Abstract
We proposed a scan-RNA model for genome rearrangement based on finding small RNAs that hybridized preferentially to micronuclear-specific sequences and on the properties of Twi1p, a PPD protein required for both sequence elimination and small RNA accumulation in Tetrahymena. Here we show that Twi1p interacts with the small RNAs in both the old and the developing macronucleus, and is required for their stability. We show that the specificity of the small RNAs for micronuclear-limited sequences increases during conjugation. These results indicate that the small RNAs observed in conjugating cells have the properties predicted for scan RNAs.
Keywords: Tetrahymena, genome rearrangement, RNAi, PPD protein, small RNA
Like most ciliated protozoans, Tetrahymena thermophila (referred to as Tetrahymena below) have two structurally and functionally different nuclei in a single cell (see Karrer 2000). The diploid, germ-line micronucleus and the polyploid, somatic macronucleus are derived from the same zygotic nucleus formed by fertilization of two micronucleus-derived, haploid, meiotic nuclei during the sexual process of conjugation. Concomitant with formation of a new macronucleus during conjugation, the old macronucleus is destroyed. Most, if not all, transcription required for vegetative cell growth occurs in the macronucleus.
Tetrahymena undergoes extensive programmed genome rearrangement during conjugation (see Yao et al. 2002) resulting in elimination of ∼15% of the genome. About 6000 internal eliminated sequences (IESs), varying from 0.5 to >20 kb in length, are eliminated, and flanking, macronucleus-destined sequences are ligated during macronuclear development in Tetrahymena. The precise ends of IESs can occur reproducibly at a specific site or at a limited number of alternative sites. Because no obvious consensus sequence had been found in and around IESs, it was not clear how IESs were precisely recognized until recently. We and others showed that an RNAi-related mechanism was involved in the IES elimination in Tetrahymena (Mochizuki et al. 2002; Yao et al. 2003). A PPD (PAZ-Piwi Domain) protein, Twi1p, was required for IES elimination, and for accumulation of the siRNA-like small RNAs homologous to micronuclear-specific (largely IES) sequences. Twi1p was detected only during conjugation and accumulated first in the cytoplasm, then in parental (old) macronuclei and then relocalized to developing (new) macronuclei.
Based on the results above, on the presence of micronuclear transcripts from both strands of the IESs in conjugating cells (Chalker and Yao 2001), and on the demonstration that sequences in the old macronucleus could epigenetically affect IES elimination (Chalker and Yao 1996), we proposed a model to explain how IESs, lacking any consensus sequences, are recognized during macronuclear development (Mochizuki et al. 2002; Mochizuki and Gorovsky 2004). First, the micronuclear genome is transcribed bidirectionally to make double-stranded (ds) RNAs that are processed to small RNAs by a Dicer-related RNase. We named these hypothetical small RNAs scan (scn) RNAs. We proposed that the scnRNAs are localized first to the old macronucleus, along with Twi1p, and that the scnRNAs homologous to any DNA found there are degraded. We hypothesized that the remaining (i.e., micronucleus-specific) scnRNAs, still complexed with Twi1p, are then transferred to the new macronucleus, where they promote elimination of the IES sequences to which they are homologous.
Here we test three critical predictions of the scnRNA hypothesis: (1) that the small RNAs are associated with Twi1p; (2) that the small RNAs are transferred from old to new macronuclei; and (3) that macronuclear-specific sequences are eliminated from the small RNA population as conjugation proceeds. We show that a significant fraction of the ∼28-nt RNA can be coimmunoprecipitated with Twi1p when this PPD protein is localized in either the old or the new macronucleus. Thus, much of the small RNAs is associated with Twi1p and probably is transferred with it from the old to the new macronucleus. We also show that small RNAs present early in conjugation are much less specific for micronuclear DNA than for macronuclear DNA, suggesting that the enrichment for IES sequences predicted by the scnRNA hypothesis actually occurs. These results argue strongly that the small, ∼28-nt RNAs specifically expressed during conjugation have the predicted functions of the hypothetical scnRNAs.
Results and Discussion
Expression of small RNA is greatly reduced but not eliminated in TWI1 knockout cells
Previously we reported that small RNAs specifically expressed during conjugation in wild-type cells could not be detected in TWI1 (somatic) knockout cells in which all somatic (macronuclear) copies of TWI1 were replaced with a drug resistance marker (Mochizuki et al. 2002). However, low and asynchronous mating of these TWI1 somatic knockout cells prevented us from determining whether the small RNAs were absent or just greatly reduced in the absence of TWI1. Although the cause of this low and asynchronous mating was not clear, we suspected that senescence (Nanney 1974) or cellular damage occurred during the long period (∼40 d = 300∼400 generations) of culture and treatment with high concentration of the drug (up to 50 mg/mL of paromomycin sulfate) required for phenotypic assortment to produce somatic knockouts. To avoid these problems, we used a different strategy to make TWI1 knockout cells. One of the two copies of the TWI1 genes in the diploid micronucleus was replaced by the drug resistance marker and homozygous micronuclear knockout strains were made by uniparental pronuclear transfer during conjugation (Hai et al. 2000). Cells created by this process are homozygous TWI1 germ-line knockout heterokaryons, homozygous for disrupted TWI1 genes in their micronucleus and containing wild-type TWI1 genes in their macronucleus (see Fig. 1A). Two homozygous heterokaryon strains were crossed and homozygous TWI1 knockout homokaryon progeny cells, referred to as ΔTWI1 cells were obtained. These cells contained disrupted TWI1 genes in both macro- and micronuclei and have not been subjected to long periods of drug selection.
Figure 1.
Construction and characterization of TWI1 germ-line knockout strains and Flag-TWI1 strains. (A) Genotyping of TWI1 germ-line knockout strains and Flag-TWI1 strains. The genotypes of the TWI1 loci in macro- and micronuclei of each cell type are illustrated at the top. TWI1 and its targeted loci are schematically drawn on the left. Arrows indicate the locations of primers used for genotyping PCR. (Lanes O1,O2) Some homozygous TWI1 knockout homokaryon strains had deletions in the neo3 cassette and gave both intact (4.3 kb) and deleted (∼2.5 kb) products. (Lanes O1′,O2′) Because macronuclear chromosomes are randomly segregated when macronuclei divide, intact targeted loci can be sorted out and knockout cells containing only deleted targeted loci can be obtained. (Lane F) The Flag-TWI1 strain was constructed in a homozygous TWI1 knockout homokaryon background and complete disruption of endogenous TWI1 loci in this strain was confirmed. Note that the region corresponding to the downstream primer was absent in pD5H8-Flag-TWI1, and thus the Flag-TWI1 gene could not be detected by PCR. (B) IES elimination assay. Double horizontal lines indicate DNA retained in the macronucleus and the filled box indicates an IES. Four primers (arrows on the horizontal lines) were used for nested PCR. The sizes of the processed (macronuclear form) and unprocessed (micronuclear form) of the M region are marked by arrowheads with “a” and “i”, respectively. (M) Molecular weight marker. (C) Expression of small RNA in wild-type and ΔTWI1 homozygous homokaryon cells. Total RNA was extracted from mating wild-type or ΔTWI1 homozygous homokaryon cells at 0, 4, 8, and 12 h postmixing and the RNA corresponding to 5 × 104 cells was analyzed. The RNA was fractionated in 12% acrylamide-urea gels and stained with ethidium bromide. In vitro transcribed, 17-, 26-, and 51-nt RNAs were used as markers (M). The gel was partially destained and tRNAs amounts were observed to normalize the loading. The amount of the small RNAs in wild-type cells at 4 h postmixing was adjusted as 1.00 and the relative ratio of the small RNAs at other times is indicated at the bottom. Total RNA was used for Northern hybridization with a probe for PDD1 (631–943 bp, GenBank TTU66363) mRNA which is specifically expressed in conjugating cells.
Complete replacement of TWI1 loci with the drug resistance markers was confirmed by PCR analyses (Fig. 1A). In contrast to the original somatic knockout strains, two ΔTWI1 strains mated almost normally and without delay (data not shown) and expressed the conjugation-specific PDD1 gene normally (see Fig. 1C). Thus, the low and asynchronous matings of TWI1 somatic knockout strains were not due to the absence of TWI1. As seen for somatic TWI1 knockouts (Mochizuki et al. 2002), when two ΔTWI1 strains were mated, no progeny grew upon refeeding of >10 million progeny tested (see Materials and methods). Again, like TWI1 somatic knockout cells, elimination of the M-region IES (Austerberry and Yao 1987) in the progeny of the ΔTWI1 strains also failed to occur (Fig. 1B), confirming that TWI1 is required for genome rearrangement.
Next, total RNA was extracted from mating wild-type or ΔTWI1 cells at different stages of conjugation and analyzed by acrylamide-urea gel electrophoresis (Fig. 1C). The small RNAs could be detected in ΔTWI1 cells although the amount was greatly reduced compared with wild-type cells. In early conjugation (4 h), the amount of the small RNA in TWI1 knockout cells was about one-fourth of that of wild-type cells but these RNAs did not persist into mid- to late stages of conjugation (Fig. 1C). These results suggest that TWI1 is dispensable for production of the small RNAs but is required for their stability. This view is supported by two additional observations. First, a Dicer-like protein (Dcl1p), required for small RNA production, is localized in the meiotic micronucleus (K. Mochizuki and M.A. Gorovsky, unpubl.) but localization of Twi1p in the micronucleus was not observed (Mochizuki et al. 2002). Second, Twi1p and the small RNAs are associated with each other in early to late stages of conjugation (see Fig. 2). These results indicate that formation of the small RNAs in the micronucleus precedes and is independent of its association with Twi1p. Because IES elimination was not observed in the progeny of TWI1 germ-line knockout cells (Fig. 1B), it is likely that either the amount of the small RNAs detected in ΔTWI1 cells is insufficient to function in genome rearrangement or that the small RNAs can not act in genome rearrangement without Twi1p.
Figure 2.
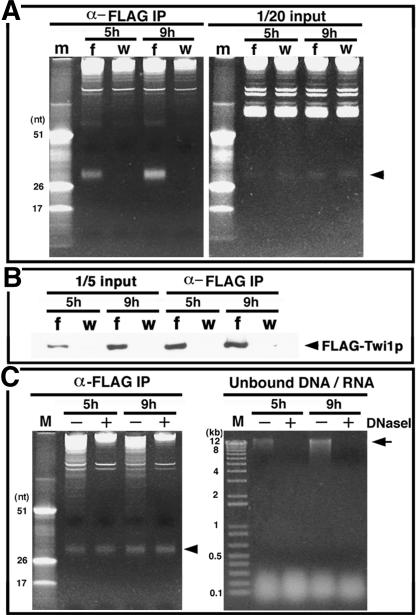
Coimmunoprecipitation of small RNAs with Flag-Twi1p. (A) Whole cell lysates were prepared from the mating of Flag-TWI1 × CU428 (f) or B2086 × CU428 (w) and the Flag-Twi1p-containing complex was immunoprecipitated using anti-Flag antibody. RNA extracted from one-twentieth of the lysates (right panel) and from the fraction eluted from the immunoprecipitate with Flag peptide (left panel) was analyzed by 12% acrylamide-urea gel electrophoresis followed by ethidium bromide staining. The marker (m) was the same as in Figure 1. The arrowhead shows the position of the small RNAs. (B) Flag-Twi1p in the elution (right) and the lysate (left, corresponding to one-fifth of total protein used for immunoprecipitation), was analyzed on Western blot using anti-Flag antibody. (C, Left) Immunoprecipitation was performed with (+) or without (-) DNase treatment and RNA was analyzed. The arrowhead shows the position of the small RNA. (Right) To confirm that genomic DNA in lysates had been digested, nucleic acids were extracted from the unbound fraction after immunoprecipitation with phenol/chloroform and fractionated in a 1% agarose gel followed by ethidium bromide staining. The arrow indicates the position of high-molecular-weight (genomic) DNA.
Twi1p interacts with small RNA
Previously, we proposed that the small RNAs travel from old to new macronuclei in association with Twi1p (Mochizuki et al. 2002). However, there was no direct evidence for the association of the small RNAs with Twi1p or for their transfer. Thus, we wished to determine if Twi1p interacts with the small RNAs in old and new macronuclei. To address this, we constructed strains expressing Flag-tagged Twi1p. The Flag-tag-encoding sequence was added just after the methionine initiation codon of TWI1 and a genomic DNA sequence including the whole coding and the promoter region driving TWI1 transcription was introduced into the transformation vector, pD5H8 (Godiska et al. 1993), resulting in pD5H8-Flag-TWI1. Two homozygous TWI1 germ-line knockout heterokaryons were crossed and their progeny were transformed with pD5H8-Flag-TWI1. In the transformed progeny (referred to as Flag-TWI1 strains), all endogenous TWI1 loci were disrupted in both macro- and micronuclei (confirmed by PCR, see Fig. 1A) and Flag-TWI1 expressed from pD5H8-Flag-TWI1 was the only source of TWI1 in these strains. As shown in Table 1, Flag-TWI1 strains, as well as homozygous TWI1 germ-line knockout heterokaryon strains, produced viable progeny when mated with ΔTWI1 cells. By contrast, as described above, two ΔTWI1 strains could not produce viable progeny (Table 1). Thus, Flag-Twi1p expressed from pD5H8-Flag-TWI1 can replace the essential function of Twi1p during conjugation, arguing that it retains normal physical interactions with other molecules required for the function of Twi1p.
Table 1.
Viability of the progeny
| Types of mating cells | Pairs tested | Progeny |
|---|---|---|
| heΔTWII-F3-1 × heΔTWII-C4-4-3 | 288 | 30 |
| ΔTWII-5-1 × heΔTWII-F3-1 | 288 | 26 |
| ΔTWII-5-1 × ΔTWII-WG7-1 | 287 | 0 |
| ΔTWII-5-1 × FLAG-TWII-18 | 288 | 35 |
At ∼10 h postmixing, single mating pairs were placed into drops of SPP medium to allow postconjugation growth. Completion of conjugation was confirmed by testing for expression of a drug resistance marker specific either for parental macronuclei or newly developed macronuclei (see Materials and Methods).
Next, a Flag-TWI1 strain was crossed with a wild-type strain and a whole cell lysate was prepared from the mating cells at 5 h (when Twi1p is localized mainly in the old macronucleus) and 9 h (when it is mainly in the new macronucleus) postmixing. Anti-Flag antibody was used to immunoprecipitate Flag-Twi1p and molecules associated with it and the immunoprecipitated components were eluted. RNA was extracted from the eluted products and analyzed by acrylamide-urea gel electrophoresis followed by ethidium bromide staining. As a control, two wild-type strains were crossed and processed in parallel. The ∼28-nt small RNA coimmunoprecipitated with Flag-Twi1p both in 5 h and 9 h postmixing samples (Fig. 2A). It was estimated that ∼39% (5 h) and 41% (9 h) of the small RNA in the original lysate was coimmunoprecipitated. In contrast, when two wild-type strains were crossed, the small RNAs were not precipitated. Based on the Western blot (Fig. 2B), it was estimated that ∼50% (5 h) and 41% (9 h) of Flag-Twi1p in the original lysate were collected in the elution. Assuming that unimmunoprecipitated Flag-Twi1p was also complexed with the small RNA in the same manner as the precipitated Flag-Twi1p, we calculate that 78% (5 h) and almost 100% (9 h) of the small RNAs in the cell were complexed with Twi1p. These numbers were unexpectedly high because wild-type, nontagged Twi1p expressed from the wild-type partner, also should be associated with the small RNAs, should not be immunoprecipitated in this experiment. Two explanations are possible for this unexpected result. First, because Flag-Twi1p was overexpressed from the rDNA based pD5H8 vector, the majority of Twi1p in mating cells should be Flag-tagged. Alternatively, Twi1p–small RNA complexes could contain multiple Twi1p molecules. We repeated this experiment, using mating of ΔTWI1 and Flag-TWI1 strains. In this case, all Twi1p molecules should be tagged. We also ran quantitative standards to ensure that the measured signals for small RNAs and for Twi1p were in the linear range of quantification. In this experiment, ∼27% (5 h) and 51% (9 h) of the small RNA and 41% (5 h) and 52% (9 h) of Flag-Twi1p in the original lysate was coimmunoprecipitated (data not shown). Again, if immunoprecipitated and unimmunoprecipitated Flag-Twi1p are similarly complexed with the small RNA, it is estimated that ∼66% (5 h) and 97% (9 h) of the small RNAs were in the Twi1p complex.
Because Twi1p and the small RNAs were proposed to function in genome rearrangements controlled by DNA–RNA interactions (Mochizuki et al. 2002), it was possible that the association of Twi1p and the small RNAs was bridged by DNA. To test this, immunoprecipitation was performed with or without DNase (Fig. 2C) treatment. Similar amounts of small RNAs were precipitated in the presence or absence of DNase, suggesting that the interaction of Twi1p and the small RNAs was not mediated by DNA.
As noted, at 5 and 9 h postmixing, the most Twi1p is present in old and new macronuclei, respectively (Mochizuki et al. 2002). Thus, a large fraction of the small RNAs is likely to be associated with Twi1p, and travel with it from old to new macronuclei, as predicted.
In the experiments described here, we did not detect any proteins that interacted with Flag-Twi1p by silver staining although Flag-Twi1p was clearly detected (data not shown). In addition the PAZ domains of five different PPD proteins in fly and human have been shown to interact directly with RNA in vitro (Lingel et al. 2003; Song et al. 2003; Yan et al. 2003). Thus, Twi1p probably interacts with the small RNA directly.
Small RNAs homologous to micronucleus-specific sequences are gradually enriched as conjugation proceeds
The scnRNA model hypothesizes that the whole micronuclear genome is transcribed bidirectionally and processed to the small RNAs, and that the small RNAs then localize to the old macronucleus where those homologous to the macronuclear DNA are degraded (Mochizuki et al. 2002). If this is correct, small RNAs homologous to micronuclear-specific sequences should be enriched as conjugation proceeds. To test this prediction, purified macro- and micronuclear DNAs were probed with radioactively labeled small RNAs purified from conjugating cells at different time points. When small RNAs isolated at 2 h postmixing were used as probe, hybridization to micronuclear DNA was approximately three times greater than to macronuclear DNA (Fig. 3). This ratio gradually increased to >20 times at 8 h postmixing, when macronuclear development starts (Fig. 3). These results argue that small RNAs homologous to micronucleus-specific DNA sequences are enriched from early to midconjugation. The observation that small RNAs isolated even at early stages (2 h postmixing) of conjugation already hybridize approximately three times more to micronuclear DNA than to macronuclear DNA has four possible explantations. First, micronuclear IESs might be preferentially transcribed in early conjugation. Second, the whole micronuclear genome could be transcribed but the micronuclear sequences, many of which are repeated, might preferentially form the dsRNA substrate required for production of scnRNAs. Third, “selection” might occur so rapidly that some of small RNAs corresponding to the macronuclear sequences were already eliminated by 2 h. Fourth, it is possible that the repeated nature of many IESs causes their transcripts to be enriched and their concentration on the blot to be high, favoring their detection. Regardless of the explanation, it is clear that the specificity of the small RNAs for micronuclear sequences increases dramatically from early to mid-conjugation. At later stages (10 and 12 h postmixing), the ratio decreased (Fig. 3). We speculate that small RNAs are continuously produced until late stages of conjugation (either by transcription or by amplification by RNA-dependent RNA polymerase [RdRP]) but, after new macronuclear development and old macronuclear degradation are initiated, the small RNAs cannot enter the “selection pathway” that eliminates small RNAs homologous to macronuclear sequences. At present, the detailed mechanism of “selective enrichment” of IES-derived sequences is not clear. We previously proposed that a RNaseH-like activity, able to degrade RNA in RNA–DNA hybrid, may be involved in degradation of small RNAs homologous to macronucleus-destined sequences. Alternatively, because the absence of Twi1p prevents accumulation of the small RNAs (Fig. 1C), it is possible that DNA–RNA pairing displaces Twi1p from the small RNA–Tw1ip complex, allowing the small RNA to be degraded by a nonspecific RNase. Selective amplification, but not degradation, of small RNAs homologous to the micronucleus-specific DNA by RdRP could occur.
Figure 3.
Hybridization of macro- and micronuclear DNA with labeled small RNAs. (A) Macronuclear (a) or micronuclear (i) DNA was isolated and digested with EcoRI. The DNA was separated in a 1% agarose gel and transferred to a membrane. To prepare the probe, total RNA from wild-type conjugating cells at 2, 4, 6, 8, 10, and 12 h postmixing was fractionated in a 12% acrylamide-urea gel and the small RNA was extracted from the gel. The purified small RNA was end-labeled with 32P and hybridized to macro- and micronuclear DNAs on a Southern blot. As a loading control, a probe for the HHO1 gene encoding the histone H1 was hybridized to the same blot. Note that approximately three times more DNA was loaded on lanes containing macronuclear DNA than on micronuclear DNA lanes. (B) The ratio of the small RNAs hybridized to micronuclear DNA versus macronuclear DNA from different time points is shown as a bar graph. The loading was normalized by using the signal obtained with the HHO1 probe.
The small RNAs act as scnRNAs
In the present study, we show that Twi1p is associated with ∼28-nt small RNA in both old and new macronuclei (Fig. 2) and is probably required for its stability (Fig. 1C). Thus, Twi1p might function as a molecular “chaperone” of the small RNAs. Because, Twi1p relocalizes from old to new macronuclei (Mochizuki et al. 2002) and a large fraction of the small RNAs is associated with Twi1p, the movement of the small RNAs from old to new macronuclei also is likely to occur. The increase in specificity of small RNAs for micronuclear sequences during conjugation (Fig. 3), also argues for the existence of a post-transcriptional mechanism that selects the small RNAs homologous to the micronuclear-specific sequences. These results argue that the small RNAs observed in conjugating Tetrahymena cells have functions like those hypothesized for scnRNAs (Mochizuki et al. 2002; Mochizuki and Gorovsky 2004). Thus, we propose to name the ∼28-nt small RNAs specifically expressed during conjugation in Tetrahymena scnRNAs.
Materials and methods
Strains and culture conditions
Wild-type B2086 and CU428 strains of Tetrahymena thermophila (provided by Dr. P.J. Bruns, Cornell University, Ithaca, NY) were grown in SPP medium (Gorovsky et al. 1975) at 30°C. For conjugation, logarithmically growing cells of different mating types were washed, starved (16–24 h at 30°C), and mixed in 10 mM Tris (pH 7.5).
Construction of TWI1 germ-line knockout
The targeting construct, with the whole TWI1 coding sequence replaced by the neo3 cassette conferring paromomycin (pm) resistance, was described previously (Mochizuki et al. 2002). B2086 and CU428 cells were mated and the targeting construct was introduced at 2.5–3.5 h postmixing as described (Cassidy-Hanley et al. 1997). Two transformants that had neo3 in the TWI1 loci of both the macro- and the micronucleus were obtained. The TWI1 disrupted loci in the macronucleus were eliminated by phenotypic assortment in the absence of selection. These heterozygous transformants were crossed with “star” strain B*VII to make homozygous germ-line knockout heterokaryon strains (heΔTWI1-C4-4-3 and heΔTWI1-F3-1) by uniparental micronuclear transfer (Hai et al. 2000). Then two homozygous germ-line knockout heterokaryons were crossed to obtain homozygous homokaryon strains, referred to as ΔTWI1. Complete elimination of TWI1 genes was confirmed by PCR. In some ΔTWI1 strains, the neo3 cassette was partially eliminated from the macronucleus probably by a mechanism similar to that used to eliminate IESs (Yao et al. 2003). The remaining neo3 cassettes were eliminated by phenotypic assortment to make pm-sensitive ΔTWI1 strains (ΔTWI1-5-1 and ΔTWI1-WG7-1). These strains can be distinguished from their progeny in the viability test and IES elimination analyses by their drug sensitivity.
Construction of Flag-TWI1 strain
Flag-encoding sequence was added immediately after the initiation codon of the TWI1 gene by overlapping PCR. The primers used for the PCR were 5′-TWI1–NotI, 5′-CAGCGGCCGCCGATCCTTTCTCTAT GTGTCCAC-3′; Flag-TWI1-RV, 5′-CTTATCGTCGTCATCCTTGTAAT CcatGGATGTTAATTATATCGCTT-3′; Flag-TWI1-FW, 5′-GATTACA AGGATGACGACGATAAGTCTAACAAAGGCCTTGTCTA-3′; 3′-TWI1–NotI, 5′-CAGCGGCCGCACATAAAACGATAGCTAT-3′. NotI site is underlined, Flag-encoding sequence is in italics, and the TWI1 start codon is in lower case. 5′-TWI1 and Flag-TWI1 RV were used to amplify ∼800 bp upstream from 5′UTR, 5′UTR, and the codon for the first methionine followed by the Flag sequence. Also, Flag-TWI1 FW and 3′-TWI1 were used to amplify the Flag-encoding sequence followed by the second amino acid coding codon, the rest of the coding sequence, 3′UTR and ∼1.1 kbp 3′ of noncoding sequence. Then these two fragments were combined by overlapping PCR using primers 5′-TWI1 and 3′-TWI1. The amplified product was digested with NotI and inserted into the NotI site of the ribosomal DNA vector, pD5H8 (provided by Dr. M.C. Yao, Fred Hutchinson Cancer Research Center, Seattle, WA; Godiska et al. 1993), resulting pFlag-TWI1. pD5H8 confers pm resistance in Tetrahymane cells.
Homozygous germ-line knockout heterokaryon strains heΔTWI1C4-4-3 and heΔTWI1F3-1 were mated and pFlag-TWI1 was introduced at 10 h postmixing as above. Cells were incubated in 10 mM Tris (pH 7.5) for 24 h at 30°C and then refed by adding an equal amount of 2× SPP. One hundred micrograms per milliliter of pm (paromomycin sulfate, Sigma) was added 3 h after refeeding and cells were aliquoted to the microtiter plates. The cells were incubated until resistant clones were grown up. Although the progeny of heΔTWI1C4-4-3 and heΔTWI1F3-1 had pm resistance genes in the disrupted TWI1 loci, these were driven by the cadmium inducible MTT1 promoter and thus were not expressed in the absence of Cd2+ in the medium. To select cells with increasing numbers of the pFlag-TWI1-derived rDNA, the transformants were subjected to step-wise selection in increasing concentrations of pm, starting from 100 μg/mL to a final of 3–4 mg/mL, above which the cells failed to grow. One of these strains ΔTWI1-Flag-TWI1-18 was used for further experiments.
Viability test
To test the viability of the progeny of ΔTWI1 cells, starved ΔTWI1-5-1 and ΔTWI1-WG7-1 cells (1 × 107 cells each) were mixed to initiate mating and refed at 24 h postmixing by adding an equal amount of 2× SPP. CdCl2 was added (0.5 μg/mL final) at 3 h after refeeding and cells were cultured for 1 h. Then CdCl2 (to 1 μg/mL total) and pm (120 μg/mL final) were added and the culture was aliquoted to microtiter plates. Microscopic analyses indicated that ∼65% of the cells were exconjugants at 24 h postmixing. No pm resistant progeny were obtained. Thus, the viability was estimated as less than one out of 1.3 × 107.
To determine the functionality of Flag-Twi1p, ΔTWI1-Flag-TWI1-18 and ΔTWI1-5-1 were crossed, individual pairs were placed into SPP drops at 8–10 h postmixing, and cultured at 30°C. At 48–60 h after cloning, drops were examined for growth of cells. Then, the phenotypes of growing cells were tested to determine whether cells had completed or aborted conjugation. These cells were incubated with 120 μg/ml pm with or without 1 μg/mL CdCl2. Clones that showed pm resistance with CdCl2 but were sensitive in its absence were scored as progeny. This assay depends on the fact that ΔTWI1-Flag-TWI1-18 had pD5H8 derived rDNA gene that conferred pm resistance in Tetrahymena without CdCl2 in the macronucleus while both ΔTWI1-Flag-TWI1-18 and ΔTWI1-5-1 had neo3 that conferred pm resistance in Tetrahymena with CdCl2 in the micronucleus. As controls, conjugations of ΔTWI1-5-1 × heΔTWI1-F3-1 or ΔTWI1-5-1 × ΔTWI1-WG7-1 were also tested. To determine the completion of conjugation in these control experiment, cells were cultured in 1× SPP including 120 μg/mL pm and 1 μg/mL CdCl2 and scored for cells resistant to pm.
Small RNA and IES elimination analysis
Total RNA extraction and analysis of the small RNAs were performed as described (Mochizuki et al. 2002). IES elimination was analyzed as described (Mochizuki et al. 2002) except cells were analyzed at 48 h post-mixing.
Coimmunoprecipitation
CU428 or ΔTWI1-WG7-1 and ΔTWI1-Flag-TWI1-18 (2 × 105cells/mL of each) were mated. At 5 and 9 h postmixing, 2 × 106 cells were collected and homogenized in 1 mL lysis solution (50 mM Tris at pH 7.5, 150 mM NaCl, 2 mM MgCl2, 1% Tween 20, 1× Complete Proteinase Inhibitor (Roche), 2 mM phenyl methyl sulfonylfluoride (Roche), 0.2 mg/mL Ribonucleoside Vanadyl Complex (New England Biolabs). Insoluble materials were sedimented at 10,000 × g for 15 min and the supernatant was used for immunoprecipitation. Anti-Flag M2-agarose affinity gel (Sigma) was mixed with the supernatant to precipitate the Flag-Twi1p complex. After overnight incubation at 4°C, the gel was rinsed in wash buffer (50 mM Tris at pH 7.5, 150 mM NaCl, 2 mM MgCl2) and the complex was eluted in 150 μg/mL 3× Flag peptide (Sigma) in wash buffer. Nucleic acids were extracted from the eluted fraction with phenol/chloroform and then ethanol precipitated. The small RNAs were analyzed as described (Mochizuki et al. 2002). The intensities of bands were measured using NIH Image version 1.59. For DNase treatment, 0.1 mg/mL of DNase I (RNase free, Sigma) was added to the initial lysate. After immunoprecipitation, nucleic acid was extracted from the unbound fraction and analyzed on 1% agarose gels to check DNA digestion.
Southern hybridization with labeled small RNAs
The small RNAs were purified from acrylamide urea gels and end-labeled as described (Mochizuki et al. 2002). Southern hybridization to isolated macro- and micronuclear DNA was performed as described (Mochizuki et al. 2002). To normalize the loading of DNA, the membranes were stripped and reprobed with HHO1 probe (random primed DNA probe, 2.4-kb EcoRI–HindIII fragment of pTtDR1, Wu et al. 1986). The radioactivity on the blot was analyzed using a PhosphorImager (Molecular Dynamics/Amersham).
Acknowledgments
We thank Josephine Bowen for critical reading of the manuscript. This work was supported by grant GM21793 from the National Institutes of Health.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article published online ahead of print. Article and publication date are at http://www.genesdev.org/cgi/doi/10.1101/gad.1219904.
References
- Austerberry C.F. and Yao, M.C. 1987. Sequence structures of two developmetally regulated, alternative DNA deletion junctions in Tetrahymena thermophila. Mol. Cell. Biol. 8: 3947-3950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassidy-Hanley D., Bowen, J., Lee, J.H., Cole, E., VerPlank, L.A., Gaertig, J., Gorovsky, M.A., and Bruns, P.J. 1997. Germline and somatic transformation of mating Tetrahymena thermophila by particle bombardment. Genetics 146: 135-147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalker D.L. and Yao, M.C. 1996. Non-Mendelian, heritable blocks to DNA rearrangement are induced by loading the somatic nucleus of Tetrahymena thermophila with germ line-limited DNA. Mol. Cell. Biol. 16: 3658-3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ____. 2001. Nongenic, bidirectional transcription precedes and may promote developmental DNA deletion in Tetrahymena thermophila. Genes & Dev. 15: 1287-1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godiska R., James, C., and Yao, M.C. 1993. A distant 10-bp sequence specifies the boundaries of a programmed DNA deletion in Tetrahymena. Genes & Dev. 7: 2357-2365. [DOI] [PubMed] [Google Scholar]
- Gorovsky M.A., Yao, M.C., Keevert, J.B., and Pleger, G.L. 1975. Isolation of micro- and macronuclei of Tetrahymena pyriformis. Meth. Cell Biol. 9: 311-327. [DOI] [PubMed] [Google Scholar]
- Hai B., Gaertig, J., and Gorovsky, M.A. 2000. Knockout heterokaryons enable facile mutagenic analysis of essential genes in Tetrahymena. Meth. Cell Biol. 62: 513-531. [DOI] [PubMed] [Google Scholar]
- Karrer K.M. 2000. Tetrahymena genetics: Two nuclei are better than one. Meth. Cell Biol. 62: 127-186. [DOI] [PubMed] [Google Scholar]
- Lingel A., Simon, B., Izaurralde, E., and Sattler, M. 2003. Structure and nucleic-acid binding of the Drosophila Argonaute 2 PAZ domain. Nature 426: 465-469. [DOI] [PubMed] [Google Scholar]
- Mochizuki K. and Gorovsky, M.A. 2004. Small RNAs in genome rearrangement in Tetrahymena. Curr. Opin. Genet. Dev. 14: 181-187. [DOI] [PubMed] [Google Scholar]
- Mochizuki K., Fine, N.A., Fujisawa, T., and Gorovsky, M.A. 2002. Analysis of a piwi-related gene implicates small RNAs in genome rearrangement in Tetrahymena. Cell 110: 689-699. [DOI] [PubMed] [Google Scholar]
- Nanney D.L. 1974. Aging and long-term temporal regulation in ciliated protozoa. A critical review. Mech. Ageing Dev. 3: 81-105. [DOI] [PubMed] [Google Scholar]
- Song J.J., Liu, J., Tolia, N.H., Schneiderman, J., Smith, S.K., Martienssen, R.A., Hannon, G.J., and Joshua-Tor, L. 2003. The crystal structure of the Argonaute2 PAZ domain reveals an RNA binding motif in RNAi effector complexes. Nat. Struct. Biol. 10: 1026-1032. [DOI] [PubMed] [Google Scholar]
- Wu M., Allis, C.D., Richman, R., Cook, R.G., and Gorovsky, M.A. 1986. An intervening sequence in an unusual histone H1 gene of Tetrahymena thermophila. Proc. Natl. Acad. Sci. 83: 8674-8678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan K.S., Yan, S., Farooq, A., Han, A., Zeng, L., and Zhou, M.M. 2003. Structure and conserved RNA binding of the PAZ domain. Nature 426: 468-474. [DOI] [PubMed] [Google Scholar]
- Yao M.C., Duharcourt, S., and Chalker, D.L. 2002. Genome-wide rear-rangements of DNA in ciliates. In Mobile DNA II (eds. N. Craig et al.), pp. 730-758. Academic Press, New York.
- Yao M.C., Fuller, P., and Xi, X. 2003. Programmed DNA deletion as an RNA-guided system of genome defense. Science 300: 1581-1584. [DOI] [PubMed] [Google Scholar]