Abstract
Different RNA species are exported from the nucleus by distinct mechanisms. Among the different RNAs, mRNAs and major spliceosomal U snRNAs share several structural similarities, yet they are exported by distinct factors. We previously showed that U1 snRNAs behaved like an mRNA in nuclear export if various ∼300-nucleotide fragments were inserted in a central position. Here we show that this export switch is dependent on the length of the insertion but independent of its position, indicating unequivocally that this switch is indeed the result of RNA length. We also show that intronless mRNAs can be progressively converted to use the U snRNA export pathway if the mRNAs are progressively shortened by deletion. In addition, immunoprecipitation experiments show that the protein composition of export RNPs is influenced by RNA length. These findings indicate that RNA length is one of the key determinants of the choice of RNA export pathway. Based on these results and previous observations, a unified model of how an RNA is committed to a specific export pathway is proposed.
Keywords: RNA export, mRNA, U snRNA, RNA length
The nuclear envelope separates the eukaryotic cells into two major compartments, the nucleus and the cytoplasm. This separation necessitates transport through nuclear pore complexes (NPCs), which form gateways across the nuclear envelope barrier (Macara 2001; Lei and Silver 2002; Suntharalingam and Wente 2003). All the nuclear proteins that are made in the cytoplasm must be imported to the nucleus. In contrast, the vast majority of the RNA species, following their synthesis and processing in the nucleus, are exported to the cytoplasm. It has been shown that different RNA species are exported from the nucleus via distinct export pathways, that is, by distinct sets of export factors (Jarmolowski et al. 1994; Komeili and O'Shea 2001; Cullen 2003). Accumulating evidence shows that the pathway of RNA export can influence the fate of an RNA in the cytoplasm (Ohno et al. 2002; Jin et al. 2003; Wiegand et al. 2003; Kuersten et al. 2004; Nott et al. 2004; Swanson et al. 2004), indicating the biological importance of the choice of RNA export pathway. This means that the cellular export machinery must be able to infallibly discriminate distinct RNA species.
The discrimination mechanism for tRNA export seems simple. The major export factor for tRNAs is Xpo-t (Los1 in yeast), a member of the family of nuclear transport receptors, the prototype of which is importin β. Xpo-t binds directly to tRNAs cooperatively with RanGTP, and this trimeric complex formation facilitates transit of the RNA to the cytoplasm (Arts et al. 1998a; Hellmuth et al. 1998; Kutay et al. 1998). Xpo-t requires major features of the higher-order structure of tRNAs in order to bind them efficiently, and these features are not present in other classes of RNA (Arts et al. 1998b; Lipowsky et al. 1999).
By contrast, how the cellular export machinery can discriminate between U snRNAs and mRNAs is not so easy to understand because these two classes of RNA share several similarities. Both classes of RNA are transcribed by RNA polymerase II, and therefore, both initially acquire a seven-methyl guanosine cap (m7G-cap) structure. In addition, neither class of RNA has conserved RNA sequences or structures, with the exception of the poly(A) tail for mRNAs and the Sm-binding site for U snRNAs. Neither of these two elements is required for nuclear export (Hamm and Mattaj 1990; Jarmolowski et al. 1994). Despite these similarities, export of these two classes of RNA is mediated by different factors.
Major spliceosomal U snRNAs such as U1, U2, U4, and U5 are initially exported from the nucleus in Metazoa (Mattaj 1988; Will and Luhrmann 2001). U snRNA export is mediated by CRM1, another member of the importin β family (Fornerod et al. 1997). CRM1 is also known to be the export receptor for proteins carrying a leucine-rich nuclear export signal or NES (Fischer et al. 1995; Wen et al. 1995; Fornerod et al. 1997; Fukuda et al. 1997; Ossareh-Nazari et al. 1997; Stade et al. 1997). CRM1 binds directly to NES but indirectly to U snRNAs. Two adaptor proteins bridge the interaction between CRM1 and the U snRNAs. One is the heterodimeric cap-binding complex (CBC), which binds specifically to the essential export signal of U snRNAs, the m7G-cap structure (Ohno et al. 1990; Izaurralde et al. 1994, 1995; Kataoka et al. 1994, 1995). The other adaptor is PHAX, which bridges the interaction between CRM1 and the CBC/RNA complex (Ohno et al. 2000). PHAX has a leucine-rich NES to which CRM1 binds cooperatively with RanGTP. In this way these five proteins and a U snRNA assemble into the export complex in the nucleus, and this complex subsequently transits to the cytoplasm.
On the other hand, the major export receptor for mRNAs is a protein called TAP or NXF1 (and its related proteins) in vertebrates and Mex67p in yeast (Segref et al. 1997; Grüter et al. 1998; Katahira et al. 1999). TAP is one of the few non-importin β family transport receptors identified to date. Several RNA-binding proteins are also implicated in mRNA export (Dreyfuss et al. 2002; Stutz and Izaurralde 2003; Dimaano and Ullman 2004), and at least some of them, such as REF/Aly (Yral in yeast), mediate the interaction of TAP with mRNAs (Strässer and Hurt 2000; Stutz et al. 2000; Zhou et al. 2000; Rodrigues et al. 2001; Huang et al. 2003).
In the course of attempts to search for distinguishing features between mRNAs and U snRNAs that can be recognized by the cellular export machinery, we previously found that the presence of introns in mRNA precursors is one such feature. If a pre-mRNA intron was artificially inserted into U1 snRNA, the spliced U1 snRNA was exported via the mRNA export pathway instead of the U snRNA pathway, indicating that introns can function as an identity element for mRNA export (Ohno et al. 2002). It was very likely that this identity element functions through splicing-dependent deposition of a protein complex termed the exon–exon junction complex (EJC), in which mRNA-specific export factor REF is included (Zhou et al. 2000; Kim et al. 2001; Le Hir et al. 2001).
The presence of introns, however, cannot be the only mRNA identity element because there are many mRNAs that do not contain introns. We previously found that if U1 snRNA was elongated by insertion of ∼300-nucleotide (nt) fragments of various sequences, the resultant artificial U1 snRNAs were also exported via the mRNA pathway (Ohno et al. 2002), suggesting that RNA length, sequence, or structure is another distinguishing feature of mRNAs and U snRNAs in nuclear export.
In this study, we have focused on the RNA length issue and obtained several novel findings that unequivocally show that RNA length is an extremely important determinant of RNA export pathway choice. The obtained results have allowed us to propose a unified model of how an RNA is committed to a specific RNA export pathway.
Results
Elongated U1 RNAs behaves like an mRNA in nuclear export.
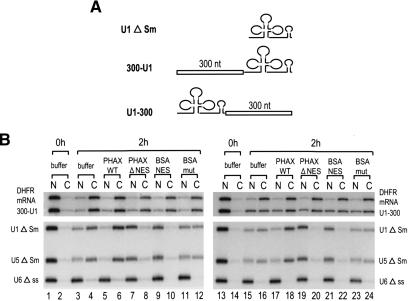
It was previously shown that U1 snRNAs in which ∼300 nt fragments of various sequences had been inserted at the central Sm-site position behaved like an mRNA in export (Ohno et al. 2002). Although this result already suggested that RNA length might be an important determinant of the choice of RNA export pathway, it was still possible that the insertion might have disrupted a hypothetical U snRNA identity element(s) required for committing the RNA to the U snRNA export pathway. To check this possibility, a 300-nt fragment derived from ftz cDNA (see Supplementary Fig. 1) was inserted at two novel positions of U1ΔSm RNA, the very 5′ end (just downstream of the cap structure, Fig. 1A; 300-U1) or the very 3′ end (Fig. 1A; U1-300), and the export pathway of the resultant artificial U1 RNAs was analyzed in Xenopus oocytes (Fig. 1B). We used U1ΔSm RNA as the parental RNA because it has mutations in the Sm-binding site and therefore is defective in reimport into the nucleus (Hamm and Mattaj 1990).
Figure 1.
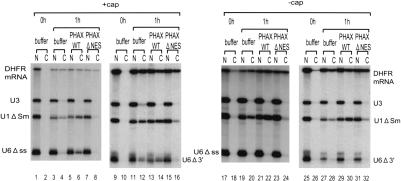
Positional effect of insertion into U1 RNA on RNA export. (A) Schematic representation of U1 RNA derivatives used for the export analysis. A 300-nt sequence from ftz mRNA was inserted at either the 5′ end (300-U1) or the 3′ end (U1-300) of U1 ΔSm RNA. (B) 32P-labeled 300-U1 or U1-300 RNA was microinjected into Xenopus oocyte nuclei together with 32P-labeled DHFR mRNA, U1 ΔSm, U5 ΔSm, and U6 Δss RNAs, with either 0.4 mg/mL PHAX wild-type recombinant protein (PHAX WT; lanes 5,6,17,18), 0.4 mg/mL PHAX ΔNES mutant protein (PHAX ΔNES; lanes 7,8,19,20), 4 mg/mL BSA–NES (lanes 9,10,21,22), or BSA-mut (lanes 11,12,23,24), or without proteins (buffer; lanes 1–4,13–16). All the RNAs were m7G-capped with the exception of U6, which was uncapped. RNA was extracted from nuclear (N) and cytoplasmic (C) fractions, immediately (0 h; lanes 1,2,13,14) or 2 h (2 h; lanes 3–12,15–24) after injection and analyzed by 8% denaturing PAGE.
Two different mixtures of 32P-labeled RNAs were generated: one containing 300-U1 RNA together with DHFR mRNA, U1ΔSm, U5ΔSm, and U6Δss RNAs (Fig. 1B, left); and the other containing U1-300 RNA in place of 300-U1 RNA (Fig. 1B, right). All the RNAs except U6Δss were m7G-capped. Immediately after injection, all the RNAs were nuclear (Fig. 1B, lanes 1,2,13,14). After 2 h, 300-U1, U1-300, and the reference RNAs had been exported partially to the cytoplasm, except for the nonexported U6 RNA control (Fig. 1B, lanes 3,4,15,16). To determine the export pathway of the two U1 derivatives, two inhibitors of CRM1-mediated export of U snRNAs were used. The first was a conjugate of NES peptides coupled to BSA (BSA–NES) that saturates CRM1 export (Fischer et al. 1995), and the second was a dominant-negative mutant of a U snRNA export factor, PHAX, that lacks the NES sequence (PHAX ΔNES) and thus blocks U snRNA–CRM1 complex formation (Ohno et al. 2000).
When BSA–NES was injected together with the RNAs, export of U1 and U5 control RNAs was inhibited as expected, whereas the CRM1-independent export of DHFR mRNA was unaffected (Fig. 1B, lanes 9,10,21,22). Export of the two U1 derivatives (300-U1 and U1-300) was also not affected by BSA–NES (Fig. 1B, lanes 9,10,21,22). When the control conjugate BSA-mut was used, the RNA export pattern was very similar to that without the conjugate (Fig. 1B, lanes 11,12,23,24). Similarly, when PHAX ΔNES was coinjected with the RNAs, export of U1 and U5 RNAs was severely inhibited, whereas export of mRNA was not inhibited (Fig. 1B, lanes 7,8,19,20). Export of the two U1 derivatives (300-U1 and U1-300) was also not inhibited by PHAX ΔNES (Fig. 1B, lanes 7,8,19,20). When wild-type PHAX (PHAX WT) was used, export of U1 and U5 was stimulated whereas export of mRNA and the two U1 derivatives was essentially unaffected (Fig. 1B, lanes 5,6,17,18). These results indicated that the two U1 derivatives (300-U1 and U1-300) were exported via the mRNA export pathway. Thus, the effect of fragment insertion is likely to be position-independent, and therefore, the above-mentioned possibility that the insertion might have disrupted a hypothetical U snRNA identity element(s) is unlikely (see Fig. 3, below).
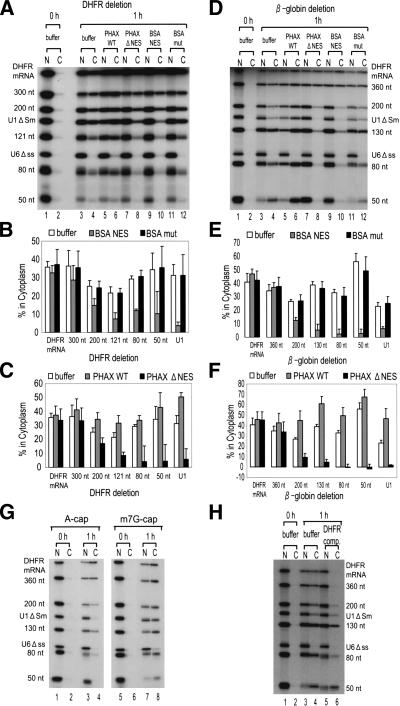
Figure 3.
Effect of mRNA shortening on RNA export. (A) A 32P-labeled RNA mixture containing shortened DHFR mRNAs (300 nt, 200 nt, 121 nt, 80 nt, and 50 nt), full-length DHFR mRNA, U1 ΔSm, and U6 Δss RNAs was microinjected with either PHAX WT (lanes 5,6), PHAX ΔNES (lanes 7,8), BSA–NES (lanes 9,10), or BSA-mut (lanes 11,12), or without proteins (buffer; lanes 3,4). All the RNAs except U6 were m7G-capped. RNA was extracted immediately (0 h; lanes 1,2) or 1 h (1 h; lanes 3–12) after injection, and analyzed as in Figure 1. (B,C) Quantitation of RNA export from three independent experiments carried out as in A. (D–F) The same as in A–C except that β-globin mRNA derivatives (360 nt, 300 nt, 200 nt, 130 nt, 80 nt, and 50 nt) were used. (G) Effect of the cap structure on export of the shortened β-globin mRNAs. The same RNA mixture as in D was prepared by primed-transcription with either dinucleotide ApppG (A-cap; lanes 1–4) or m7GpppG (m7G-cap; lanes 5–8) and RNA export was analyzed as in D. Note that U6 Δss RNA was always uncapped. (H) Effect of an mRNA competitor on RNA export. The same RNA mixture as in D was microinjected either with 0.5 pmole of unlabeled uncapped DHFR mRNA (DHFR comp., 500 times excess over labeled RNAs) or without it (buffer). RNA was extracted and analyzed as in D.
RNA export pathway gradually shifts as the length of U1 RNA increases.
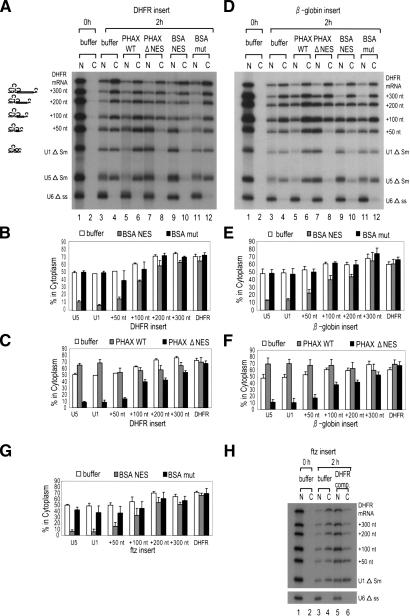
To examine how much longer U1 RNA must be to behave like an mRNA, fragments of various lengths (50, 100, 200, and 300 nt) derived from the DHFR cDNA sequence (see Supplementary Fig. 1) were inserted into U1ΔSm RNA at the central position, and export pathway of the resultant elongated U1 RNAs was examined as in Figure 1 (Fig. 2A–C). RNA export gradually shifted from U snRNA export to mRNA export as the insert became longer, as judged by the requirement for CRM1 and PHAX proteins (Fig. 2A [lanes 3–12], B,C for quantitation). The longer the insert, the more like an mRNA the RNA behaved, and if the insert was as long as 300 nt, the elongated U1 RNAs behaved indistinguishably from an mRNA in export (Fig. 2A–C). When the full-length DHFR mRNA sequence (720 nt) was inserted, the resultant U1 RNA also behaved identically to an mRNA (data not shown). Similar results were obtained with other mRNA fragments derived from β-globin and ftz cDNAs (Fig. 2D–F,G, respectively; see Supplementary Fig. 1).
Figure 2.
Effect of U1 RNA elongation on RNA export. (A) A 32P-labeled mixture of U1 RNA derivatives, containing 50 nt (+50 nt), 100 nt (+100 nt), 200 nt (+200 nt), or 300 nt (+300 nt) sequence from DHFR mRNA, was microinjected into Xenopus oocyte nuclei together with DHFR mRNA, U1 ΔSm, U5 ΔSm, and U6 Δss RNAs. All the RNAs except U6 were m7G-capped. The effects of PHAX and BSA–NES proteins were examined as in Figure 1B. (B) Quantitation of RNA export from three independent experiments as in A. Averages and standard deviations with BSA–NES (gray bars) or BSA-mut (black bars), or without proteins (buffer; white bars) are shown. (C) The same as in B except that PHAX WT (gray bars) or PHAX ΔNES (black bars) proteins were used. (D–F) The same as in A–C except that β-globin mRNA sequences were inserted instead of DHFR sequences. (G) The same as B and E except that ftz mRNA were used as the insert. Only quantitation is shown. (H) Effect of an mRNA competitor on RNA export. The same RNA mixture as in G was microinjected either with 0.5 pmole of unlabeled uncapped DHFR mRNA (DHFR comp., 500 times excess over labeled RNAs) or without it (buffer).
To further confirm that the elongated U1 RNAs used the mRNA export pathway, another criterion was examined. A characteristic of mRNA export is its high sensitivity to uncapped mRNA competitors (Jarmolowski et al. 1994). Therefore, the effect of coinjection of excess uncapped DHFR mRNA on the export of elongated U1 RNAs with ftz fragments was examined (Fig. 2H). Export of DHFR mRNA was severely inhibited as expected, whereas export of U1 RNA was hardly affected (Fig. 2H, lanes 3–6). RNA export gradually shifted from U snRNA export to mRNA export as the insert became longer, as judged by the sensitivity to the competitor (Fig. 2H, lanes 3–6). Similar results were obtained with elongated U1 RNAs with β-globin fragments (data not shown). These results, together with the previous data (Ohno et al. 2002), indicated that U1 RNAs elongated by the insertion of >300-nt fragments were induced to use the mRNA export pathway and that the nucleotide sequence or the position of the inserted fragments was not important for this effect.
Shortened mRNAs behave like a U snRNA in nuclear export.
The results described so far strongly suggested that RNA length has an important role in the choice of RNA export pathway. Moreover, the results implied an intriguing possibility that intronless mRNAs might be induced to behave like a U snRNA in export if the mRNAs were sufficiently shortened. We therefore tested this possibility. An m7G-capped DHFR mRNA (720 nt) was progressively shortened by deletion from the 3′-end, and export of the shortened DHFR-derived mRNAs was examined. If the mRNAs were >300 nt, they did not require CRM1 or PHAX for export and thus used the mRNA pathway (Fig. 3A [lanes 3–12], B,C for quantitation). By contrast, if the mRNAs were <120 nt, they used the U snRNA export pathway exclusively, (Fig. 3A [lanes 3–12], B,C). If the mRNA had an intermediate length (200 nt), it showed intermediate behavior between mRNA and U snRNA (Fig. 3A–C). Very similar results were obtained with another intronless mRNA derived from β-globin cDNA (Fig. 3D [lanes 3–12], E,F for quantitation). Moreover, shortening of an intronless mRNA derived from ftz cDNA from the 5′-end gave similar results (data not shown), indicating the effect is independent of the location of the deletion.
To further confirm that the shortened mRNAs used the U snRNA export pathway, two additional criteria were examined. As previously reported (Hamm and Mattaj 1990) U snRNA export is dependent on the m7G-cap structure, whereas mRNA export is not. Export of m7G-capped RNAs and A-capped RNAs, the latter of which are hardly recognized by CBC (Ohno et al. 1990; Izaurralde et al. 1994; Kataoka et al. 1994), was therefore compared (Fig. 3G). Export of DHFR mRNA and β-globin mRNA (360 nt) was independent of m7G-cap as expected (Fig. 3G, lanes 3,4,7,8). Export of the shortened β-globin mRNAs, however, became increasingly dependent on the m7G-cap as the RNA length decreased, and RNAs <130 nt behaved very similarly to U1 RNA, the export of which was dependent on m7G-cap (Fig. 3G, cf. lanes 3,4 and 7,8). In addition, the effect of coinjection of excess uncapped DHFR mRNA on the export of shortened m7G-capped β-globin mRNAs was examined (Fig. 3H). Export of DHFR mRNA and β-globin mRNA (360 nt) was abolished as expected, whereas export of U1 RNA and shortened β-globin mRNAs was not at all or was less affected (Fig. 3H, lanes 3–6). Export of the mRNAs <130 nt was as insensitive to the competitor as that of U1. These results confirmed that mRNAs longer than ∼300 nt were still using the mRNA export pathway but that mRNAs <130 nt were induced to use the U snRNA export pathway.
Behavior of highly structured RNAs
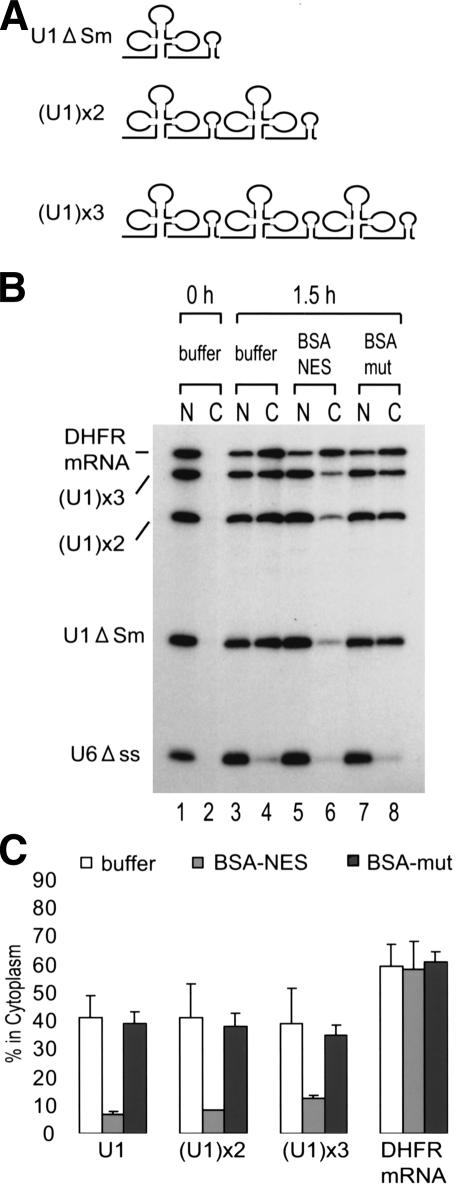
The results so far indicated that RNA length is a very important element for discriminating between mRNA and U snRNA export. m7G-capped RNAs longer than ∼300 nt use the mRNA export pathway, whereas m7G-capped RNAs shorter than ∼130 nt use the U snRNA export pathway. RNAs with intermediate length show intermediate phenotypes. However, U1 snRNA has an intermediate length (∼170 nt) but does not at all show an intermediate phenotype. U2 snRNA is even longer (∼190 nt), but export of this RNA follows the U snRNA export pathway (Jarmolowski et al. 1994). U snRNAs such as U1 and U2 are not homologous to each other in RNA sequence or structure per se, but U snRNAs are generally highly structured. Therefore, we hypothesized that highly structured RNAs are not recognized by the mRNA export machinery independent of their real length. To test this hypothesis, we generated artificial RNAs with the U1 RNA sequence tandemly repeated two or three times [Fig. 4A; (U1)x2 and (U1)x3, respectively]. The latter of these, (U1)x3 RNA, is ∼500 nt long and should be long enough to behave like an mRNA in export. However, (U1)x3 RNA, as well as (U1)x2, still behaved like a U snRNA as judged by the requirement for CRM1 (Fig. 4B,C for quantitation). When the anti-sense U1 RNA sequence was tandemly repeated, similar results were obtained, although export of these antisense U1 derivatives was significantly slower than that of the sense derivatives (data not shown), indicating that the cognate U1 sequence or structure was not important for this phenomenon. These results taken together support the hypothesis that highly structured RNAs are not effective in exerting the RNA length effect on the choice of export pathways.
Figure 4.
Export of tandemly repeated U1 RNAs. (A) Schematic representation of tandemly repeated U1 RNAs. U1 RNA repeated two or three times was named as (U1)x2 or (U1)x3, respectively. (B) A 32P-labeled RNA mixture of U1 ΔSm, (U1)x2 and (U1)x3 RNAs, DHFR mRNA, and U6 Δss RNA was injected and analyzed. (C) Quantitation of three independent experiments carried out as in B.
U6 snRNA is also highly structured, but this RNA is not exported. This is because U6 RNA naturally does not have an m7G-cap structure (Singh and Reddy 1989) and also because U6 is retained in the nucleus possibly by the action of La protein, which binds near the 3′ end of U6 (Stefano 1984; Terns et al. 1992). Even if U6Δss RNA was artificially m7G-capped, the RNA was hardly exported (Fig. 5, lanes 3,4). However, coinjection of PHAX WT recombinant protein could induce export of U6 (Fig. 5, lanes 5,6). If eight nucleotides were deleted from the 3′-end of U6 to which La protein binds (Terns et al. 1993; Boelens et al. 1995), export of U6 was observed even without PHAX WT recombinant protein (Fig. 5, lanes 11,12). Furthermore, export of this RNA was stimulated by PHAX WT but was blocked by PHAX ΔNES (Fig. 5, lanes 11–16), indicating that this artificial U6 RNA was exported via the U snRNA pathway. If U6 RNA was uncapped, it stayed in the nucleus in all the tested conditions (Fig. 5, lanes 17–32). These results, together with the results shown in Figure 3, strongly suggest that the presence of the m7G-cap structure and shortness of RNA length are the only essential determinants of U snRNA export, and the specific RNA sequence or structure per se is not important.
Figure 5.
Export of m7G-capped U6 RNA derivatives. Either m7G-capped (+cap) or uncapped (-cap) 32P-labeled U6 Δss RNA (lanes 1–8,17–24) or 3′-tail-deleted U6 RNA (U6 Δ3′; lanes 9–16,25–32) was injected with a 32P-labeled reference RNA mixture containing m7G-capped DHFR mRNA, uncapped U3 RNA, and m7G-capped U1 ΔSm RNA, and RNA export was analyzed as in Figure 1A.
Remodeling of RNA–protein complex in response to the RNA length
The results so far suggested that RNAs were associated with different export factors in an RNA-length-dependent manner. It was previously shown that U1 RNAs in which ∼300-nt fragments of several different sequences had been inserted into the central Sm-site position were associated with mRNA specific factor Y14 (Kataoka et al. 2000) but not with U snRNA export factor PHAX, as judged by immunoprecipitation (Ohno et al. 2002). However, because the role of Y14 in mRNA export is not clarified, use of Y14 as a marker protein of mRNA export was inappropriate. Here we extended the previous analysis by using REF/Aly (Zhou et al. 2000; Kim et al. 2001; Rodrigues et al. 2001) as a marker of mRNA export. Proteins associated with the elongated U snRNAs and the shortened mRNAs were analyzed by immunoprecipitation.
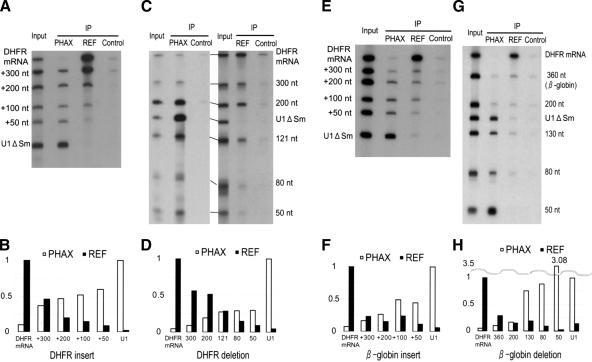
Control U1ΔSm RNA was efficiently precipitated by an antibody against PHAX, an export factor for U snR-NAs, but the same U1 RNA was hardly precipitated by an antibody against REF, an export factor for mRNAs (Fig. 6A–H, cf. PHAX lanes and REF lanes), indicating that U1 RNA was bound by PHAX but not by REF in the nucleus. Conversely, control DHFR mRNA behaved oppositely, indicating that DHFR mRNA was bound by REF but not by PHAX (Fig. 6A–H). In the case of U1 RNAs elongated with the DHFR fragments, less PHAX but more REF was bound as the RNA length increased (Fig. 6A,B for quantitation). Similar results were obtained with the shortened DHFR mRNAs. More PHAX but less REF was bound to the RNA as the RNA length decreased (Fig. 6C,D). When U1 RNAs elongated with β-globin fragments (Fig. 6E,F) and shortened β-globin mRNAs (Fig. 6G,H) were used, comparable results were obtained. These results were in good agreement with the gradual shift of export pathway in response to RNA length. These results suggest that there is constant competition between U snRNA export factors and mRNA export factors for binding to the RNA and also that there is an active RNP remodeling mechanism that depends upon RNA length.
Figure 6.
Effect of RNA length on RNP compositions. (A) A 32P-labeled RNA mixture containing U1 RNAs elongated with DHFR mRNA sequence, together with DHFR mRNA itself and U1 ΔSm RNA was injected into the nucleus. The nuclear fraction was prepared after 1 h, and immunoprecipitation (IP) was performed with either anti-PHAX antibody (PHAX), anti-Aly/REF antibody (REF), or anti-mouse IgG antibody (control) that had been prebound to Protein A-Sepharose beads. RNA precipitated with each antibody was recovered and analyzed. The input lane contains 10% of the input used in each IP lane. (B) Quantitation of IP shown in A. Relative precipitation of elongated U1 RNAs is shown. When anti-PHAX antibody (PHAX; white bars) or anti-Aly/REF antibody (REF; black bars) was used, precipitation of U1 RNA or DHFR mRNA was used as the standard (1.0), respectively. (C,D) Results of immunoprecipitation analysis with shortened DHFR mRNAs, performed as in A and B. (E,F) Immunoprecipitation analysis with U1 RNAs elongated with β-globin mRNA sequence, performed as in A and B. (G,H) Immunoprecipitation analysis with shortened β-globin mRNAs.
Discussion
In this study, the effect of RNA length on the choice of RNA export pathway was investigated. We found that U1 snRNA can be induced to use the mRNA export pathway if the U1 RNA is elongated by insertion of various RNA sequences of ≥300 nt. Conversely, intronless mRNAs can be induced to use the U snRNA export pathway if the mRNAs are shortened to ∼130 nt or shorter. RNAs of intermediate length exhibit intermediate export behavior. Immunoprecipitation experiments suggest that RNA length can define the protein composition of pre-export RNPs in the nucleus. Thus, RNA length is an extremely important determinant of RNA export pathway.
A model of RNA export
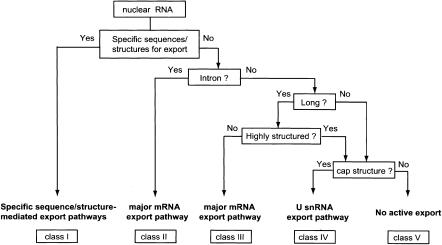
Based on the results obtained in this study together with those reported in previous publications, a model of how an RNA is committed to a specific export pathway is presented (Fig. 7). In this model, RNA export is divided into five classes. We discuss the classes one by one.
Figure 7.
A model of RNA export. The RNA export pathway is determined through several discrimination steps in the nucleus. This flowchart shows the major criteria that determine the RNA export pathway. See the text for details.
First of all, if the RNA contains specific RNA sequences or structures for export, the RNA is committed to the corresponding specific export pathways (Fig. 7, class I). Most, if not all, RNA polymerase I and III transcripts are in this category, and therefore, this class is heterogeneous in terms of RNA export pathways. As was already mentioned, proper recognition of tRNAs by Xpo-t requires major features of the higher-order structure of tRNAs. Although the details of the export mechanisms of large and small rRNAs remain unknown, it is clear that these RNAs have to be properly processed and subsequently have to be assembled into ribosomal subunits in order to be exported to the cytoplasm (Johnson et al. 2002; Tschochner and Hurt 2003). The proper processing and subunit assembly obviously require major structural features of the rRNAs. It has been recently shown that some short double-stranded RNAs such as pre-miRNAs and adenovirus VAI are exported by exportin-5 (Gwizdek et al. 2003; Yi et al. 2003; Lund et al. 2004). These RNAs should be also categorized in this class.
A specialized fraction of RNA polymerase II transcripts are also categorized in class I. Export of some viral mRNAs requires virus-specific RNA sequences and also often virus-specific proteins. One of the best-characterized examples is HIV-1 mRNAs. Export of partially spliced and unspliced HIV-1 mRNAs requires virus-encoded protein Rev and the virus-specific RNA structure Rev Response Element or RRE (Malim et al. 1989; Pollard and Malim 1998). Interestingly, simpler retroviruses such as simian type D retroviruses use a comparable system to export their unspliced RNAs. In this case, however, only a virus-specific RNA element, the constitutive transport element (CTE), is required. The cellular export factor TAP binds directly to the CTE without the aid of viral proteins (Pasquinelli et al. 1997; Grüter et al. 1998). Export of some specific cellular mRNAs also requires a specific RNA sequence. Histone H2A mRNA, which is naturally devoid of introns, has a specific RNA sequence termed the intronless transport element, which was shown to stimulate RNA export through binding to specific cellular factors (Huang and Carmichael 1997; Huang and Steitz 2001; Huang et al. 2003).
If the RNA does not contain any of the above features, then the next point of selectivity is whether the RNA contains a pre-mRNA intron (Fig. 7). If the answer is no, the next question is whether the RNA is long or not. If the RNA is longer than ∼300 nt, it is committed to the major mRNA pathway mediated by Tap (Fig. 7, class III). If the RNA is shorter than ∼130 nt and contains an m7G-cap, it will be committed to the U snRNA export pathway (Fig. 7, class IV). It was previously shown that export of mRNAs became inefficient when intronless mRNAs were shortened to ∼200 nt (Rodrigues et al. 2001). In light of our results, ∼200 nt is an intermediate length that is too short for efficient export as an mRNA and too long for efficient export as a U snRNA, and therefore, the overall kinetics of export could be slower with this length. Although we did not observe a severe decrease or cessation of export of the RNAs we examined here at the range of 200 nt (Fig. 3), kinetic changes in export may depend on the specific sequence of the RNA substrate.
Another important issue regarding RNA length is whether the poly(A) tail of mRNAs can be counted in as part of the RNA length. The average length of the poly(A) tail is ∼200 nt, and it is possible that this length may contribute to make the RNA an mRNA. However, it is equally possible that the poly(A) tail does not contribute to the length effect because the poly(A) tail must be covered by poly(A)-binding proteins in the nucleus. So far we have not succeeded in testing these possibilities because short RNAs that were artificially poly(A)-tailed in vitro were found to be unstable in Xenopus oocytes (data not shown).
It was previously reported that three RNA elements in U1 snRNA, the m7G-cap and 5′- and 3′-stem loop structures, are important for the export of U1 snRNA (Terns et al. 1993). However, our present results show that there is no absolute requirement for internal RNA sequences or structures to commit an RNA to the U snRNA export pathway. This conclusion was also implicit in Hamm et al.'s (1989, 1990) analysis of U1 and U2 mutants in splicing, in which all the mutants except ΔSm were able to associated with the Sm proteins and reimported into the nucleus. The only requirements are the presence of the m7G-cap and the shortness of the RNA length. Our results are not necessarily contradictory with the results of Terns et al. (1993), because we are dealing with essential identity elements of U snRNA whereas Terns et al. (1993) dealt with efficiency of RNA export. It is also possible that stem-loop RNA structures apparently stimulate RNA export simply because such structures increase RNA stability.
In contrast to capped RNAs, uncapped short RNAs in general would not be actively exported but rather stay in the nucleus unless they diffuse through the NPCs (Figs. 7 [class V], 3). Exceptions to the above-mentioned length rule include highly structured RNAs, which are not effective in exerting the RNA length effect on the choice of export pathway (Fig. 4). Strong secondary structures may be sufficient to prevent interaction of mRNA-specific RNA-binding proteins with the RNAs.
Finally, if the RNA contains a pre-mRNA intron(s), the RNA is committed to the major mRNA export pathway regardless of the length and regardless of the presence of the cap structure (Fig. 7, class II). Short capped RNAs that are exported via the U snRNA pathway or short uncapped RNAs that stay in the nucleus are both induced to be exported via the mRNA pathway if they are produced via mRNA splicing reactions (Ohno et al. 2002). It is highly likely that this phenomenon is dependent on the deposition of the EJC on the spliced RNAs (Dreyfuss et al. 2002; Reed and Hurt 2002; Le Hir et al. 2003).
Nuclear RNA retention mechanisms are not discussed in this model, although such mechanisms are clearly important for understanding RNA export. There are specific retention mechanisms for certain RNAs, such as U6, and for unspliced RNA precursors (Terns et al. 1993; Boelens et al. 1995; Rutz and Seraphin 2000; Galy et al. 2004), but such retention mechanisms may also influence the export of many RNAs. RNA export in general might be achieved under constant competition between export and retention mechanisms.
Other factors that may influence the choice of RNA export pathway
The results presented here indicate that a relatively unstructured RNA region of certain length (>300 nt) can induce mRNA export. However, does the RNA sequence itself not at all influence this effect? We have observed small but reproducible differences between different RNA fragments in the activity to induce mRNA export when the fragments were inserted into U1 RNA. For instance, ftz fragments are slightly more effective than β-globin fragments of the same length in inducing mRNA export. This indicates that the sequence of the RNA fragments can influence the inducing activity. However, we do not know the cause of this difference at the moment. It is to be clarified whether this difference is due to some specific RNA sequence properties that may actively influence the choice of RNA export pathway or due to the difference in the stability of general RNA secondary structure that counteracts the RNA length effect.
In this study, we analyze RNA export by microinjecting in vitro transcribed RNAs in Xenopus oocyte nuclei. In this system, RNA export is uncoupled from transcription and 3′-end processing. Despite this, we can observe the expected specificity of export factors, indicating that the export factors can be loaded onto RNAs posttranscriptionally. However, we cannot rule out the possibility that in vivo transcription and/or 3′-end processing may contribute to the specificity of export because it has been shown that these processes contribute at least to the association of export factors (Huang and Carmichael 1996; Strässer et al. 2002).
Factors that recognize the RNA length and remodeling of export RNPs
Our results show that relatively unstructured RNAs of >300 nt are considered to be mRNAs by the cellular export machinery. This indicates that RNA length is some-how measured by the cellular export machinery. This probably means that a certain length of unstructured RNA would be required for stable binding to mRNA export factors. Consistent with this notion is our finding that the mRNA-specific RNA-binding protein REF is recruited to RNA in an RNA-length-dependent manner (Fig. 6). However, this does not necessarily mean that REF itself is the factor that measures RNA length. The length could be measured by other nuclear RNA-binding protein(s) because it is not clear whether REF binding is the cause or the result of the determination of mRNA identity. Regarding this issue, it is enigmatic why a relatively long region (>300 nt) is necessary for this effect. The length of 300 nt seems to be too long only for a single protein factor to bind. It is possible that binding of multiple copies of a single factor is required for determining mRNA identity and that the minimal length that allows the binding of the minimal necessary copies is ∼300 nt. A more intriguing alternative possibility is that efficient binding of a large protein complex that imprints mRNA identity requires 300 nt. In either scenario, it is interesting to consider that very long mRNAs may need multiple TAP molecules along the RNA for efficient translocation through the NPCs just like the centipedes use many pairs of legs for walking. The length of 300 nt might be the unit to which one TAP molecule, that is, a pair of legs in centipedes, is recruited.
As a result of the binding of mRNA identity factors, an RNA is committed to the mRNA export pathway. Moreover, our results indicate that U snRNA-specific export factors such as PHAX are not found on the RNA if the RNA is committed to the mRNA export pathway. We previously observed similar RNP remodeling in the case of U1 RNA that had been produced via pre-mRNA splicing (Ohno et al. 2002). Thus, it may be a general rule that if the RNA is committed to one pathway, association of the export factors of other pathways is prevented. It will be interesting to elucidate the molecular mechanisms that operate to achieve such remodeling of export RNPs.
Materials and methods
DNA constructs
For U1 derivative plasmids, corresponding PCR-amplified DNA fragments, designed for XhoI and SalI digestion, were inserted into the XhoI site of Xenopus T7 U1 ΔSm plasmid (Hamm et al. 1987). For the U3 RNA plasmid, U3A (Savino et al. 1992) gene was PCR-amplified from Xenopus laevis genomic DNA. The T7 promoter was then added by a second PCR, and the amplified product was directly used as a template for in vitro transcription. The deletion of β-globin cDNA was done by PCR from a human β-globin cDNA fragment cloned into pSP64 (Promega; Mayeda and Ohshima 1990). DHFR deletion was similarly done with the mouse DHFR gene inserted into pBluescribe (Kambach and Mattaj 1992). For the latter, a termination codon was introduced at the end of each deletion point. For the tandemly repeated U1 constructs, a BglII site was introduced at the 5′ end of the U1 sequence in T7 U1ΔSm plasmid (pBgl-U1). The U1 fragment was prepared by BglII/BamHI digestion of pBgl-U1 and cloned into the BamHI site of T7 U1ΔSm. For the 300-U1 and U1-300 constructs, a ftz cDNA fragment of 300 nt was inserted into the BglII or BamHI site of pBgl-U1 plasmid, respectively. For the U6 Δ3′ construct, 8 nt (AUAUUUUU) were deleted from the 3′ end of the U6 sequence in the U6 Dgr;ss RNA by PCR, and the PCR product was directly used for in vitro transcription.
In vitro transcription
32P-labeled RNAs were transcribed as described previously (Jarmolowski et al. 1994) except that only [α-32P]UTP was used. The transcription reaction was usually performed for 45–60 min at 37°C. Uncapped DHFR competitor RNA was synthesized by using MEGAscript (Ambion) according to the manufacturer's instructions.
Recombinant proteins
Mouse PHAX WT or NES mutant proteins with a his-tag were expressed as described previously (Ohno et al. 2000) except that Escherichia coli BL21 (DE3) codon plus (Stratagene) with pRep4 plasmid was used. His-tagged proteins were purified by using a His Bind kit (Novagen) according to the manufacturer's protocol.
RNA microinjection into Xenopus oocytes
RNA microinjection into Xenopus oocytes was performed as previously described (Jarmolowski et al. 1994). For preparation of BSA–NES and BSA-mut, PKI NES peptide (CELALKLAGLDIN) or a mutant peptide (CELALKAAGADIN) was conjugated to BSA as described previously (Fischer et al. 1995). Analysis and quantitation of RNA bands were performed with BAS-2500 (FujiFilm) and Image Gauge version 3.45 (FujiFilm).
Immunoprecipitation experiments
Different antibodies were individually bound to Protein A-Sepharose beads (Amersham Biosciences) on a rotating platformfor 1 h at 4°C. In thecase of mouse monoclonal antibodies, rabbit antimouse IgG antibody was also used as a bridging antibody. After RNA injection into Xenopus oocyte nuclei, the injected nuclei were isolated and resuspended in RSB100N buffer (10 mM Tris at pH 7.4, 100 mM NaCl, 2.5 mM MgCl2, 0.1% NP-40) supplemented with complete protease inhibitor cocktail (Roche) and 1000 U/mL RNasin (Promega). The mixture was centrifuged at 14,000 rpm for 15 min at 4°C, and the resultant supernatant was incubated with the antibody-bound beads for 1 h at 4°C. After the beads were washed five times with RSB100N buffer, they were incubated in HomoMix (50 mM Tris at pH 7.4, 5 mM EDTA, 1.5% SDS, 300 mM NaCl, 1.5 mg/mL Proteinase K; Nakalai Tesque) for 30 min at 50°C to elute RNA in the supernatant. RNA was recovered from the supernatant by phenol/chloroform extraction and ethanol precipitation. The recovered RNA was analyzed by denaturing PAGE.
Acknowledgments
We are grateful to Drs. Iain Mattaj and Gideon Dreyfuss for anti-PHAX antibody and anti-Aly/REF antibody, respectively. We thank Dr. Iain Mattaj for helpful comments on this manuscript. We also thank members of our laboratory, especially Drs. Naoto Mabuchi and Makoto Kitabatake, for suggestions and criticisms of this manuscript. This work was supported by CREST, JST, and grants from the Ministry of Education, Science, Sports and Culture of Japan. K.M. and I.T. were supported by the 21st Century COE Program of the Ministry of Education, Culture, Sports, Science and Technology. K.M. is a JSPS Research Fellow.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article published online ahead of print. Article and publication date are at http://www.genesdev.org/cgi/doi/10.1101/gad.1216204.
Supplemental material is available at http://www.genesdev.org.
References
- Arts G.J., Fornerod, M., and Mattaj, I.W. 1998a. Identification of a nuclear export receptor for tRNA. Curr. Biol. 8: 305-314. [DOI] [PubMed] [Google Scholar]
- Arts G.J., Kuersten, S., Romby, P., Ehresmann, B., and Mattaj, I.W. 1998b. The role of exportin-t in selective nuclear export of mature tRNAs. EMBO J. 17: 7430-7441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boelens W.C., Palacios, I., and Mattaj, I.W. 1995. Nuclear retention of RNA as a mechanism for localization. RNA 1: 273-283. [PMC free article] [PubMed] [Google Scholar]
- Cullen B.R. 2003. Nuclear RNA export. J. Cell. Sci. 116: 587-597. [DOI] [PubMed] [Google Scholar]
- Dimaano C. and Ullman, K.S. 2004. Nucleocytoplasmic transport: Integrating mRNA production and turnover with export through the nuclear pore. Mol. Cell. Biol. 24: 3069-3076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreyfuss G., Kim, V.N., and Kataoka, N. 2002. Messenger-RNA-binding proteins and the messages they carry. Nat. Rev. Mol. Cell. Biol. 3: 195-205. [DOI] [PubMed] [Google Scholar]
- Fischer U., Huber, J., Boelens, W.C., Mattaj, I.W., and Luhrmann, R. 1995. The HIV-1 Rev activation domain is a nuclear export signal that accesses an export pathway used by specific cellular RNAs. Cell 82: 475-483. [DOI] [PubMed] [Google Scholar]
- Fornerod M., Ohno, M., Yoshida, M., and Mattaj, I.W. 1997. CRM1 is an export receptor for leucine-rich nuclear export signals. Cell 90: 1051-1060. [DOI] [PubMed] [Google Scholar]
- Fukuda M., Asano, S., Nakamura, T., Adachi, M., Yoshida, M., Yanagida, M., and Nishida, E. 1997. CRM1 is responsible for intracellular transport mediated by the nuclear export signal. Nature 390: 308-311. [DOI] [PubMed] [Google Scholar]
- Galy V., Gadal, O., Fromont-Racine, M., Romano, A., Jacquier, A., and Nehrbass, U. 2004. Nuclear retention of unspliced mRNAs in yeast is mediated by perinuclear Mlp1. Cell 116: 63-73. [DOI] [PubMed] [Google Scholar]
- Grüter P., Tabernero, C., von Kobbe, C., Schmitt, C., Saavedra, C., Bachi, A., Wilm, M., Felber, B.K., and Izaurralde, E. 1998. TAP, the human homolog of Mex67p, mediates CTE-dependent RNA export from the nucleus. Mol. Cell 1: 649-659. [DOI] [PubMed] [Google Scholar]
- Gwizdek C., Ossareh-Nazari, B., Brownawell, A.M., Doglio, A., Bertrand, E., Macara, I.G., and Dargemont, C. 2003. Exportin-5 mediates nuclear export of minihelix-containing RNAs. J. Biol. Chem. 278: 5505-5508. [DOI] [PubMed] [Google Scholar]
- Hamm J. and Mattaj, I.W. 1990. Monomethylated cap structures facilitate RNA export from the nucleus. Cell 63: 109-118. [DOI] [PubMed] [Google Scholar]
- Hamm J., Kazmaier, M., and Mattaj, I.W. 1987. In vitro assembly of U1 snRNPs. EMBO J. 6: 3479-3485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamm J., Dathan, N.A., and Mattaj, I.W. 1989. Functional analysis of mutant Xenopus U2 snRNAs. Cell 59: 159-169. [DOI] [PubMed] [Google Scholar]
- Hamm J., Dathan, N.A., Scherly, D., and Mattaj, I.W. 1990. Multiple domains of U1 snRNA, including U1 specific protein binding sites, are required for splicing. EMBO J. 9: 1237-1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellmuth K., Lau, D.M., Bischoff, F.R., Kunzler, M., Hurt, E., and Simos, G. 1998. Yeast Los1p has properties of an exportin-like nucleocytoplasmic transport factor for tRNA. Mol. Cell. Biol. 18: 6374-6386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y. and Carmichael, G.C. 1996. Role of polyadenylation in nucleocytoplasmic transport of mRNA. Mol. Cell. Biol. 16: 1534-1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ____. 1997. The mouse histone H2a gene contains a small element that facilitates cytoplasmic accumulation of intronless gene transcripts and of unspliced HIV-1-related mRNAs. Proc. Natl. Acad. Sci. 94: 10104-10109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y. and Steitz, J.A. 2001. Splicing factors SRp20 and 9G8 promote the nucleocytoplasmic export of mRNA. Mol. Cell 7: 899-905. [DOI] [PubMed] [Google Scholar]
- Huang Y., Gattoni, R., Stevenin, J., and Steitz, J.A. 2003. SR splicing factors serve as adapter proteins for TAP-dependent mRNA export. Mol. Cell 11: 837-843. [DOI] [PubMed] [Google Scholar]
- Izaurralde E., Lewis, J., McGuigan, C., Jankowska, M., Darzynkiewicz, E., and Mattaj, I.W. 1994. A nuclear cap binding protein complex involved in pre-mRNA splicing. Cell 78: 657-668. [DOI] [PubMed] [Google Scholar]
- Izaurralde E., Lewis, J., Gamberi, C., Jarmolowski, A., McGuigan, C., and Mattaj, I.W. 1995. A cap-binding protein complex mediating U snRNA export. Nature 376: 709-712. [DOI] [PubMed] [Google Scholar]
- Jarmolowski A., Boelens, W.C., Izaurralde, E., and Mattaj, I.W. 1994. Nuclear export of different classes of RNA is mediated by specific factors. J. Cell. Biol. 124: 627-635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin L., Guzik, B.W., Bor, Y.C., Rekosh, D., and Hammarskjold, M.L. 2003. Tap and NXT promote translation of unspliced mRNA. Genes & Dev. 17: 3075-3086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson A.W., Lund, E., and Dahlberg, J. 2002. Nuclear export of ribosomal subunits. Trends Biochem. Sci. 27: 580-585. [DOI] [PubMed] [Google Scholar]
- Kambach C. and Mattaj, I.W. 1992. Intracellular distribution of the U1A protein depends on active transport and nuclear binding to U1 snRNA. J. Cell. Biol. 118: 11-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katahira J., Strässer, K., Podtelejnikov, A., Mann, M., Jung, J.U., and Hurt, E. 1999. The Mex67p-mediated nuclear mRNA export pathway is conserved from yeast to human. EMBO J. 18: 2593-2609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kataoka N., Ohno, M., Kangawa, K., Tokoro, Y., and Shimura, Y. 1994. Cloning of a complementary DNA encoding an 80 kilodalton nuclear cap binding protein. Nucleic Acids Res. 22: 3861-3865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kataoka N., Ohno, M., Moda, I., and Shimura, Y. 1995. Identification of the factors that interact with NCBP, an 80 kDa nuclear cap binding protein. Nucleic Acids Res. 23: 3638-3641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kataoka N., Yong, J., Kim, V.N., Velazquez, F., Perkinson, R.A., Wang, F., and Dreyfuss, G. 2000. Pre-mRNA splicing imprints mRNA in the nucleus with a novel RNA-binding protein that persists in the cytoplasm. Mol. Cell 6: 673-682. [DOI] [PubMed] [Google Scholar]
- Kim V.N., Yong, J., Kataoka, N., Abel, L., Diem, M.D., and Dreyfuss, G. 2001. The Y14 protein communicates to the cytoplasm the position of exon–exon junctions. EMBO J. 20: 2062-2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komeili A. and O'Shea, E.K. 2001. New perspectives on nuclear transport. Annu. Rev. Genet. 35: 341-364. [DOI] [PubMed] [Google Scholar]
- Kuersten S., Segal, S.P., Verheyden, J., LaMartina, S.M., and Goodwin, E.B. 2004. NXF-2, REF-1, and REF-2 affect the choice of nuclear export pathway for tra-2 mRNA in C. elegans. Mol. Cell 14: 599-610. [DOI] [PubMed] [Google Scholar]
- Kutay U., Lipowsky, G., Izaurralde, E., Bischoff, F.R., Schwarzmaier, P., Hartmann, E., and Görlich, D. 1998. Identification of a tRNA-specific nuclear export receptor. Mol. Cell 1: 359-369. [DOI] [PubMed] [Google Scholar]
- Le Hir H., Gatfield, D., Izaurralde, E., and Moore, M.J. 2001. The exon–exon junction complex provides a binding platform for factors involved in mRNA export and nonsensemediated mRNA decay. EMBO J. 20: 4987-4997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Hir H., Nott, A., and Moore, M.J. 2003. How introns influence and enhance eukaryotic gene expression. Trends Biochem. Sci. 28: 215-220. [DOI] [PubMed] [Google Scholar]
- Lei E.P. and Silver, P.A. 2002. Protein and RNA export from the nucleus. Dev. Cell. 2: 261-272. [DOI] [PubMed] [Google Scholar]
- Lipowsky G., Bischoff, F.R., Izaurralde, E., Kutay, U., Schafer, S., Gross, H.J., Beier, H., and Görlich, D. 1999. Coordination of tRNA nuclear export with processing of tRNA. RNA 5: 539-549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund E., Guttinger, S., Calado, A., Dahlberg, J.E., and Kutay, U. 2004. Nuclear export of microRNA precursors. Science 303: 95-98. [DOI] [PubMed] [Google Scholar]
- Macara I.G. 2001. Transport into and out of the nucleus. Microbiol. Mol. Biol Rev. 65: 570-594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malim M.H., Hauber, J., Le, S.Y., Maizel, J.V., and Cullen, B.R. 1989. The HIV-1 rev trans-activator acts through a structured target sequence to activate nuclear export of unspliced viral mRNA. Nature 338: 254-257. [DOI] [PubMed] [Google Scholar]
- Mattaj I.W. 1988. U snRNP assembly and transport. In: Structure and function of major and minor small nuclear ribonucleoprotein particles (ed. M.L. Birnstiel), pp. 100-114. Springer-Verlag, Berlin, Germany.
- Mayeda A. and Ohshima, Y. 1990. β-Globin transcripts carrying a single intron with three adjacent nucleotides of 5′ exon are efficiently spliced in vitro irrespective of intron position or surrounding exon sequences. Nucleic Acids Res. 18: 4671-4676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nott A., Le Hir, H., and Moore, M.J. 2004. Splicing enhances translation in mammalian cells: An additional function of the exon junction complex. Genes & Dev. 18: 210-222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno M., Kataoka, N., and Shimura. Y. 1990. A nuclear cap binding protein from HeLa cells. Nucleic Acids Res. 18: 6989-6995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno M., Segref, A., Bachi, A., Wilm, M., and Mattaj, I.W. 2000. PHAX, a mediator of U snRNA nuclear export whose activity is regulated by phosphorylation. Cell 101: 187-198. [DOI] [PubMed] [Google Scholar]
- Ohno M., Segref, A., Kuersten, S., and Mattaj, I.W. 2002. Identity elements used in export of mRNAs. Mol. Cell 9: 659-671. [DOI] [PubMed] [Google Scholar]
- Ossareh-Nazari B., Bachelerie, F., and Dargemont, C. 1997. Evidence for a role of CRM1 in signal-mediated nuclear protein export. Science 278: 141-144. [DOI] [PubMed] [Google Scholar]
- Pasquinelli A.E., Ernst, R.K., Lund, E., Grimm, C., Zapp, M.L., Rekosh, D., Hammarskjold, M.L., and Dahlberg, J.E. 1997. The constitutive transport element (CTE) of Mason-Pfizer monkey virus (MPMV) accesses a cellular mRNA export pathway. EMBO J. 16: 7500-7510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard V.W. and Malim, M.H. 1998. The HIV-1 Rev protein. Annu. Rev. Microbiol. 52: 491-532. [DOI] [PubMed] [Google Scholar]
- Reed R. and Hurt, E. 2002. A conserved mRNA export machinery coupled to pre-mRNA splicing. Cell 108: 523-531. [DOI] [PubMed] [Google Scholar]
- Rodrigues J.P., Rode, M., Gatfield, D., Blencowe, B.J., Carmo-Fonseca, M., and Izaurralde, E. 2001. REF proteins mediate the export of spliced and unspliced mRNAs from the nucleus. Proc. Natl. Acad. Sci. 98: 1030-1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutz B. and Seraphin, B. 2000. A dual role for BBP/ScSF1 in nuclear pre-mRNA retention and splicing. EMBO J. 19: 1873-1886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savino R., Hitti, Y., and Gerbi, S.A. 1992. Genes for Xenopus laevis U3 small nuclear RNA. Nucleic Acids Res. 20: 5435-5442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segref A., Sharma, K., Doye, V., Hellwig, A., Huber, J., Luhrmann, R., and Hurt, E. 1997. Mex67p, a novel factor for nuclear mRNA export, binds to both poly(A)+ RNA and nuclear pores. EMBO J. 16: 3256-3271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh R. and Reddy, R. 1989. Gamma-monomethyl phosphate: A cap structure in spliceosomal U6 small nuclear RNA. Proc. Natl. Acad. Sci. 86: 8280-8283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stade K., Ford, C.S., Guthrie, C., and Weis, K. 1997. Exportin 1 (Crm1p) is an essential nuclear export factor. Cell 90: 1041-1050. [DOI] [PubMed] [Google Scholar]
- Stefano J.E. 1984. Purified lupus antigen La recognizes an oligouridylate stretch common to the 3′ termini of RNA polymerase III transcripts. Cell 36: 145-154. [DOI] [PubMed] [Google Scholar]
- Strässer K.A. and Hurt, E. 2000. Yra1p, a conserved nuclear RNA-binding protein, interacts directly with Mex67p and is required for mRNA export. EMBO J. 19: 410-420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strässer K., Masuda, S., Mason, P., Pfannstiel, J., Oppizzi, M., Rodriguez-Navarro, S., Rondon, A.G., Aguilera, A., Struhl, K., Reed, R., et al. 2002. TREX is a conserved complex coupling transcription with messenger RNA export. Nature 417: 304-308. [DOI] [PubMed] [Google Scholar]
- Stutz F. and Izaurralde, E. 2003. The interplay of nuclear mRNP assembly, mRNA surveillance and export. Trends Cell Biol. 13: 319-327. [DOI] [PubMed] [Google Scholar]
- Stutz F., Bachi, A., Doerks, T., Braun, I.C., Seraphin, B., Wilm, M., Bork, P., and Izaurralde, E. 2000. REF, an evolutionary conserved family of hnRNP-like proteins, interacts with TAP/Mex67p and participates in mRNA nuclear export. RNA 6: 638-650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suntharalingam M. and Wente, S.R. 2003. Peering through the pore: Nuclear pore complex structure, assembly, and function. Dev. Cell. 4: 775-789. [DOI] [PubMed] [Google Scholar]
- Swanson C.M., Puffer, B.A., Ahmad, K.M., Doms, R.W., and Malim, M.H. 2004. Retroviral mRNA nuclear export elements regulate protein function and virion assembly. EMBO J. 23: 2632-2640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terns M.P., Lund, E., and Dahlberg, J.E. 1992. 3′-End-dependent formation of U6 small nuclear ribonucleoprotein particles in Xenopus laevis oocyte nuclei. Mol. Cell. Biol. 12: 3032-3040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terns M.P., Dahlberg, J.E., and Lund, E. 1993. Multiple cis-acting signals for export of pre-U1 snRNA from the nucleus. Genes & Dev. 7: 1898-1908. [DOI] [PubMed] [Google Scholar]
- Tschochner H. and Hurt, E. 2003. Pre-ribosomes on the road from the nucleolus to the cytoplasm. Trends Cell Biol. 13: 255-263. [DOI] [PubMed] [Google Scholar]
- Wen W., Meinkoth, J.L., Tsien, R.Y., and Taylor, S.S. 1995. Identification of a signal for rapid export of proteins from the nucleus. Cell 82: 463-473. [DOI] [PubMed] [Google Scholar]
- Wiegand H.L., Lu, S., and Cullen, B.R. 2003. Exon junction complexes mediate the enhancing effect of splicing on mRNA expression. Proc. Natl. Acad. Sci. 100: 11327-11332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Will C.L. and Luhrmann, R. 2001. Spliceosomal UsnRNP biogenesis, structure and function. Curr. Opin. Cell. Biol. 13: 290-301. [DOI] [PubMed] [Google Scholar]
- Yi R., Qin, Y., Macara, I.G., and Cullen, B.R. 2003. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes & Dev. 17: 3011-3016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z., Luo, M.J., Strässer, K., Katahira, J., Hurt, E., and Reed, R. 2000. The protein Aly links pre-messenger-RNA splicing to nuclear export in metazoans. Nature 407: 401-405. [DOI] [PubMed] [Google Scholar]