Abstract
We report on the function of the human ortholog of Saccharomyces cerevisiae Rif1 (Rap1-interacting factor 1). Yeast Rif1 associates with telomeres and regulates their length. In contrast, human Rif1 did not accumulate at functional telomeres, but localized to dysfunctional telomeres and to telomeric DNA clusters in ALT cells, a pattern of telomere association typical of DNA-damage-response factors. After induction of double-strand breaks (DSBs), Rif1 formed foci that colocalized with other DNA-damage-response factors. This response was strictly dependent on ATM (ataxia telangiectasia mutated) and 53BP1, but not affected by diminished function of ATR (ATM- and Rad3-related kinase), BRCA1, Chk2, Nbs1, and Mre11. Rif1 inhibition resulted in radiosensitivity and a defect in the intra-S-phase checkpoint. The S-phase checkpoint phenotype was independent of Nbs1 status, arguing that Rif1 and Nbs1 act in different pathways to inhibit DNA replication after DNA damage. These data reveal that human Rif1 contributes to the ATM-mediated protection against DNA damage and point to a remarkable difference in the primary function of this protein in yeast and mammals.
Keywords: 53BP1, ATM, checkpoint, Nbs1, Rif1, telomere
The ability of mammalian cells to survive chromosomal damage depends on the coordinated activity of DNA-response pathways that halt cell cycle progression prior to DNA replication or mitosis. The ataxia telangiectasia mutated (ATM) kinase plays a crucial function as a transducer of the DNA-damage signal (for review, see Kastan and Lim 2000; Zhou and Elledge 2000; Abraham 2001; Shiloh 2003). Following the generation of doublestrand breaks (DSBs) by IR or clastogens, inactive ATM kinase dimers are converted into active monomers through autophosphorylation on Ser 1981 (Bakkenist and Kastan 2003). ATM has a number of direct targets, including Nbs1, Chk2, Mdm2, 53BP1, BRCA1, Rad17, Smc1, FANCD2, and H2AX (for review, see Kastan and Lim 2000; D'Andrea and Grompe 2003; Shiloh 2003). Many ATM targets and other proteins involved in the DNA-damage response physically accumulate at or near DNA lesions forming ionizing radiation-induced foci (IRIF; Haaf et al. 1995; for review, see Petrini and Stracker 2003).
The ATM- and Rad3-related kinase, ATR, responds to replication stress and UV damage as well as to DSBs (for review, see Abraham 2001). ATR requires an interacting partner, ATRIP, and the loading of RPA on single-stranded DNA for its activation (Cortez et al. 2001; Zou and Elledge 2003). There is considerable redundancy in the ATM and ATR-signaling pathways, in part because many of the effectors in these pathways can be phosphorylated by both kinases.
A hallmark of A-T cells is their diminished ability to survive ionizing radiation (IR) or other genotoxic treatments that create DSBs (Taylor et al. 1975; for review, see Shiloh 2003). The ATM-dependent DNA-damage response can block progression through the cell cycle before, during, and after DNA replication. The intra-S-phase checkpoint involves ATM-mediated phosphorylation of Nbs1, Smc1, and BRCA1 (Gatei et al. 2000; Lim et al. 2000; Wu et al. 2000; Zhao et al. 2000; Xu et al. 2001, 2002b; Kim et al. 2002; Yazdi et al. 2002). ATM controls a second parallel intra-S-phase pathway that results in degradation of Cdc25A and reduced Cdk2 activity (Falck et al. 2001, 2002). Radioresistant DNA synthesis (RDS), a second feature of A-T cells, occurs when any of these intra-S-phase checkpoint pathways is compromised.
Here, we describe a novel component of the ATM pathway, Rif1 (Rap1-interacting factor 1), the human ortholog of yeast Rif1. Rif1 was identified in Saccharomyces cerevisiae as one of two Rap1-binding partners required for the regulation of telomere length (Hardy et al. 1992), and a similar function has been ascribed to Rif1 from Schizosaccharomyces pombe (Kanoh and Ishikawa 2001). The maintenance of yeast telomeres is controlled by a negative feedback loop, in which the telomere length regulator Rap1 binds to the telomeric repeat tract (Marcand et al. 1997; for review, see Smogorzewska and de Lange 2004). Telomere-bound Rap1 recruits Rif1 and Rif2, which both limit extension of the telomeric tract by telomerase (Hardy et al. 1992; Marcand et al. 1997; Wotton and Shore 1997). Our data show that human Rif1 functions as a DNA-damage-response factor required for cell survival after radiation damage and proper execution of the intra-S-phase checkpoint. These findings underscore the close relationship between the DNA-damage response and telomere function and further highlight the substantial changes that have taken place during the evolution of the telomeric complex.
Results
Human Rif1 does not accumulate at functional telomeres
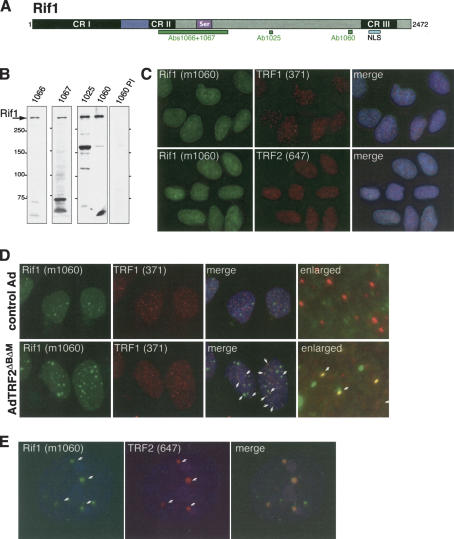
Human Rif1 was identified on the basis of sequence similarity to the RIF1 genes from S. pombe and S. cerevisiae (BLAST search; P = 9 × 10-5; Hardy et al. 1992; Kanoh and Ishikawa 2001; Rif1 GenBank accession no. AY585745). A mouse Rif1 ortholog was recently reported, but the function of murine Rif1 was not addressed (Adams and McLaren 2004). Alignment of human, mouse, and fugu Rif1 ORFs revealed three conserved regions, CRI–III (Fig. 1A; Supplementary Fig. 1A). Comparison to S. cerevisiae Rif1 indicated that 20%–25% of the amino acids in CRI–III are conserved (Supplementary Fig. 1B). The most notable feature of the mammalian Rif1 proteins is the predicted presence of eight HEAT- or ARMADILLO-type repeats in the N-terminal 340 amino acids (see Supplemental Material for details). HEAT/ARMADILLO repeats are helical folds that typically occur in long arrays, creating an extended curved protein or RNA interaction surface.
Figure 1.
Human Rif1 accumulates preferentially at uncapped telomeres and ALT cell telomeres. (A) Schematic of human Rif1 indicating the approximate position of the regions conserved in vertebrate Rif1 proteins (CRI–III), a predicted bipartite NLS, and a serine-rich region. The positions of antibody antigens are indicated (green boxes). (B) Detection of human Rif1 protein. Immunoblots of HeLa cell extract probed with the indicated antibodies or a preimmune serum (1060 PI). (C) Rif1 is present throughout the nucleoplasm. IF of paraformaldehyde fixed HeLa1.2.11 cells with mouse Rif1 serum (1060; green) and antibodies against TRF1 and TRF2 (371 and 647, respectively; both red), along with DAPI staining (blue). (D) Rif1 foci at uncapped telomeres. hTERT-BJ cells were infected with the indicated adenoviruses (control: β-galactosidase virus) fixed 48 h postinfection and processed for IF, Rif1, and TRF1 as described in Takai et al. (2003). DAPI (blue) DNA stain was added in the merged images. White arrows highlight a subset of the Rif1 foci at telomeres. The enlarged images are derived from different cells in the same experiment. (E) Association of Rif1 with ALT cell APBs. Costaining of Rif1 with TRF2 on APBs in the ALT cell line GM847.
Antisera raised against Rif1 peptides and a GST–Rif1 fusion reacted with the same large (>250 kDa) polypeptide in immunoblots of human cell lines (Fig. 1A,B). Rif1 siRNAs diminished the abundance of this protein (see below), confirming that this polypeptide is encoded by the Rif1 gene. Human Rif1 was detectable in different human cell lines and strains (e.g., IMR90, BJ, WI38, GM847, VA-13, HeLa, U-2OS, HCT15, HT1080) and Rif1 mRNA was present in a wide variety of tissues (data not shown), suggesting ubiquitous expression.
Indirect immunofluorescence (IF) with Rif1 antibodies revealed a diffuse nuclear localization in interphase cells (Fig. 1C). Rif1 did not show significant colocalization with telomeric proteins, such as TRF1, TRF2, Rap1, and TIN2, suggesting that Rif1 does not accumulate on chromosome ends (Fig. 1C; data not shown). The same result was obtained with different methods for cell fixation (paraformaldehyde and methanol), and after extraction of soluble nucleoplasmic proteins with Triton-X 100 (data not shown). Furthermore, Rif1 was not recovered in immunoprecipitates of the telomeric proteins TRF1, TRF2, and Rap1 (data not shown), and chromatin immunoprecipitations with Rif1 antiserum did not bring down telomeric DNA (J. Donigian, R. Wang, D. Loayza, J. Silverman, and T. de Lange, unpubl.).
Telomeres protect natural chromosome ends from being recognized as sites of DNA damage. This function depends on the presence of the telomeric-binding protein TRF2 (for review, see de Lange 2002). Removal of TRF2 using a dominant negative allele results in telomere dysfunction (van Steensel et al. 1998; Karlseder et al. 1999). The resulting uncapped telomeres are recognized as sites of DNA damage, leading to the accumulation of DNA-damage-response factors, such as Nbs1, 53BP1, ATM, Rad17, and γ-H2AX at chromosome ends (d'Adda di Fagagna et al. 2003; Takai et al. 2003). The foci formed at uncapped telomeres resemble DNA-damage-response foci and are referred to as telomere dysfunction-induced foci or TIFs. IF revealed that Rif1 colocalized with a subset of the telomeres in cells infected with a dominant negative allele of TRF2, suggesting that Rif1 can be a component of TIFs (Fig. 1D).
Rif1 was also associated with unusual telomeric structures present in human cell lines that maintain telomeric DNA in absence of telomerase (alternative lengthening of telomeres, or ALT cells; for review, see Henson et al. 2002). ALT cells can contain large foci of telomeric DNA that contain a number of protein factors not normally present at telomeres, including the PML protein, Rad52, and RPA (Yeager et al. 1999). These structures are referred to as ALT-associated PML Bodies, or APBs. IF analysis showed that Rif1 was detectable in most of the APBs in the ALT cell line GM847 (Fig. 1E).
Rif1 accumulates at DSBs
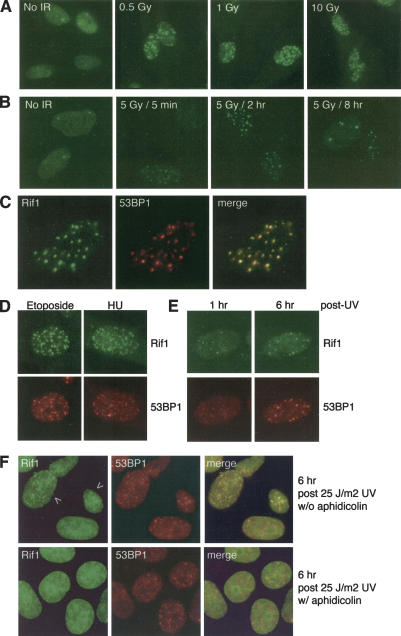
The association of Rif1 with dysfunctional telomeres and with the APBs of ALT cells suggested a role for Rif1 in the DNA-damage response. To test this, primary human fibroblasts were irradiated, and changes in the Rif1 pattern were determined by IF. After IR, all cells showed numerous discrete Rif1 foci that colocalized with 53BP1 (Fig. 2A–C). This response was uniform, rapid, persisted for many hours, and was dependent on radiation dose (Fig. 2A,B). At 8 h after irradiation, the number of Rif1 foci per cell had diminished (Fig. 2B). Rif1 also formed foci after treatment with etoposide and prolonged incubation in the presence of hydroxyurea (Fig. 2D), representing conditions that can lead to formation of DSBs. UV light also led to a Rif1 response (Fig. 2E), but these foci only formed in a subset (∼20%–30%) of the cells, and their appearance was delayed by several hours. Aphidicolin-treated cells failed to form Rif1 foci after UV (Fig. 2F), suggesting that the Rif1 response to UV light depends on DNA replication that could convert the primary UV damage into DSBs. In contrast, the 53BP1 foci induced by UV treatment did not require S-phase progression (Fig. 2F). Collectively, these findings suggest that Rif1 responds primarily to DSBs.
Figure 2.
Rif1 foci in response to DSBs. (A) Rif1 foci after IR treatment. Rif1 IF (Ab 1060) on IMR90 primary fibroblasts, exposed to the indicated levels of IR and fixed after 30 min. (B) Time course of the Rif1 response. IF as in A, but after 5 Gy IR and with fixation at the indicated time points. (C) Colocalization of Rif1 with 53BP1 foci. IF of IMR90 cells, exposed to 5 Gy IR and fixed 2 h post-IR. Cells were then stained with Rif1 (green) and 53BP1 (red) antibodies for IF. (D) Rif1 foci in response to etoposide and hydroxyurea treatment. IMR90 cells were treated for 1 h with 50 μg/mL etoposide. For HU treatment, cells were incubated for 18 h with 2 mM hydroxyurea prior to fixation. Rif1 and 53BP1 were detected as in C. (E) Response of Rif1 to UV irradiation. IMR90 cells were exposed to UV light (25 J/m2), fixed at the indicated time points, and processed for Rif1 and 53BP1 IF as in C. (F) Rif1 foci in UV-treated cells are DNA replication dependent. HeLa1.2.11 cells were incubated with 5 μg aphidicolin/mL during the 6-h period after the indicated UV treatment. Cells were processed for IF as in C. A total of 20%–30% of the cells showed Rif1 foci in cells not treated with aphidicolin.
In addition to 53BP1, the IR-induced Rif1 foci colocalized with several other DNA-damage-response factors, including γ-H2AX, Chk1 phosphorylated on S317, activated ATM (phosphorylated on S1981), Rad17 phosphorylated on S645, Mre11, and BRCA1 (Supplementary Fig. 2). As a control, TRF1 and other telomeric proteins did not localize to sites of DNA damage (data not shown; Zhu et al. 2000). We conclude that Rif1 behaves similarly to previously described factors that accumulate at sites of DSBs.
Rif1 controlled by ATM not ATR
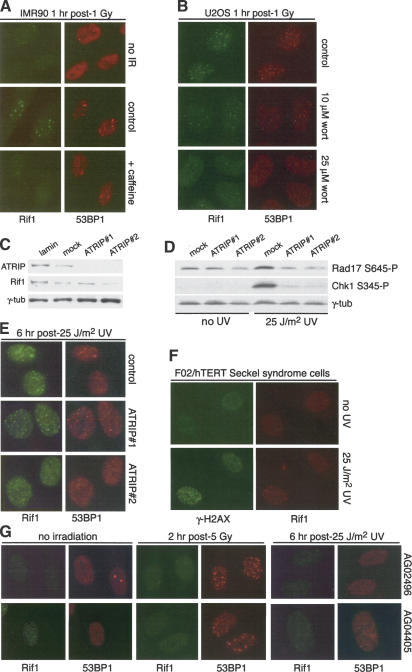
Treatment with caffeine or wortmannin, inhibitors of the PI3 kinase-related family of kinases (PIKKs), severely diminished Rif1 response to DNA damage (Fig. 3A,B; data not shown), suggesting that Rif1 is regulated by ATR and/or ATM. To test the dependence of Rif1 on the ATR kinase, we examined the response of Rif1 to UV in cells treated with siRNAs to ATRIP, an essential partner for ATR (Cortez et al. 2001). Two ATRIP siRNAs resulted in nearly complete depletion of the ATRIP protein (Fig. 3C) and strongly reduced the phosphorylation of Chk1 and Rad17 in response to UV (Fig. 3D). Despite the diminished ATR signaling, UV treatment continued to induce Rif1 foci (Fig. 3E). Furthermore, the Rif1 response to UV was preserved in cells derived from a Seckel syndrome patient with a severely reduced ATR protein level due to a splicing defect (Fig. 3F; O'Driscoll et al. 2003).
Figure 3.
Rif1 dependence on ATM, not ATR. (A) Effect of caffeine on Rif1 foci. IMR90 cells were treated with caffeine 2 h prior to 1 Gy IR and fixed 30 min post-IR. IF for Rif1 and 53BP1 as in Figure 2C. (B) Effect of wortmannin on Rif1 foci. U-2OS cells were treated with the indicated concentrations of wortmannin 1 h prior to 5 Gy IR, fixed 30 min post-IR, and processed for IF as in Figure 2C. (C) Immunoblotting analysis of the effect of ATRIP siRNAs. HeLa1.2.11 cells were transfected twice with the indicated siRNAs and analyzed by immunoblotting 72 h after the first transfection for expression of the indicated proteins. Rif1 was detected with mouse serum 1060. (D) Immunoblotting analysis of the effects of ATRIP siRNAs on Rad17 S345 and Chk1 S345 phosphorylation. HeLa1.2.11 cells were treated as in C and exposed to 25 J/m2 UV 72 h after siRNA transfection. Cells were collected 1 h post-UV and immunoblotting for the indicated proteins. (E) UV-induced Rif1 foci occur in cells treated with ATRIP siRNA. HeLa1.2.11 cells were exposed to 25 J/m2 UV 72 h after siRNA transfection. Cells were fixed after 6 h and processed for IF as in A. (F) UV-induced Rif1 foci in F02-98/hTERT cells. F02-98/hTERT Seckel syndrome fibroblasts were exposed to 0 or 25 J/m2 UV, fixed after 6 h, and processed for IF as in Figure 2C. After UV treatment, but not without UV, 10%–20% cells showed Rif1 foci that colocalized with γ-H2AX. (G) Absence of Rif1 foci in A-T cells. A-T fibroblasts (AG02496 and AG04405) were exposed to either 5 Gy IR or 25 J/m2 UV. Cells were fixed at the indicated time points and processed for IF as in Figure 2C.
To test the dependence of Rif1 on ATM, fibroblasts from two unrelated A-T patients lacking the ATM kinase were treated with IR. No Rif1 foci were observed in these cells (Fig. 3G), even though the cells expressed normal amounts of Rif1 protein (data not shown). Even prolonged (8 h) incubation after 5 Gy or treatment with 20 Gy failed to induce Rif1 foci (data not shown). In contrast, IR-treated A-T cells formed 53BP1 foci (Fig. 3G), albeit at diminished levels as previously noted (Rappold et al. 2001). A-T cells also failed to form Rif1 foci after UV treatment, whereas 53BP1 foci still were formed (Fig. 3G). These data indicate that the ability of Rif1 to form foci after the induction of DSBs requires the function of ATM, but not ATR.
Dependence of Rif1 on 53BP1
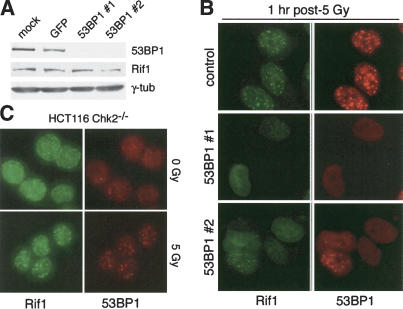
Inhibition of 53BP1 completely abolished the Rif1 response to DNA damage. Reduction in 53BP1 expression was achieved with two different siRNAs as demonstrated by immunoblotting (Wang et al. 2002; Fig. 4A). Rif1 protein levels were not affected (Fig. 4A). As expected, the induction of 53BP1 foci by IR was strongly reduced in cells treated with the 53BP1 siRNAs (Fig. 4B). Cells without 53BP1 foci also lacked Rif1 foci, indicating a functional relationship between these factors. Normal Rif1 and 53BP1 foci occurred in the few cells that probably had not taken up the siRNAs providing an internal control in the experiments (Fig. 4B). Thus, Rif1 appears to be regulated by two components of the ATM pathway, 53BP1 and ATM itself. As 53BP1 forms foci in A-T cells, but Rif1 does not (Fig. 3), it appears that Rif1 requires an ATM-regulated event in addition to normal levels of 53BP1 at sites of DNA damage. An alternative interpretation is that 53BP1 acts upstream of ATM and controls Rif1 in this way (Mochan et al. 2003). In contrast to 53BP1, several other components of the ATM-signaling pathway did not have an effect on the Rif1 response to DSBs. The Rif1 response was unaffected in Chk2-/- cells and cells lacking normal function of Mre11, Nbs1, and BRCA1 (Fig. 4C; Supplementary Fig. 3).
Figure 4.
Regulation of Rif1 by 53BP1 but not Chk2. (A) Immunoblotting analysis of the effect of 53BP1 siRNAs. HeLa cells were transfected twice with the indicated siRNAs and analyzed by immunoblotting 72 h after the first transfection for expression of the indicated proteins. Rif1 was detected with mouse 1060. (B) IR-induced Rif1 foci in cells treated with 53BP1 siRNA. Cells were exposed to 5 Gy 72 h after mock transfection or transfection with either control (GFP) or 53BP1 siRNAs. Cells were fixed 1 h after IR and processed as in Figure 2C. (C) IR-induced Rif1 foci in Chk2-/- HCT116 cells. HCT116 cells lacking functional Chk2 were treated with 5 Gy and processed for Rif1 and 53BP1 IF after 1 h as in Figure 2C.
Increased radiosensitivity upon Rif1 inhibition
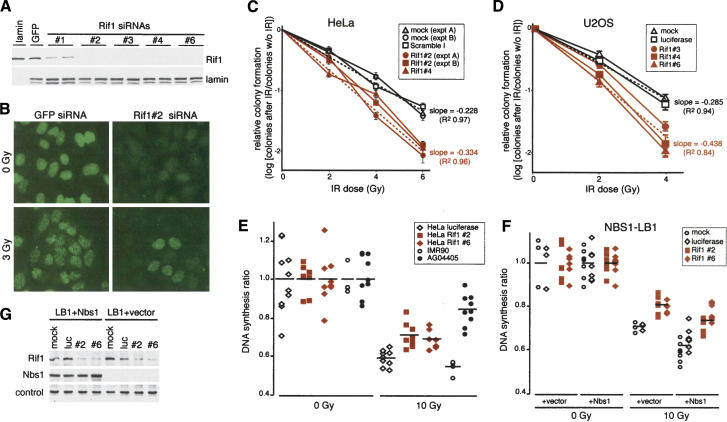
The consequences of diminished Rif1 function were assessed using several siRNAs to Rif1, which strongly reduced Rif1 protein levels (Fig. 5A) and blocked the formation of Rif1 foci after IR (Fig. 5B). Diminished Rif1 levels did not affect the autophosphorylation of ATM at Ser 1981 or the phosphorylation of Nbs1, Chk1, BRCA1, and p53 after irradiation (Supplementary Fig. 4A) or IR-induced focus formation by 53BP1 (Supplementary Fig. 4B).
Figure 5.
Rif1 inhibition increases radiosensitivity and affects the intra-S-phase checkpoint. (A) Immunoblotting analysis of Rif1 reduction using siRNAs. HeLa cells were transfected twice with the indicated siRNAs and analyzed by immunoblotting 72 h after the first transfection for expression of the indicated proteins. Rif1 was detected with Ab 1060. (B) Rif1 siRNA abrogates Rif1 ionizing radiation-induced foci. Rif1 siRNA treatment as in A. Cells were treatment with 0 or 3 Gy. IF for Rif1 (green) was performed using Ab 1060. (C) Effect on Rif1 siRNAs on clonogenic survival after IR. Cells treated with Rif1 siRNA #2 as in A were brought to the same cell density and irradiated with the indicated dose of IR at 72 h after transfection. Colonies on Coomassie-stained plates were counted 2 wk after plating. Bars indicate S.D.s of triplicate experiments. Broken lines indicate the least squared linear regression through either mock and Scramble I (black) or two independent Rif1 siRNAs (#2 and #4; red) data points. (D) Effect on Rif1 siRNAs on clonogenic survival after IR in U-2OS cells. Method as in C.(E) Effect of Rif1 siRNAs on the intra-S-phase checkpoint. Cells treated with siRNA as in A were treated with either 0 or 10 Gy IR at 72 h posttransfection. The DNA synthesis ratio (see Materials and Methods) were normalized to untreated cultures. (F) Rif1 siRNAs induced RDS in NBS1-LBI cells. NBS1-LB1 cells stably infected with control vector or wild-type Nbs1 retroviruses were treated as in E.(G) Immunoblot analysis of NBS1-LB1 cells treated with Rif1 siRNA. Rif1 siRNA does not affect ectopically expressed NBS1 protein level in NBS1-LB1 reconstituted wild-type NBS1. The loading control is a nonspecific band detected with the Nbs1 antibody.
However, cells with diminished Rif1 levels displayed increased radiosensitivity, a phenotype also seen for deficiency in ATM or 53BP1 (Taylor et al. 1975; Ward et al. 2003). Three tumor cell lines (HeLa, U-2OS, A549) showed a significant decrease in clonogenic survial upon treatment with Rif1 siRNAs compared with control cultures (Fig. 5C,D; data not shown). For instance, after 6 Gy, HeLa cells treated with Rif1 siRNAs showed a 4.3-fold reduction in clonogenic survival (P < 0.0001; Student's t-test). This effect is comparable to the radiosensitivity due to knockdown of BRCA1 with siRNA (Ganesan et al. 2002) or inhibition of ATM and SMC1 function with mutated alleles (Kim et al. 2002), but modest compared with the radiosensitivity of A-T cells, which typically show 10-fold lower survival rates. The RNAi experiments may underestimate the importance of Rif1 for survival after DNA damage, as siRNA effects are incomplete and short-lived.
Hypersensitivity to IR can be due to a deficiency in the Fanconi anemia (FA) pathway (D'Andrea and Grompe 2003), a target of ATM signaling (Taniguchi et al. 2002). To determine whether Rif1 contributes to the FA pathway, we determined whether Rif1 siRNA treatment resulted in hypersensitivity to the cross-linking agent MMC, a hallmark of FA cells, but not seen in A-T cells (Jaspers et al. 1982). The results show that Rif1 siRNAs did not diminish the ability of cells to survive in the presence of MMC (Supplementary Fig. 5), arguing against a role for Rif1 in the FA pathway.
Rif1 functions in the intra-S-phase checkpoint
A second consequence of ATM or 53BP1 deficiency is a defect in the intra-S-phase checkpoint (for review, see Osborn et al. 2002). Rif1 depletion led to a defect in the intra S-phase checkpoint as evidenced by RDS (Fig. 5E; Supplementary Table 1). In HeLa cells, knockdown of Rif1 with two siRNAs led to a partial RDS phenotype when compared with cells treated with luciferase siRNA or mock-treated cells (p = 0.0014; Scheffe's test; Supplementary Table 1). However, the RDS defect after Rif1 knockdown was significantly less prominent than the phenotype of A-T cells (p = 0.0007; Scheffe's test; Fig. 5E; Supplementary Table 1), consistent with the view that there are multiple ATM-regulated intra-S-phase checkpoint pathways.
The Mre11/Rad50/Nbs1 complex defines one of the ATM-regulated intra-S-phase checkpoints (Gatei et al. 2000; Lim et al. 2000; Wu et al. 2000; Zhao et al. 2000; Xu et al. 2001, 2002b; Kim et al. 2002; Yazdi et al. 2002). To determine whether Rif1 acts in this pathway, we tested the RDS phenotype of Rif1 inhibition in NBS1 mutant LB1 cells. These cells showed the expected effect of Nbs1 deficiency in terms of RDS (Fig. 6F; Supplementary Table 2) and retroviral expression of wild-type Nbs1 in these cells improved the intra-S-phase checkpoint (p = 0.0074; Scheffe's test; Fig. 5F,G; Supplementary Table 2), as reported previously (Zhao et al. 2000). Inhibition of Rif1 in LB1 cells resulted in an RDS phenotype (p = 0.0017; Scheffe's test; Supplementary Table 2), regardless of NBS1 status (Fig. 5F,G). Furthermore, the effects of Rif1 and Nbs1 deficiency were additive. This result would suggest that Rif1 acts in an intra-S-phase checkpoint pathway that is separate from the Nbs1 pathway.
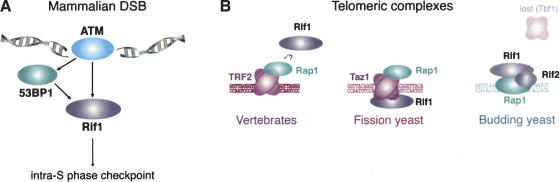
Figure 6.
Distinct functions of Rif1 in mammals and fungi (see Discussion for details).
One known Nbs1-independent intra-S-phase checkpoint pathway involves Chk2 (Falck et al. 2001, 2002). The phosphorylation state of Chk2 is in part under control of 53BP1 (Mochan et al. 2003), which also regulates Rif1. We therefore examined the effect of Rif1 inhibition on the phosphorylation of Chk2 on T68 after IR. Rif1 siRNA treatment did not diminish this phosphorylation event (Supplementary Fig. 4C). Rif1 also did not affect Chk2 phosphorylation in cells lacking functional Nbs1. In this regard, Rif1 is notably different from 53BP1, which is necessary for Chk2 phosphorylation in absence of normal Nbs1 function (Mochan et al. 2003; Supplementary Fig. 4C). In addition, we found that Rif1 siRNA treatment did not affect the degradation of Cdc25A after IR (data not shown). The simplest interpretation of these data is that Rif1 is not required for Chk2 activation. Conversely, Chk2 is not required for Rif1 focus formation after IR (Fig. 4C).
ATM and 53BP1 are also required for the rapid arrest of cells before mitosis after IR in G2 (Painter and Young 1980; DiTullio et al. 2002; Wang et al. 2002; Xu et al. 2002a). We monitored the G2/M checkpoint by assessing the propensity of cells to enter mitosis (on the basis of the presence of phosphorylated histone H3) within 1 h after IR. HeLa and U-2OS cells treated with Rif1 siRNA arrested prior to mitosis to the same extend as control cells, (Supplementary Fig. 6A,B; data not shown), indicating that the ATM-regulated G2/M checkpoint does not require full Rif1 function. Finally, the G1/S checkpoint appeared to be unaffected by Rif1 siRNAs, as both Chk2 phosphorylation and p53 activation occurred normally after IR (Supplementary Figs. 4, 6C).
Discussion
Human Rif1 is the ortholog of a fungal protein that associates with telomeres and regulates telomere length. We report that human Rif1 does not accumulate detectably at functional telomeres. Although a role for human Rif1 at telomeres is not excluded, our data show that the primary function of Rif1 is in the DNA-damage response. Rif1 localizes to DSBs in an ATM- and 53BP1-dependent manner and functions in the intra-S-phase checkpoint that serves to slow down DNA synthesis when DNA damage has occurred.
Rif1 as a new component of the ATM and 53BP1 pathway
Rif1 has a unique pattern of regulation, requiring functional 53BP1 and ATM for its accumulation at sites of DNA damage (Fig. 6A). Rif1 is unusual in its absolute dependence on ATM signaling. Because of this dependency, Rif1 foci are a useful indicator of ATM activity, providing a cell-based assay for ATM that is not confounded by activation of ATR. For example, the ATM-dependent UV response of Rif1 confirms earlier reports that UV treatment can activate both ATR and ATM (Jaspers et al. 1982; Oakley et al. 2001; Hannan et al. 2002; Heinloth et al. 2003). The UV-induced Rif1 foci were not formed when S-phase progression was blocked, indicating that the ATM pathway can be activated indirectly by UV lesions after they are processed during DNA replication. The strict dependence of Rif1 on ATM is also informative in the context of the Rif1 foci formed at a subset of uncapped telomeres. The Rif1 response to uncapped telomeres implicates that telomere damage signaling is in part mediated by ATM, a conclusion consistent with previous data (Karlseder et al. 1999; d'Adda di Fagagna et al. 2003; Takai et al. 2003; Wong et al. 2003).
Distinct functions for human and yeast Rif1: Intra-S-phase checkpoint and telomere length control
Inhibition human Rif1 recapitulates the two phenotypic hallmarks of ATM deficiency, radiosensitivity and RDS. The latter phenotype reflects inappropriate DNA synthesis after cells have been damaged with IR. The ability of Rif1 to modulate DNA synthesis after damage is independent of the Nbs1 component of the Mre11 complex, a previously identified mediator of the ATM intra-Sphase checkpoint. The budding yeast intra-S-phase checkpoint is largely similar to the human pathway, involving ATM/ATR orthologs and the Mre11 complex, as well as the yeast orthologs of Chk1 and Chk2. The conservation of this pathway raises the possibility that budding yeast Rif1 has a role in the yeast intra-S-phase checkpoint as well. It is less likely that Rif1 plays a role in the second S-phase checkpoint that responds to replication damage, as deficiencies in this pathway generally lead to gross chromosomal rearrangements, whereas strains lacking the RIF1 gene do not have this phenotype (Myung et al. 2001a,b).
The only known function of budding yeast Rif1 is at telomeres where it controls the frequency of telomere elongation by telomerase (Teixeira et al. 2004). Short telomeres contain less Rif1, and therefore, have a higher probability to be in a state that is permissive for telomerase action, resulting in telomere length homeostasis. Telomere length control by Rif1 is part of a pathway governed by the budding yeast ortholog of ATM (Craven and Petes 1999; Chan et al. 2001). This parallel with the control of mammalian Rif1 by ATM raises the possibility that telomere length regulation and the inhibition of DNA synthesis share important regulatory and/or mechanistic aspects.
Telomeres ensure that natural chromosome ends are not inappropriately processed by DNA-repair pathways and guarding against the activation of the ATM and ATR. Yet, telomeres contain numerous DNA repair factors and associate with components of the damage-signaling pathway. The emerging view is that telomeres deploy the DNA-damage-response machinery for the processing and maintenance of the telomeric complex, but do so in a highly controlled manner, such that the detrimental outcomes are avoided (see for instance, de Lange 2004). Examples are the nucleotide excision repair endonuclease ERCC1/XPF and nonhomologous end-joining factors Ku70/80 and DNA-PKcs, which contribute to the protection of human telomeres (Bailey et al. 1999; Zhu et al. 2003). These same factors have been implicated in the generation of telomere fusions, and must therefore be controlled by the telomeric complex in its functional state (Smogorzewska et al. 2002; Zhu et al. 2003). The role of Rif1 at yeast telomeres may be another example of this principle.
Evolutionary changes in the telomere complex
Budding yeast Rif1 is an interacting partner of Rap1, a DNA-binding protein that binds directly to yeast telomeres. Human Rap1 is distinct in that it does not bind DNA but associates with a telomeric protein, TRF2 (Li et al. 2000). Similarly, fission yeast telomeres contain a TRF-like protein (Taz1; for review, see Ferreira et al. 2004) that recruits Rap1 to chromosome ends, and a TRF1/2 ortholog was recently identified in Trypnosoma brucei (B. Li and G. Cross, pers. comm.) These findings corroborate the view that budding yeast telomeres have undergone a considerable alteration, accompanied by the loss of the TRF1/2-like telomere-binding proteins and associated rearrangements in other telomeric proteins (Fig. 6B).
The analysis of human Rif1 reveals further changes in the architecture of the telomeric protein complex (Fig. 6B). Whereas budding yeast Rif1 is recruited to telomeres through its interaction with Rap1, human Rif1 does not bind to Rap1 and does not accumulate strongly at telomeres unless they are dysfunctional. Fission yeast Rif1 is present at functional telomeres due to an interaction with Taz1, but binds more prominently when Rap1 is absent (Kanoh and Ishikawa 2001). As loss of Rap1 results in partial telomere dysfunction (M. Godinho, M.G. Fereira, and J. Cooper, pers. comm.), the strong binding of Rif1 to uncapped telomeres suggests that fission Rif1 also acts in the DNA-damage response. Therefore, human and fission yeast Rif1 may be similar in function both at telomeres and in the DNA-damage response.
Materials and methods
Cell culture and DNA-damaging agents
IMR90 human fibroblasts (ATCC, passage 20 or less), AG02496 and AG04405 A-T fibroblasts (Coriell Cell Repository), GM847 (ATCC), BJ/hTERT, ATLD-3 and ATLD-4 cells (Stewart et al. 1999; a gift from J.H.J. Petrini, Memorial Sloan-Kettering Cancer Center, New York, NY) were grown in DMEM/15% FBS. HeLa cells and NBS-1 LB1 cells (Kraakman-van der Zwet et al. 1999; a gift from M. Zdziencka, University of Leiden, Leiden, The Netherlands) infected with pLPC-Nbs1 or pLPC were grown in DMEM/10% BCS. U-2OS cells (ATCC) and HCT116 Chk2-/-; cells (a gift from P. Jallipalli, Memorial Sloan-Kettering Cancer Center, New York, NY; Jallepalli et al. 2003) were grown in McCoy's 5A/10% FBS. HCC1937 cells (ATCC) were grown in RPMI/10% FBS and 1 mM sodium pyruvate. All media were supplemented with 2 mM L-glutamine, 0.1 mM nonessential amino acids, 100 U of penicillin, and 0.1 μg of streptomycin/mL. For IR treatment, cells were exposed to a Cs137 source at a rate of 7.7 Gy/min in 6-cm dishes. For UV radiation, cells were washed with PBS and exposed to 25 J/m2 UV (265 nm) in a Stratalinker (Stratagene) without lid.
Immunoblotting and IF
Immunoblotting and immunofluorescence analysis were performed as described previously (Li et al. 2000; Zhu et al. 2000; Mirzoeva and Petrini 2001; Takai et al. 2003). Abs 1025 and 1060 were affinity purified from rabbit serum immunized with KLH-conjugated Rif1 peptides NSESDSSEAKEEGSRKKRSGK WKNK-Cys and EEGIIDANKTETNTEYSKSEEKLDN-Cys. Ab 1060 was also used without affinity purification and the same antigen was used to raise mouse serum to Rif1. Rabbit antibodies 1066 and 1067 were raised against GST fused to amino acids 906–1326 and used crude. Additional Abs used were as follows: TRF1, 371 (van Steensel and de Lange 1997); TRF2, 647 (Smogorzewska et al. 2000) or a monoclonal from Upstate; ATM 2C1 (GeneTex); phospho-ATM Ser 1981 (Bakkenist and Kastan 2003; a gift from M. Kastan, St. Jude's Children's Research Hospital, Memphis, TN); ATRIP-N (Cortez et al. 2001; a gift from S. Elledge, Harvard Medical School, Boston, MA); BRCA1 Ab-1 (Oncogene Research Products); phospho-BRCA1 Ser 1524 (Oncogene Research Products); phospho-Chk1 Ser 317 and phospho-Chk1 Ser 345 (Cell Signaling); γ-H2AX Ser 139 (JBW301, Upstate); Nbs1 (Maser et al. 2001; a gift from J.H.J. Petrini); phospho-p53 Ser15 (Cell Signaling); anti-p53 (DO-1; Santa Cruz); anti-p21 (F-5; Santa Cruz); phospho-Rad17 Ser 645 (Cell Signaling); 53BP1 (Ouellette et al. 2000; a gift from T. Halazonetis, Wistar Institute, Philadelphia, PA), or Novus Biologicals 100-305A; anti-phospho Chk2 (Cell Signaling #2661); α-tubulin clone B-5-1-2 (Sigma); lamin A/C (Santa Cruz sc-7292); and γ-tubulin clone GTU-88 (Sigma).
RNAi
Rif1 siRNAs (Dharmacon): #1, AACAGCAAGAAAUAGCAC CUAdTdT; #2, AAUGAGACUUACGUGUUAAAAdTdT; #3, AAGAGAAACCAGGUUCUGAAGdTdT; #4, AAGAAUGAG CCCCUAGGGAAAdTdT; #5, AAGAGGAAAAGUCUACUG ACUdTdT; #6, AAGAGCAUCUCAGGGUUUGCUdTdT. 53BP1 siRNAs 1 and 2 (Wang et al. 2002); ATRIP siRNAs 1 and 2 (Cortez et al. 2001); BRCA1 (Ganesan et al. 2002); GFP (Novina et al. 2002); lamin A/C and luciferase GL2 (Elbashir et al. 2001); and scramble I (Dharmacon, Scramble I duplex) were used. HeLa2, U-2OS, or NBS-1 LB1 cells were transfected using Oligofectamine (Invitrogen). Briefly, 1.0 × 105 of HeLa or 0.8 × 105 of U-2OS or NBS1-LB1 cells/well of a six-well plate were plated 18–24 h prior to transfection. Cells were transfected twice with a 24-h interval and processed 48 h later.
Clonogenic survival assay
Cells transfected with Rif1 and control siRNAs were harvested and counted in parallel 72 h after transfection. The cells diluted to the same cell density (5 × 104 cells/mL), irradiated, and subsequently plated in triplicate at a range of cell densities. After 10–14 d, plates were gently washed with PBS and stained with 50% MeOH/7% glacial acetic acid/43% H2O/0.1% Coomassie Brilliant Blue G for 10 min, rinsed with water, and air-dried. Colonies were counted for the various treatments on dilution plates yielding 50–200 colonies.
RDS assay
Inhibition of DNA synthesis after ionizing radiation was assessed on cells treated with siRNAs as previously described (Lim et al. 2000). Briefly, 4 h after the second siRNA transfection were placed into DMEM/10% FCS and 10 nCi of [methyl-14C]-Thymidine (Amersham) per milliliter, and incubated for 24 (HeLa and NBS1-LB1) or 48 h (AG04405 and IMR90). Cells were brought into medium without label, incubated for 24 h, and irradiated with either 0 or 10 Gy from a 137Cs source (dose rate: 3 Gy/min). After IR, cells were incubated for 30 min at 37°C and pulse-labeled with 2.5 mCi of [methyl-3H]Thymidine (Amersham Biosciences) per milliliter for 20 min at 37°C, followed by rinsing (two times) with DMEM containing 2.5 mM unlabeled thymidine, and cells were collected by trypsinization. After transfer to GF/C filters (Whatman), cells were washed with 10% trichloroacetic acid. The filters were rinsed with 100% ethanol, air-dried, and assayed in a liquid scintillation counter. The resulting ratios of 3H counts/min to 14C counts/min (3H/14C ratio) were corrected for channel crossover. DNA synthesis ratio after exposure to ionizing radiation was calculated as 3H/14C ratio in irradiated cells/3H/14C ratio in unirradiated cells.
G2/M checkpoint assay
G2/M checkpoint assay was performed as described (Xu et al. 2002b). Briefly, HeLa and U-2OS cells were transfected with control or Rif1 siRNA twice as described above. Cells were replated at a concentration of 4 × 105 cells/6-cm dish 24 h after the second transfection, and were cultured another 24 h. One hour after IR, cells were harvested, washed with PBS, fixed in 70% ethanol, resuspended in PBS containing 0.1% Triton-X and 0.5% BSA, and stained with anti-phospho-Histone H3 (Ser 10) antibody (Upstate) and propidium iodide for FACS analysis.
Acknowledgments
We are very grateful for antibodies and cell lines provided by J. Petrini, M. Zdziencka, S. Elledge, M. Kastan, T. Halazonetis, D. Livingston, and P. Jallipalli. J. Petrini, S. Elledge, and T. Halazonetis are thanked for discussion of this work in its early stages. We thank Jeff Ye, Richard Wang, Xu-Dong Zhu, and Jan Theunissen for technical advice and discussion. Xu-Dong Zhu and Richard Wang are also thanked for retrovirally infected NBS1-LB1 cells. This work was supported by a grant from the NCI to T.dL. (CA76027) and by the Breast Cancer Research Foundation. H.T. was supported by a Charles H. Revson fellowship. S.B.C.B. is supported by the William D. Hassett Fellowship of the Damon Runyon Cancer Research Foundation. (DRG-1753-03). F.E. is grateful for support from Boehringer Ingelheim, from the Gen-AU BIN program, from FWF (P15037), and from the Austrian Ministry for Economics. J.S. was supported by an NIH MSTP grant (GM07739) to the Cornell/RU/MSK Tri-Institutional MD/PhD program and by the Abby Rockefeller Mauze Medical Scientist Fellowship.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication date are at http://www.genesdev.org/cgi/doi/10.1101/gad.1216004.
Supplemental material is available at http://www.genesdev.org.
References
- Abraham R.T. 2001. Cell cycle checkpoint signaling through the ATM and ATR kinases. Genes & Dev. 15: 2177-2196. [DOI] [PubMed] [Google Scholar]
- Adams I.R. and McLaren, A. 2004. Identification and characterization of mRif1: A mouse telomere-associated protein highly expressed in germ cells and embryo-derived pluripotent stem cells. Dev. Dyn. 229: 733-744. [DOI] [PubMed] [Google Scholar]
- Bailey S.M., Meyne, J., Chen, D.J., Kurimasa, A., Li, G.C., Lehnert, B.E., and Goodwin, E.H. 1999. DNA double-strand break repair proteins are required to cap the ends of mammalian chromosomes. Proc. Natl. Acad. Sci. 96: 14899-14904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakkenist C.J. and Kastan, M.B. 2003. DNA damage activates ATM through intermolecular autophosphorylation and dimer dissociation. Nature 421: 499-506. [DOI] [PubMed] [Google Scholar]
- Chan S.W., Chang, J., Prescott, J., and Blackburn, E.H. 2001. Altering telomere structure allows telomerase to act in yeast lacking ATM kinases. Curr. Biol. 11: 1240-1250. [DOI] [PubMed] [Google Scholar]
- Cortez D., Guntuku, S., Qin, J., and Elledge, S.J. 2001. ATR and ATRIP: Partners in checkpoint signaling. Science 294: 1713-1716. [DOI] [PubMed] [Google Scholar]
- Craven R.J. and Petes, T.D. 1999. Dependence of the regulation of telomere length on the type of subtelomeric repeat in the yeast Saccharomyces cerevisiae. Genetics 152: 1531-1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- d'Adda di Fagagna F., Reaper, P.M., Clay-Farrace, L., Fiegler, H., Carr, P., Von Zglinicki, T., Saretzki, G., Carter, N.P., and Jackson, S.P. 2003. A DNA damage checkpoint response in telomere-initiated senescence. Nature 426: 194-198. [DOI] [PubMed] [Google Scholar]
- D'Andrea A.D. and Grompe, M. 2003. The Fanconi anaemia/BRCA pathway. Nat. Rev. Cancer 3: 23-34. [DOI] [PubMed] [Google Scholar]
- de Lange T. 2002. Protection of mammalian telomeres. Oncogene 21: 532-540. [DOI] [PubMed] [Google Scholar]
- ____. 2004. T-loops and the origin of telomeres. Nat. Rev. Mol. Cell Biology 5: 323-329. [DOI] [PubMed] [Google Scholar]
- DiTullio Jr R.A., Mochan, T.A., Venere, M., Bartkova, J., Sehested, M., Bartek, J., and Halazonetis, T.D. 2002. 53BP1 functions in an ATM-dependent checkpoint pathway that is constitutively activated in human cancer. Nat. Cell Biol. 4: 998-1002. [DOI] [PubMed] [Google Scholar]
- Elbashir S.M., Harborth, J., Lendeckel, W., Yalcin, A., Weber, K., and Tuschl, T. 2001. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature 411: 494-498. [DOI] [PubMed] [Google Scholar]
- Falck J., Mailand, N., Syljuasen, R.G., Bartek, J., and Lukas, J. 2001. The ATM-Chk2-Cdc25A checkpoint pathway guards against radioresistant DNA synthesis. Nature 410: 842-847. [DOI] [PubMed] [Google Scholar]
- Falck J., Petrini, J.H., Williams, B.R., Lukas, J., and Bartek, J. 2002. The DNA damage-dependent intra-S phase checkpoint is regulated by parallel pathways. Nat. Genet. 30: 290-294. [DOI] [PubMed] [Google Scholar]
- Ferreira M.G., Miller, K.M., and Cooper, J.P. 2004. Indecent exposure: When telomeres become uncapped. Mol. Cell 13: 7-18. [DOI] [PubMed] [Google Scholar]
- Ganesan S., Silver, D.P., Greenberg, R.A., Avni, D., Drapkin, R., Miron, A., Mok, S.C., Randrianarison, V., Brodie, S., Salstrom, J., et al. 2002. BRCA1 supports XIST RNA concentration on the inactive X chromosome. Cell 111: 393-405. [DOI] [PubMed] [Google Scholar]
- Gatei M., Young, D., Cerosaletti, K.M., Desai-Mehta, A., Spring, K., Kozlov, S., Lavin, M.F., Gatti, R.A., Concannon, P., and Khanna, K. 2000. ATM-dependent phosphorylation of nibrin in response to radiation exposure. Nat. Genet. 25: 115-119. [DOI] [PubMed] [Google Scholar]
- Haaf T., Golub, E.I., Reddy, G., Radding, C.M., and Ward, D.C. 1995. Nuclear foci of mammalian Rad51 recombination protein in somatic cells after DNA damage and its localization in synaptonemal complexes. Proc. Natl. Acad. Sci. 92: 2298-2302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannan M.A., Hellani, A., Al-Khodairy, F.M., Kunhi, M., Siddiqui, Y., Al-Yussef, N., Pangue-Cruz, N., Siewertsen, M., Al-Ahdal, M.N., and Aboussekhra, A. 2002. Deficiency in the repair of UV-induced DNA damage in human skin fibroblasts compromised for the ATM gene. Carcinogenesis 23: 1617-1624. [DOI] [PubMed] [Google Scholar]
- Hardy C.F., Sussel, L., and Shore, D. 1992. A RAP1-interacting protein involved in transcriptional silencing and telomere length regulation. Genes & Dev. 6: 801-814. [DOI] [PubMed] [Google Scholar]
- Heinloth A.N., Shackelford, R.E., Innes, C.L., Bennett, L., Li, L., Amin, R.P., Sieber, S.O., Flores, K.G., Bushel, P.R., and Paules, R.S. 2003. ATM-dependent and -independent gene expression changes in response to oxidative stress, γ irradiation, and UV irradiation. Radiat. Res. 160: 273-290. [DOI] [PubMed] [Google Scholar]
- Henson J.D., Neumann, A.A., Yeager, T.R., and Reddel, R.R. 2002. Alternative lengthening of telomeres in mammalian cells. Oncogene 21: 598-610. [DOI] [PubMed] [Google Scholar]
- Jallepalli P.V., Lengauer, C., Vogelstein, B., and Bunz, F. 2003. The Chk2 tumor suppressor is not required for p53 responses in human cancer cells. J. Biol. Chem. 278: 20475-20479. [DOI] [PubMed] [Google Scholar]
- Jaspers N.G., de Wit, J., Regulski, M.R., and Bootsma, D. 1982. Abnormal regulation of DNA replication and increased lethality in ataxia telangiectasia cells exposed to carcinogenic agents. Cancer Res. 42: 335-341. [PubMed] [Google Scholar]
- Kanoh J. and Ishikawa, F. 2001. spRap1 and spRif1, recruited to telomeres by Taz1, are essential for telomere function in fission yeast. Curr. Biol. 11: 1624-1630. [DOI] [PubMed] [Google Scholar]
- Karlseder J., Broccoli, D., Dai, Y., Hardy, S., and de Lange, T. 1999. p53- and ATM-dependent apoptosis induced by telomeres lacking TRF2. Science 283: 1321-1325. [DOI] [PubMed] [Google Scholar]
- Kastan M.B. and Lim, D.S. 2000. The many substrates and functions of ATM. Nat. Rev. Mol. Cell. Biol. 1: 179-186. [DOI] [PubMed] [Google Scholar]
- Kim S.T., Xu, B., and Kastan, M.B. 2002. Involvement of the cohesin protein, Smc1, in Atm-dependent and independent responses to DNA damage. Genes & Dev. 16: 560-570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraakman-van der Zwet M., Overkamp, W.J., Friedl, A.A., Klein, B., Verhaegh, G.W., Jaspers, N.G., Midro, A.T., Eckardt-Schupp, F., Lohman, P.H., and Zdzienicka, M.Z. 1999. Immortalization and characterization of Nijmegen Breakage syndrome fibroblasts. Mutat. Res. 434: 17-27. [DOI] [PubMed] [Google Scholar]
- Li B., Oestreich, S., and de Lange, T. 2000. Identification of human Rap1: Implications for telomere evolution. Cell 101: 471-483. [DOI] [PubMed] [Google Scholar]
- Lim D.S., Kim, S.T., Xu, B., Maser, R.S., Lin, J., Petrini, J.H., and Kastan, M.B. 2000. ATM phosphorylates p95/nbs1 in an S-phase checkpoint pathway. Nature 404: 613-617. [DOI] [PubMed] [Google Scholar]
- Marcand S., Gilson, E., and Shore, D. 1997. A protein-counting mechanism for telomere length regulation in yeast Science 275: 986-990. [DOI] [PubMed] [Google Scholar]
- Maser R.S., Bressan, D.A., and Petrini, J. 2001. The Mre11–Rad50 complex: Diverse functions in the cellular DNA damage response. In DNA damage and repair (eds. M.F. Hoekstra and J.A. Nickoloff), pp. 147-172. Humana Press, Totowa, NJ.
- Mirzoeva O.K. and Petrini, J.H. 2001. DNA damage-dependent nuclear dynamics of the Mre11 complex. Mol. Cell. Biol. 21: 281-288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochan T.A., Venere, M., DiTullio Jr., R.A., and Halazonetis, T.D. 2003. 53BP1 and NFBD1/MDC1-Nbs1 function in parallel interacting pathways activating ataxia-telangiectasia mutated (ATM) in response to DNA damage. Cancer Res. 63: 8586-8591. [PubMed] [Google Scholar]
- Myung K., Chen, C., and Kolodner, R.D. 2001a. Multiple pathways cooperate in the suppression of genome instability in Saccharomyces cerevisiae. Nature 411: 1073-1076. [DOI] [PubMed] [Google Scholar]
- Myung K., Datta, A., and Kolodner, R.D. 2001b. Suppression of spontaneous chromosomal rearrangements by S phase checkpoint functions in Saccharomyces cerevisiae. Cell 104: 397-408. [DOI] [PubMed] [Google Scholar]
- Novina C.D., Murray, M.F., Dykxhoorn, D.M., Beresford, P.J., Riess, J., Lee, S.K., Collman, R.G., Lieberman, J., Shankar, P., and Sharp, P.A. 2002. siRNA-directed inhibition of HIV-1 infection. Nat. Med. 8: 681-686. [DOI] [PubMed] [Google Scholar]
- Oakley G.G., Loberg, L.I., Yao, J., Risinger, M.A., Yunker, R.L., Zernik-Kobak, M., Khanna, K.K., Lavin, M.F., Carty, M.P., and Dixon, K. 2001. UV-induced hyperphosphorylation of replication protein a depends on DNA replication and expression of ATM protein. Mol. Biol. Cell 12: 1199-1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Driscoll M., Ruiz-Perez, V.L., Woods, C.G., Jeggo, P.A., and Goodship, J.A. 2003. A splicing mutation affecting expression of ataxia-telangiectasia and Rad3-related protein (ATR) results in Seckel syndrome. Nat. Genet. 33: 497-501. [DOI] [PubMed] [Google Scholar]
- Osborn A.J., Elledge, S.J., and Zou, L. 2002. Checking on the fork: The DNA-replication stress-response pathway. Trends Cell. Biol. 12: 509-516. [DOI] [PubMed] [Google Scholar]
- Ouellette M.M., McDaniel, L.D., Wright, W.E., Shay, J.W., and Schultz, R.A. 2000. The establishment of telomerase-immortalized cell lines representing human chromosome instability syndromes. Hum. Mol. Genet. 9: 403-411. [DOI] [PubMed] [Google Scholar]
- Painter R.B. and Young, B.R. 1980. Radiosensitivity in ataxiatelangiectasia: A new explanation. Proc. Natl. Acad. Sci. 77: 7315-7317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrini J. and Stracker, T. 2003. The cellular response to DNA double strand breaks: Defining the sensors and mediators. Trends Cell. Biol. 13: 458-462. [DOI] [PubMed] [Google Scholar]
- Rappold I., Iwabuchi, K., Date, T., and Chen, J. 2001. Tumor suppressor p53 binding protein 1 (53BP1) is involved in DNA damage-signaling pathways. J. Cell. Biol. 153: 613-620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiloh Y. 2003. ATM and related protein kinases: Safeguarding genome integrity. Nat. Rev. Cancer 3: 155-168. [DOI] [PubMed] [Google Scholar]
- Smogorzewska A. and de Lange, T. 2004. Regulation of telomerase by telomeric proteins. Annu. Rev. Biochem. 73: 177-208. [DOI] [PubMed] [Google Scholar]
- Smogorzewska A., van Steensel, B., Bianchi, A., Oelmann, S., Schaefer, M.R., Schnapp, G., and de Lange, T. 2000. Control of human telomere length by TRF1 and TRF2. Mol. Cell. Biol. 20: 1659-1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smogorzewska A., Karlseder, J., Holtgreve-Grez, H., Jauch, A., and de Lange, T. 2002. DNA ligase IV-dependent NHEJ of deprotected mammalian telomeres in G1 and G2. Curr. Biol. 12: 1635. [DOI] [PubMed] [Google Scholar]
- Stewart G.S., Maser, R.S., Stankovic, T., Bressan, D.A., Kaplan, M.I., Jaspers, N.G., Raams, A., Byrd, P.J., Petrini, J.H., and Taylor, A.M. 1999. The DNA double-strand break repair gene hMRE11 is mutated in individuals with an ataxia-telangiectasia-like disorder. Cell 99: 577-587. [DOI] [PubMed] [Google Scholar]
- Takai H., Smogorzewska, A., and de Lange, T. 2003. DNA damage foci at dysfunctional telomeres. Curr. Biol. 13: 1549-1556. [DOI] [PubMed] [Google Scholar]
- Taniguchi T., Garcia-Higuera, I., Xu, B., Andreassen, P.R., Gregory, R.C., Kim, S.T., Lane, W.S., Kastan, M.B., and D'Andrea, A.D. 2002. Convergence of the fanconi anemia and ataxia telangiectasia signaling pathways. Cell 109: 459-472. [DOI] [PubMed] [Google Scholar]
- Taylor A.M., Harnden, D.G., Arlett, C.F., Harcourt, S.A., Lehmann, A.R., Stevens, S., and Bridges, B.A. 1975. Ataxia telangiectasia: A human mutation with abnormal radiation sensitivity. Nature 258: 427-429. [DOI] [PubMed] [Google Scholar]
- Teixeira M.T., Arneric, M., Sperisen, P., and Lingner, J. 2004. Telomere length homeostasis is achieved via a switch between telomerase-extendible and -nonextendible states. Cell 117: 323-335. [DOI] [PubMed] [Google Scholar]
- van Steensel B. and de Lange, T. 1997. Control of telomere length by the human telomeric protein TRF1 Nature 385: 740-743. [DOI] [PubMed] [Google Scholar]
- van Steensel B., Smogorzewska, A., and de Lange, T. 1998. TRF2 protects human telomeres from end-to-end fusions. Cell 92: 401-413. [DOI] [PubMed] [Google Scholar]
- Wang B., Matsuoka, S., Carpenter, P.B., and Elledge, S.J. 2002. 53BP1, a mediator of the DNA damage checkpoint. Science 298: 1435-1438. [DOI] [PubMed] [Google Scholar]
- Ward I.M., Minn, K., van Deursen, J., and Chen, J. 2003. p53 binding protein 53BP1 is required for DNA damage responses and tumor suppression in mice. Mol. Cell. Biol. 23: 2556-2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong K.K., Maser, R.S., Bachoo, R.M., Menon, J., Carrasco, D.R., Gu, Y., Alt, F.W., and DePinho, R.A. 2003. Telomere dysfunction and Atm deficiency compromises organ homeostasis and accelerates ageing. Nature 421: 643-648. [DOI] [PubMed] [Google Scholar]
- Wotton D. and Shore, D. 1997. A novel Rap1p-interacting factor, Rif2p, cooperates with Rif1p to regulate telomere length in Saccharomyces cerevisiae. Genes & Dev. 11: 748-760. [DOI] [PubMed] [Google Scholar]
- Wu X., Ranganathan, V., Weisman, D.S., Heine, W.F., Ciccone, D.N., O'Neill, T.B., Crick, K.E., Pierce, K.A., Lane, W.S., Rathbun, G., et al. 2000. ATM phosphorylation of Nijmegen breakage syndrome protein is required in a DNA damage response. Nature 405: 477-482. [DOI] [PubMed] [Google Scholar]
- Xu B., Kim, S., and Kastan, M.B. 2001. Involvement of Brca1 in S-phase and G(2)-phase checkpoints after ionizing irradiation. Mol. Cell. Biol. 21: 3445-3450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu B., Kim, S.T., Lim, D.S., and Kastan, M.B. 2002a. Two molecularly distinct G(2)/M checkpoints are induced by ionizing irradiation. Mol. Cell. Biol. 22: 1049-1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu B., O'Donnell, A.H., Kim, S.T., and Kastan, M.B. 2002b. Phosphorylation of serine 1387 in Brca1 is specifically required for the Atm-mediated S-phase checkpoint after ionizing irradiation. Cancer Res. 62: 4588-4591. [PubMed] [Google Scholar]
- Yazdi P.T., Wang, Y., Zhao, S., Patel, N., Lee, E.Y., and Qin, J. 2002. SMC1 is a downstream effector in the ATM/NBS1 branch of the human S-phase checkpoint. Genes & Dev. 16: 571-582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeager T.R., Neumann, A.A., Englezou, A., Huschtscha, L.I., Noble, J.R., and Reddel, R.R. 1999. Telomerase-negative immortalized human cells contain a novel type of promyelocytic leukemia (PML) body. Cancer Res. 59: 4175-4179. [PubMed] [Google Scholar]
- Zhao S., Weng, Y.C., Yuan, S.S., Lin, Y.T., Hsu, H.C., Lin, S.C., Gerbino, E., Song, M.H., Zdzienicka, M.Z., Gatti, R.A., et al. 2000. Functional link between ataxia-telangiectasia and Nijmegen breakage syndrome gene products. Nature 405: 473-477. [DOI] [PubMed] [Google Scholar]
- Zhou B.B. and Elledge, S.J. 2000. The DNA damage response: Putting checkpoints in perspective. Nature 408: 433-439. [DOI] [PubMed] [Google Scholar]
- Zhu X.D., Kuster, B., Mann, M., Petrini, J.H., and de Lange, T. 2000. Cell-cycle-regulated association of RAD50/MRE11/NBS1 with TRF2 and human telomeres. Nat. Genet. 25: 347-352. [DOI] [PubMed] [Google Scholar]
- Zhu X.D., Niedernhofer, L., Kuster, B., Mann, M., Hoeijmakers, J.H., and de Lange, T. 2003. ERCC1/XPF removes the 3′ overhang from uncapped telomeres and represses formation of telomeric DNA-containing double minute chromosomes. Mol. Cell 12: 1489-1498. [DOI] [PubMed] [Google Scholar]
- Zou L. and Elledge, S.J. 2003. Sensing DNA damage through ATRIP recognition of RPA–ssDNA complexes. Science 300: 1542-1548. [DOI] [PubMed] [Google Scholar]