Abstract
Background
We and others have shown four distinct and presumably related effects of mammalian proliferating cell nuclear antigen (PCNA) on DNA synthesis catalyzed by mammalian DNA polymerase δ(pol δ). In the presence of homologous PCNA, pol δ exhibits 1) increased absolute activity; 2) increased processivity of DNA synthesis; 3) stable binding of synthetic oligonucleotide template-primers (t1/2 of the pol δ•PCNA•template-primer complex ≥2.5 h); and 4) enhanced synthesis of DNA opposite and beyond template base lesions. This last effect is potentially mutagenic in vivo. Biochemical studies performed in parallel with in vivo genetic analyses, would represent an extremely powerful approach to investigate further, both DNA replication and repair in eukaryotes.
Results
Drosophila PCNA, although highly similar in structure to mammalian PCNA (e.g., it is >70% identical to human PCNA in amino acid sequence), can only substitute poorly for either calf thymus or human PCNA (~10% as well) in affecting calf thymus pol δ. However, by mutating one or only a few amino acids in the region of Drosophila PCNA thought to interact with pol δ, all four effects can be enhanced dramatically.
Conclusions
Our results therefore suggest that all four above effects depend at least in part on the PCNA-pol δ interaction. Moreover unlike mammals, Drosophila offers the potential for immediate in vivo genetic analyses. Although it has proven difficult to obtain sufficient amounts of homologous pol δ for parallel in vitro biochemical studies, by altering Drosophila PCNA using site-directed mutagenesis as suggested by our results, in vitro biochemical studies may now be performed using human and/or calf thymus pol δ preparations.
Background
Many Drosophila melanogaster homologs of the proteins required for both DNA replication and repair have been identified and in several cases purified to apparent homogeneity. These include DNA polymerase α holoenzyme [1,2], DNA polymerase δ(pol δ) [2-4], replication protein A (RP-A; [5]), replication factor C (RF-C; e.g., see [6-9]) and various origin recognition complex (ORC) subunits (see e.g., [10,11]). Moreover, complete replication of DNA containing the SV40 origin of replication has been reconstituted in vitro using purified SV40 T-antigen and Drosophila cell-free extracts [7].
A protein about which much information has been obtained is proliferating cell nuclear antigen (PCNA). Drosophila PCNA was first identified both as a highly purified protein able to substitute, albeit poorly, for human PCNA in a cell-free SV40 DNA replication system reconstituted from purified proteins [12] and by Yamaguchi et al. [13] who used an oligonucleotide probe to detect the Drosophila PCNA cDNA and gene, express the protein in E. coli and deduce its complete amino acid sequence. Further results indicated that in flies, PCNA was encoded by a single gene located at position 56F5-15 on the right arm of chromosome 2. This was subsequently identified as the Drosophila mus209 locus [14]. Recently, a second Drosophila PCNA gene of limited homology to the original and of unknown biological function has also been found [15].
Protocols have been established for purification of wild-type human PCNA from tissue culture cells [16,17], unmodified wild-type human PCNA after regulated expression in E. coli [18] and NH2-terminally his-tagged but otherwise wild-type human PCNA, also engineered for bacterial expression [19]. All were comparably effective at stimulating mammalian pol δ. Similar protocols have been developed for Drosophila PCNA and strategies for site-directed mutagenesis have been devised and implemented [20].
Recently, Zhang et al. [21] (see also [22]) as well as others (e.g., see [23]) identified the interdomain connector loop of PCNA (amino acids 119-133 of human PCNA) as crucial for binding pol δ. Of note, relative to wild-type PCNA, mutations of the molecule within this region such as glutamine at position 125 changed to glutamic acid (Q125E) promoted increased pol δ-processivity [21]. In human PCNA, residues 123, 126, 127 and 128 were defined as being essential for interaction with pol δ [21]. Comparison of human with Drosophila PCNA sequences in this region indicated that of these four amino acids, three (residues 126, 127 and 128) are identical. The fourth, residue 123, is glutamine (Q123) in wild-type Drosophila PCNA. The corresponding residue in human PCNA is valine (V).
To investigate the role of the interdomain connector loop of PCNA on the effects of PCNA on pol δ, we mutagenized residues within this region of Drosophila PCNA so that they more nearly resembled human amino acids. After bacterial expression and purification, we tested the effects of these site-specifically modified ("humanized") Drosophila PCNA molecules on purified calf thymus pol δ (two-subunit form; see [17,24]). Calf thymus and human pol δ are highly similar in amino acid sequence [25-27] and can, for our purposes, be used interchangeably. "Humanization" of a single Drosophila PCNA residue, conversion of Q123 to V (Q123V), conferred upon it, enhanced ability to affect several properties of calf thymus pol δ. More extensive mutagenesis, in which the entire interdomain connector loop of Drosophila PCNA (amino acids 119-133) was replaced by the corresponding human residues, was still more effective at stimulation of calf thymus pol δ, than either wild-type or Q123V Drosophila PCNA. However, it was considerably less effective than wild-type human PCNA at altering the properties of calf thymus pol δ. These results therefore suggest that in addition to the interdomain connnector loop, other regions of PCNA are also important effectors of pol δ activity. They also provide a means to couple operationally, the considerable power of in vivo genetic analyses performed in Drosophila with the sophistication of mammalian biochemistry.
Results
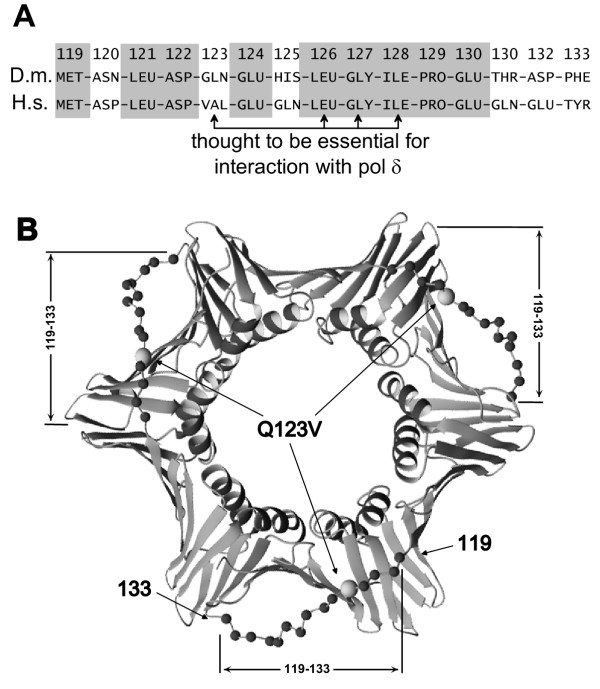
To study the role of the interdomain connector loop of PCNA (amino acids 119-133), we compared human and Drosophila homologs. Of the 15 interdomain connector loop residues, nine are identical between the two; identical residues are shaded (Fig. 1A). Overall, Drosophila PCNA is >70% identical to that from mammals (e.g., humans; see [13]). Others showed that PCNA residues 123, 126, 127 and 128 were essential for interaction with pol δ [28]. Of these four, only one (residue 123) differs between flies and humans. Also shown is a model constructed from the X-ray crystallographically determined structure of PCNA indicating the locations of the sites to be mutated in Drosophila PCNA (Fig. 1B). Shown (Fig. 1B) is the X-ray crystal structure of human PCNA. The Drosophila homolog is assumed to be similar.
Figure 1.
Structure and structural rationale for mutating Drosophila PCNA. A: amino acid sequences of the interdomain loops of Drosophila (designated D.m.) and human (designated H.s.) PCNA. Gray boxes indicate amino acids identical for both organisms; arrows show amino acids thought essential for interaction of human PCNA with human pol δ. Amino acid 123 is the only one which is both essential and different in Drosophila versus human PCNA. B: the "front" side of the human PCNA trimer. Amino acids 119-133 of the interdomain loops are highlighted by showing their α-carbon atoms as black spheres. The α-carbon atom of Val123 is shown as a larger gray sphere.
Purification of wild-type and site-specifically mutated PCNA
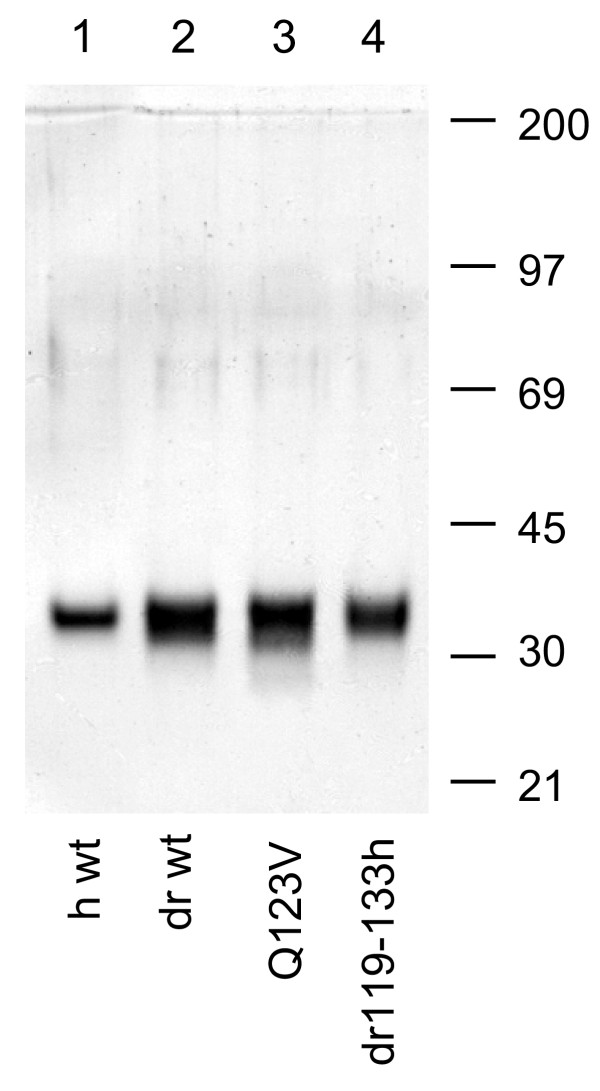
Four NH2-terminally his-tagged PCNA variants were highly purified; purity for each is shown (Fig. 2). First constructs were prepared encoding 1) NH2-terminally his-tagged wild-type human PCNA; 2) NH2-terminally his-tagged wild-type Drosophila PCNA (dPCNA) and two dPCNA derivatives; 3) one in which amino acid 123 was mutated from glutamine to valine (Q123V dPCNA); and 4) the other, in which Drosophila amino acids 119-133 were replaced by the corresponding human sequence (dr119-133h dPCNA). Then all four were transformed separately into E. coli (strain M15 [pREP4]) and respective proteins were expressed. Finally bacteria were lysed and his-tagged proteins were purified using various procedures including Ni2+-IDA Sepharose chromatography. The purity of each was determined by SDS-PAGE and is shown as indicated (Fig. 2). The identity of wild-type human PCNA was confirmed using mouse monoclonal anti-mammalian PCNA antibody PC10; the identity of wild-type Drosophila PCNA was confirmed using affinity purified polyclonal anti-Drosophila PCNA antibodies prepared in rabbits [12] (not shown).
Figure 2.
SDS-PAGE analysis of his-tagged PCNA purified from E. coli extracts after regulated bacterial expression. Purification and SDS-PAGE were as described (Experimental Procedures). Lane 1, 0.4 μg wild-type human PCNA was subjected to electrophoresis. Lane 2, 0.8 μg wild-type Drosophila PCNA was subjected to electrophoresis. Lane 3, 0.8 μg Drosophila PCNA containing valine substituted for glutamine at position 123 was subjected to electrophoresis. Lane 4, 0.45 μg Drosophila PCNA containing amino acids 119-133 substituted with the corresponding human PCNA amino acids was subjected to electrophoresis. Migration positions of molecular mass standards are indicated to the right of the figure.
Stimulation of calf thymus pol δ activity by highly purified wild-type versus selected mutant PCNA fractions
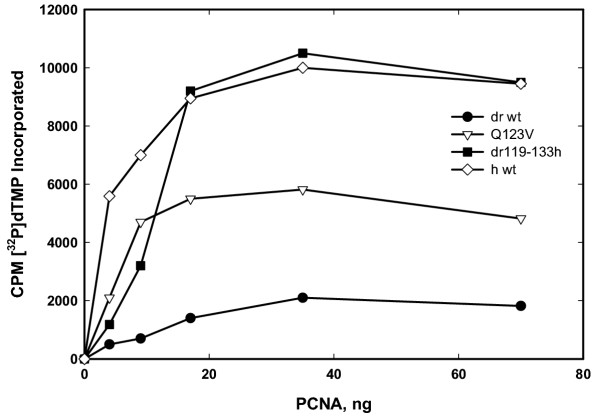
Calf thymus pol δ (apparently homogeneous two-subunit form; see [24]) was purified and assayed for polymerase activity in the presence of varying concentrations of both highly purified wild-type and specific mutant PCNA molecules. We showed previously that either calf thymus or human PCNA could be used interchangeably as stimulatory co-factors for calf thymus pol δ [29] (see also [12,18,19]). Assays were performed using poly(dA)-oligo(dT) as described (Experimental Procedures). As can be seen, human PCNA resulted in robust stimulation of calf thymus pol δ; much less stimulation was observed for wild-type Drosophila PCNA (Fig. 3). Mutation of Drosophila PCNA resulted in substantially increased stimulation of calf thymus pol δ; both substitution of a single amino acid (Q123V dPCNA) and replacement of the entire fly interdomain connector loop with corresponding human amino acids (dr119-133h dPCNA) had demonstrable effects. Of note, at relatively high concentrations, Drosophila PCNA but with the entire fly interdomain connector loop replaced by corresponding human amino acids (dr119-133h dPCNA) was similarly effective to wild-type human PCNA at stimulating the activity of calf thymus pol δ; however, it was considerably less effective at lower concentrations (Fig. 3). This suggests an effect on binding of PCNA to pol δ and/or on mutant PCNA multimerization.
Figure 3.
Effect of various purified PCNA fractions on the DNA polymerase activity of calf thymus pol δ. Calf thymus pol δ was incubated in a reaction mixture as described (see Materials and Methods) for 5 min at room temperature. Each incubation contained 10 ng of pol δ. DNA product synthesized was determined after placing 5-μl aliquots on Whatman DE-81 filters and subsequently washing with a 5% (w/v) solution of Na2HPO4•12H2O. Radioactivity retained on filters was then determined by liquid scintillation counter. Reaction mixtures contained increasing amounts, as indicated on the abscissa, of various PCNA samples, also as indicated.
The effects of highly purified wild-type versus selected mutant PCNA fractions on the processivity of incorporation by calf thymus pol δ
To examine further, the stimulation of calf thymus pol δ by both wild-type and specific mutant PCNA molecules, we examined effects on processivity of nucleotide incorporation. Processivity is defined as the number of deoxyribonucleotides incorporated each time a DNA polymerase binds its template-primer. As can be seen, without PCNA (Fig. 4 lane 1), pol δ is essentially a distributive enzyme incorporating only a few nucleotides as a result of each binding event. With increasing concentrations of wild-type human PCNA (concentrations increasing from right to left as indicated), processivity of incorporation increases dramatically (Fig. 4 lanes 2–4). This correlates quite closely with the PCNA-mediated activity increase (see Fig. 3). Wild-type Drosophila PCNA had relatively much less effect on the processivity of calf thymus pol δ (Fig. 4 lanes 5–7; concentrations again increasing from right to left as indicated). This is also consistent with activity data presented herein (Fig. 3) as well as with results reported previously [12]. When mutants of Drosophila PCNA were tested, both Q123V dPCNA (Fig. 4 lanes lanes 8–10; concentrations again increasing from right to left as indicated) and dr119-133h dPCNA (Fig. 4 lanes lanes 11–13; concentrations again increasing from right to left as indicated), promoted increased pol δ processivities, again consistent with increased activities (Fig. 3). Increases were concentration-dependent, also as expected.
Figure 4.
Effect of various purified PCNA fractions on the processivity of nucleotide incorporation by calf thymus pol δ. Incorporation of [α32P]dTMP by calf thymus pol δ was monitored by standard denaturing PAGE. The substrates used were (dA)~500-(dT)12–18 as template-primer and [α-32P]dTTP. Concentrations of PCNA, both wild-type and mutant proteins, are as indicated. h, human; dr, Drosophila melanogaster. NH2-terminally his-tagged-PCNA fractions are as indicated; wt, wild-type; Q123V, recombinant Drosophila PCNA containing a single amino acid, glutamine at position 123, changed to valine; dr119-133h, recombinant Drosophila PCNA containing the entire interdomain connector loop (amino acids 119-133) replaced with the corresponding human PCNA amino acids.
Stable complex formation among pol δ, 32P-labeled oligonucleotide template-primer and highly purified wild-type versus selected mutant PCNA fractions
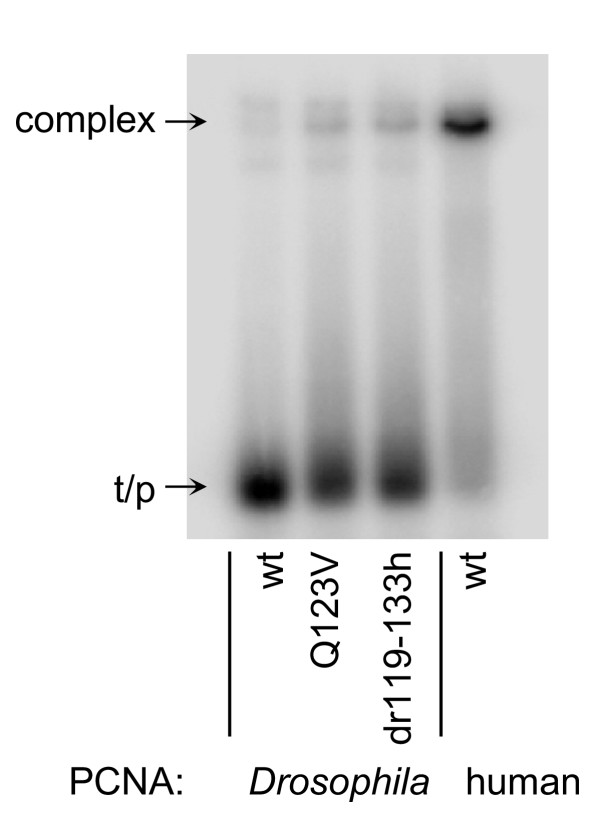
PAGE band mobility shift assays were used to evaluate, in an essentially qualitative manner, the stability of complex formation among calf thymus pol δ, labeled template-primer and highly purified wild-type versus selected mutant PCNA molecules. As can be seen, wild-type Drosophila PCNA promoted almost no pol δ•PCNA•template-primer complex formation (Fig. 5). In contrast, complex-formation with both Drosophila PCNA mutants (Q123V dPCNA and dr119-133h dPCNA) was readily detectable but neither gave results as robust as those seen with wild-type human PCNA (Fig. 5).
Figure 5.
Effect of various purified PCNA fractions on calf thymus pol δ•PCNA•32P-labeled oligonucleotide template-primer complex formation. Complex formation among pol δ, various purified PCNA fractions and 32P-labeled synthetic oligonucleotide template-primers (30-21-mers) was monitored by standard non-denaturing PAGE-band-mobility-shift assays [32]. Each incubation contained 10 ng of pol δ, 70 ng of PCNA and 0.1 pmol/reaction (useable 3'-OH) of annealed template-primer. NH2-terminally his-tagged-PCNA fractions are as indicated; wt, wild-type; Q123V, recombinant Drosophila PCNA containing a single amino acid, glutamine at position 123, changed to valine; dr119-133h, recombinant Drosophila PCNA containing the entire interdomain connector loop (amino acids 119-133) replaced with the corresponding human PCNA amino acids.
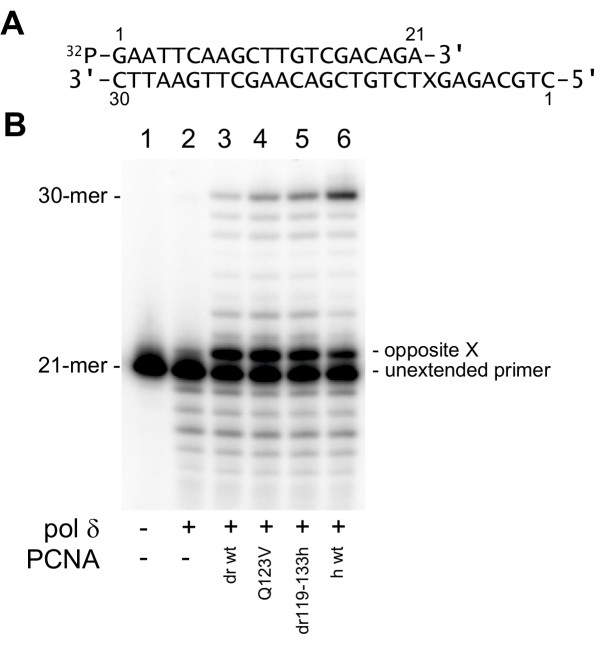
DNA synthesis beyond chemically defined template base lesions promoted by highly purified wild-type versus selected mutant PCNA fractions
As a final test, we examined the abilities of various PCNA fractions to promote pol δ-dependent DNA synthesis beyond template base lesions (TLS). PCNA-dependent TLS by pol δ was first reported by O'Day et al. [30] and subsequently analyzed in detail biochemically [29]. The structure of the synthetic oligonucleotide used for evaluation is shown in Fig. 6A. For the data shown (Fig. 6B), X represents the model abasic site (hereafter termed the abasic site [31]) used previously for many of our studies (e.g., see [29]). The mobility of the labeled 21-mer primer, PAGE-purified but without any subsequent enzymatic incubation is shown (Fig. 6B lane 1). When calf thymus pol δ alone was added, primer extension opposite the template abasic site was detected but there was no discernible elongation of the resulting 22-mer primer and no full-length product (30-mer) was observed; some degradation of the 21-mer primer, presumably resulting from the activity of the intrinsic pol δ 3'-5' exonuclease, was seen (Fig. 6B lane 2). Addition to incubations of wild-type Drosophila PCNA resulted in slight but readily detectable DNA synthesis beyond the template abasic site; this included some full-length 30-mer (Fig. 6B lane 3). Relatively more full-length 30-mer was seen when Q123V mutant Drosophila PCNA was included in addition to calf thymus pol δ (Fig. 6B lane 4) and still more full-length 30-mer was seen when dr119-133h Drosophila PCNA was added (Fig. 6B lane 5). Clearly, the greatest amount of full-length 30-mer product was seen when wild-type human PCNA was incubated with calf thymus pol δ (Fig. 6B lane 6). Of note, wild-type human PCNA also promotes the tightest complex formation between calf thymus pol δ and 32P-labeled template-primer DNA (see Fig. 5).
Figure 6.
Effect of various purified PCNA fractions to promote nucleotide incorporation by calf thymus pol δ beyond chemically defined template base lesions. A: the structure of the 5'-32P-labeled 30-21-mer template-primer; only the primer (21-mer) was radiolabeled and X indicates the position of a modified tetrahydrofuran moiety (model abasic site) on the 30-mer template. B: lane 1, gel-purified primer alone was subjected to electrophoresis; lanes 2–6, incubations were formulated as indicated with the template-primer shown in A followed by standard denaturing PAGE. h, human; dr, Drosophila melanogaster. For lanes 2–6, each incubation contained 0.5 pmol of labeled primer (3'-OH ends) annealed to 0.5 pmol of template (3'-OH ends), 10 ng pol δ and 70 ng PCNA as indicated. NH2-terminally his-tagged-PCNA fractions are as indicated; wt, wild-type; Q123V, recombinant Drosophila PCNA containing a single amino acid, glutamine at position 123, changed to valine; dr119-133h, recombinant Drosophila PCNA containing the entire interdomain connector loop (amino acids 119-133) replaced with the corresponding human PCNA amino acids.
Discussion
Although human PCNA and Drosophila PCNA are more than 70% identical at the level of primary amino acid sequence, wild-type Drosophila PCNA is only a very poor substitute for human PCNA in cell-free reactions with calf thymus pol δ. This is documented both in this report and previously [12,32]. However, mutating only a single Drosophila PCNA amino acid, glutamine at position 123 (Q123) to valine (V), leads to a dramatic enhancement in the abilities of Drosophila PCNA to stimulate calf thymus pol δ. Effects were shown on total activity (Fig. 3), processivity (Fig. 4), pol δ•PCNA•template-primer complex formation (Fig. 5) and extended DNA synthesis beyond a template abasic site (Fig. 6). Replacing the entire interdomain connector loop of Drosophila PCNA (amino acids 119-133) with the corresponding residues from human PCNA resulted in additional enhancement (Figs. 3,4,5,6), but in neither case were the mutants of Drosophila PCNA (Q123V dPCNA or dr119-133h dPCNA) equivalent to wild-type human PCNA in the stimulation of calf thymus pol δ.
Our data indicate that although a single Drosophila PCNA amino acid at position 123 (in addition to conserved residues 126–128) is very important for pol δ-stimulation, the further enhancement of stimulation seen when the entire interdomain connector loop of Drosophila PCNA (amino acids 119-133) was replaced with the corresponding residues from human PCNA suggests that other residues in this loop are also involved directly in binding pol δ. Alternatively, it is possible that loop residues other than 123 and 126–128 play a secondary or indirect (e.g., conformational) role in positioning crucial amino acids so as to optimize their direct binding to pol δ.
In this context, we would like to call attention to the fact that at relatively low concentrations, dr119-133h dPCNA is considerably less effective than wild-type human PCNA in stimulating the activity of calf thymus pol δ; at higher concentrations, dr119-133h dPCNA and wild-type human PCNA stimulate calf thymus pol δ similarly. This implies complex protein-protein interactions between PCNA and pol δ such that biochemical properties recorded in dilute solutions in vitro may not accurately predict properties manifest at much different and generally much higher intranuclear concentrations present in vivo. Alternatively, PCNA must be present as a trimer (three-subunit ring) in order to function. Since the equilibrium among monomer, dimer and trimer was shown to depend on PCNA protein concentration [33], it is certainly possible that the difference observed between dr119-133h dPCNA and wild-type human PCNA actually reflects differences in the Keq for PCNA multimerization. These two possibilities, concerning both complicated pol δ•PCNA interactions and PCNA multimerization, are not mutually exclusive.
Similarly, the fact that replacement of the entire interdomain connector loop of Drosophila PCNA (amino acids 119-133) with the corresponding residues from human PCNA did not result in a molecule as effective in stimulating calf thymus pol δ as human PCNA suggests that regions other than the interdomain connector loop are important for pol δ-stimulation. Our data do not address the question of whether these putative "other regions" affect pol δ directly (e.g., like the interdomain loop) or indirectly (e.g., through conformational effects on other regions of the molecule that do bind pol δ directly). Additional mutagenesis studies may shed light on this issue. For example, based on experiments of others, it seems likely that the extreme C-terminus of PCNA also interacts directly with pol δ (see [23,34-36]). Hence it may be of interest to perform similar mutagenesis experiments to those reported here, focusing instead on the C-terminal region of Drosophila PCNA, rather than the interdomain connector loop.
We think it should also be noted that both Oku et al. [35] and Ola et al. [36] prepared hybrid proteins between human and S. cerevisiae PCNA. As in our studies, Ola et al. [36] found that regions other than the interdomain connector loop of PCNA were important for interaction with pol δ. These authors suggested that additional interacting regions were likely to exist both in the PCNA C-terminus and N-terminus.
It may also be of interest to prepare double-mutants, first in the interdomain connector loop of Drosophila PCNA, thereby allowing efficient in vitro function with purified calf thymus pol δ, and then elsewhere in the PCNA molecule corresponding to interesting sites defined phenotypically by in vivo genetic studies of others. For example, it might be possible to determine if particular mus209 mutations leading to enhanced mutagen sensitivity among affected organisms (see [37] and references therein) alter any functional interactions between PCNA and pol δ in vitro. Results of such studies could lead to novel biochemical insights regarding the mechanism(s) by which point mutations in the Drosophila PCNA gene lead to enhanced mutagen sensitivity among animals bearing these mutations.
The strategy taken here will presumably allow study of interactions between PCNA and other proteins with which it interacts. In this context, we think it important to note that partial effects on pol δ-stimulation have been recorded. This suggests that our methodology will also allow detection of partial rather than complete effects on the binding of other proteins. Interactions between PCNA and many of the molecules with which it interacts have recently been mapped [23] and for example, one might immediately compare interactions between several mammalian proteins (e.g., human RF-C, DNA ligase I, FEN I and/or p21) and both various wild-type and mutant PCNA molecules described in this paper. Functional (e.g., effects on pol δ activity) as well as direct binding measurements may be made. As with PCNA•pol δ interactions, it may ultimately be feasible to correlate interesting PCNA molecules defined phenotypically using genetic analyses performed in living animals and biochemical studies of specific PCNA•protein binding. For example, do mutagen sensitive mus209 animals bear mutations in a region of PCNA responsible for MSH binding? Both MSH3 and MSH6 were reported to possess a consensus motif for binding to the interdomain connector loop of PCNA [38].
Finally, we think it important to note that pol δ has most recently been reported to contain at least four subunits (see e.g., [39,40]) yet all experiments performed here were with the two-subunit form of the enzyme purified from calf thymus. We and others have shown that the larger subunit, p125, is catalytic while the smaller, p50, does not seem to contact the DNA closely (see e.g, [41]), but instead, is required for processivity-stimulation by PCNA (e.g., see [42]) to which it apparently binds. It is also clear that PCNA binds to what has been termed, the third pol δ subunit, p68 or p66 in mammalian systems [39,43,44], Cdc27p in S. pombe [40] and Pol32p in S. cerevisiae [45,46]. Clearly the physiologically important interaction between PCNA (either mutant or wild-type) and this third pol δ subunit was omitted from our analyses, but could markedly affect any or all of the responses of polymerase to PCNA that we reported here.
Conclusions
Through our experiments, we showed that Drosophila PCNA could be "humanized" and that "humanization" (mutation of key Drosophila residues to human ones) increased effects on mammalian pol δ. The highly purified two-subunit form of pol δ was used for all of our studies. It is possible, though we think it unlikely, that different conclusions would be reached if a different form of pol δ (three-or four-subunit) was used. Nevertheless two of the effects we observed could be considered beneficial. They were enhancement of polymerase activity and processivity. A third effect seems likely to be detrimental, at least over the long term, that is increased synthesis opposite and beyond a chemically defined template base lesion (TLS). Our data suggest that all three of these effects result from enhancement of PCNA-dependent stability of the pol δ•PCNA•template-primer complex. In other words, in the range that we have studied, the more tightly pol δ binds to DNA, the greater its activity, the greater its processivity, but also the more likely it is to catalyze TLS. Our results provide an explicit approach to correlate in vivo genetic studies with rigorous in vitro biochemistry.
Methods
Materials
Unlabeled deoxyribonucleoside triphosphates (dNTPs) were from Boehringer-Mannheim; [α-32P]ATP and [α-32P]dTTP were from Amersham Corp. E. coli DNA polymerase I Klenow fragment without 3'-5' exonuclease activity (exo-), was expressed and purified according to standard protocols [47]. Terminal deoxynucleotidyl transferase (TdT) was from Sigma. Micrococcal nuclease was from Boehringer-Mannheim. Pfu DNA polymerase was from Stratagene. Ni2+-IDA Sepharose was from Pharmacia (Piscataway, NJ). Acrylamide and methylene bis-acrylamide were from Eastman Organic Chemicals and for protein SDS-PAGE, were further purified by adsorption of impurities to activated charcoal. For PAGE of nucleic acids, they were purified by adsorption to an ion exchange resin. All other materials were of reagent grade and were used without additional purification.
Proteins
PCNA was purified to apparent homogeneity from calf thymus [17] as was pol δ [24,48]. Human PCNA cDNA was cloned into a bacterial expression vector and human PCNA was purified from an E. coli extract, also to apparent homogeneity [18]. D. melanogaster PCNA was purified to apparent homogeneity identically after bacterial expression [13]. A his-tag was added to the NH2-termini of both human and Drosophila PCNA by cDNA insertion into pQE30 (Qiagen, Valencia, CA) using BamH1 and HindIII restriction endonuclease sites.
Nucleic acids
Templates and primers, all of defined sequence, were synthesized conventionally by Dr. F. Johnson and colleagues (Stony Brook). Before use, they were purified by standard denaturing PAGE [49]. All other DNA manipulations were performed according to standard techniques [49].
Methods
Much of the methodology was described in detail previously [12,19,20,24,29,32,41,50,51]. SDS-PAGE was according to Laemmli [52] as modified [53] on minigels or as reported previously [54]. For immunoblots, proteins were transferred electrophoretically to nitrocellulose [55] and resulting replicas were probed with antibodies. Reactivity was visualized colorimetrically [56] with alkaline phosphatase-conjugated goat anti-IgG antibodies [57,58] and a one-solution phosphatase substrate (Kirkegaard and Perry, Gaithersburg, MD). Immunologic detection of human PCNA was with mouse monoclonal antibody (mAb) PC10 (Oncogene Sciences, Uniondale, NY). Detection of Drosophila PCNA was with affinity purified polyclonal rabbit anti-Drosophila PCNA antibodies [12]. Restriction endonucleases were from Boehringer (Indianapolis, IN) and were used according to the vendor's instructions. DNA sequencing performed in both directions was according to Sanger et al. [59] using a fluorescence-based method and an ABI 373 (Applied Biosystems, Foster City, CA) automated DNA sequencer.
Site-directed mutagenesis of Drosophila PCNA
Site-directed mutagenesis of NH2-terminally his-tagged Drosophila PCNA was performed exactly as described [20] to generate either the Q123V protein or chimeric molecules containing the entire Drosophila PCNA sequence except for amino acids 119-133 which were replaced by the corresponding residues from human PCNA.
Purification of his-tagged PCNA
Purification of his-tagged PCNA to apparent homogeneity was performed exactly as previously described [20]. Characterization was by SDS-PAGE (Fig. 2) and immunoblot analysis.
DNA polymerase δ incubations
Assays of pol δ on synthetic oligonucleotide template-primers were performed essentially as previously described [24]. Primers were 5' end-labeled with T4 polynucleotide kinase in the presence of [γ-32P]ATP. Afterward, labeled primer was annealed to an unlabeled template. The standard reaction mixture for pol δ contained 40 mM Bis-Tris, pH 6.7, 6 mM MgCl2, 1 mM dithiothreitol, 10% glycerol and 40 μg/ml bovine serum albumin. Additional details are provided in the figure legends. Incubations were terminated by addition of standard stop solution and aliquots were subjected to 12% PAGE in the presence of 7 M urea and 15% formamide. After electrophoresis, gels were subjected to autoradiography and/or Molecular Dynamics 445 SI PhosphorImager analyses.
Pol δ processivity
Processivity was evaluated qualitatively using (dA)~500 annealed to (dT)12–18 (both from Pharmacia) in a final volume of 5 μl containing 6 nmol poly(dA) (nucleotide), 0.2 nmol (dT)12–18 (nucleotide), 10 μM dTTP, 100 μCi [α-32P]dTTP, 40 mM Bis-Tris, pH 6.7, 6 mM MgCl, 1 mM dithiothreitol, 10% glycerol, 40 μg/ml bovine serum albumin, 10 ng of highly purified pol δ and various quantities of different PCNA samples as indicated. Assays were for 5 min at room temperature and were stopped by addition of standard PAGE stop solution and PAGE in the presence of 7 M urea. After electrophoresis, gels were subjected to autoradiography and/or Molecular Dynamics 445 SI PhosphorImager analyses.
Nondenaturing PAGE band mobility shift assays
Nondenaturing PAGE band mobility shift assays were performed essentially as previously described [32] but without MgCl2 and otherwise as detailed in the figure legend. EDTA was included in each incubation and in the gel electrophoresis buffer at a final concentration of 3 mM.
Authors' contributions
DJuM performed all enzymologic and mobility shift assays with DNA polymerase δ in combination with both wild-type and various mutant PCNA molecules. He also designed, engineered and characterized all recombinant PCNA molecules. DJuM expressed several recombinant proteins in bacteria and purified them. Finally, he participated in DNA polymerase purification and drafted the original manuscript. MM expressed some recombinant proteins in bacteria and purified them. She also purified and characterized most DNA polymerase substrates. HM participated in DNA polymerase purification and manuscript preparation. PAF advised DJuM on execution and interpretation of experiments and assisted both in figure design and all other aspects of manuscript preparation. All authors read and approved the final manuscript.
Acknowledgments
Acknowledgements
These studies were supported by NIH Research Grant ES04068.
Contributor Information
Dmitry Ju Mozzherin, Email: dim@pharm.sunysb.edu.
Maeve McConnell, Email: mcconnell@pharm.sunysb.edu.
Holly Miller, Email: miller@pharm.sunysb.edu.
Paul A Fisher, Email: paul@pharm.sunysb.edu.
References
- Kaguni LS, Rossignol J-M, Conaway RC, Lehman IR. Isolation of an intact DNA polymerase-primase from embryos of Drosophila melanogaster. Proc Natl Acad Sci USA. 1983;80:2221–2225. doi: 10.1073/pnas.80.8.2221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsis PG, Chiang CS, Lehman IR. Purification of DNA polymerase-primase (DNA polymerase alpha) and DNA polymerase delta from embryos of Drosophila melanogaster. Methods Enzymol. 1995;262:62–77. doi: 10.1016/0076-6879(95)62009-5. [DOI] [PubMed] [Google Scholar]
- Aoyagi N, Matsuoka S, Furunobu A, Matsukage A, Sakaguchi K. Drosophila DNA polymerase delta. Purification and characterization. J Biol Chem. 1994;269:6045–6050. [PubMed] [Google Scholar]
- Chiang CS, Mitsis PG, Lehman IR. DNA polymerase delta from embryos of Drosophila melanogaster. Proc Natl Acad Sci U S A. 1993;90:9105–9109. doi: 10.1073/pnas.90.19.9105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsis PG, Kowalczykowski SC, Lehman IR. A single-stranded DNA binding protein from Drosophila melanogaster: characterization of the heterotrimeric protein and its interaction with single-stranded DNA. Biochemistry. 1993;32:5257–5266. doi: 10.1021/bi00070a038. [DOI] [PubMed] [Google Scholar]
- Bluyssen HA, Naus NC, van Os RI, Jaspers I, Hoeijmakers JH, de Klein A. Human and mouse homologs of the Schizosaccharomyces pombe rad17+ cell cycle checkpoint control gene. Genomics. 1999;55:219–228. doi: 10.1006/geno.1998.5642. [DOI] [PubMed] [Google Scholar]
- Kamakaka RT, Kaufman PD, Stillman B, Mitsis PG, Kadonaga JT. Simian virus 40 origin- and T-antigen-dependent DNA replication with Drosophila factors in vitro. Mol Cell Biol. 1994;14:5114–5122. doi: 10.1128/mcb.14.8.5114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller RC, Mossi R, Maga G, Wellinger RE, Hubscher U, Sogo JM. Electron microscopic analysis reveals that replication factor C is sequestered by single-stranded DNA. Nucleic Acids Res. 1999;27:3433–3437. doi: 10.1093/nar/27.17.3433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mossi R, Jonsson ZO, Allen BL, Hardin SH, Hubscher U. Replication factor C interacts with the C-terminal side of proliferating cell nuclear antigen. J Biol Chem. 1997;272:1769–1776. doi: 10.1074/jbc.272.3.1769. [DOI] [PubMed] [Google Scholar]
- Chesnokov I, Gossen M, Remus D, Botchan M. Assembly of functionally active Drosophila origin recognition complex from recombinant proteins. Genes Dev. 1999;13:1289–1296. doi: 10.1101/gad.13.10.1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gossen M, Pak DT, Hansen SK, Acharya JK, Botchan MR. A Drosophila homolog of the yeast origin recognition complex. Science. 1995;270:1674–1677. doi: 10.1126/science.270.5242.1674. [DOI] [PubMed] [Google Scholar]
- Ng L, Prelich G, Anderson CW, Stillman B, Fisher PA. Drosophila proliferating cell nuclear antigen. Structural and functional homology with its mammalian counterpart. J Biol Chem. 1990;265:11948–11954. [PubMed] [Google Scholar]
- Yamaguchi M, Nishida Y, Moriuchi T, Hirose F, Hui CC, Suzuki Y, Matsukage A. Drosophila proliferating cell nuclear antigen (cyclin) gene: structure, expression during development, and specific binding of homeodomain proteins to its 5'-flanking region. Mol Cell Biol. 1990;10:872–879. doi: 10.1128/mcb.10.3.872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson DS, Banga SS, Grigliatti TA, Boyd JB. Mutagen sensitivity and suppression of position-effect variegation result from mutations in mus209, the Drosophila gene encoding PCNA. EMBO J. 1994;13:1450–1459. doi: 10.1002/j.1460-2075.1994.tb06399.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams MD, Celniker SE, Holt RA, Evans CA, Gocayne JD, Amanatides PG, Scherer SE, Li PW, Hoskins RA, Galle RF, George RA, Lewis SE, Richards S, Ashburner M, Henderson SN, Sutton GG, Wortman JR, Yandell MD, Zhang Q, Chen LX, Brandon RC, Rogers YH, Blazej RG, Champe M, Pfeiffer BD, Wan KH, Doyle C, Baxter EG, Helt G, Nelson CR, Gabor GL, Abril JF, Agbayani A, An HJ, Andrews-Pfannkoch C, Baldwin D, Ballew RM, Basu A, Baxendale J, Bayraktaroglu L, Beasley EM, Beeson KY, Benos PV, Berman BP, Bhandari D, Bolshakov S, Borkova D, Botchan MR, Bouck J, Brokstein P, Brottier P, Burtis KC, Busam DA, Butler H, Cadieu E, Center A, Chandra I, Cherry JM, Cawley S, Dahlke C, Davenport LB, Davies P, de Pablos B, Delcher A, Deng Z, Mays AD, Dew I, Dietz SM, Dodson K, Doup LE, Downes M, Dugan-Rocha S, Dunkov BC, Dunn P, Durbin KJ, Evangelista CC, Ferraz C, Ferriera S, Fleischmann W, Fosler C, Gabrielian AE, Garg NS, Gelbart WM, Glasser K, Glodek A, Gong F, Gorrell JH, Gu Z, Guan P, Harris M, Harris NL, Harvey D, Heiman TJ, Hernandez JR, Houck J, Hostin D, Houston KA, Howland TJ, Wei MH, Ibegwam C, Jalali M, Kalush F, Karpen GH, Ke Z, Kennison JA, Ketchum KA, Kimmel BE, Kodira CD, Kraft C, Kravitz S, Kulp D, Lai Z, Lasko P, Lei Y, Levitsky AA, Li J, Li Z, Liang Y, Lin X, Liu X, Mattei B, McIntosh TC, McLeod MP, McPherson D, Merkulov G, Milshina NV, Mobarry C, Morris J, Moshrefi A, Mount SM, Moy M, Murphy B, Murphy L, Muzny DM, Nelson DL, Nelson DR, Nelson KA, Nixon K, Nusskern DR, Pacleb JM, Palazzolo M, Pittman GS, Pan S, Pollard J, Puri V, Reese MG, Reinert K, Remington K, Saunders RD, Scheeler F, Shen H, Shue BC, Siden-Kiamos I, Simpson M, Skupski MP, Smith T, Spier E, Spradling AC, Stapleton M, Strong R, Sun E, Svirskas R, Tector C, Turner R, Venter E, Wang AH, Wang X, Wang ZY, Wassarman DA, Weinstock GM, Weissenbach J, Williams SM, WoodageT. Worley KC, Wu D, Yang S, Yao QA, Ye J, Yeh RF, Zaveri JS, Zhan M, Zhang G, Zhao Q, Zheng L, Zheng XH, Zhong FN, Zhong W, Zhou X, Zhu S, Zhu X, Smith HO, Gibbs RA, Myers EW, Rubin GM, Venter JC. The genome sequence of Drosophila melanogaster. Science. 2000;287:2185–2195. doi: 10.1126/science.287.5461.2185. [DOI] [PubMed] [Google Scholar]
- Prelich G, Kostura M, Marshak DR, Mathews MB, Stillman B. The cell-cycle regulated proliferating cell nuclear antigen is required for SV40 DNA replication in vitro. Nature. 1987;326:471–475. doi: 10.1038/326471a0. [DOI] [PubMed] [Google Scholar]
- Tan CK, Castillo C, So AG, Downey KM. An auxiliary protein for DNA polymerase-delta from fetal calf thymus. J Biol Chem. 1986;261:12310–12316. [PubMed] [Google Scholar]
- Fien K, Stillman B. Identification of replication factor C from Saccharomyces cerevisiae: a component of the leading-strand DNA replication complex. Molecular Cell Biol. 1992;12:155–163. doi: 10.1128/mcb.12.1.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaika A, Mozzherin DJ, Tan CK, Downey KM, Fisher PA. A two-dimensional support for selective binding of polyhistidine-tagged proteins: identification of a proliferating cell nuclear antigen point mutant with altered function in vitro. Anal Biochem. 1999;268:193–200. doi: 10.1006/abio.1998.3074. [DOI] [PubMed] [Google Scholar]
- Mozzherin DJ, McConnell M, Fisher PA. Drosophila replication and repair proteins: proliferating cell nuclear antigen (PCNA) Methods: A Companion to Methods in Enzymology. 1999;18:401–406. doi: 10.1006/meth.1999.0798. [DOI] [PubMed] [Google Scholar]
- Zhang P, Sun Y, Hsu H, Zhang L, Zhang Y, Lee MY. The interdomain connector loop of human PCNA is involved in a direct interaction with human polymerase delta. J Biol Chem. 1998;273:713–719. doi: 10.1074/jbc.273.2.713. [DOI] [PubMed] [Google Scholar]
- Roos G, Jiang Y, Landberg G, Nielsen NH, Zhang P, Lee MY. Determination of the epitope of an inhibitory antibody to proliferating cell nuclear antigen. Exp Cell Res. 1996;226:208–213. doi: 10.1006/excr.1996.0220. [DOI] [PubMed] [Google Scholar]
- Jonsson ZO, Hindges R, Hubscher U. Regulation of DNA replication and repair proteins through interaction with the front side of proliferating cell nuclear antigen. EMBO Journal. 1998;17:2412–2425. doi: 10.1093/emboj/17.8.2412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng L, Tan CK, Downey KM, Fisher PA. Enzymologic mechanism of calf thymus DNA polymerase delta. J Biol Chem. 1991;266:11699–11704. [PubMed] [Google Scholar]
- Chung DW, Zhang JA, Tan CK, Davie EW, So AG, Downey KM. Primary structure of the catalytic subunit of human DNA polymerase delta and chromosomal location of the gene. Proc Natl Acad Sci U S A. 1991;88:11197–11201. doi: 10.1073/pnas.88.24.11197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Tan CK, McMullen B, Downey KM, So AG. Cloning of the cDNAs for the small subunits of bovine and human DNA polymerase delta and chromosomal location of the human gene (POLD2) Genomics. 1995;29:179–186. doi: 10.1006/geno.1995.1229. [DOI] [PubMed] [Google Scholar]
- Zhang J, Chung DW, Tan CK, Downey KM, Davie EW, So AG. Primary structure of the catalytic subunit of calf thymus DNA polymerase delta: sequence similarities with other DNA polymerases. Biochemistry. 1991;30:11742–11750. doi: 10.1021/bi00115a002. [DOI] [PubMed] [Google Scholar]
- Zhang SJ, Zeng XR, Zhang P, Toomey NL, Chuang RY, Chang LS, Lee MY. A conserved region in the amino terminus of DNA polymerase delta is involved in proliferating cell nuclear antigen binding. J Biol Chem. 1995;270:7988–7992. doi: 10.1074/jbc.270.14.7988. [DOI] [PubMed] [Google Scholar]
- Mozzherin DJ, Shibutani S, Tan CK, Downey KM, Fisher PA. Proliferating cell nuclear antigen promotes DNA synthesis past template lesions by mammalian DNA polymerase delta. Proc Natl Acad Sci U S A. 1997;94:6126–6131. doi: 10.1073/pnas.94.12.6126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Day CL, Burgers PMJ, Taylor J-S. PCNA-induced DNA synthesis past cis-syn and trans-syn-l thymine dimers by calf thymus DNA polymerase d in vitro. Nucleic Acids Res. 1992;20:5403–5406. doi: 10.1093/nar/20.20.5403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeshita M, Chang CN, Johnson F, Will S, Grollman AP. Oligodeoxynucleotides containing synthetic abasic sites. Model substrates for DNA polymerases and apurinic/apyrimidinic endonucleases. J Biol Chem. 1987;262:10171–10179. [PubMed] [Google Scholar]
- Ng L, McConnell M, Tan CK, Downey KM, Fisher PA. Interaction of DNA polymerase delta, proliferating cell nuclear antigen, and synthetic oligonucleotide template-primers. Analysis by polyacrylamide gel electrophoresis-band mobility shift assay. J Biol Chem. 1993;268:13571–13576. [PubMed] [Google Scholar]
- Zhang P, Zhang SJ, Zhang Z, Woessner J. F., Jr., Lee MY. Expression and physicochemical characterization of human proliferating cell nuclear antigen. Biochemistry. 1995;34:10703–10712. doi: 10.1021/bi00034a002. [DOI] [PubMed] [Google Scholar]
- Fukuda K, Morioka H, Imajou S, Ikeda S, Ohtuska E, Tsurimoto T. Structure-function relationship of the eukaryotic DNA replication factor, proliferating cell nuclear antigen. Journal of Biological Chemistry. 1995;270:22527–22534. doi: 10.1074/jbc.270.38.22527. [DOI] [PubMed] [Google Scholar]
- Oku T, Ikeda S, Sasaki H, Fukuda K, Morioka H, Ohtsuka E, Yoshikawa H, Tsurimoto T. Functional sites of human PCNA which interact with p21 (Cip1/Waf1), DNA polymerase delta and replication factor C. Genes Cells. 1998;3:357–369. doi: 10.1046/j.1365-2443.1998.00199.x. [DOI] [PubMed] [Google Scholar]
- Ola A, Waga S, Ellison V, Stillman B, McGurk M, Leigh IM, Waseem NH, Waseem A. Human-Saccharomyces cerevisiae proliferating cell nuclear antigen hybrids: oligomeric structure and functional characterization using in vitro DNA replication. J Biol Chem. 2001;276:10168–10177. doi: 10.1074/jbc.M008929200. [DOI] [PubMed] [Google Scholar]
- Henderson DS. DNA repair defects and other mustakes in Drosophila melanogaster. Methods: A Companion to Methods in Enzymology. 1999;18:377–400. doi: 10.1006/meth.1999.0797. [DOI] [PubMed] [Google Scholar]
- Kleczkowska HE, Marra G, Lettieri T, Jiricny J. hMSH3 and hMSH6 interact with PCNA and colocalize with it to replication foci. Genes Dev. 2001;15:724–736. doi: 10.1101/gad.191201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie B, Mazloum N, Liu L, Rahmeh A, Li H, Lee MY. Reconstitution and characterization of the human DNA polymerase delta four-subunit holoenzyme. Biochemistry. 2002;41:13133–13142. doi: 10.1021/bi0262707. [DOI] [PubMed] [Google Scholar]
- Bermudez VP, MacNeill SA, Tappin I, Hurwitz J. The influence of the Cdc27 subunit on the properties of the Schizosaccharomyces pombe DNA polymerase delta. J Biol Chem. 2002;277:36853–36862. doi: 10.1074/jbc.M202897200. [DOI] [PubMed] [Google Scholar]
- Mozzherin DJ, Tan CK, Downey KM, Fisher PA. Architecture of the active DNA polymerase delta.proliferating cell nuclear antigen.template-primer complex. J Biol Chem. 1999;274:19862–19867. doi: 10.1074/jbc.274.28.19862. [DOI] [PubMed] [Google Scholar]
- Zhou JQ, He H, Tan CK, Downey KM, So AG. The small subunit is required for functional interaction of DNA polymerase delta with the proliferating cell nuclear antigen. Nucleic Acids Res. 1997;25:1094–1099. doi: 10.1093/nar/25.6.1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mo J, Liu L, Leon A, Mazloum N, Lee MY. Evidence that DNA polymerase delta isolated by immunoaffinity chromatography exhibits high-molecular weight characteristics and is associated with the KIAA0039 protein and RPA. Biochemistry. 2000;39:7245–7254. doi: 10.1021/bi0000871. [DOI] [PubMed] [Google Scholar]
- Podust VN, Chang LS, Ott R, Dianov GL, Fanning E. Reconstitution of human DNA polymerase delta using recombinant baculoviruses: the p12 subunit potentiates DNA polymerizing activity of the four-subunit enzyme. J Biol Chem. 2002;277:3894–3901. doi: 10.1074/jbc.M109684200. [DOI] [PubMed] [Google Scholar]
- Burgers PM, Gerik KJ. Structure and processivity of two forms of Saccharomyces cerevisiae DNA polymerase delta. J Biol Chem. 1998;273:19756–19762. doi: 10.1074/jbc.273.31.19756. [DOI] [PubMed] [Google Scholar]
- Gerik KJ, Li X, Pautz A, Burgers PM. Characterization of the two small subunits of Saccharomyces cerevisiae DNA polymerase delta. J Biol Chem. 1998;273:19747–19755. doi: 10.1074/jbc.273.31.19747. [DOI] [PubMed] [Google Scholar]
- Joyce CM, Grindley ND. Construction of a plasmid that overproduces the large proteolytic fragment (Klenow fragment) of DNA polymerase I of Escherichia coli. Proc Natl Acad Sci U S A. 1983;80:1830–1834. doi: 10.1073/pnas.80.7.1830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MYWT, Tan C-K, Downey KM, So AG. Further studies on calf thymus DNA polymerase d purified to homogeneity by a new procedure. Biochemistry. 1984;23:1906–1913. doi: 10.1021/bi00304a003. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY, Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- McConnell M, Miller H, Mozzherin DJ, Quamina A, Tan CK, Downey KM, Fisher PA. The mammalian DNA polymerase delta--proliferating cell nuclear antigen--template-primer complex: molecular characterization by direct binding. Biochemistry. 1996;35:8268–8274. doi: 10.1021/bi9530649. [DOI] [PubMed] [Google Scholar]
- Mozzherin DJ, McConnell M, Jasko MV, Krayevsky AA, Tan CK, Downey KM, Fisher PA. Proliferating cell nuclear antigen promotes misincorporation catalyzed by calf thymus DNA polymerase delta. J Biol Chem. 1996;271:31711–31717. doi: 10.1074/jbc.271.49.31711. [DOI] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Fisher PA, Berrios M, Blobel G. Isolation and characterization of a proteinaceous subnuclear fraction composed of nuclear matrix peripheral lamina and nuclear pore complexes from embryos of Drosophila melanogaster. J Cell Biol. 1982;92:674–686. doi: 10.1083/jcb.92.3.674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schagger H, von Jagow G. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal Biochem. 1987;166:368–379. doi: 10.1016/0003-2697(87)90587-2. [DOI] [PubMed] [Google Scholar]
- Harlow E, Lane D. Antibodies: a laboratory manual. Cold Spring Harbor, NY, Cold Spring Harbor Laboratory; 1988. [Google Scholar]
- McGadey J. A tetrazolium method for non-specific alkaline phosphatase. Histochemie. 1970;23:180–184. doi: 10.1007/BF00305851. [DOI] [PubMed] [Google Scholar]
- Blake MS, Johnston KH, Russell-Jones GJ, Gotschlich EC. A rapid sensitive method for detection of alkaline phosphatase conjugated anti-antibody on Western blots. Anal Biochem. 1984;136:175–179. doi: 10.1016/0003-2697(84)90320-8. [DOI] [PubMed] [Google Scholar]
- Smith DE, Fisher PA. Identification developmental regulation and response to heat shock of two antigenically related forms of a major nuclear envelope protein in Drosophila embryos. Application of an improved method for affinity purification of antibodies. J Cell Biol. 1984;99:20–28. doi: 10.1083/jcb.99.1.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F, Nicklen S, Coulson AR. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]