Abstract
Environmental cues transduced by an endocrine network converge on Caenorhabditis elegans nuclear receptor DAF-12 to mediate arrest at dauer diapause or continuous larval development. In adults, DAF-12 selects long-lived or short-lived modes. How these organismal choices are molecularly specified is unknown. Here we show that coregulator DIN-1 and DAF-12 physically and genetically interact to instruct organismal fates. Homologous to human corepressor SHARP, DIN-1 comes in long (L) and short (S) isoforms, which are nuclear localized but have distinct functions. DIN-1L has embryonic and larval developmental roles. DIN-1S, along with DAF-12, regulates lipid metabolism, larval stage-specific programs, diapause, and longevity. Epistasis experiments reveal that din-1S acts in the dauer pathways downstream of lipophilic hormone, insulin/IGF, and TGFβ signaling, the same point as daf-12. We propose that the DIN-1S/DAF-12 complex serves as a molecular switch that implements slow life history alternatives in response to diminished hormonal signals.
Keywords: Nuclear receptor, coregulator, aging, dauer, heterochrony
Nuclear hormone receptors orchestrate diverse aspects of metazoan development, metabolism, and homeostasis, often coordinating programs throughout the body via lipophilic hormones (Mangelsdorf et al. 1995). Some nuclear receptors, such as retinoic acid and thyroid receptors, turn on target genes in response to hormone, and turn them off in the absence, by assembling coactivator and corepressor complexes, respectively (Glass and Rosenfeld 2000). Identified coactivators such as SRC-1, GRIP, CBP/p300 and PCAF promote histone acetylation and potentiate transcription. Conversely, corepressors such as NCoR and SMRT, recruit histone deacetylases and catalyze transcriptional silencing. Although intensely studied biochemically, physiological descriptions of such complexes are just beginning (Yao et al. 1998; Jepsen et al. 2000; Xu and Li 2003). Governed by systemic signals, these complexes are well poised to act as molecular switches that can work at the level of the whole organism, specifying metabolic states, developmental alternatives, or transitions from juvenile to adult forms.
A genetic analysis of the Caenorhabditis elegans nuclear hormone receptor DAF-12 reveals the remarkable intricacy of receptor signaling in a simple in vivo model. DAF-12 is homologous to vertebrate vitamin D, pregnane, and androstane receptors (Antebi et al. 1998, 2000). In harsh environments, DAF-12 promotes dauer diapause, a stress-resistant, long-lived alternate third larval stage (Riddle et al. 1981; Antebi et al. 1998). In favorable environments, DAF-12 promotes reproductive development, regulating third stage (L3) and later programs within the heterochronic circuit, a regulatory pathway that controls developmental timing (Antebi et al. 1998, 2000). Moreover, it influences lipid metabolism, fertility, and adult life span (Larsen et al. 1995; Gems et al. 1998; Hsin and Kenyon 1999; Gerisch et al. 2001; Jia et al. 2002). Alleles fall into at least six classes based on dauer and heterochronic phenotypes, reflecting daf-12 functional complexity (Antebi et al. 1998). Notably, DNA-binding domain mutants fail to form dauer larvae (Daf-d), whereas some ligand-binding domain mutants constitutively form dauer larvae (Daf-c).
Many organisms, including mammals, couple environmental and physiological signals, such as nutrient availability to metabolism, development, and life span. For example, dietary restriction can delay puberty and prolong life in rodents (Tatar et al. 2003). The role of daf-12 and functionally related genes in C. elegans dauer formation gives an unprecedented view of how environmental signals transduced by endocrine networks influence similar processes (Riddle and Albert 1997; Finch and Ruvkun 2001). In abundant food, C. elegans transits rapidly through four larval stages (L1–L4) to short-lived reproductive adults (fast life history). Conditions signaling starvation, overcrowding, and thermal stress drive juveniles into long-lived dauer (slow life history). Molecular genetic and cellular studies of dauer formation reveal that specific neurons integrate sensory information and secrete insulin/IGF and TGFβ peptides in favorable environments (Ren et al. 1996; Riddle and Albert 1997; Finch and Ruvkun 2001; Li et al. 2003). Their respective signal transduction pathways ultimately converge on cytochrome P450, DAF-9, which is postulated to produce a lipophilic hormone, possibly a sterol, in endocrine cells (Gerisch et al. 2001; Jia et al. 2002; Gerisch and Antebi 2004; Mak and Ruvkun 2004). Acting through the nuclear receptor DAF-12, this hormone antagonizes diapause, promotes reproductive development, and short life. In unfavorable environments, peptide signaling pathways are suppressed; it is proposed that no hormone or an alternate hormone is made, and DAF-12 promotes dauer formation and long life. In higher metazoans, insulin/IGF signaling regulates sexual maturation, reproduction, and life span, and could do so in part by modulating lipophilic hormones (Tatar et al. 2003).
How are these activities specified? To elucidate how signaling circuits modulate daf-12, we investigated interacting proteins by the yeast two-hybrid method and identified DIN-1. It belongs to a family of large nuclear proteins typified by human SHARP and fly SPEN, which are corepressors for several transcription factors (Wiellette et al. 1999; Chen and Rebay 2000; Kuang et al. 2000; Shi et al. 2001; Oswald et al. 2002). By using genetic epistasis, we show that a small isoform, DIN-1S, works in an in vivo context for nuclear hormone signaling. We propose that as sterol hormone production declines in unfavorable environments, the DIN-1S/DAF-12 complex arrests development and specifies slow life history alternatives. This study reveals that coregulators can instruct alternate organismal fates.
Results
DIN-1 and DAF-12 interact
Nuclear receptors consist of an N-terminal ligand-independent activation function, a DNA-binding domain (DBD), a hinge region, and a ligand-binding domain (LBD) that binds corepressors, as well as fat soluble hormones and coactivators dependent on a C-terminal transactivation helix called AF-2 (Mangelsdorf et al. 1995). To identify DAF-12 complexes, we sought interacting proteins by the yeast two-hybrid method. With hinge and LBD of DAF-12 (amino acids 281–753) as bait, (Fig. 1B) we screened against an oligo-dT primed C. elegans mixed-stage cDNA library and obtained three independent clones corresponding to the C terminus of F07A11.6, dubbed DIN-1 (DAF-12-interacting protein 1; Fig. 1A, amino acids 1–567).
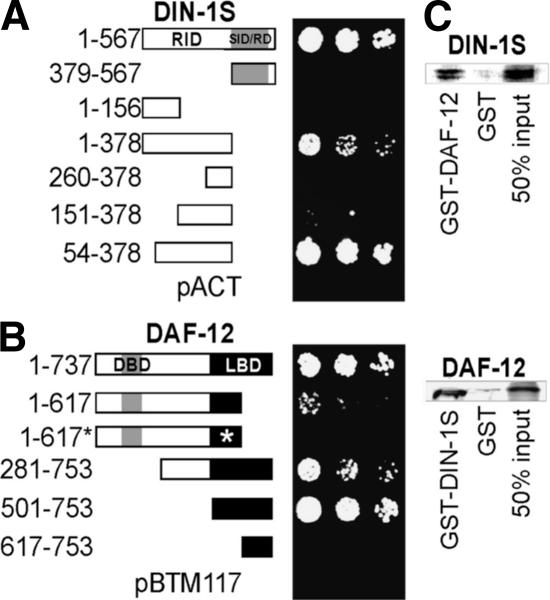
Figure 1.
DAF-12 and DIN-1 interact. (A) Yeast two-hybrid interaction of DAF-12 (281–753) bait with DIN-1S constructs. Dilution series of the yeast strain L40ccu transformed with the indicated constructs, growth selected on media lacking leucine, tryptophan, (for bait [pBTM117] and prey [pACT] plasmids), histidine, and uracil (for two-hybrid interaction). (B) Yeast two-hybrid interaction of DIN-1S (1–567) bait with DAF-12 constructs. Asterisk indicates the dh115 derivative of the DAF-12 (1–617) rh61 mutant protein. Numbering is according to isoform DAF-12A1 except 1–737, isoform A3 (Antebi et al. 2000). (C, Top) Radiolabeled DIN-1S (61 kd) interacts in vitro with GST::DAF-12. (Bottom) Radiolabeled DAF-12 (84 kd) interacts with GST::DIN-1S. Fifty percent of the radiolabeled input used for incubations is shown.
din-1 structure
The din-1 locus contains a coding region composed of 23 exons spanning 18.3 kb (Fig. 2A), and a 13-kb predicted promoter. The cDNA structures, determined by RT–PCR and from existent ESTs, revealed at least five isoforms. Exons 3 and 4 were joined in some isoforms, and exons 15 and 19 were alternately spliced. Notably, DIN-1L long isoforms (A–D) lacked exon 19, whereas DIN-1S short isoform uniquely contained it. One DIN-1S product was trans-spliced to SL1 at the exon 19 junction, another 438 bp upstream of exon 19. Other EST clones (yk814c08 and yk38b7) had even longer 5′ UTRs (452 and 688 bp, respectively), suggesting din-1S is transcribed as a discrete functional unit. Indeed, the analysis described below shows that din-1L and din-1S have distinct functions, and that din-1S specifically interacted with daf-12.
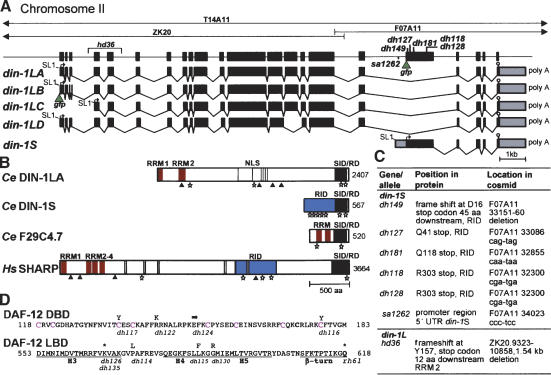
Figure 2.
din-1 structure. (A) Cosmids from the din-1 region, gene structure with mutations and din-1S::gfp insertion site (green triangle) are shown. (Bottom) mRNA isoforms containing SL1 trans-splice leader (line), start codon (arrow), stop codon (stop sign), and polyadenylation site (poly A). gfp insertion site of din-1LB is shown. (B) Domain structure of DIN-1 and homologs. (RRM) RNA recognition motif (red); (NLS) Nuclear localization sites (gray); (RID) nuclear receptor interaction domain (blue); (SID/RD) SMRT interaction domain/repression domain (black); (stars) L/IXXL/I/VL/I motifs; (triangles) AKT sites. (C) din-1 mutations. (D) daf-12(rh61) intragenic revertants. (Top) DAF-12 DBD, with conserved cysteines (magenta). (Bottom) LBD helices (H3–5). Amino acid changes are shown above the sequence, allele names below. (Arrow) Splice mutation; (asterisk) stop codon.
DIN-1 is evolutionarily conserved
DIN-1L belongs to a taxonomically conserved protein family, which includes founding member SPEN, implicated in multiple signaling pathways in flies (Wiellette et al. 1999; Kuang et al. 2000; Lane et al. 2000; Rebay et al. 2000; Lin et al. 2003); human corepressor SHARP (Shi et al. 2001); and mouse MINT (Newberry et al. 1999). DIN-1 and homologs are large nuclear proteins that contain two to four N-terminal RNA recognition motifs (RRMs); a long hinge of low complexity, predicted nuclear localization and AKT phosphorylation sites (RXRXXS/T); and a C-terminal conserved domain known as SID/RD (SMRT interaction domain/repressor domain; Fig. 2B). With respect to SHARP, there is 29% identity within the SID/RD domain and 22% and 26% identity within RRM1 and RRM2. DIN-1S uniquely contains exon 19, comprising the DAF-12 receptor interaction domain (RID), fused only to the SID/RD (Fig. 2A,B). SHARP binds SMRT/NCoR corepressors through the SID/RD (Shi et al. 2001; Ariyoshi and Schwabe 2003). Additionally, SHARP binds unliganded RAR and PPARδ through its RID (Shi et al. 2001, 2002), and several motifs resembling a hydrophobic corepressor helix (L/IXXI/VI) identified in SMRT (Hu and Lazar 1999) are postulated to mediate this interaction (Shi et al. 2001). DIN-1 isoforms contain variants of this motif (L/IXXI/V/L), as well as LXXLL motifs found in several nuclear coregulators (Heery et al. 1997; Fernandes et al. 2003), though the overall identity in the RID is weak. A smaller C. elegans DIN-1 paralog, F29C4.7 (Fig. 2B) containing the RRMs and SID/RD, better resembles the human OTT/RBM15 protein implicated in acute megakaryocytic leukemia (Ma et al. 2001; Mercher et al. 2001) and fly CG2910.
Analysis of interacting regions
To further define interacting regions, we analyzed various DIN-1 peptides for their association with the DAF-12 bait (Fig. 1A). The interaction of short isoform DIN-1S (amino acids 1–567) was delimited to the N-terminal half encoded by exon 19 (amino acids 1–378) and defined by mutation as functionally crucial (see below). Further truncations within this region led to observable (amino acids 54–378), reduced (amino acids 151–378) or no interaction (amino acids 1–156 and 260–378), suggesting the interacting domains may be distributed. In contrast, the SID/RD, common to DIN-1S and DIN-1L showed no interaction (amino acids 379–567). We also examined the association of DIN-1S with several DAF-12 peptides (Fig. 1B). Full-length isoform DAF-12A1 was unstable, but isoform DAF-12A3 (amino acids 1–737), which also contains DBD and LBD but differs in the hinge by 16 amino acids, interacts. So does a fragment roughly corresponding to isoform B (amino acids 501–753), composed of LBD only. A truncation of the LBD C terminus corresponding to the daf-12(rh61) mutant protein (amino acids 1–617) produced visible but reduced interaction. Finally, removal of the LBD N terminus (amino acids 617–753) abolished interaction, revealing that amino acids 501–617 are crucial. Interestingly, this region comprises helices H1–H5, which in vertebrate receptors interact with corepressors NCoR and SMRT (Horlein et al. 1995; Hu and Lazar 1999; Perissi et al. 1999). Pull-down experiments showed that short isoform DIN-1S specifically associated with DAF-12::GST in vitro but not with GST alone (Fig. 1C). Similarly, radiolabeled DAF-12 interacted with DIN-1S::GST.
din-1S alleles
RNAi against exon 19 of din-1S potently suppressed the Daf-c and heterochronic defects of daf-12 LBD mutants. We used this observation to obtain bonafide din-1 alleles by selecting for revertants of daf-12(rh61) and daf-12(rh274) gonadal migration (Mig) mutants (below). Such screens yielded five din-1 alleles: dh118, dh128, dh149, dh127, and dh181. Another allele, sa1262, suppresses the Daf-c phenotypes of ncr-1ncr-2 mutants (J. Li, G. Brown, and J. Thomas, pers. comm.). ncr-1 and ncr-2 encode homologs of Niemann-Pick C1 (Sym et al. 2000), a neurodegenerative disease locus implicated in cholesterol trafficking. Remarkably, all six din-1 alleles clustered around exon 19 (Fig. 2A,C) and selectively affected din-1S. Since the DAF-12 receptor interaction domain localized to this region (Fig. 1A, amino acids 1–378), association is likely largely disrupted.
Mapping farthest upstream, dh149 deleted 10 bp, causing a frameshift at D16 that results in a stop codon 45 amino acids downstream (Fig. 2C). Stop codons dh127, dh181, dh118/dh128 truncated the protein at Q41, Q118, and R303, respectively. Finally, sa1262 harbored a point mutation in the promoter/5′ UTR, 808 nucleotides before exon 19, suggesting that din-1S comprises a distinct transcriptional unit.
We also obtained nine daf-12(rh61) intragenic revertants from these genetic screens. Interestingly, five mutations affected the LBD between helices 3 and 5 (Fig. 2D), a region crucial for DIN-1 interaction (above), and thus might affect binding. Accordingly, we found that while the rh61 fragment, DAF-12 (amino acids 1–617), interacted with DIN-1S in the yeast two-hybrid system, a derivative containing the dh115 (L587F) lesion did not (Fig. 1B, amino acids 1–617*). Finally, four mutations affected the DBD (Fig. 2D), suggesting that loss of DNA binding abolishes the unregulated activity of daf-12(rh61) mutant protein.
din-1S regulates dauer formation
Based on the homology of DIN-1 with nuclear coregulators, we hypothesized that DIN-1 and DAF-12 work together in C. elegans dauer formation. To test this, we specifically reduced din-1S expression by using an exon 19 fragment in RNAi feeding experiments. Alleles isolated in genetic screens gave similar phenotypes. Candidate molecular null alleles (dh149, dh127, and dh181) were partially dauer-formation defective (Daf-d) forming fewer dauer larvae, which abnormally resumed pharyngeal pumping and gonadal development (Table 1). dh181 had the most severe dauer formation defects, whereas dh128 was actually non-Daf.
Table 1.
Interactions of din-1 with daf-12 and daf-9
| Genotype | % Normal dauer formationa | Daf phenotype | % L3 seamb | % adult seamc | % Reflexed distal tip celld | Brood sizee |
|---|---|---|---|---|---|---|
| N2 | 100, 100f | non-Daf | 100 | 100 | 100 | 303 ± 26, 190 ± 42f |
| din-1(dh149) | 19, 0f | Daf-d | 100 | 100 | 100 | N.D. |
| dh127 | 41, 0f | Daf-d | 100 | 100 | 100 | 235 ± 50 |
| dh181 | 7, 0f | Daf-d | 100 | 100 | 100 | N.D. |
| dh128 | 100, 100f | non-Daf | 100 | 100 | 100 | 270 ± 27 |
| hd36 | 100 | non-Daf | 100 | 100 | 89g | 76 ± 52, 4.6 ± 5.6f |
| daf-12(rh61) | 0 | Daf-d | 10 | 80 | 0 | 37 ± 29 |
| dh149 rh61 | 0 | Daf-d | 50 | 99 | 100 | N.D. |
| dh127 rh61 | 0 | Daf-d | 62 | 99 | 100 | 134 ± 33 |
| dh181 rh61 | 0 | Daf-d | 77 | 99 | 100 | N.D. |
| dh128 rh61 | 0 | Daf-d | 82 | 100 | 100 | 191 ± 40 |
| din-1SRNAi rh61 | 0 | Daf-d | 87 | 98 | 100 | N.D. |
| daf-12(rh61dh115) | 0 | Daf-d | 80 | N.D. | 100 | N.D. |
| din-1SRNAi rh61dh115 | 0 | Daf-d | 74 | N.D. | 100 | N.D. |
| daf-9(rh50) | 100 | non-Daf | 100 | 100 | 11 | N.D. |
| din-1SRNAi rh50 | N.D. | N.D. | N.D. | N.D. | 91 | N.D. |
N.D. not determined.
Percentage of dauer larvae that have normal developmental arrest in all tissues and do not pump the pharynx, n ≥ 25 dauers (from starved out plates)
n ≥ 10 animals (110 cells)
n ≥ 25 animals
n ≥ 25 animals (50 cells), distal tip cells that reach the dorsal side
n ≥ 5 animals
Measured at 25°C (phenotypes measured at 20°C except where noted)
11% Mig includes any mismigrated gonadal arm
To place din-1S in the dauer pathways, we performed genetic epistasis experiments with various dauer constitutive (Daf-c) mutants. din-1S RNAi or dh127 suppressed Daf-c phenotypes of conditional mutants that impede signaling from Insulin/IGF, TGFβ, and cGMP pathways at 25°C (Fig. 3A). Suppression of daf-2/Insulin/IGF receptor was seen with weak alleles (e1368, e1369, and m41), but not strong ones (e1370 and e1391). Similarly, suppression of the daf-7(m62) null was incomplete.
Figure 3.
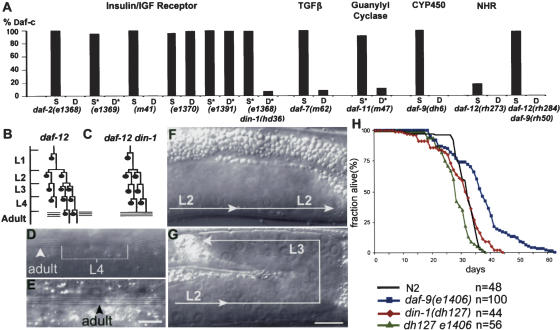
din-1S mutants suppress Daf-c, heterochronic, and long-lived phenotypes. (A) din-1S suppresses Daf-c mutants. daf-c single (S) mutant; daf-c din-1(dh127) double (D) mutants; daf-c mutant fed L4440 control plasmid (S*); daf-c fed din-1S RNAi (D*). n > 100 animals except for e1370D (88), e1391S* (48), and e1391D* (44). (B) daf-12(rh61) seam cell lineage defect. Seam cells repeat L2 programs of proliferative division at the L3 stage, and L4 programs of stem cell division at the adult stage giving rise to gaps in adult alae, indicated by a break in the three lines. (C) din-1(dh128) suppresses rh61 seam cell lineage defects, restoring a wild-type-like pattern. (D) Discontinuous alae of young adult rh61 hermaphrodite. Adult program, alae (arrowhead). L4 program, no alae (bracket). (E) Continuous alae in rh61dh128, which resembles wild type. (F) Unreflexed gonad arm of rh61, repeating L2 programs. (G) Reflexed gonad arm of rh61dh128 expressing normal L3 programs. Bar, 10 μm. (H) din-1 suppresses daf-9 longevity (15°C). N2 (mean [m] = 30 ± 1 d, Max [M] = 39 ± 1 d); din-1(dh127) (P = 0.3, m = 30 ± 0 d, M = 44 ± 1 d); daf-9(e1406) (p < 0.001, m = 33 ± 5 d, M = 56 ± 8 d); e1406dh127 (P < 6 × 10-8 compared with e1406, m = 27 ± 2 d, M = 38 ± 1 d). P values are calculated using the Student's t-test for survival curves relative to N2 unless indicated.
We next performed genetic epistasis on Daf-c mutants acting further downstream. Mutations in daf-9/cytochrome P450 are thought to block production of a lipophilic hormone for DAF-12, whereas mutations in the DAF-12 LBD (e.g., rh273, rh274) presumably diminish hormone binding itself (Antebi et al. 2000; Gerisch et al. 2001; Jia et al. 2002). Notably, din-1(dh127) suppressed Daf-c phenotypes of such mutants completely (Fig. 3A). Moreover, although hypomorphic Mig mutants daf-12(rh284) and daf-9(rh50) alone are not visibly Daf-c, the rh50rh284 double displayed a penetrant synthetic Daf-c phenotype (97%; n = 489), similar to the daf-9 null. din-1(dh127) completely suppressed the Daf-c phenotype of this double (0%, n > 500; Fig. 3A), consistent with action in a singular pathway. Because daf-12 and din-1S nulls behave similarly, suppression of Daf-c mutants could arise from DAF-12 destabilization. However, the daf-12::gfp expression pattern was unchanged in din-1(dh127) (n = 20). We conclude that din-1S acts partially downstream or parallel to insulin/IGF and TGFβ signaling, but fully downstream from lipophilic hormonal regulation of nuclear receptor daf-12.
din-1S influences larval stage-specific programs
The heterochronic loci are stage selectors that regulate C. elegans developmental timing. Mutants repeat or delete larval stage-specific programs in different tissues. Alone, din-1S alleles or din-1S RNAi had no observable heterochrony, but suppressed the delayed heterochronic phenotypes of daf-12 LBD and daf-9 hypomorphic mutants. In particular, daf-12(rh61) mutants fail to execute L3 seam cell divisions, adult cuticle programs, and gonadal cell migrations (Mig phenotype) on schedule and instead inappropriately repeat earlier larval programs (Fig. 3B,D,F; Table 1; Antebi et al. 1998). daf-9(rh50) mutants display gonadal Mig phenotypes. Significantly, din-1S mutants restored L3 stem cell division, adult cuticle formation, gonadal cell migration, and brood size in daf-12(rh61), and gonadal cell migration in daf-9(rh50) (Fig. 3C,E,G; Table 1). Moreover, din-1(dh127) suppressed the Mig phenotypes of several other daf-12 mutants including rh193, rh285, and rh284 (Supplementary Table S1), which are predicted to disrupt ligand binding or transactivation, suggesting that DIN-1S works with the unliganded receptor. In addition, dh127rh61 double mutants were similar in their residual heterochronic phenotypes to the daf-12(rh61dh115) intragenic revertant (Table 1), which affects DAF-12 association with DIN-1S (Fig. 1B, amino acids 1–617*). Finally, din-1 RNAi in the rh61dh115 background made no difference in phenotype, further supporting the idea that DIN-1 acts principally through the DAF-12 LBD.
Although a ligand for DAF-12 is unidentified, current evidence hints at a derivative of cholesterol, the precursor to many lipophilic hormones. Cholesterol deprivation of wild-type phenocopies Mig and Daf-c defects of daf-9 and daf-12 LBD mutants and enhances hypomorphic alleles (Gerisch et al. 2001), suggesting that loss of hormone production or binding causes these severe phenotypes. Importantly, we found that din-1(dh149) suppressed these induced phenotypes. Whereas 21% and 28% of wild type were Mig and Daf-c (n = 352) on low cholesterol media, 0% of dh149 mutants exhibited either phenotype (n = 73), revealing a critical function downstream of a cholesterol sensitive step. Similarly, such suppression was also seen with daf-12 nulls (Gerisch et al. 2001), suggesting din-1 and daf-12 may work together in this process.
din-1S is required for longevity
Lipophilic hormones may also influence nematode life span. daf-9 Daf-c animals that have recovered to adult live at least 25% longer than wild type, particularly at 15°C (Gerisch et al. 2001; Jia et al. 2002). In addition, they are thermotolerant (B. Gerisch and A. Antebi, unpubl.), often a correlate of longevity (Lithgow et al. 1995). din-1(dh127) alone had little effect on life span, but fully suppressed daf-9(e1406) longevity (Fig. 3H). Similarly, daf-9 longevity largely depends on daf-12(+) (Gerisch et al. 2001; Jia et al. 2002). These results suggest that in the absence of a daf-9-produced hormone, the DIN-1/DAF-12 complex specifies programs that promote longer life. dh127e1406 animals actually lived shorter than dh127 alone, perhaps reflecting a synthetic phenotype. Indeed, they also displayed impenetrant heterochronic phenotypes in L3 seams (17% mutant, n = 121 cells) not seen in either single mutant, but reminiscent of daf-12 nulls (21% mutant; Antebi et al. 1998).
DIN-1S has intrinsic repressor activity
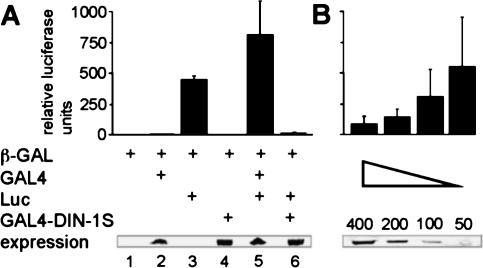
Given the similarity to the SHARP corepressor, we asked whether DIN-IS displayed any repressor activity. To address this, we fused DIN-1S to the yeast GAL4 DBD and assayed transcription from a GAL4 UAS-luciferase reporter in HEK293 cells. As expected, the luciferase reporter displayed a basal level of expression that was transactivated by GAL4 (Fig. 4A, lanes 3,5). Transactivation was potently inhibited by the GAL4–DIN-IS fusion protein far below basal levels of transcription (Fig. 4A, lane 6) and was dose dependent (Fig. 4B). Moreover, a GAL-4-DIN-1S (amino acids 1–378) construct lacking the SID/RD domain abolished repression (data not shown). These experiments reveal that DIN-1S possesses intrinsic repressor activity and suggest DIN-1S can recruit human components that mediate transcriptional repression.
Figure 4.
DIN-1S has intrinsic repressor activity. (A) The GAL4–UAS::luciferase reporter construct (Luc) was cotransfected into HEK293 cells with GAL4 or the GAL4–DIN-1S fusion protein. Relative luciferase activity was measured and normalized to β-galactosidase activity from the transfection control (β-GAL). Expression levels of GAL4 fusions are shown below. (B) Dose response curve of GAL4–DIN-1S. Nanograms of DNA cotransfected with luciferase reporter are shown.
din-1L is required for embryonic and larval development
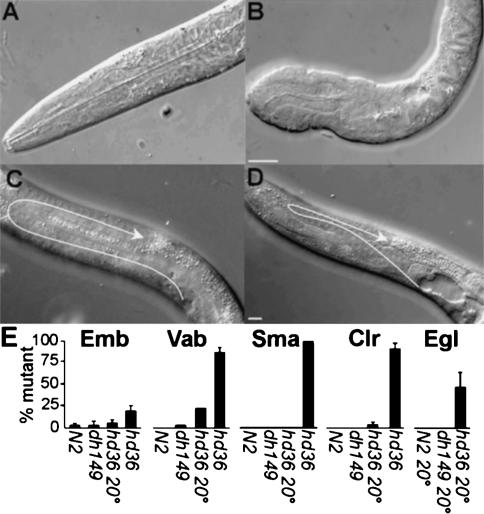
Because long isoforms were mutationally untouched and had no visible RNAi phenotype, we screened for a deletion among mutagenized pools using PCR (McKay and Jones 2002). hd36 deleted 1.54 kb, removing exons 5–6 (RRM2) and creating a frameshift after Y157 that truncates the protein 12 amino acids downstream (Fig. 2A,C). hd36 displayed temperature sensitive phenotypes wholly different from din-1S. At 20°C half the animals were egg-laying defective (Egl) with occasional mispositioned gonadal arms (Fig. 5C–E; Table 1). At 25°C, 18% arrested as embryos (Emb). Those animals that hatched usually displayed variable morphology defects in body and pharynx (Vab; Fig. 5A,B,E). Nearly all animals that reached adult were small (Sma), clear (Clr), slightly uncoordinated, constipated, and virtually sterile (Fig. 5E; Table 1), phenotypes not seen in din-1S mutants. hd36 was non-Daf, failed to suppress the Daf-c phenotype of daf-2(e1368) and had little effect on the ability of din-1S to do so (Fig. 3A). Moreover, hd36 din-1SRNAi animals had no additional discernible phenotype (not shown).
Figure 5.
din-1L(hd36) mutants display embryonic and larval defects. (A) Wild-type L1 head region. (B) hd36 L1 head region. (C) Wild-type gonad L4, arrow traces the path of gonadal outgrowth. (D) hd36 gonad L4. Bar, 10 μm. (E) Percentage embryonic lethal (Emb), variable body morphology defect (Vab), small (Sma), clear (Clr), or egg laying defective (Egl). n ≥ 50 animals, except hd36 Vab phenotypes (n = 27 at 20°C and 46 at 25°C). Phenotypes were measured at 25°C unless noted.
din-1L and din-1S are functionally independent
Although linked, din-1S and din-1L behaved essentially as two independent genes. Notably, they complemented one another: din-1S(dh127)/din-1L(hd36) transheterozygotes looked normal (100% non-Clear, non-Small, n = 68) and did not suppress daf-2 Daf-c phenotypes. In all, 62% of the F1 progeny of din-1S/din-1L daf-2(e1368) mothers formed Daf-c dauer larvae at 25°C (n = 872), approximating the expected 75% frequency had hd36 fully complemented dh127.
As another test of separability, we examined genomic constructs for rescue of din-1 phenotypes. First, cosmid T14A11, which encodes the entire locus, rescued both din-1L and din-1S. From a hd36 strain segregating T14A11 extrachromosomal array dhEx282, 100% of the animals scored as wild type (non-Clear, non-Small, non-Sterile, n = 50) contained the array, whereas 70% scored as mutants lacked it (n = 50). Thus, hd36 phenotypes arise from a disruption of din-1L, and not from a linked locus. From din-1S(dh127)daf-2(e1368) dhEx282 animals, 24% (n = 38) of those carrying the transgene showed antisuppression, restoring the daf-2 Daf-c phenotype. Moreover, extrachromosomal array dhEx235, comprising a mere 8.5 kb genomic fragment (exons 17–23), also rescued (39% Daf-c, n = 218), showing that a minigene encoding din-1S suffices to restore function.
din-1 influences lipid metabolism
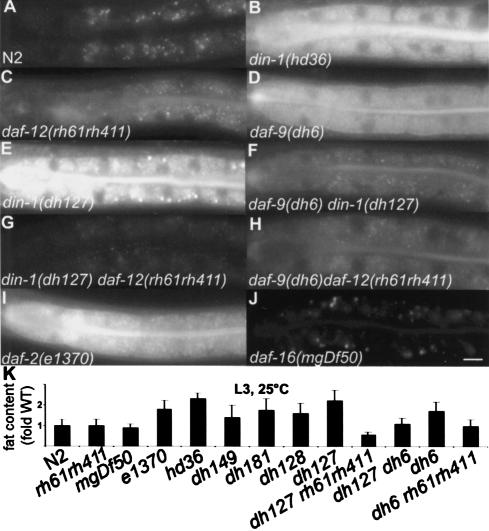
Prior to diapause, nematodes store lipid in intracellular droplets within the intestine and hypodermis. Daf-c mutants, such as daf-9 and daf-2, inappropriately accumulate excess intestinal fat in favorable conditions, a phenotype suppressed by Daf-d loci daf-12 and daf-16, respectively (Ogg et al. 1997; Gerisch et al. 2001). Surprisingly, din-1S Daf-d mutants as well as din-1L stored nearly twice the fat as wild type (Fig. 6A,B,E,K), as measured by vital staining with Nile red (Ashrafi et al. 2003). Moreover, daf-12 nulls suppressed the din-1S phenotype (Fig. 6C,E,G,K), suggesting that din-1S(+) antagonizes daf-12(+) fat storage. Paradoxically, din-1(dh127) also suppressed Nile red accumulation in daf-9 (Fig. 6D,F,K). Whereas each single mutant accumulated excess fat, din-1daf-9 did not, displaying a synthetic phenotype resembling the daf-12 null (Fig. 6C,H,K). Thus, din-1(+) antagonizes fat storage under reproductive growth but promotes it during entry to dauer, revealing a key regulatory role.
Figure 6.
Lipid deposition phenotypes. (A–J) Nile red accumulation of indicated genotypes (L3 larvae, 25°C, except e1370 and dh6, L2d larvae). Bar, 10 μm. (K) Nile red quantitation in N2 wild type, daf-12(rh61rh411), daf-16(mgDf50), daf-2(e1370), din-1S(dh149, dh128, dh181, dh127), din-1L(hd36), daf-9(dh6) and doubles (n ≥ 28).
din-1 is widely expressed in cell nuclei
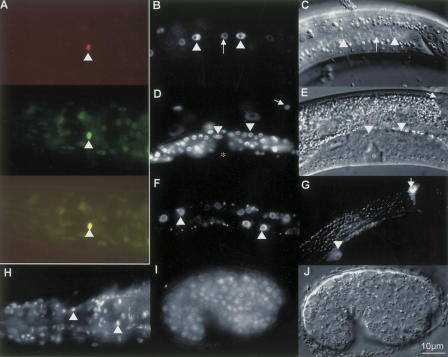
To investigate the din-1 expression pattern, we generated various GFP fusions. For din-1S, we inserted gfp in frame near the beginning of exon 19 in the 8.5 kb minimal genomic rescuing fragment (Fig. 2A). This construct was functional as measured by phenotypic rescue, and arrays dhEx285 and dhEx343 gave similar expression patterns. din-1S::gfp localized to the nuclei of most cell types. Detected in hypodermis, seam, intestine, and somatic gonad including the distal tip cells (Fig. 7B–G), din-1S was also expressed in neurons, vulval precursors, body wall muscle, and pharynx, all tissues with heterochronic phenotypes or remodeled during dauer. Expression was first detected in a few nuclei by the comma stage of embryogenesis. By hatch, din-1S was widely expressed, albeit weakly. Overall expression in most tissues was detected at various levels into adult and in dauer larvae. Interestingly, in late L1 and L2 larvae din-1S was often (75%, n = 16) highly expressed in the XXX cells (Fig. 7A), specialized neuroendocrine cells proposed to be a site of synthesis for the daf-9-produced hormone (Gerisch et al. 2001; Jia et al. 2002; Ohkura et al. 2003), suggesting control with endocrine circuits themselves. For din-1L, we placed 8 kb of the din-1 upstream region before gfp followed by din-1LB cDNA. Extrachromosomal arrays dhEx191 and dhEx194 were expressed in the nuclei of most cells from embryo to adult (Fig. 7H–J), as well as in dauer larvae. din-1L expressed more strongly in embryo and L1 larvae than din-1S, consistent with an earlier role.
Figure 7.
DIN-1 proteins are nuclear localized. (A–F) din-1S::gfp. (G) din-1Lp::gfp. (H–J) din-1LBcDNA::gfp. (A, top) rfp fusion to sdf-9/phosphatase marks XXX cell in L1 (Ohkura et al. 2003). (Middle) din-1S::gfp is highly expressed in XXX. (Bottom) Merge of both. (B) L3, din-1S::gfp in seam (arrowhead), hypodermal cells (arrow). (C) DIC image. (D) L4, distal tip (arrow), somatic gonadal cells (arrowhead). (E) DIC image. Asterisk, vulva. (F) L2, intestinal cells. (G) L4, din-1L promoter construct in seam (arrowhead) and distal tip cell (arrow). (H) Adult, din-1LBcDNA::gfp in pharynx and head neurons. (I) embryo. (J) DIC image. Bar, 10 μm.
Discussion
Nuclear receptor transcriptional complexes are key regulators of organismal biology. Our studies suggest that the transcriptional coregulator DIN-1 acts in a hormone-regulated molecular switch that specifies alternative metabolism, development, and life span in C. elegans. Because DIN-1 is evolutionarily conserved, these findings could have implications for higher metazoans. Whereas nuclear receptors are generally known for their hormonal activation, this work highlights the significance of the unliganded receptor. Moreover, a genetic approach reveals the in vivo role of DIN-1 in the context of larger endocrine networks including insulin/IGF, TGFβ, and sterol hormone signaling.
din-1 encodes functionally distinct isoforms
We found that din-1 encodes a complex locus with two largely separable functions encoded by short (S) and long (L) genes (Fig. 2). The major focus of this study was DIN-1S, which functionally interacted with DAF-12 nuclear receptor in dauer formation, stage specification, fat metabolism, and life span (see below). din-1L phenotypes included embryonic lethality and misshapen larvae. Those animals that grew were uncoordinated, small, stored excess fat, and were nearly sterile (Figs. 5, 6). Widespread expression and pleiotropic phenotypes probably indicate roles in multiple differentiation pathways that deserve future detailed exploration. Indeed, Drosophila SPEN works in EGF (Chen and Rebay 2000; Kuang et al. 2000), E2F (Lane et al. 2000), homeobox (Wiellette et al. 1999), and wingless signaling (Lin et al. 2003).
Although alternate isoforms or promoters are not uncommon in gene organization, the resultant products usually have related functions, often expressed in different tissues. Here, the two products have congruent expression patterns, but almost entirely different functions. Their diversification may reveal how coregulator complexes evolve to selectively interact with some transcription factors and not others. din-1 homologs in other species come in various isoforms (Kuang et al. 2000), though a short isoform is not yet described. Notably, DIN-1L homologs are highly variable in length (2407–5554 amino acids). Speculatively, interaction interfaces for different transcription factors could be modularly added or removed rapidly in evolution.
DIN-1S, a putative nuclear coregulator
Several lines of evidence strongly suggest that DIN-1S is a coregulator of DAF-12. First, DIN-1S and DAF-12 physically interacted by the yeast two-hybrid method and GST pull down experiments (Fig. 1). The interacting domain on DAF-12 corresponded to helices 1–5 of the LBD, a region in other nuclear receptors that docks coregulators (Horlein et al. 1995; Hu and Lazar 1999; Perissi et al. 1999), while the interacting region of DIN-1S was specific to this isoform and contained several putative hydophobic coregulator helices. Second, genetic experiments confirmed the in vivo relevance of these regions. Suppressors of the daf-12(rh61) LBD mutant either fell within the interacting region of DIN-1S (exon 19 RID), or of DAF-12 (helix 3–5), suggesting an in vivo interaction map (Fig. 2A,C,D). Particularly revealing was DAF-12(L587F) (dh115), which has conserved equivalents in PPARγ (L318) and PPARα (L309) that interact directly with the hydrophobic helices of coregulators, as shown by X-ray crystallography (Nolte et al. 1998; Xu et al. 2002). Direct contacts with DIN-1S are perturbed by dh115, since two-hybrid interactions were abolished (Fig. 1B, cf. amino acids 1–617 and 1–617*). Accordingly, dh115rh61 animals grown on din-1 RNAi were indistinguishable from dh115rh61 alone (Table 1). Third, din-1S mutants or RNAi potently suppressed nearly all daf-12 LBD mutants, presumably deficient in ligand binding as well as daf-9 mutants, deficient in ligand production (Table 1; Fig. 3; Supplementary Fig. S1), suggesting DIN-1S works with the unliganded receptor. Similarly, vertebrate coregulators are typically ligand regulated. Fourth, both din-1S and daf-12 null alleles were Daf-d and suppressed the Daf-c phenotypes of mutants in insulin, TGFβ, cGMP, and sterol signaling (Table 1; Fig. 3A). Thus they have similar phenotypes and patterns of epistasis. Finally, both were widely expressed and nuclear localized (Fig. 7).
Another important clue to DIN-1's potential biochemical activity comes from studies on the closest Homo sapiens homolog, SHARP. It functions as a corepressor with CBF-1 in the absence of intracellular Notch-IC (Oswald et al. 2002), as well as with unliganded nuclear receptors RAR and PPARδ (Shi et al. 2001, 2002), suggesting that interaction with nuclear receptors is phyletically ancient. What DIN-1S and SHARP share in common is a receptor interaction domain and the C-terminal SID/RD domain, which has been shown to coprecipitate SMRT corepressor and five members of the NURD complex, HDAC1, HDAC2, MTA2, MBD3, and RbAp48 causing target gene repression (Shi et al. 2001; Ariyoshi and Schwabe 2003). Importantly, unliganded receptors such as RAR or TR typically act as transcriptional repressors by recruiting such histone deacetylase complexes to promoter sites (Glass and Rosenfeld 2000).
As mentioned earlier, nearly all din-1 phenotypes were revealed with daf-12 mutants predicted to be unable to bind ligand, consistent with a corepressor function. For example, DIN-1 physically interacted with the rh61 mutant protein DAF-12 (amino acids 1–617; Fig. 1B), and din-1S mutants suppressed rh61 heterochrony (Fig. 3B–G). Truncated upstream of the C-terminal AF-2 transactivation helix, the rh61 encoded protein is likely ligand insensitive and predicted to repress transcription, based on analogous lesions in vertebrates (Hu and Lazar 2000). Similarly, din-1S mutants suppressed the phenotypes of several other daf-12 LBD mutants, including rh273 (Fig. 3A), which affects a predicted ligand contact site and is thought to diminish hormone binding to the receptor, and rh284 (Supplementary Table S1), which affects the AF-2 transactivation helix (Antebi et al. 2000). Moreover, din-1S suppressed daf-9/CYP450 mutants (Fig. 3A,H; Table 1) presumably deficient in DAF-12 hormone production (Gerisch et al. 2001; Jia et al. 2002). Consistent with a postulated role as corepressor, DIN-1S demonstrated intrinsic repressor activity when tethered to GAL4 in mammalian cell culture system (Fig. 4). Arguably, DIN-1S could still function as a coactivator, as it contains several LXXLL motifs typical of coactivators such as SRC-1 (Heery et al. 1997), and even classical corepressors coactivate in some promoter contexts (Tagami et al. 1999).
In either scenario, a simple hypothesis is that DIN-1S and DAF-12 form an active coregulator complex that is inhibited by ligand. In the absence of ligand, this complex could repress reproductive genes, activate dauerspecific genes, or both. This view not only helps clarify daf-12 phenotypic complexity, but helps explain how loss of ligand binding can cause more severe phenotypes than loss of DNA binding (Antebi et al. 1998; Antebi et al. 2000), as in cases of thyroid resistance (Hu and Lazar 2000). Such complexes likely inhibit the activity of other transcription factors resident at those promoters, thus silencing expression even below the basal level. In contrast, DBD mutants can neither transactivate nor repress. Therefore their phenotypes are milder because basal transcription is unimpeded. Further exploration of this mechanism will be facilitated by the identification of target genes and ligand. Below we discuss the physiological role of din-1S in more detail.
din-1S acts in the dauer pathways
For the first time in any organism, we placed a SHARP-like coregulator in an in vivo context for nuclear receptor signaling. In particular, because din-1S was separable from din-1L, we could focus specifically upon this pathway. Our studies showed that din-1S was crucial for dauer formation and variably suppressed Daf-c mutants (Fig. 3A). Importantly, complete suppression was seen only with daf-9 nulls and daf-12 LBD mutants, suggesting a proximal role within the nuclear receptor branch. Altogether, our findings suggest that din-1S acts partly downstream or parallel to insulin/IGF and TGFβ signaling, but fully downstream from hormonal regulation of the nuclear receptor, a genetic position consistent with the detected DAF-12/DIN-1S physical interaction. Partial suppression of insulin/IGF and TGFβ signaling mutants, however, does not exclude a direct role within these pathways either. Whereas daf-12 is absolutely essential for diapause, din-1S is not, arguing that other complexes could substitute. These could include DAF-3/SMAD and DAF-16/FOXO, which mediate the transcriptional output of TGFβ and insulin/IGF signaling, respectively (Finch and Ruvkun 2001), since their homologs are coregulators of nuclear receptors (Yanagisawa et al. 1999; Zhao et al. 2001; Dowell et al. 2003).
din-1S mediates daf-12 heterochrony
Evidently, hormonal signals also control the timing of developmental events including gonadal distal tip cell outgrowth (Gerisch et al. 2001; Gerisch and Antebi 2004; Mak and Ruvkun 2004). Mutations that perturb these signals delay or arrest migration; in daf-9(rh50) and daf-12 LBD mutants, distal tip cells fail their L3 migratory turns. din-1S mutants potently suppressed these phenotypes (Fig. 3B–G; Table 1), suggesting that din-1 works with daf-12 to regulate larval stage-specific programs. Interestingly, fly TAIMAN, a homolog of the AIB1 coactivator, also influences ecdysone dependent migrations of ovarian border cells (Bai et al. 2000), suggesting that hormonal control of gonadal outgrowth may be conserved. Delayed heterochrony in L3 and adult epidermal seam cells was also rescued, revealing that din-1S functions in multiple tissues. Accordingly, din-1S was nuclear in most stages and cells (Fig. 7), including those exhibiting heterochronic phenotypes as well as tissues remodeled during dauer formation. Homologs in other species are also nuclear and expressed widely (Newberry et al. 1999; Chen and Rebay 2000; Kuang et al. 2000; Shi et al. 2001).
din-1 perturbs lipid metabolism
While din-1S and daf-12 mutants had similar phenotypes for many processes, din-1S mutants influenced lipid metabolism in an unexpected way. Animals amassed excess lipid as measured by Nile red accumulation (Fig. 6). This is surprising because daf-12 mutants, as well as other Daf-d loci, have the opposite phenotype. Moreover, excess lipid accumulation in din-1(dh127) was suppressed by daf-12 nulls. This implies a novel function for the DIN-1/DAF-12 complex in the presence of the DAF-12 ligand during reproductive growth. Perhaps din-1S(+) prevents daf-12(+) from assembling with competing complexes that promote lipid accumulation. Both din-1 and daf-12 mutants also suppressed excess fat storage of daf-9, suggesting that upon dauer entry, din-1(+) has the opposite function, to promote fat storage. These results indicate a key role of receptor and coregulator in controlling organismal lipid metabolism.
din-1S mediates daf-9 longevity
daf-9 longevity is proposed to arise from a heterochronic re-expression of dauer-like programs in the adult (Gerisch et al. 2001). din-1S(+) was required for longer life, as daf-9 longevity was suppressed by din-1S mutants (Fig. 3H). Conceivably, in the absence of a daf-9-produced hormone, the DIN-1S/DAF-12 complex specifies dauer-like programs that promote somatic endurance, stress resistance, and survival. Whereas Insulin/IGF signal transduction is clearly established as a central regulator of metazoan aging, recent work suggests that nuclear receptor signaling and silencing complexes may mediate or modulate some of the effects (Tatar et al. 2003). Mutations that attenuate the fly ecdysone receptor (Simon et al. 2003) and histone deacetylase rpd3 (Rogina et al. 2002) result in increased longevity. rpd3 loss augments the NAD-dependent deacetylase, sir2, whose increased activity extends life span in yeast and worms (Guarente 2000; Tissenbaum and Guarente 2001). Intriguingly, human sir2 works in a corepressor complex with the PPARγ nuclear receptor to promote fat mobilization from white adipocytes (Picard et al. 2004). Conceivably, sir2 influences longevity in part through PPARγ.
Because din-1Sdaf-9 double mutants were slightly shorter-lived than either single mutant, din-1S is not strictly epistatic. Intriguingly, in other respects, din-1Sdaf-9 animals displayed synthetic phenotypes, such as weak heterochronic defects in L3 seam cells or fat storage phenotypes, not seen in either single mutant, but resembling daf-12 nulls. Presumably daf-12 null mutants can neither activate nor repress transcription. Similarly, the double might be deficient in both daf-9(+) dependent hormonal activation and din-1(+) mediated repression.
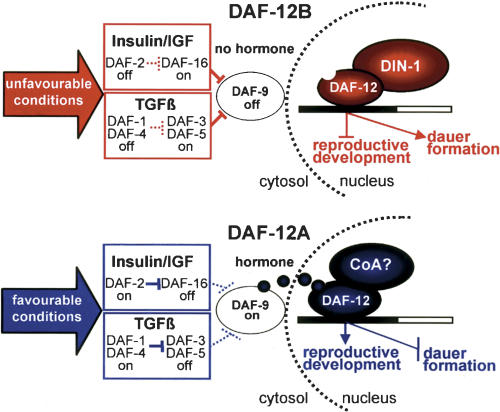
din-1S instructs slow life history traits
An emergent model for C. elegans' life history regulation by the nuclear receptor DAF-12 suggests that it has two activities: DAF-12A, which specifies reproductive development and short life span, and DAF-12B, which specifies dauer development and slow aging (Antebi et al. 1998; Gerisch et al. 2001). Genetic data show that DAF-12 is essential for diapause but only partly required for reproductive growth. Several lines of evidence suggest that A and B activities are specified by the presence and absence of a DAF-12 ligand, respectively. First, daf-9 encodes a cytochrome P450 related to steroidogenic and fatty acid hydroxylases that regulates DAF-12 and has phenotypes similar to DAF-12 ligand-binding domain mutants, notably Daf-c and Mig defects (Gerisch et al. 2001; Jia et al. 2002). Second, cholesterol deprivation phenocopies daf-12 LBD and daf-9 mutants, suggesting the hormone could be a sterol. Third, daf-9/CYP450 acts cell nonautonomously from select endocrine tissues and is regulated by environmental and genetic inputs through a DAF-12 feedback loop (Gerisch and Antebi 2004; Mak and Ruvkun 2004), all features characteristic of an endocrine mechanism.
In this work, we extend this model by showing the coregulator din-1 specifically works with the unliganded receptor to specify daf-12B slow life history traits (Fig. 8). We propose that when worms sense environmental stress, insulin/IGF, TGFβ, and cGMP pathways are suppressed, resulting in down-regulation of a DAF-9-produced hormone. Without hormone, DAF-12B/DIN-1S coregulator complexes inhibit programs of reproductive development, specify diapause, fat storage, and long life (slow life history). In favorable environments, sensory cues stimulate insulin/IGF, TGFβ, and cGMP signaling to activate DAF-9, which then produces a putative sterol hormone for DAF-12. With hormone, DAF-12A assembles postulated coactivator complexes to promote rapid reproductive growth and short life, whereas DIN-1S antagonizes fat storage (fast life history). It is highly adaptive for animals to adjust their physiology attuned to their environment. In particular, the capacity to shift metabolism, delay or arrest development and reproduction, and invest in maintenance and stress resistance to outlast adversity, could consequently delay somatic decline and aging. Endocrine coordination of these traits by lipophilic hormone and Insulin/IGF signaling, and their associated transcriptional complexes, could prove a common feature in metazoan evolution (Tatar et al. 2003).
Figure 8.
Life history model (see Discussion). Nondotted lines are active; dotted lines are inactive.
Materials and methods
Nematode culture, mutant isolation, and phenotypic analysis
Except where noted, nematodes were cultured at 20°C on NGM media inoculated with Escherichia coli strain OP50. Low cholesterol media was prepared as described (Gerisch et al. 2001). din-1 alleles dh118, dh127, dh128, and dh149 were isolated after ethyl methane sulfonate (EMS) mutagenesis in F2 screens of 168,000 haploid genomes for reversion of the daf-12(rh61) Mig phenotype (Antebi et al. 1998). din-1(dh181) was isolated in a screen of 112,000 haploid genomes for reversion of daf-12(rh274) Mig and slow growth phenotypes. Revertants were outcrossed to +/dpy-10 males. Mig cross males indicated extragenic suppressors, whereas non-Mig cross males indicated X-linked intragenic suppressors. Linkage to dpy-10(II) was established to identify din-1 candidates. hd36 was isolated in PCR-based screens for gene deletions using the poison primer protocol (McKay and Jones 2002). To identify mutations, genomic DNA from mutants was PCR amplified and sequenced on both strands. Mutants were outcrossed at least three times.
Brood size, L3 and adult seam, dauer formation, and life span assays were carried out as described (Antebi et al. 1998; Gerisch et al. 2001). Statistical analyses were performed with the Excel 98 Student's t-test. RNAi feeding experiments were carried out according to published procedures (Timmons et al. 2001). Nile red accumulation was carried out according to Ashrafi et al. (2003). Stained animals were observed by epifluorescent microscopy using Rhodamine filters on a Zeiss Axioplan microscope. Photographs were taken with a Hamamatsu Orca ER CCD digital camera, using Zeiss Axiovision software to quantitate pixels, and relative values normalized to N2.
Yeast two-hybrid screens
Yeast two-hybrid screens were performed as described (Wanker et al. 1997) using strain L40ccu. A LexA-DAF-12 hybrid containing daf-12 cDNA fragment corresponding to amino acids 281–753, was cloned into the KpnI site of LexA bait vector, pBTM117. The LexA–DAF-12 construct was used to screen a C. elegans oligo-dT primed cDNA library cloned into the GAL4 activation domain in pACT2 (R. Barstead, Oklahoma Medical Research Center, Oklahoma City). Independent transformants (108) were plated on minimal medium lacking histidine and uracil. His+, Ura+ colonies were tested for β-galactosidase activity. Counter selection on canavanine (15 μg/mL) induced loss of the bait plasmid. Positive clones were selected for Ampr in E. coli and sequenced. For DAF-12/DIN-1 interaction, daf-12 fragments using gene specific primers were subcloned into pBTM117, transformed into L40ccu expressing pACT-DIN-1S-31, and assayed for growth on selective media. Similarly, din-1S fragments were cloned into pACT2 and transformed into L40ccu expressing DAF-12 bait. Fusion proteins in yeast were detected by Western blot using anti-LexA or GAL4 antibody, respectively.
GST pull-down experiments
daf-12A cDNA and din-1S cDNA were cloned into the pGEX4T-1 vector. GST fusion proteins were induced in bacterial cultures in the presence of 10 μM ZnSO4. Recombinant protein was bound to glutathione-agarose beads (Sigma). 35S-methionine labeled protein was produced in vitro using the TnT expression kit (Promega), incubated with GST fusions or GST alone, and washed with 20 mM TrisHCl (pH 7.4), 200 mM NaCl, 1 mM EDTA. Proteins were resolved by SDS-page, blotted to nitrocellulose membranes, and followed by autoradiography.
Repressor assays
din-1S cDNA was cloned in frame downstream from GAL4 in pCMX-GAL4 to make pCMX-GAL4–DIN-1S. Various plasmid constructs (Shi et al. 2001) were cotransfected into HEK293 cells grown on 3-cm dishes at the following levels: pCMX-GAL4 (600 ng), pCMX-GAL4–DIN-1S (600 ng), and pMH100X4-TK-LUC (200 ng). pCMX-βGAL (200 ng) was used as a transfection control for normalization. Luciferase and β-galactosidase were assayed with the Enhanced Luciferase Assay Kit (BD Pharmingen) and Gal-Screen System, (Applied Biosystems) respectively, and quantitated by scintillation counting. At least three independent trials were performed. Expression of constructs was confirmed by Western blotting with anti-GAL4 antibody.
cDNA clones
YK501h2, YK297g11, YK132c3, YK96e12, YK38b7, and YK1121c03 din-1 cDNA clones from Y. Kohara (National Institute of Genetics, Mishima, Shizuoka, Japan) were sequenced on both strands. The 5′ ends of the L and S isoforms were determined essentially as described (Antebi et al. 2000). To obtain full-length din-1LB cDNA overlapping 5′ (ZK20.12055-4845) and 3′ fragments (ZK20.5090-F07A11.29280) were amplified from nematode cDNA and spliced together by their common XbaI site in TOPO-Blunt II, resulting in the plasmid Topo-din-1LB-cDNA.
Constructs and transgenics
For RNAi feeding constructs, a BglII fragment containing exon 19 (F07A11.32391-33145) was excised from pACT-DIN-1S-31 and ligated into the BglII site of RNAi expression vector L4440 (Timmons et al. 2001). To make the din-1Lp::gfp promoter construct, an 8-kb fragment of the din-1 promoter region (ZK20.20003-12055) flanked by NotI (5′) and AgeI (3′) sites was PCR amplified and cloned into gfp vector L3781 (A. Fire, Stanford University, Palo Alto, CA). To construct the din-1LB::gfp translational fusion, the 7.7-kb din-1LB cDNA flanked by NheI and ApaI sites was cloned into din-1Lp::gfp, placing GFP at the N terminus. For the din-1S genomic construct, an 8.5 kb genomic fragment (ZK20.1081-F07A11.28147) was cloned into TOPO blunt (Topo-din-1S). For the din-1 genomic construct tagged with gfp, a 3.4-kb din-1S promoter region (ZK20.1081-F07A11.33195) flanked by KpnI sites was amplified and cloned in front of gfp in vector L3781 (called din-1Sp::gfp). The 5-kb coding region, (F07A11.33195-28230) flanked by NheI and SpeI sites was amplified and cloned into din-1Sp::gfp to give din-1S::gfp. T14A11 cosmid, gfp, and plasmid constructs were injected at 10–20 ng/μL into the gonads of lin-15(n765) hermaphrodites, using 75–90 ng/μL of lin-15(+) pL15.EK or sur-5::gfp (pTG96) as a coinjectible marker.
Acknowledgments
We thank A. Fire for gfp vectors; the CGC for worm strains; Y. Kohara for cDNAs; A. Coulson for cosmids; B. Barstead for two-hybrid libraries; J. Lie, G. Brown, and J. Thomas for sa1262; I. Katsura for sdf-9::rfp; R. Evans for plasmids; A. Stein, P. Dowell, and A. Gruenweller for technical help; and A. Ehrenhofer-Murray for comments on the manuscript. This work was supported by the MPG and EC grant QLK6CT199902071.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article published online ahead of print. Article and publication date are at http://www.genesdev.org/cgi/doi/10.1101/gad.312604.
Supplemental material is available at http://www.genesdev.org.
References
- Antebi A., Culotti, J.G., and Hedgecock, E.M. 1998. daf-12 regulates developmental age and the dauer alternative in C. elegans. Development 125: 1191-1205. [DOI] [PubMed] [Google Scholar]
- Antebi A., Yeh, W.H., Tait, D., Hedgecock, E.M., and Riddle, D.L. 2000. daf-12 encodes a nuclear receptor that regulates the dauer diapause and developmental age in C. elegans. Genes & Dev. 14: 1512-1527. [PMC free article] [PubMed] [Google Scholar]
- Ariyoshi M. and Schwabe, J.W. 2003. A conserved structural motif reveals the essential transcriptional repression function of Spen proteins and their role in developmental signaling. Genes & Dev. 17: 1909-1920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashrafi K., Chang, F.Y., Watts, J.L., Fraser, A.G., Kamath, R.S., Ahringer, J., and Ruvkun, G. 2003. Genome-wide RNAi analysis of C. elegans fat regulatory genes. Nature 421: 268-272. [DOI] [PubMed] [Google Scholar]
- Bai J., Uehara, Y., and Montell, D.J. 2000. Regulation of invasive cell behavior by taiman, a Drosophila protein related to AIB1, a steroid receptor coactivator amplified in breast cancer. Cell 103: 1047-1058. [DOI] [PubMed] [Google Scholar]
- Chen F. and Rebay, I. 2000. split ends, a new component of the Drosophila EGF receptor pathway, regulates development of midline glial cells. Curr. Biol. 10: 943-946. [DOI] [PubMed] [Google Scholar]
- Dowell P., Otto, T.C., Adi, S., and Lane, M.D. 2003. Convergence of PPARγ and Foxo1 signaling pathways. J. Biol. Chem. 278: 45485-45491. [DOI] [PubMed] [Google Scholar]
- Fernandes I., Bastien, Y., Wai, T., Nygard, K., Lin, R., Cormier, O., Lee, H.S., Eng, F., Bertos, N.R., Pelletier, N., et al. 2003. Ligand-dependent nuclear receptor corepressor LCoR functions by histone deacetylase-dependent and -independent mechanisms. Mol. Cell 11: 139-150. [DOI] [PubMed] [Google Scholar]
- Finch C.E. and Ruvkun, G. 2001. The genetics of aging. Annu. Rev. Genomics Hum. Genet. 2: 435-462. [DOI] [PubMed] [Google Scholar]
- Gems D., Sutton, A.J., Sundermeyer, M.L., Albert, P.S., King, K.V., Edgley, M.L., Larsen, P.L., and Riddle, D.L. 1998. Two pleiotropic classes of daf-2 mutation affect larval arrest, adult behavior, reproduction and longevity in C. elegans. Genetics 150: 129-155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerisch B. and Antebi, A. 2004. Hormonal signals produced by DAF-9/cytochrome P450 regulate C. elegans dauer diapause in response to environmental cues. Development 131: 1765-1776. [DOI] [PubMed] [Google Scholar]
- Gerisch B., Weitzel, C., Kober-Eisermann, C., Rottiers, V., and Antebi, A. 2001. A hormonal signaling pathway influencing C. elegans metabolism, reproductive development, and life span. Dev. Cell 1: 841-851. [DOI] [PubMed] [Google Scholar]
- Glass C.K. and Rosenfeld, M.G. 2000. The coregulator exchange in transcriptional functions of nuclear receptors. Genes & Dev. 14: 121-141. [PubMed] [Google Scholar]
- Guarente L. 2000. Sir2 links chromatin silencing, metabolism, and aging. Genes & Dev. 14: 1021-1026. [PubMed] [Google Scholar]
- Heery D.M., Kalkhoven, E., Hoare, S., and Parker, M.G. 1997. A signature motif in transcriptional co-activators mediates binding to nuclear receptors. Nature 387: 733-736. [DOI] [PubMed] [Google Scholar]
- Horlein A.J., Naar, A.M., Heinzel, T., Torchia, J., Gloss, B., Kurokawa, R., Ryan, A., Kamei, Y., Soderstrom, M., Glass, C.K., et al. 1995. Ligand-independent repression by the thyroid hormone receptor mediated by a nuclear receptor corepressor. Nature 377: 397-404. [DOI] [PubMed] [Google Scholar]
- Hsin H. and Kenyon, C. 1999. Signals from the reproductive system regulate the lifespan of C. elegans. Nature 399: 362-366. [DOI] [PubMed] [Google Scholar]
- Hu X. and Lazar, M.A. 1999. The CoRNR motif controls the recruitment of corepressors by nuclear hormone receptors. Nature 402: 93-96. [DOI] [PubMed] [Google Scholar]
- ____. 2000. Transcriptional repression by nuclear hormone receptors. Trends Endocrinol. Metab. 11: 6-10. [DOI] [PubMed] [Google Scholar]
- Jepsen K., Hermanson, O., Onami, T.M., Gleiberman, A.S., Lunyak, V., McEvilly, R.J., Kurokawa, R., Kumar, V., Liu, F., Seto, E., et al. 2000. Combinatorial roles of the nuclear receptor corepressor in transcription and development. Cell 102: 753-763. [DOI] [PubMed] [Google Scholar]
- Jia K., Albert, P.S., and Riddle, D.L. 2002. DAF-9, a cytochrome P450 regulating C. elegans larval development and adult longevity. Development 129: 221-231. [DOI] [PubMed] [Google Scholar]
- Kuang B., Wu, S.C., Shin, Y., Luo, L., and Kolodziej, P. 2000. split ends encodes large nuclear proteins that regulate neuronal cell fate and axon extension in the Drosophila embryo. Development 127: 1517-1529. [DOI] [PubMed] [Google Scholar]
- Lane M.E., Elend, M., Heidmann, D., Herr, A., Marzodko, S., Herzig, A., and Lehner, C.F. 2000. A screen for modifiers of cyclin E function in Drosophila melanogaster identifies Cdk2 mutations, revealing the insignificance of putative phosphorylation sites in Cdk2. Genetics 155: 233-244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen P.L., Albert, P.S., and Riddle, D.L. 1995. Genes that regulate both development and longevity in C. elegans. Genetics 139: 1567-1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Kennedy, S.G., and Ruvkun, G. 2003. daf-28 encodes a C. elegans insulin superfamily member that is regulated by environmental cues and acts in the DAF-2 signaling pathway. Genes & Dev. 17: 844-858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin H.V., Doroquez, D.B., Cho, S., Chen, F., Rebay, I., and Cadigan, K.M. 2003. Splits ends is a tissue/promoter specific regulator of Wingless signaling. Development 130: 3125-3135. [DOI] [PubMed] [Google Scholar]
- Lithgow G.J., White, T.M., Melov, S., and Johnson, T.E. 1995. Thermotolerance and extended life-span conferred by singlegene mutations and induced by thermal stress. Proc. Natl. Acad. Sci. USA 92: 7540-7544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Z., Morris, S.W., Valentine, V., Li, M., Herbrick, J.A., Cui, X., Bouman, D., Li, Y., Mehta, P.K., Nizetic, D., et al. 2001. Fusion of two novel genes, RBM15 and MKL1, in the t(1;22)(p13;q13) of acute megakaryoblastic leukemia. Nat. Genet. 28: 220-221. [DOI] [PubMed] [Google Scholar]
- Mak H.Y. and Ruvkun, G. 2004. Intercellular signaling of reproductive development by the C. elegans DAF-9 cytochrome P450. Development 131: 1777-1786. [DOI] [PubMed] [Google Scholar]
- Mangelsdorf D.J., Thummel, C., Beato, M., Herrlich, P., Schutz, G., Umesono, K., Blumberg, B., Kastner, P., Mark, M., Chambon, P., et al. 1995. The nuclear receptor superfamily: The second decade. Cell 83: 835-839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKay S.J. and Jones, S.J. 2002. AcePrimer: Automation of PCR primer design based on gene structure. Bioinformatics 18: 1538-1539. [DOI] [PubMed] [Google Scholar]
- Mercher T., Coniat, M.B., Monni, R., Mauchauffe, M., Khac, F.N., Gressin, L., Mugneret, F., Leblanc, T., Dastugue, N., Berger, R., et al. 2001. Involvement of a human gene related to the Drosophila spen gene in the recurrent t(1;22) translocation of acute megakaryocytic leukemia. Proc. Natl. Acad. Sci. USA 98: 5776-5779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newberry E.P., Latifi, T., and Towler, D.A. 1999. The RRM domain of MINT, a novel Msx2 binding protein, recognizes and regulates the rat osteocalcin promoter. Biochemistry 38: 10678-10690. [DOI] [PubMed] [Google Scholar]
- Nolte R.T., Wisely, G.B., Westin, S., Cobb, J.E., Lambert, M.H., Kurokawa, R., Rosenfeld, M.G., Willson, T.M., Glass, C.K., and Milburn, M.V. 1998. Ligand binding and co-activator assembly of the peroxisome proliferator-activated receptor-γ. Nature 395: 137-143. [DOI] [PubMed] [Google Scholar]
- Ogg S., Paradis, S., Gottlieb, S., Patterson, G.I., Lee, L., Tissenbaum, H.A., and Ruvkun, G. 1997. The Fork head transcription factor DAF-16 transduces insulin-like metabolic and longevity signals in C. elegans. Nature 389: 994-999. [DOI] [PubMed] [Google Scholar]
- Ohkura K., Suzuki, N., Ishihara, T., and Katsura, I. 2003. SDF-9, a protein tyrosine phosphatase-like molecule, regulates the L3/dauer developmental decision through hormonal signaling in C. elegans. Development 130: 3237-3248. [DOI] [PubMed] [Google Scholar]
- Oswald F., Kostezka, U., Astrahantseff, K., Bourteele, S., Dillinger, K., Zechner, U., Ludwig, L., Wilda, M., Hameister, H., Knochel, W., et al. 2002. SHARP is a novel component of the Notch/RBP-Jκ signalling pathway. Embo. J. 21: 5417-5426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perissi V., Staszewski, L.M., McInerney, E.M., Kurokawa, R., Krones, A., Rose, D.W., Lambert, M.H., Milburn, M.V., Glass, C.K., and Rosenfeld, M.G. 1999. Molecular determinants of nuclear receptor–corepressor interaction. Genes & Dev. 13: 3198-3208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picard F., Kurtev, M., Chung, N., Topark-Ngarm, A., Senawong, T., Oliveira, R.M., Leid, M., McBurney, M.W., and Guarente, L. 2004. Sirt1 promotes fat mobilization in white adipocytes by repressing PPAR-γ. Nature 429: 771-776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebay I., Chen, F., Hsiao, F., Kolodziej, P.A., Kuang, B.H., Laverty, T., Suh, C., Voas, M., Williams, A., and Rubin, G.M. 2000. A genetic screen for novel components of the Ras/Mitogen-activated protein kinase signaling pathway that interact with the yan gene of Drosophila identifies split ends, a new RNA recognition motif-containing protein. Genetics 154: 695-712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren P., Lim, C.S., Johnsen, R., Albert, P.S., Pilgrim, D., and Riddle, D.L. 1996. Control of C. elegans larval development by neuronal expression of a TGF-β homolog. Science 274: 1389-1391. [DOI] [PubMed] [Google Scholar]
- Riddle D.L. and Albert, P.S. 1997. Genetic and environmental regulation of dauer larva development. In C. elegans II (eds. D.L. Riddle et al.), pp. 739-768. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [PubMed]
- Riddle D.L., Swanson, M.M., and Albert, P.S. 1981. Interacting genes in nematode dauer larva formation. Nature 290: 668-671. [DOI] [PubMed] [Google Scholar]
- Rogina B., Helfand, S.L., and Frankel, S. 2002. Longevity regulation by Drosophila Rpd3 deacetylase and caloric restriction. Science 298: 1745. [DOI] [PubMed] [Google Scholar]
- Shi Y., Downes, M., Xie, W., Kao, H.Y., Ordentlich, P., Tsai, C.C., Hon, M., and Evans, R.M. 2001. Sharp, an inducible cofactor that integrates nuclear receptor repression and activation. Genes & Dev. 15: 1140-1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y., Hon, M., and Evans, R.M. 2002. The peroxisome proliferator-activated receptor δ, an integrator of transcriptional repression and nuclear receptor signaling. Proc. Natl. Acad. Sci. USA 99: 2613-2618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon A.F., Shih, C., Mack, A., and Benzer, S. 2003. Steroid control of longevity in Drosophila melanogaster. Science 299: 1407-1410. [DOI] [PubMed] [Google Scholar]
- Sym M., Basson, M., and Johnson, C. 2000. A model for Niemann-Pick type C disease in the nematode C. elegans. Curr. Biol. 10: 527-530. [DOI] [PubMed] [Google Scholar]
- Tagami T., Park, Y., and Jameson, J.L. 1999. Mechanisms that mediate negative regulation of the thyroid-stimulating hormone α gene by the thyroid hormone receptor. J. Biol. Chem. 274: 22345-22353. [DOI] [PubMed] [Google Scholar]
- Tatar M., Bartke, A., and Antebi, A. 2003. The endocrine regulation of aging by insulin-like signals. Science 299: 1346-1351. [DOI] [PubMed] [Google Scholar]
- Timmons L., Court, D.L., and Fire, A. 2001. Ingestion of bacterially expressed dsRNAs can produce specific and potent genetic interference in C. elegans. Gene 263: 103-112. [DOI] [PubMed] [Google Scholar]
- Tissenbaum H.A. and Guarente, L. 2001. Increased dosage of a sir-2 gene extends lifespan in C. elegans. Nature 410: 227-230. [DOI] [PubMed] [Google Scholar]
- Wanker E.E., Rovira, C., Scherzinger, E., Hasenbank, R., Walter, S., Tait, D., Colicelli, J., and Lehrach, H. 1997. HIP-I: A huntingtin interacting protein isolated by the yeast two-hybrid system. Hum. Mol. Genet. 6: 487-495. [DOI] [PubMed] [Google Scholar]
- Wiellette E.L., Harding, K.W., Mace, K.A., Ronshaugen, M.R., Wang, F.Y., and McGinnis, W. 1999. spen encodes an RNP motif protein that interacts with Hox pathways to repress the development of head-like sclerites in the Drosophila trunk. Development 126: 5373-5385. [DOI] [PubMed] [Google Scholar]
- Xu J. and Li, Q. 2003. Review of the in vivo functions of the p160 steroid receptor coactivator family. Mol. Endocrinol. 17: 1681-1692. [DOI] [PubMed] [Google Scholar]
- Xu H.E., Stanley, T.B., Montana, V.G., Lambert, M.H., Shearer, B.G., Cobb, J.E., McKee, D.D., Galardi, C.M., Plunket, K.D., Nolte, R.T., et al. 2002. Structural basis for antagonist-mediated recruitment of nuclear co-repressors by PPARα. Nature 415: 813-817. [DOI] [PubMed] [Google Scholar]
- Yanagisawa J., Yanagi, Y., Masuhiro, Y., Suzawa, M., Watanabe, M., Kashiwagi, K., Toriyabe, T., Kawabata, M., Miyazono, K., and Kato, S. 1999. Convergence of transforming growth factor-β and vitamin D signaling pathways on SMAD transcriptional coactivators. Science 283: 1317-1321. [DOI] [PubMed] [Google Scholar]
- Yao T.P., Oh, S.P., Fuchs, M., Zhou, N.D., Ch'ng, L.E., Newsome, D., Bronson, R.T., Li, E., Livingston, D.M., and Eckner, R. 1998. Gene dosage-dependent embryonic development and proliferation defects in mice lacking the transcriptional integrator p300. Cell 93: 361-372. [DOI] [PubMed] [Google Scholar]
- Zhao H.H., Herrera, R.E., Coronado-Heinsohn, E., Yang, M.C., Ludes-Meyers, J.H., Seybold-Tilson, K.J., Nawaz, Z., Yee, D., Barr, F.G., Diab, S.G., et al. 2001. Forkhead homologue in rhabdomyosarcoma functions as a bifunctional nuclear receptor-interacting protein with both coactivator and corepressor functions. J. Biol. Chem. 276: 27907-27912. [DOI] [PubMed] [Google Scholar]