (Sigma-Aldrich, cat. no. P9416)
Urea (VWR Chemicals, cat. no. 28877.292)
Xylene cyanol FF (Sigma-Aldrich, cat. no. X-4126)
Software
Cutadapt (https://cutadapt.readthedocs.org/en/stable/)
Fastqc (http://www.bioinformatics.babraham.ac.uk/projects/fastqc)
-
mNET_SNR (https://github.com/tomasgomes/mNET_snr)
▲ CRITICAL Requires Python v2.7 (https://www.python.org/) and pysam v0.8.3 (https://github.com/pysam-developers/pysam)
Samtools v0.1.19 (http://samtools.sourceforge.net/)
TopHat v2.0.13 (http://ccb.jhu.edu/software/tophat/index.shtml)
Equipment
▲ CRITICAL Nuclease-free equipment is required before starting mNET-seq experiment: spray pipettes, pipette aids and bench with RNaseZap. Also use an RNase-free centrifuge and thermomixer if possible.
Cell scrapers (Fisher Scientific, cat. no. 08-100-240)
Cell culture dishes, 150 mm × 25 mm (Corning, cat. no. 430599)
Cell culture hood
Centrifuge (Thermo scientific, Heraeus)
CO2 incubator (CO2 at 5%, humidified, 37 °C)
Developer for X-ray film (Konica Minolta, cat. no. SRX-101A)
Dynamag2, magnetic rack for microcentrifuge tubes, 1.5 ml (Life technologies, cat. no. 12321D)
Filter tips (Fisher)
Gel tank for PAGE (Engineering and Design Plastics Ltd., no. EV400)
Glass filter, 1 cm (Whatman, cat. no. 1823-010)
MILLEX GP 0.22 μm (Millipore, cat. no. SLGP033RS)
Nanodrop 2000 spectrophotometer (Thermo Scientific, cat. no. ND-2000)
Needle, 19G (Thermo)
Novex Tris-borate-EDTA (TBE) gel, 6%, 10 well (Life Technologies, cat. no. EC6265BOX)
Novex TBE running buffer, 5× (Life technologies, cat. no. LC6675)
Nuclease-free microcentrifuge tubes, 1.5 ml (Axygen, cat. no. MCT-175-C)
Nuclease-free tubes, 0.2 ml (Qiagen, cat. no. 981005)
Nuclease-free tubes, 0.5 ml (Life Technologies, cat. no. AM12300)
Pipetman (Gilson)
Pipette aid (Corning)
Qubit 3.0 fluorometer (Thermo Scientific, cat. no. Q33216)
Radioactive monitor
Sarogold wrap
Spin-X centrifuge tube, filter 0.22 μm (Coster, cat. no. 8160)
Super RX, Fuji medical X-ray film (Fuji film, cat. no. 47410 19237)
Syringe, 1 ml (Thermo, cat. no. SS+01T1)
Thermomixer C (Eppendorf, cat. no. 035963)
Thermal cycler (Biometra)
Tube rotator (Stuart)
Vortex (Fisons)
XCell SureLock Mini-Cell, Norvex gel tank (Life Technologies, cat. no. E10001)
Reagent Setup
HLB+N HLB+N is 10mM Tris-HCl (pH 7.5), 10 mM NaCl, 2.5 mM MgCl2 and 0.5% (vol/vol) NP-40. 4 ml of HLB+N is required for each preparation; for 12 cell fractionations, prepare 50 ml as follows. Add 0.5 ml of 1 M Tris-HCl (pH 7.5), 0.1 ml of 5 M NaCl, 0.125 ml of 1 M MgCl2 and 2.5 ml of 10% (vol/vol) NP-40 to 46.725 ml of autoclaved distilled water. ▲ CRITICAL Always use autoclaved distilled water and after mixing filter using a MILLEX GP 0.22 μm. Just before use, add 1× Complete proteinase inhibitor cocktail and 1× PhosSTOP. Store it at 4 °C for up to 1 year; once opened, use it within 1 week.
HLB+NS HLB+NS is 10 mM Tris-HCl (pH 7.5), 10 mM NaCl, 2.5 mM MgCl2, 0.5% (vol/vol) NP-40 and 10% (wt/vol) sucrose. 1 ml of HLB+NS is required for each preparation; for 30 cell fractionations, prepare 50 ml as follows. Add 0.5 ml of 1 M Tris-HCl (pH 7.5), 100 μl of 5 M NaCl, 125 μl of 1 M MgCl2, 2.5 ml of 10% (vol/vol) NP-40 and 10 ml of 50% (wt/vol) sucrose to 36.725 ml of autoclaved distilled water. ▲ CRITICAL Always use autoclaved distilled water, and after mixing filter the solution using a MILLEX GP 0.22 μm. Just before use, add 1× Complete proteinase inhibitor cocktail and 1× PhosSTOP. Store the solution at 4 °C for up to 1 year; once opened, use it within 1 week.
NUN1 buffer NUN1 buffer is 20 mM Tris-HCl (pH 7.9), 75 mM NaCl, 0.5 mM EDTA and 50% (vol/vol) glycerol. 125 μl of NUN1 is required for each preparation; for 400 cell fractionations, prepare 50 ml as follows. Add 1 ml of 1 M Tris-HCl (pH 7.9), 750 μl of 5 M NaCl, 125 μl of 0.2 M EDTA and 25 ml of 100% (vol/vol) glycerol to 23.125 ml of autoclaved distilled water. ▲ CRITICAL Always use autoclaved distilled water, and after mixing filter using a MILLEX GP 0.22 μm. Just before use, add 1× Complete proteinase inhibitor cocktail and 1× PhosSTOP. Store the solution at 4 °C for up to 1 year; once opened, use it within 1 week.
NUN2 buffer NUN2 buffer is 20 mM HEPES-KOH (pH 7.6), 300 mM NaCl, 0.2 mM EDTA, 7.5 mM MgCl2, 1% (vol/vol) NP-40 and 1 M urea. 1.2 ml of NUN2 is required for each preparation; for 40 cell fractionations, prepare 50 ml as follows. Add 1 ml of 1 M HEPES-KOH (pH 7.6), 3 ml of 5 M NaCl, 50 μl of 0.2 M EDTA, 375 μl of 1 M MgCl2, 5 ml of 10% (vol/vol) NP-40 and 3 g of urea, and then fill it up to 50 ml with autoclaved distilled water. ▲ CRITICAL Always use autoclaved distilled water, and after mixing filter the solution using a MILLEX GP 0.22 μm. Just before use, add 1× Complete proteinase inhibitor cocktail and 1× PhosSTOP. Store the solution at 4 °C for up to 1 year; once opened, use it within 1 week.
NET-2 buffer NET-2 buffer is 50 mM Tris-HCl (pH 7.4, 150 mM NaCl and 0.05% (vol/vol) NP-40. 1 ml of NET-2 is required for each preparation; for 1,000 washes, prepare 1 liter as follows. Add 50 ml of 1 M Tris-HCl (pH 7.4) and 30 ml of 5 M NaCl to 915 ml of distilled water. ▲ CRITICAL 5 ml of 10% (vol/vol) NP-40 must be added after autoclaving. Store it at room temperature (25 °C) for up to 1 year.
RNA gel elution buffer RNA gel elution buffer is 1 M NaOAc and 1 mM EDTA. 400 μl of RNA gel elution buffer is required for each preparation; for 75 RNA elutions, prepare 30 ml as follows. Add 10 ml of 3 M NaOAc and 150 μl of 0.2 M EDTA to 19.85 ml of autoclaved distilled water. ▲ CRITICAL After mixing, filter the solution using a MILLEX GP 0.22 μm. Store it at room temperature for up to 1 year.
MNase buffer MNase buffer is 1× MNase buffer and 1× BSA. 100 μl of MNase buffer is required for each wash; for nine washes, prepare 1 ml as follows. Add 100 μl of 10× MNase buffer and 10 μl of 100× BSA to 890 μl of nuclease-free water. 10× MNase buffer and 100× BSA come with MNase enzyme. ▲ CRITICAL Freshly prepare the buffer for each experiment.
PNKT PNKT is 1× T4 PNK buffer and 0.1% (vol/vol) Tween. 300 μl of PNKT is required for each wash; for 30 washes, prepare 10 ml as follows. Add 1 ml of 10 × T4 PNK buffer and 100 μl of 10% (vol/vol) Tween to 8.9 ml of nuclease-free water. 10× PNK buffer comes with T4 PNK enzyme. ▲ CRITICAL Store it at −20 °C for up to 1 year.
10× TBE 10× TBE is 890 mM Tris, 890 mM boric acid and 20 mM EDTA. Prepare 1 liter as follows. Add 107.81 g of Tris, 55.02 g of boric acid and 7.43 g of EDTA and then fill up to 1 liter with distilled water. ▲ CRITICAL Autoclave the solution after mixing. Store it at room temperature for up to 1 year.
Urea dye Urea dye is 7 M urea, 0.05% (wt/vol) xylene cyanol and 0.05% (wt/vol) bromophenol blue. 10 μl of urea dye is required for each sample; for 1,000 samples, prepare 10 ml as follows. Add 4.2 g of urea, 0.5 ml of 1% (wt/vol) xylene cyanol and 0.5 ml of 1% (wt/vol) bromophenol blue and then fill up to 10 ml with nuclease-free water. ▲ CRITICAL Store it at room temperature for up to 1 year.
5× Ficoll dye 5× Ficoll dye is 15% (wt/vol) Ficoll-400 and 0.05% (wt/vol) bromophenol blue. 12 μl of 5× Ficoll dye is required for each sample; for 800 samples, prepare 10 ml as follows. Add 1.5 g of Ficoll-400 and 0.5 ml of 1% (wt/vol) bromophenol blue, and then fill up to 10 ml with nuclease-free water. ▲ CRITICAL Store it at room temperature for up to 1 year.
Procedure
cell culture ● TIMING 16 h
1| Seed cells of choice onto 150-mm dishes at 40% confluency, and grow them overnight in medium appropriate for the cell line being used (e.g., for HeLa cells, use DMEM with 10% (vol/vol) FBS). Prepare 6–8 dishes for each mNET-seq experiment sample or control.
Preparation of antibody-conjugated beads ● TIMING 1.5 h (or overnight)
▲ CRITICAL In Steps 2–7, volumes given provide sufficient beads for one plate of cells from Step 1. For six plates, 300 μl of beads (in a final volume of 600 μl of NET-2 buffer at Step 7) will need to be prepared. For IP control, the same amount of beads will need to be prepared separately, which at Step 4 can be left unconjugated (no antibody) or can be conjugated to the appropriate control antibody (e.g., mouse IgG).
2| Wash 50 μl of Dynabeads protein G (for 8WG16) or Dynabeads sheep anti-mouse IgG (for CMA601, CMA602 and CMA603; see Table 2) with 1 ml of ice-cold NET-2 buffer by vortexing at room temperature: repeat this step twice (total three times).
3| Resuspend the Dynabeads in 50 μl of ice-cold NET-2 buffer.
4| Add 5 μg of the appropriate Pol II antibody to washed beads. For IP control, no antibody or 5 μg of mouse IgG can be used.
▲ CRITICAL STEP Always use fresh antibody aliquots. The appropriate amount of antibody should be optimized if a previously untested Pol II antibody is used.
5| Incubate the mixture on a tube rotator (12 r.p.m.) in a cold room for at least 1 h or overnight.
6| Wash the Dynabeads with 1 ml of ice-cold NET-2 buffer by vortexing at room temperature: repeat this step twice (total three times).
7| Replace the wash buffer with 100 μl of ice-cold NET-2 buffer. Place on ice until use (see Step 25).
cell fractionation ● TIMING 1 h
▲ CRITICAL Steps 8–16 should be performed in the cold room.
8| Wash the cells from Step 1 with 20 ml of ice-cold PBS twice and, using a scraper, collect the cells into a 10-ml tube.
▲ CRITICAL STEP Use one 10-ml tube for each dish from Step 1.
9| Centrifuge the cells at 420g for 5 min at 4 °C, and then discard the supernatant to collect the cell pellets.
10| Resuspend the cells with 4 ml of ice-cold HLB+N and incubate the samples on ice for 5 min.
11| By using a 1-ml pipette, underlay the cell pellet with 1 ml of ice-cold HLB+NS.
▲ CRITICAL STEP Prepare ∼1.5 ml of HLB+NS for each sample. Add the solution under the resuspended cells slowly so as not to disturb the layer.
12| Centrifuge the cells at 420g for 5 min at 4 °C, and then discard the supernatant to collect the nuclear pellets.
▲ CRITICAL STEP Remove white debris in the supernatant, as that is possibly cytoplasmic membrane.
13| Add 125 μl of ice-cold NUN1 buffer to the nuclear pellet, resuspend by pipetting and transfer the sample to a new 1.5-ml microcentrifuge tube.
14| Add 1.2 ml of ice-cold NUN2 buffer and then vortex it at maximum speed to mix.
15| Incubate the samples on ice for 15 min.
▲ CRITICAL STEP Vortex the samples every 3–4 min. You should see white chromatin pellets in this step.
16| Centrifuge the samples at 16,000g for 10 min at 4 °C, and then discard the supernatant to collect the chromatin pellets.
Chromatin and RNA digestion ● TIMING ∼5 min
17| Add 100 μl of MNase buffer to the chromatin pellets from Step 16.
▲ CRITICAL STEP Do not try to resuspend, as the pellet cannot be broken until MNase is added.
? TROUBLESHOOTING
18| Centrifuge the samples at 16,000g for 10 s at 4 °C, and then discard the supernatant to collect the chromatin pellets.
19| Add 100 μl of MNase reaction mixture to the pellet. The volumes per sample are given below.
| Reagents | Volume (μl) per reaction | Final concentration |
|---|---|---|
| Nuclease-free water | 87 | |
| MNase buffer, 10× | 10 | 1× |
| BSA, 100× | 1 | 1× |
| MNase (2,000 gel units/μl) | 2 | 40 gel unit/μl |
| Mix total volume | 100 |
20| Incubate the samples at 37 °C for 90 s at 1,400 r.p.m. on a thermomixer.
Immunoprecipitation ● TIMING ~ 1.5 h
21|Add 10 μl of 250 mM EGTA to the samples from Step 20; this inactivates MNase.
22| Centrifuge the samples at 16,000g for 5 min at 4 °C.
23| Take 100 μl of supernatant from each sample and mix them together (total 600–800 μl).
▲ CRITICAL STEP Be careful not contaminate with the pellet, as this may result in background signals.
24| Dilute the pooled sample from Step 23 with 6–8 ml of ice-cold NET-2 buffer.
▲ CRITICAL STEP This tenfold dilution helps prevent protein aggregation and nonspecific binding.
25| Add Pol II antibody–conjugated beads (600–800 μl for each sample pooled at Step 23) from Step 7 and mix immediately.
▲ CRITICAL STEP Add mouse IgG or no antibody beads to separate samples for negative control.
26| Incubate the samples on a tube rotator (12 r.p.m.) for 1 h in the cold room.
▲ CRITICAL STEP Do not incubate for longer than 1 h, as this may cause RNA degradation.
27| Spin the samples at 300g for 5 min at 4 °C and discard the supernatant.
28| Transfer the beads to a 1.5-ml microcentrifuge tube.
▲ CRITICAL STEP ~1 ml of supernatant should be retained to enable this transfer.
29| Place the tube on a magnetic rack and leave it for 1 min. Remove the remaining supernatant.
30| Wash the beads with 1 ml of ice-cold NET-2 buffer by inverting tube, and repeat Steps 29 and 30 seven times (total eight times) in the cold room, taking care to remove the supernatant after the final wash.
RNA phosphorylation with T4 PNK ● TIMING 10 min
31| Resuspend the beads in 300 μl of ice-cold PNKT buffer.
▲ CRITICAL STEP Transfer 15 μl of sample to a new tube for monitoring RNA size by the incorporation of radioactivity during phosphorylation (Step 33); this sample is processed alongside the experimental sample in Steps 32–49.
32| Place the tubes on a magnetic rack and leave them for 1 min. Remove the supernatant.
33| Resuspend the beads in 200 μl of PNK reaction mixture, as tabulated in the first table below. For the size-monitoring sample, resuspend the beads in 10 μl of the reaction mix tabulated in the second table below.
| Reagents | Volume (μl) per reaction | Final concentration |
|---|---|---|
| PNKT (buffer and Tween) | 160 | |
| PNK (20 units/μl) | 10 | 1 unit/μl |
| ATP, 10 mM | 30 | 1.5 mM |
| Mix total volume | 200 |
| Reagents | Volume (μl) per reaction | Final concentration |
|---|---|---|
| PNKT | 8.5 | |
| PNK (20 units/μl) | 0.5 | 1 unit/μl |
| 32P–γ-ATP, fresh | 1 | 1 μCi/μl |
| Mix total volume | 10 |
! CAUTION Always be careful when working with radioactive materials. Consult local and institutional rules for detailed guidelines.
34| Incubate the samples at 37 °C for 6 min at 1,400 r.p.m. on a thermomixer.
35| Place the tubes on a magnetic rack and leave them for 1 min. Remove the supernatant.
36| Wash the beads with 1 ml of ice-cold NET-2 buffer by inverting the tube. Place the tubes on a magnetic rack and leave them for 1 min. Remove the supernatant.
Purification of RNA ● TIMING 45 min
37| Add 1 ml of TRIzol reagent to the beads and mix them well by vortexing.
38| Incubate the mixture for 5 min at room temperature.
■ PAUSE POINT The samples can be stored for a few weeks at −20 °C.
39| Add 200 μl of chloroform and mix well by vortexing.
40| Incubate the samples for 2 min at room temperature.
41| Spin the samples at 16,000g for 15 min at 4 °C.
42| Place the new tubes on the magnetic rack and add 1 μl of GlycoBlue co-precipitant.
43| Transfer 500 μl of upper phase from tubes in Step 41 into a new tube on the rack.
44| Add 500 μl of isopropanol and mix it well by vortexing.
45| Incubate the samples for 10 min at room temperature.
46| Spin the samples at 16,000g for 10 min at 4 °C.
47| Discard the supernatants completely using a pipette.
48| Dry the samples with the lid open for 3 min at room temperature.
49| Dissolve the pellets in 10 μl of urea dye.
RNA size fractionation ● TIMING 8 h
50| Apply the samples from Step 49 to a denaturing 8% (wt/vol) polyacrylamide gel containing 7 M urea.
▲ CRITICAL STEP Use a sequencing gel (200 × 380 mm, 0.5 mm) to ensure proper separation of bands.
▲ CRITICAL STEP The radiolabeled sample (usually >2,000 c.p.s.) should be applied to the same gel. There must be at least two empty lanes between radiolabeled and unlabeled samples to avoid radioactive cross-contamination of the mNET-seq sample. It is also useful to include a radiolabeled DNA size marker (~2,000 c.p.s.) for size calibration; we recommend MspI-digested pBR322 for this purpose.
51| Run the gel in 1×TBE buffer for 30 min at 30 W.
▲ CRITICAL STEP Regulate by wattage, as voltage will change during the electrophoresis.
52| Take the plates apart and cover the gel with Sarogold wrap.
53| Expose the gel to Super RX, Fuji medical X-ray film for 5 min at room temperature, and then develop the film.
? TROUBLESHOOTING
54| Place the developed film on the gel and mark the 35- to 100-nt size on Sarogold wrap with a marker pen.
▲ CRITICAL STEP Normally, 35- to 100-nt RNAs lie between the two dyes (xylene cyanol and bromophenol blue) in 8% (wt/vol) urea gel. Signals should be a smear (Fig. 2a).
55| Cut the gel and place the gel slices into a new 1.5-ml microcentrifuge tube.
■ PAUSE POINT The gel fragments can be frozen for several weeks at −80 °C.
56| Add 400 μl of RNA elution buffer to the gel slices from Step 55, and then crush the gel with a 1-ml plastic syringe.
57| Elute the RNA by rotating it for 2 h at room temperature.
58| Centrifuge the sample at 16,000g for 10 s at room temperature.
59| Transfer 400 μl of supernatants to a spin-X column, which has two glass filters on the membrane.
60| Centrifuge the tube at 16,000g for 1 min at room temperature.
61| Transfer the flow-through to a new 1.5-ml microcentrifuge tube.
62| Add 0.5 μl of GlycoBlue co-precipitant and 1 ml of 100% ethanol.
■ PAUSE POINT The samples can be stored at −20 °C.
63| Repeat the elution (Steps 57–62).
▲ CRITICAL STEP The first- and second-round elutes should not be combined in this step: keep them in different tubes.
64| Centrifuge the samples at 16,000g for 20 min at 4 °C.
65| Wash the samples with 1 ml of 70% (vol/vol) ethanol twice.
66| Air-dry the pellets for 3 min at room temperature.
▲ CRITICAL STEP The first-round pellet should be bigger than the second-round pellet.
67| Dissolve the pellets in 6 μl of fresh nuclease-free water, and combine the first- and second-round elution samples. Now the total volume is 12 μl. Place the samples on ice.
68| Measure the RNA concentration of samples by Nanodrop or Qubit assay.
▲ CRITICAL STEP Nanodrop gives an overestimated concentration. Normally, 30–60 ng/μl RNA will be collected by this method (from HeLa cells).
? TROUBLESHOOTING
■ PAUSE POINT The RNA samples can be stored for several weeks at −80 °C.
RNA library preparation: 3′ adapter ligation ● TIMING 1.5 h
▲ CRITICAL RNA library preparation (Steps 69–95) is performed using the TrueSeq small RNA preparation kit, mostly in accordance with the manufacturer’s instructions. Reagents are supplied with the TrueSeq kit, with the exception of T4 RNA ligase (deletion mutant) and the Superscript III reverse transcriptase kit.
69| Mix 1 μl of RNA 3′ adapter (RA3) and RNA sample as follows, in a nuclease-free 0.2-ml tube by pipetting.
| Reagents | Volume (μl) per reaction | Final concentration |
|---|---|---|
| RA3 | 1 | |
| RNA sample (30–100 ng/μl) | 5 | ~50 ng/μl |
| Mix total volume | 6 |
70| Heat the sample for 2 min at 70 °C on a thermal cycler. The lid temperature should be 90 °C.
71| Add 4 μl of ligation reaction mixture to the sample from Step 70, as tabulated below. Mix it well by pipetting.
| Reagents | Volume (μl) per reaction | Final concentration |
|---|---|---|
| Ligation buffer (HML) | 2 | 1× |
| RNase inhibitor | 1 | 20 unit/μl |
| T4 RNA ligase, deletion mutant | 1 | |
| Mix total volume | 4 |
72| Incubate the sample for 1 h at 28 °C on a thermal cycler. The lid temperature should be 90 °C.
73| To inactivate the RNA ligation reaction, add 1 μl of stop solution (Now a total of 11 μl).
74| Incubate the sample for 15 min at 28 °C, and then place the tube on ice.
RNA library preparation: preparation of 5′ adapter ligation reaction mixture ● TIMING 5 min
75| Add 1 μl of RNA 5′ adapter into a new nuclease-free 0.2-ml tube.
76| Heat the tube for 2 min at 70 °C; immediately after heating, place it on ice.
77| Add 1 μl of 10 mM ATP and mix by pipetting.
78| Add 1 μl of T4 RNA ligase and mix by pipetting, and then place the tube on ice.
▲ CRITICAL STEP Use wild-type T4 RNA ligase, as provided with the Illumina kit.
RNA library preparation: 5′ adapter ligation ● TIMING 1 h
79| Mix 11 μl of the RA3-ligated RNA sample (Step 74) and 3 μl of 5′ adapter ligation mixture (Step 78).
80| Incubate the mixture for 1 h at 28 °C on a thermal cycler. The lid temperature should be 90 °C. After incubation, place the tube on ice.
RNA library preparation: reverse transcription ● TIMING 1 h 15 min
81| Transfer 6 μl of ligated RNA from Step 80 to a new nuclease-free 0.2-ml tube. Add 1 μl of RNA reverse transcription primer (RTP) and mix it well by pipetting.
▲ CRITICAL STEP Do not use all of the ligated RNA; the remaining sample (~8 μl) can be frozen at −80 °C for several weeks. If overamplification or no amplification is detected at Step 95, the number of PCR cycles can be changed by returning to this step and repeating the procedure with the remaining sample.
82| Heat the tube for 2 min at 70 °C, and immediately after heating place it on ice.
83| Prepare the reverse transcription reaction mixture tabulated below, mix it well by pipetting, and add 5.5 μl of the mix to each tube from Step 82.
| Reagents | Volume (μl) per reaction | Final concentration |
|---|---|---|
| First-strand buffer, 5× | 2 | 1× |
| dNTP mix, 12.5 mM | 0.5 | 0.5 mM |
| DTT, 0.1 M | 1 | 8 μM |
| RNase inhibitor | 1 | 16 unit/μl |
| Superscript III reverse transcriptase | 1 | |
| Mix total volume | 5.5 |
84| Incubate the mixture for 1 h at 50 °C on a thermal cycler. The lid temperature should be 90 °C.
RNA library preparation: PCR amplification of the cDNA library ● TIMING 1 h
85| To amplify cDNA library, prepare 50 μl of PCR mixture for each sample from Step 84 as follows. Use nuclease-free 0.2-ml tubes.
| Reagents | Volume (μl) per reaction | Final concentration |
|---|---|---|
| Ultrapure water | 8.5 | |
| PCR mix, 2× | 25 | 1× |
| RNA PCR primer (RP1) | 2 | |
| RNA PCR primer Index | 2 | |
| cDNA library (from Step 84) | 12.5 | |
| Mix total volume | 50 |
86| PCR-amplify the sample, using the following program. The lid temperature should be 98 °C.
| Number of cycles | Denature (98 °C) | Anneal (60 °C) | Extend (72 °C) |
|---|---|---|---|
| 1 | 30 s | ||
| 13–15 | 10 s | 30 s | 20 s |
| 1 | 10 min |
The number of PCR cycles depends on the concentration of the RNA sample (at Step 68) as follows.
| RNA concentration (ng/μl) at Step 68 | PCR cycle |
|---|---|
| ~30 | 15 |
| 30–60 | 14 |
| 60–100 | 13 |
▲ CRITICAL STEP To avoid PCR duplication problems in sequencing, do not perform more than 15 PCR cycles. If PCR needs repetition because of problems such as overamplification, you can restart from Step 81 (see TROUBLESHOOTING for Step 91).
■ PAUSE POINT PCR samples can be stored for several months at −20 °C.
RNA library preparation: size purification of PCR library ● TIMING 6 h
87| Add 12 μl of 5× Ficoll dye to the PCR mixture from Step 86. Mix the total volume to 62 μl.
88| Split the mix across 2 lanes of a 6% (wt/vol) TBE polyacrylamide gel, and run the gel in 1× TBE buffer for 25 min at 250 V.
▲ CRITICAL STEP Use one gel for one sample to avoid cross-contamination. Leave two empty lanes between the DNA marker and the sample to avoid cross-contamination. Precast gels are recommended. Stop running the sample when the bromophenol blue reaches the bottom of the gel.
89| Soak the gel in fresh EtBr buffer (0.5–1 μg/ml) for 3 min.
90| Make a hole at the bottom of a nuclease-free 0.5-ml tube with a sterile 19G needle, and place the 0.5-ml tube into a nuclease-free 1.5-ml microcentrifuge tube.
91| Visualize the bands on the gel from Step 89 on a UV transilluminator; a PCR product of 150–230 bp should be visible (Fig. 2b). Cut only the 150- to 230-bp PCR product from the gel, and put the gel slice into the 0.5-ml microcentrifuge tube.
▲ CRITICAL STEP Take care to exclude 120- to 125-bp PCR products when excising the PCR product. These are primer-primer ligated DNA.
? TROUBLESHOOTING
92| Crush the gel by centrifugation at 15,000g for 1 min at room temperature. Remove the 0.5-ml tube.
■ PAUSE POINT Crushed gel can be stored for several months at −20 °C.
93| Elute the PCR product as described in Steps 56–66.
94| Dissolve the pellets in 9 μl of fresh nuclease-free water, and combine the first- and second-round elution samples. Now the total volume is 18 μl.
95| Measure the concentration of PCR libraries by Nanodrop or Qubit assay. Normally, 40–80 ng/μl PCR products will be detected.
▲ CRITICAL STEP Check the purified PCR library on a TBE polyacrylamide gel to confirm the size of 150–230 bp (Fig. 2c). Use 1–2 μl. Make sure that the PCR reaction is unsaturated; primers should be detected on the gel if the PCR reaction is in the linear range.
? TROUBLESHOOTING
■ PAUSE POINT PCR products can be stored for several months at −20 °C.
High-throughput sequencing ● TIMING 16 h
96| Sequence the PCR libraries using Illumina Hiseq2500 rapid mode. Data for the paired-end reads are generated in FASTQ format.
▲ CRITICAL STEP Use Fastqc (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/) to assess the data quality. Successful mNET-seq experiments should have high read quality and low adapter contamination. A bias in k-mer and GC content might be present, which is usually a result of precipitation of co-transcriptional processing intermediates with similar sequences.
Bioinformatic analysis ● TIMING ~24 h for basic analysis
▲ CRITICAL Steps 97–101 describe a suggested pipeline for mNET-seq data analysis. Introducing changes to the protocol or using different versions of the recommended tools, even if more recent, does not guarantee identical results. Analysis duration is dependent on the computational power available for processing.
97| Trim the adapter sequences from the 3′ end of each paired read using Cutadapt (https://cutadapt.readthedocs.org/en/stable/) with an error rate of 0.05, and allow it to match ‘N’s in the reads to the adapter sequence; discard the reads that are shorter than 10 bases, and remove the adapter only once from each read.
98| During the trimming step, some read pairs may lose one of their elements. From each FASTQ file, discard the reads that do not have a corresponding pair in the other file, while maintaining the read order in both files. This can be done either by following the instructions provided in cutadapt documentation or by applying a custom script.
99| Align paired reads using TopHat v2.0.13 with library type set as second strand, a minimum anchor length of five bases and only one alignment to the reference genome for each read allowed (http://ccb.jhu.edu/software/tophat/index.shtml).
? TROUBLESHOOTING
100| Filter the resulting BAM file with the aligned read pairs using Samtools v0.1.19 (http://samtools.sourceforge.net/) to obtain those in which both elements properly aligned with the genome. Use the command view and set the required flag parameter (-f) as 3.
101| Identify the last base incorporated by the Pol II. According to the protocol, this will be the last base from read 2, but with the directionality of read 1. This can be done using the Python script hosted in https://github.com/tomasgomes/mNET_snr (Python v2.7, https://www.python.org/, requires pysam v0.8.3, https://github.com/pysam-developers/pysam).
▲ CRITICAL STEP The BAM file resulting from Step 101 only contains information about the last base incorporated. Analysis requiring information about the whole read—such as spliced read mapping—must be performed on files resulting from the filtering step (Step 100).
? TROUBLESHOOTING
Troubleshooting advice can be found in Table 3.
Table 3.
Troubleshooting table.
| Step | Problem | Possible reason | Solution |
|---|---|---|---|
| 17 | White debris on top of the supernatant | Lipid contamination | Wash the chromatin pellets again |
| 53 | Low signals on autoradiography film | Low amount of RNA | Use fresh Pol II antibody at Step 2 |
| Use more Pol II antibody at Step 2 | |||
| Use fresh reagents throughout the procedure | |||
| Old 32P−γ ATP | Use fresh radioactive ATP and PNK at Step 33 | ||
| Short exposure | Longer exposure at −80 °C at Step 54 | ||
| 68 | Low amount of RNA | RNA is degraded | Use fresh reagents throughout the PROCEDURE |
| Low IP efficiency | Use fresh Pol II antibody at Step 2 | ||
| Use more Pol II antibody at Step 2 | |||
| Use a different lot if polyclonal antibody is used at Step 2 | |||
| Low amount of cells as starting material | Use more cells at Step 1 | ||
| Low efficiency of chromatin digestion | Check Pol II release from chromatin pellet by western blotting at Step 23 | ||
| Use fresh MNase at Step 19 | |||
| 91 | No PCR product of expected size (150–230 bp) | Underamplification | Use higher PCR cycle number, up to 15, at Step 86 |
| Use fresh PCR mix at Step 85 | |||
| Longer than expected PCR product size (>300 bp) | Overamplification | Reduce PCR cycle number | |
| 95 | Low amount of PCR library | Low elution efficiency from the gel | Crush the gel properly at Step 93 |
| Elute it for a longer time at Step 93 | |||
| Freeze the sample for >20 min before ethanol precipitation at Step 93 | |||
| Use 1 μl more GlycoBlue in the ethanol precipitation at Step 93 | |||
| 99 | Low number of mappable reads | Adapter contamination | Avoid cutting 120- to 125-bp bands at Step 91 |
| Adapter degradation | Use fresh RNA adapters at Steps 69 and 75 | ||
| RNA degradation | Use fresh RNA sample for RNA library preparation at Step 68 | ||
| Use fresh reagents throughout the PROCEDURE | |||
| PCR duplication | Use more cells at Step 1 | ||
| Reduce the PCR cycle number at Step 86 |
● TIMING
Step 1, cell culture: 16 h
Steps 2–7, preparation of antibody-conjugated beads: 1.5 h
Steps 8–16, cell fractionation: 1 h
Steps 17–20, chromatin and RNA digestion: 5 min
Steps 21–30, immunoprecipitation: 1.5 h
Steps 31–36, RNA phosphorylation with T4 PNK: 10 min
Steps 37–49, purification of RNA: 45 min
Steps 50–68, RNA size fractionation: 8 h
Steps 69–74, RNA library preparation—3′ adapter ligation: 1.5 h
Steps 75–78, RNA library preparation—preparation of 5′ adapter ligation reaction mixture: 5 min
Steps 79 and 80, RNA library preparation— 5′ adapter ligation: 1 h
Steps 81–84, RNA library preparation—reverse transcription: 1 h 15 min
Steps 85 and 86, RNA library preparation—PCR amplification of the cDNA library: 1 h
Steps 87–95, RNA library preparation—size purification of PCR library: 6 h
Step 96, high-throughput sequencing: 16 h
Step 97–101, bioinformatic analysis: 24 h
Anticipated Results
mNET-seq generates paired-end sequence data corresponding to Pol II immunoprecipitated protein-protected RNA fragments from which Pol II occupancy can be determined. By using different Pol II antibodies, Pol II modification–specific nascent RNA can be profiled at single-nucleotide resolution. mNET-seq with total Pol II–specific antibody CMA601 should generate a fully engaged Pol II profile (Supplementary Fig. 1). In addition, a Ser5-P CTD–specific antibody, such as CMA603, should detect peaks at the 3′ end of exons, which are derived from splicing.
Important quality control checkpoints in the mNET-seq protocol are described below.
RNA recovery (Steps 50 and 68)
For a typical mNET-seq experiment using eight 150-mm plates of near-confluent HeLa cells (~108 cells) and immunoprecipitated with Pol II–specific antibody, the radiolabeled RNA should give a reading of >2,000 c.p.s. (Geiger reading). However, levels will depend on the number and type of cells, the type of antibody used and how fresh the γ-32P ATP is. If signals are lower than 2,000 c.p.s., then check the Pol II IP efficiency and the working environment, which should be RNase-free. For details, see Troubleshooting Table.
Typically, 0.5–2 μg of RNA can be recovered after gel purification; 150 ng of RNA is sufficient to generate mNET-seq cDNA library.
mNET-seq cDNA library preparation (Steps 69–95)
150–400 ng of RNA is recommended as a starting point for cDNA library preparation. After cDNA amplification and gel purification of 150- to 230-bp PCR product, 0.5–2 μg of PCR library can be expected. For best results from high-throughput sequencing, submit 1–2 μg per library.
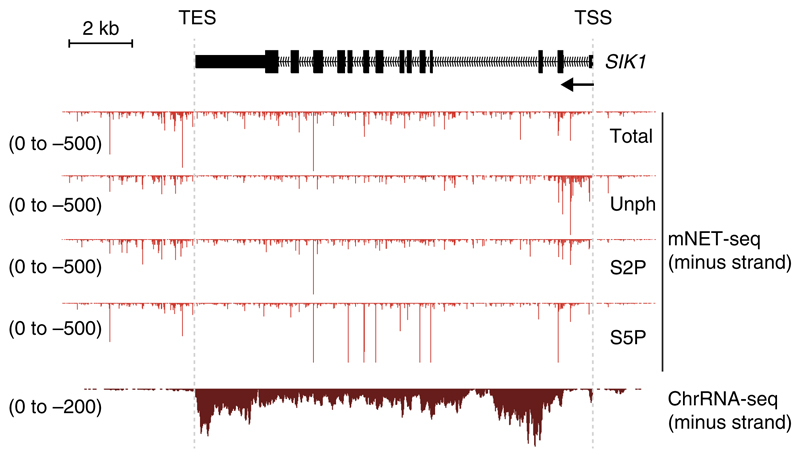
Figure 3 shows representative profiles from a successful mNET-seq experiment. This example shows mNET-seq profiles of the salt inducible kinase 1 (SIK1) gene using four different Pol II–specific antibodies that detect the Pol II active site RNA and RNA processing intermediates. The mNET-seq profiles are compared with a chromatin-bound RNA sequencing (ChrRNA-seq) profile that was used as an input for mNET-seq. Signals are observed across the gene. For 8WG16 unphosphorylated CTD (Unph), higher mNET-seq reads are detectable over the TSS, whereas for CMA603 Ser5-P CTD (S5P), specific peaks are detected at the 3′ end of exons, which are known splicing intermediates. Note that strong signals are detected downstream of the transcription end site (TES, also referred to as the polyA site), which reflect Pol II pausing associated with termination13.
Figure 3.
Example of a mNET-seq profile for the SIK1 gene. The mNET-seq minus strand reads are shown on the SIK1 gene. Four different Pol II CTD antibodies (Table 2), CMA601 (Total), 8WG16 unphosphorylated CTD (Unph), CMA602 Ser2-P CTD (S2P) and CMA603 Ser5-P CTD (S5P), were used for the mNET-seq. The RNA from the chromatin fraction was also sequenced (ChrRNA-seq, bottom). mNET-seq detects the 3′ end of nascent RNA across the SIK1 gene and also downstream of the TES. CMA603 specifically immunoprecipitated 5′SS cleaved splicing intermediates.
Unsuccessful experiments caused by low read count or quality may result in insufficient mapping of reads to the genome or incorrect read pairing. This will generate gene profiles with low read counts, and the features described above will be absent or barely detectable.
Background levels in mNET-seq analysis
To understand the ability of mNET-seq protocol to specifically target nascent Pol II transcripts, we compared the sense signal of a set of expressed protein-coding genes with that of rRNA- and tRNA-producing genes, which are transcribed by RNA polymerase I and RNA polymerase III, respectively (Supplementary Fig. 2). We show that the coverage of protein-coding genes is significantly higher when compared with the other two groups (two-sided Mann-Whitney U test: P < 2.2e–16 for both). In addition, we also compared the genome-wide coverage of total Pol II mNET-seq to GRO-seq data (Supplementary Fig. 1). By dividing the genome in 500-kb bins, the coverage of these samples shows a very high positive correlation (Pearson’s correlation coefficient = 0.93, P < 2.2e–16). Together, these results attest to the low unspecific background levels generated by the mNET-seq protocol.
Supplementary Material
Note: Any Supplementary Information and Source Data files are available in the online version of the paper.
Acknowledgments
We thank S. Murphy for critically testing this protocol, and A.R. Fialho Grosso for advice on bioinformatics analysis. This work was supported by funding to N.J.P. (Wellcome Trust Programme no. 091805/Z/10/Z and European Research Council (ERC) Advanced grant no. 339270) and to M.C-F. (Fundação Ciência e Tecnologia, Portugal).
Footnotes
Author Contributions T.N., M.C.-F. and N.J.P. designed the protocol. All authors wrote the paper. T.N. developed the protocol and performed all experiments. T.G. bioinfomatically analyzed the data.
Competing Financial Interests The authors declare no competing financial interests.
Reprints and permissions information is available online at http://www.nature.com/reprints/index.html.
References
- 1.Ozsolak F, Milos PM. RNA sequencing: advances, challenges and opportunities. Nat Rev Genet. 2011;12:87–98. doi: 10.1038/nrg2934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wilhelm BT, Landry JR. RNA-seq-quantitative measurement of expression through massively parallel RNA-sequencing. Methods. 2009;48:249–257. doi: 10.1016/j.ymeth.2009.03.016. [DOI] [PubMed] [Google Scholar]
- 3.Moore MJ, Proudfoot NJ. Pre-mRNA processing reaches back to transcription and ahead to translation. Cell. 2009;136:688–700. doi: 10.1016/j.cell.2009.02.001. [DOI] [PubMed] [Google Scholar]
- 4.Morlando M, et al. Primary microRNA transcripts are processed co-transcriptionally. Nat Struct Mol Biol. 2008;15:902–909. doi: 10.1038/nsmb.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pawlicki JM, Steitz JA. Primary microRNA transcript retention at sites of transcription leads to enhanced microRNA production. J Cell Biol. 2008;182:61–76. doi: 10.1083/jcb.200803111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Heidemann M, Hintermair C, Voss K, Eick D. Dynamic phosphorylation patterns of RNA polymerase II CTD during transcription. Biochim Biophys Acta. 2013;1829:55–62. doi: 10.1016/j.bbagrm.2012.08.013. [DOI] [PubMed] [Google Scholar]
- 7.Core LJ, Lis JT. Transcription regulation through promoter-proximal pausing of RNA polymerase II. Science. 2008;319:1791–1792. doi: 10.1126/science.1150843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kwak H, Fuda NJ, Core LJ, Lis JT. Precise maps of RNA polymerase reveal how promoters direct initiation and pausing. Science. 2013;339:950–953. doi: 10.1126/science.1229386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Weber CM, Ramachandran S, Henikoff S. Nucleosomes are context-specific, H2A.Z-modulated barriers to RNA polymerase. Mol Cell. 2014;53:819–830. doi: 10.1016/j.molcel.2014.02.014. [DOI] [PubMed] [Google Scholar]
- 10.Mayer A, et al. Native elongating transcript sequencing reveals human transcriptional activity at nucleotide resolution. Cell. 2015;161:541–554. doi: 10.1016/j.cell.2015.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Churchman LS, Weissman JS. Nascent transcript sequencing visualizes transcription at nucleotide resolution. Nature. 2011;469:368–373. doi: 10.1038/nature09652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Larson MH, et al. A pause sequence enriched at translation start sites drives transcription dynamics in vivo. Science. 2014;344:1042–1047. doi: 10.1126/science.1251871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nojima T, et al. Mammalian NET-seq reveals genome-wide nascent transcription coupled to RNA processing. Cell. 2015;161:526–540. doi: 10.1016/j.cell.2015.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wuarin J, Schibler U. Physical isolation of nascent RNA chains transcribed by RNA polymerase II: evidence for cotranscriptional splicing. Mol Cell Biol. 1994;14:7219–7225. doi: 10.1128/mcb.14.11.7219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dye MJ, Gromak N, Proudfoot NJ. Exon tethering in transcription by RNA polymerase II. Mol Cell. 2006;21:849–859. doi: 10.1016/j.molcel.2006.01.032. [DOI] [PubMed] [Google Scholar]
- 16.West S, Proudfoot NJ, Dye MJ. Molecular dissection of mammalian RNA polymerase II transcriptional termination. Mol Cell. 2008;29:600–610. doi: 10.1016/j.molcel.2007.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nojima T, Dienstbier M, Murphy S, Proudfoot NJ, Dye MJ. Definition of RNA polymerase II CoTC terminator elements in the human genome. Cell Rep. 2013;3:1080–1092. doi: 10.1016/j.celrep.2013.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Storvall H, Ramskold D, Sandberg R. Efficient and comprehensive representation of uniqueness for next-generation sequencing by minimum unique length analyses. PLoS ONE. 2013;8:e53822. doi: 10.1371/journal.pone.0053822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Martinez-Rucobo FW, et al. Molecular basis of transcription-coupled pre-mRNA capping. Mol Cell. 2015;58:1079–1089. doi: 10.1016/j.molcel.2015.04.004. [DOI] [PubMed] [Google Scholar]
- 20.Core LJ, Waterfall JJ, Lis JT. Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science. 2008;322:1845–1848. doi: 10.1126/science.1162228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Min IM, et al. Regulating RNA polymerase pausing and transcription elongation in embryonic stem cells. Genes Dev. 2011;25:742–754. doi: 10.1101/gad.2005511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Saunders A, Core LJ, Sutcliffe C, Lis JT, Ashe HL. Extensive polymerase pausing during Drosophila axis patterning enables high-level and pliable transcription. Genes Dev. 2013;27:1146–1158. doi: 10.1101/gad.215459.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kruesi WS, Core LJ, Waters CT, Lis JT, Meyer BJ. Condensin controls recruitment of RNA polymerase II to achieve nematode X-chromosome dosage compensation. Elife. 2013;2:e00808. doi: 10.7554/eLife.00808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Core LJ, et al. Analysis of nascent RNA identifies a unified architecture of initiation regions at mammalian promoters and enhancers. Nat Genet. 2014;46:1311–1320. doi: 10.1038/ng.3142. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.