Abstract
Slit proteins steer the migration of many cell types through their binding to Robo receptors, but how Robo controls cell motility is not clear. We describe the functional analysis of vilse, a Drosophila gene required for Robo repulsion in epithelial cells and axons. Vilse defines a conserved family of RhoGAPs (Rho GTPase-activating proteins), with representatives in flies and vertebrates. The phenotypes of vilse mutants resemble the tracheal and axonal phenotypes of Slit and Robo mutants at the CNS midline. Dosage-sensitive genetic interactions between vilse, slit, and robo mutants suggest that vilse is a component of robo signaling. Moreover, overexpression of Vilse in the trachea of robo mutants ameliorates the phenotypes of robo, indicating that Vilse acts downstream of Robo to mediate midline repulsion. Vilse and its human homolog bind directly to the intracellular domains of the corresponding Robo receptors and promote the hydrolysis of RacGTP and, less efficiently, of Cdc42GTP. These results together with genetic interaction experiments with robo, vilse, and rac mutants suggest a mechanism whereby Robo repulsion is mediated by the localized inactivation of Rac through Vilse.
Keywords: Robo, Slit, Rac, Drosophila, cell migration, GTPase-activating protein
Directed cell migration is a striking feature of the development and physiology of virtually all animals. A particularly dramatic example of directed cell migration takes place in the developing nervous system, as the axons and dendrites of differentiating neurons are guided toward their appropriate target regions by attractive and repulsive cues provided in the extracellular environment (Tessier-Lavigne and Goodman 1996). Several conserved families of axon guidance molecules and their receptors have recently been identified (Dickson 2002). Perhaps not surprisingly, many of these molecules act not only on neuronal growth cones, but can also direct the migration of entire cells, including both neurons and nonneuronal cells. One such guidance cue is Slit, which acts through receptors of the Robo family to direct axon and cell migration in a variety of systems and species (Kidd et al. 1998, 1999; Zallen et al. 1998; Nguyen Ba-Charvet et al. 1999; Kramer et al. 2001). In the Drosophila ventral nerve cord, some axons but not others grow across the midline. Those axons that do cross, called commissural axons, cross the midline only once. This choice between a crossing (commissural) and noncrossing (longitudinal) pathway is controlled by Slit and Robo. Slit is expressed on midline cells, and repels axons expressing its receptor Robo. Commissural axons express only very low levels of Robo, whereas longitudinal axons express high levels of Robo. Accordingly, only the former can cross (Kidd et al. 1998, 1999).
Guided cell migration is also a central process in the development of the tracheal (respiratory) network in Drosophila, which extends its branches into most tissues of the animal. Air enters the system through the spiracles and is delivered directly to internal organs through unicellular thin capillaries, which are made by specialized terminal cells that target and invade the different tissues (Uv et al. 2003). The embryonic nerve cord receives 20 ganglionic branches (GBs), which are initially guided toward the ventral side of the embryo by the expression of the Drosophila FGF homolog branchless (Sutherland et al. 1996). The tip cell of the ganglionic branch (GB1) leads the way toward the ventral nerve cord (VNC) and during embryogenesis navigates a tortuous but invariable path of 50μm tracking along distinct nerves and glia (Englund et al. 1999). Inside the VNC, GB1 becomes exposed to a highly diverse array of positional signals provided by its surrounding cells and, with its relatively large size and distinct lineage, provides an advantageous single-cell model for the genetic dissection of the signaling events that steer its migration. GB1 is guided in part by Slit acting through the two receptors Robo and Robo2. Slit appears to act as a repellent through Robo to help prevent GB1 from crossing the midline, and as an attractant through Robo2 to facilitate GB1's initial extension toward the midline (Englund et al. 2002).
Guidance receptors such as the Robo proteins are thought to direct cell or axon migration by inducing dynamic and spatially coordinated changes in the actin and microtubule network in the growth cone or migrating cell. There is compelling evidence that the Rho family of small GTPases play a critical role in signaling between guidance receptors and the cytoskeleton (Luo 2002). In particular, Rac (and possibly also Cdc42) plays a critical role downstream of Robo in midline repulsion of CNS axons by Slit and Robo. This was first suggested by the observation that some longitudinal axons aberrantly cross the midline in various mutant combinations for the three Drosophila Rac genes (Hakeda-Suzuki et al. 2002). This finding was recently extended by Bashaw and colleagues (Fan et al. 2003), who reported genetic interactions between the Rac genes and both slit and robo, as well as biochemical evidence that stimulation of Robo receptors with Slit can lead to an increase in the amount of Rac in its active GTP-bound state. These genetic and biochemical data imply that one or more of the Racs plays a positive role in transducing the Robo repulsive signal in axon guidance.
How might Robo regulate the activity of Rac and other Rho family GTPases? Like other small GTPases, these proteins are regulated by the combined activity of guanine nucleotide exchange factors (GEFs), which stimulate the release of GDP and uptake of GTP, and GTPase-activating proteins (GAPs), which stimulate the hydrolysis of the bound GTP. Because the small GTPases are only active when bound to GTP, they become activated by the GEFs and inactivated by the GAPs. In vertebrates, stimulation of Robo1 by Slit has been shown to result in the recruitment and activation of members of the srGAP family of RhoGAPs, which appear to have some specificity for Cdc42 (Wong et al. 2001). These srGAPs could thus locally inactivate Cdc42 in response to Slit stimulation of the Robo receptor. However, srGAPs neither bind to nor regulate Rac, and so are unlikely to provide a direct link between Robo and Rac (Wong et al. 2001). Moreover, the srGAPs are not found in flies and worms, where the guidance functions of Slit and Robo proteins were first defined and are best understood. Thus, additional regulatory proteins linking Robo and Rho GTPases are predicted to exist, in both invertebrates and vertebrates.
Here we report the identification of a conserved family of RhoGAPs, the Vilse proteins, with clear representatives in flies and vertebrates. We show that Drosophila Vilse is required for midline repulsion of both CNS axons and tracheal GBs. Genetic and biochemical data support a model in which Vilse provides a direct link between Robo and Rac, and perhaps also Cdc42. Somewhat surprisingly, Vilse appears to have a positive role in Robo repulsion, despite its function as a negative regulator of Rac. This mirrors the presumed action of srGAPs in signaling from Robo to Cdc42, but is perplexing in light of the genetic and biochemical data suggesting a positive role for Rac. Our data are consistent with a more complex model in which both positive and negative factors cooperate to ensure that Rac activity is regulated in the precise temporal and spatial manner required for directed cell or growth cone migration.
Results
vilse is required for tracheal and axonal pathfinding at the VNC midline
The vilse locus was identified in a P-element screen for genes with pathfinding defects in the tracheal GB (J. Hemphälä and C. Samakovlis, unpubl.). The vilse lacZ strain contained a single P[w + mC = lacW] transposon in chromosomal position 93B10-11, which caused misroutings in GB outgrowth and, at lower frequency, crossing of the VNC midline; we therefore named the gene “vilse” (which means “lost” in Swedish). We generated additional mutants and revertants of this phenotype by excision of the P-element, and chose to study the allele vilse1 because the analysis of the genomic region in this mutant and in situ hybridization indicated that it represents the zygotic null condition for the gene.
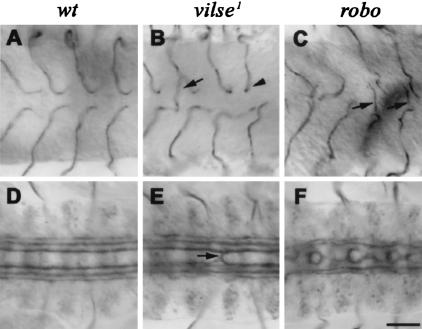
In wild-type midstage-16 embryos, the GB1 cell has reached the ventral side of the neuropil, and it turns posteriorly as it migrates in the proximity of the ventral longitudinal glia. Then, just before it reaches the midline, it abruptly turns to migrate dorsally to reach its final target on the dorsal side of the neuropil (Fig. 1A; Englund et al. 1999). In vilse1 mutants, 20% of the GBs (n = 154) migrated normally to the midline, but stalled once they reached it. An additional 14% of GBs failed to turn posteriorly; instead they extended straight toward the ventral midline, where most of them stalled (Fig. 1B, arrowhead) or, occasionally, continued to migrate across the midline (Fig. 1B, arrow). In wild-type embryos, <1% of the GBs (n = 140) migrated straight toward the midline, and none of them crossed it. Despite the low penetrance, this misguidance phenotype was interesting because it was similar to the tracheal phenotype seen in robo mutants, where GBs also migrate straight toward the midline but instead of halting there they often cross it (Fig. 1C, arrows; Englund et al. 2002).
Figure 1.
Ganglionic branch (GB) and axonal pathfinding defects in vilse1 and robo mutants. Late-stage-16 embryos stained to reveal tracheal lumen (by mAb2A12, A–C) and longitudinal fascicles (by mAb1D4, D–F). All panels show ventral views, anterior to the left. In wild-type embryos, GBs (A) and longitudinal fascicles (D) never cross the midline. (B) In vilse, a few GBs cross the midline (arrow), and several arrest upon reaching it (arrowhead). (E) Rare midline crosses are observed in vilse longitudinal fascicles. (C) GB1 midline crossing phenotypes in robo (arrows). (F) Axonal roundabouts at the midline (C–F, arrows). Bar, 20μm.
Given these similarities between vilse and robo mutants in GB1 guidance, we wondered whether vilse mutants might also show defects in CNS axon guidance similar to those in robo mutants. In wild-type embryos, Fasciclin II-positive axons project along specific pathways in the longitudinal connectives; they never cross the CNS midline (Fig. 1D). In robo mutants, many of these axons project along or across the midline (Fig. 1F; Kidd et al. 1998). A similar phenotype is also seen in vilse mutants (Fig. 1E), albeit at much lower frequency (1 in 30 embryos). This phenotype too suggested a link between vilse and the slit signaling pathway.
vilse encodes a conserved protein with WW, MyTH4, and RhoGAP domains
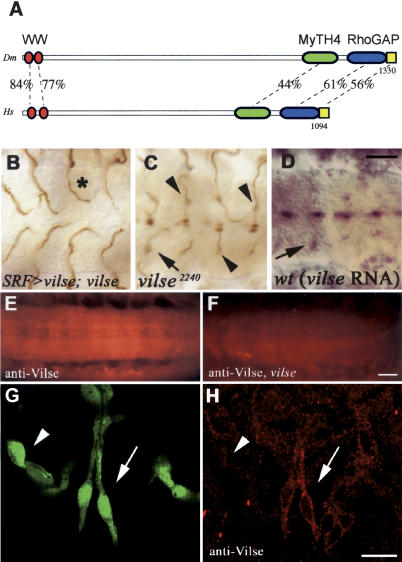
To further study the function of the affected gene in tracheal and axonal pathfinding, we cloned the genomic region surrounding the transposon insertion. Database searches with the sequence from genomic DNA flanking the vilse lacZ P-element showed that it was inserted in the 5′-untranslated region of the predicted gene CG3421 (RhoGap93B in GadFly). The search also identified several cDNAs deriving from this gene, and we sequenced the longest available clone, LD10379 (BDGP). The predicted Vilse protein contains a number of conserved domains: two N-terminal WW domains (residues 6–36 and 45–75), a more C-terminal myosin tail homology 4 (MyTH4) domain (amino acids 997–1124), a RhoGAP domain (amino acids 1154–1303), and a Pfam-B 53745 domain (amino acids 1304–1330; Fig. 2A). A predicted human protein, KIAA1688 (Nagase et al. 2000) has an identical domain structure and overall 29% identity and 51% similarity to Drosophila Vilse (Fig. 2), which is in turn the closest match to KIAA1688 in Drosophila. We therefore refer to KIAA1688 as the human Vilse protein. In addition, the human genome encodes a second Vilse homolog with an additional extensin-2 domain (48% similarity, 26% identity, GenBank accession no. gi|37574693|r). Vilse homologs can also be found in the mosquito (66% similarity, 29% identity for gi|30176853|) and mouse (50% similarity, 28% identity, gi|28380066|) genomes. We did not identify a protein with the same modular structure as Vilse in the Caenorhabditis elegans genome, the closest relative in worms (33% similarity, 13% identity) is encoded by C38D4.5 CE and contains a WW, a PH, and a RhoGAP domain.
Figure 2.
vilse encodes a RhoGAP expressed in tracheal tip cells and in the CNS. (A) Predicted domain structure of the Vilse protein (1330 amino acids), and its closest human homolog (KIAA1688, 1094 amino acids). Two WW, one MyTH4, one RhoGAP, and a conserved C-terminal region (Pfam-B 53745) were identified by Pfam. The degree of identity is shown for each domain. (B,C) Stage-16 embryos stained for the tracheal lumen (by mAb2A12; B). Expression of the vilse cDNA in the tracheal terminal cells of vilse1 mutants restores the erroneous outgrowth of GBs straight toward and across the midline (cf. C and Fig. 1B). It also causes premature turns away from the midline (*). (C) Ventral view of a vilse2240 mutant stained against β-gal and the tracheal lumen to reveal the tracheal phenotype (arrowheads) and the expression of the enhancer trap in GB1 nuclei (arrow) and midline cells. Ventral view of a wild-type stage-16 embryo. (D) Vilse RNA is visualized by in situ hybridization and is detected in the same pattern as the β-gal marker in GB terminal cells (arrow) and at the midline. (E,F) Ventral views of wild-type and vilse1 embryos stained with an anti-Vilse antiserum. Vilse protein is detected in the CNS of wild-type (E), and the signal is much reduced in the mutants (F). (G,H) Lateral view of a 1.eve-1 embryo carrying a pan-tracheal lacZ marker double-stained for β-gal (G) and Vilse (H). The terminal cells of the lateral trunk show strong cytoplasmic Vilse staining (arrow), but in the stalk cells of the ganglionic branch, Vilse is barely detectable (arrowhead). Bars: D, 20μm; F, 20μm; H, 8μm.
In situ hybridization revealed that vilse transcript is ubiquitous during the first stages of development, suggesting a robust maternal contribution (data not shown). Zygotic transcripts were prominent at stage 15 in the tip cells of all tracheal branches, the muscles, and midline cells of the VNC (Fig. 2D; data not shown). This pattern was the same as the β-gal marker expression in the enhancer trap strain (Fig. 2C), and was absent in vilse1 mutants (data not shown), indicating that vilse1 is a strong loss-of-function mutant in RhoGap93B. To analyze the expression of Vilse protein, we raised anti-Vilse antisera. Immunostainings of whole-mount wild-type embryos detected Vilse protein expression in a pattern that mirrored the pattern of the vilse transcript and that of β-gal expression in the vilse LacZ enhancer trap (data not shown). In addition, Vilse antisera stained the epidermis, the peripheral nervous system (PNS) segmental and intersegmental nerves, and the CNS longitudinal connectives and commissures (Fig. 2E; data not shown). This expression was not detected in the enhancer trap or by in situ hybridization, and may in part reflect the maternal protein. The antiserum is specific for Vilse, as the staining was much reduced in vilse1 mutants (Fig. 2F). Vilse staining showed a subcellular distribution consistent with a cytoplasmic localization of the protein (Fig. 2G,H, arrow).
The robust maternal contribution of vilse is a probable explanation for the relatively weak phenotype of the vilse1 mutant embryos, whose phenotypes are likely to be hypomorphic, given the long-lasting presence of maternal product. To analyze the function of maternal vilse and make embryos that lack Vilse completely, we generated vilse1 germ-line clones using the FLP/FRT technique (Chou and Perrimon 1996). vilse mosaic females produced only a few embryos, all of which were arrested early in embryogenesis, indicating that Vilse is also required for oogenesis (data not shown).
Next, we attempted to rescue the vilse1 GB phenotype by tracheal-directed expression of a UAS-LD10379 transgene. For this, we used the SRFGAL4 driver which selectively expresses GAL4 in the tracheal terminal cells (Jarecki et al. 1999). Only 3% of the GBs (n = 154) stalled or migrated straight to the midline in vilse1 embryos expressing LD10379 in the terminal cells (Fig. 2B), as opposed to 34% in vilse1 (see above). This rescue experiment confirms our identification of the vilse gene as RhoGAP93B, and also shows that the vilse tracheal phenotype is not a secondary consequence of the axonal defects.
We also noticed a mild gain-of-function phenotype in these experiments, in that 15% of the GBs stalled or turned away from the midline before even reaching the positions of ventral longitudinal glia (Fig. 2B, asterisk). This phenotype is even stronger in a wild-type background (25%, n = 168), indicating that it is likely due to increased levels of vilse expression. These opposing loss- and gain-of-function phenotypes resemble those for loss- and gain-of-function conditions of Robo (Englund et al. 2002), suggesting that Vilse may be a critical regulatory factor in Robo signaling during GB guidance.
vilse acts downstream of robo
To further test the idea that Robo and Vilse might act in a common signaling pathway, we examined dosage-sensitive genetic interactions between vilse and slit, robo, or robo2 mutants (Fig. 3). Specifically, we asked whether mutations in these genes would act as dominant enhancers of the vilse phenotype.
Figure 3.
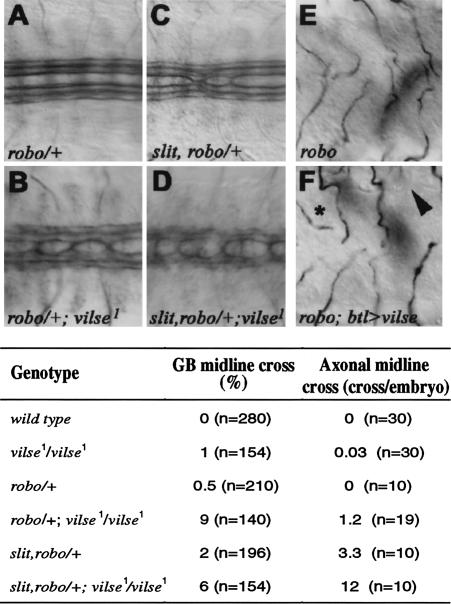
Genetic interactions of vilse with slit and robo. Ventral views of late-stage-16 embryos showing longitudinal fascicles stained by anti-FasII (A–D) and GBs (mAb2A12, E,F) of different mutant combinations. (A) In heterozygous robo embryos, longitudinal fascicles never cross the midline (anti-FasII). (B) Midline crossing is evident in all robo/+; vilse embryos. (C) Embryos lacking one copy of slit, robo exhibit three to four midline crosses per embryo. (D) This phenotype is also enhanced in slit, robo/+; vilse embryos. (E,F) Ventral views of robo and robo;BtlGAl4/UASvilse embryos. (F) Expression of UASvilse in all tracheal cells suppresses the ganglionic branch midline crossing phenotype of robo mutants. Overexpression of Vilse also causes GB premature turns (*) and stalling outside the VNC (arrowhead). The table shows the quantitation of the phenotypes.
First, we analyzed vilse1 embryos lacking one copy of robo, or of both slit and robo, and we found that the frequency of GBs crossing the midline was increased in both genotypes. In vilse1 homozygous mutant embryos, only 1% of GBs (n = 154) crossed the midline. In robo/+; vilse1 embryos, 9% of GBs (n = 140) crossed the midline. Reduction of both slit and robo in vilse1 mutants also increased midline crossing (Fig. 3), but the tracheal phenotypes of robo2/+; vilse1 embryos were indistinguishable from those vilse1 embryos. Thus, there is a selective enhancement of the vilse tracheal midline crossing phenotype when the dose of robo or slit and robo is reduced.
In parallel we analyzed the potential functional interaction between vilse, robo, and slit by looking at the longitudinal nerve fascicles. In vilse1 mutants, there was about one crossover of Fasciclin II positive axons per 30 embryos (or 0.03 crossovers/embryo, Fig. 3). This phenotype was enhanced when robo or both slit and robo function was reduced. In robo/+; vilse1 mutants, it increased to 1.2 crossovers per embryo (n = 19) and in slit, robo/+; vilse1 embryos, this phenotype was enhanced to 12 crossovers per embryo (n = 10; Fig. 3B,D). Lack of one robo copy alone caused only very weak if any defect, whereas in slit, robo heterozygous embryos only two to four longitudinal axon bundles per embryo crossed the midline (Fig. 3A,C; Kidd et al. 1999). The genetic interaction between vilse and mutations in robo and slit suggested a function of Vilse in repulsive Slit signaling. If vilse acts downstream of robo in a common pathway, we would predict that overexpression of vilse might reduce the number of tracheal branches that aberrantly cross the midline in robo mutants. To test this, we expressed UAS-vilse in the trachea of homozygous robo embryos, using the btl-GAL4 driver. UASvilse expression in robo mutants reduced the GB crossing of the midline phenotype (Fig. 3E,F) by more than half (11%, n = 154 vs. 29% in robo mutants, n = 168). In addition, an increased number of GBs failed to enter the CNS in these embryos, 21% compared with 1% in wild-type (n = 200) and 6% in robo mutants. This failure of GBs to enter the CNS was also evident when UAS-vilse was overexpressed in wild-type embryos using the btl-GAL4 driver (35%, n = 200) and suggests that vilse overexpression is sufficient to halt the early GB migration toward the CNS.
The WW domain of Vilse binds to the intracellular CC2 Robo domain
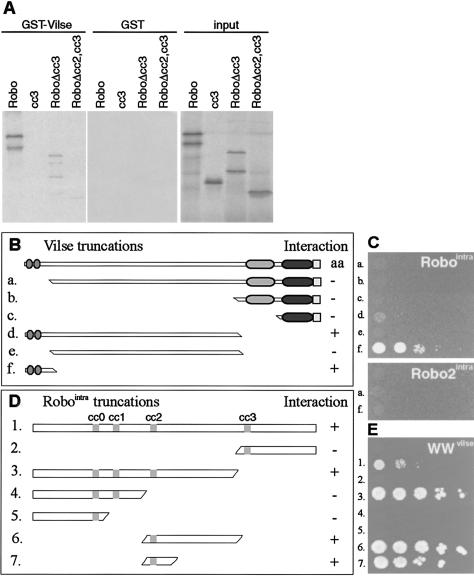
We asked whether Vilse might be a direct transducer of Slit signaling by first assessing the ability of glutathione S-transferase (GST)-Vilse-fusion proteins immobilized on glutathione beads to bind different fragments of the Robo intracellular domain, which were translated in vitro and 35S-labeled. Full-length GST-Vilse efficiently interacted with the intracellular part of the Robo receptor and a fragment containing the conserved domains CC0, CC1, and CC2. Constructs containing the CC0 and CC1 or only the CC3 domains did not bind to GST-Vilse, and none of the Robo fragments bound to beads loaded with GST alone (Fig. 4A). To further test the direct binding of Vilse to Robo and to identify the binding domains of the two proteins, we used a yeast two-hybrid assay. Variants of Vilse were expressed as LexA fusions whereas Robo deletions were expressed as fusions to the B42 activation domain. After cotransformation into yeast, only clones containing interacting proteins grow on media lacking leucine.
Figure 4.
Vilse binds to Robo in vitro and in the yeast two-hybrid assay. (A) A GST-Vilse-fusion protein or GST was used to pull down 35S-labeled in vitro translated Robointra (containing the entire intracellular domain) or truncations of it. Bound proteins were detected by fluorography. Robointra and a variant in which CC3 was deleted bind to GST-Vilse. CC2 deletion abolished the interaction. (B–E) Yeast two-hybrid assays. Drawings of Vilse truncations used as baits in C. (C) The Robointra or Robo2intra parts were cloned into the prey vector. Transformants were plated on Leu- plates. Full-length Vilse showed autoactivation (aa) and was not pursued further. (D) Vilse WW domains were necessary and sufficient for the interaction with Robointra. (E Drawings of Robo truncations used as prey in an independent experiment, where the WW domains of Vilse were the bait). The CC2 domain of Robo was necessary and sufficient for interaction with the Vilse WW domains and growth on Leu- plates.
Also in this assay, Vilse interacted with the intracellular part of the Robo receptor (Fig. 4B,C). The WW domains of Vilse were necessary and sufficient to mediate binding to Robo. In contrast, Vilse did not interact with the intracellular part of the Robo2 receptor. To pinpoint the Robo domain that confers Vilse binding, different deletions of Robo were tested. All Robo variants containing the CC2 motif could bind to the WW domain of Vilse, whereas variants lacking CC2 showed no binding (Fig. 4E). Moreover, a small 25-amino acid residue fragment of Robo containing CC2 was sufficient to bind to the WW domain of Vilse. This domain is absent in Robo2, providing an explanation for the specificity of Vilse for Robo.
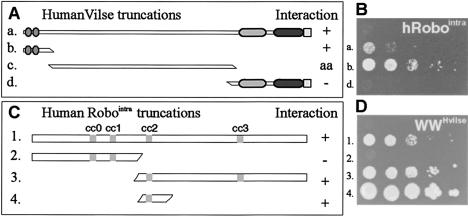
Given the extensive homology between the fly and human Vilse, we tested whether the human proteins could also bind to the human receptor Robo1. Human Vilse and Robo1 variants were tested in the yeast two-hybrid system. Full-length Vilse and the WW domain alone both showed binding to the intracellular domain of human Robo1 (Fig. 5). Again, the domain in human Robo1 involved in the binding to Vilse contained the proline-rich CC2-motif (Fig. 5), suggesting that the molecular interaction between Robo and Vilse is conserved.
Figure 5.
The WW domains of hVilse bind to the hRobo1 CC2 domain. (A) Drawings of the Vilse fragments used as bait for the Robo1intra domain in the yeast two-hybrid assay in B. (B) The full-length hVilse construct and the WW domains were necessary for growth of transformants on Leu- plates. The construct containing the middle part of Vilse was autoactivating in this assay. (C) Drawings of Robo1intra truncations used as prey in an experiment with the WW domains of Vilse as bait. (D) The CC2 region of Robo was necessary and sufficient for the interaction with the Vilse WW domains and growth on Leu-.
Vilse is a Rac/cdc42 GAP
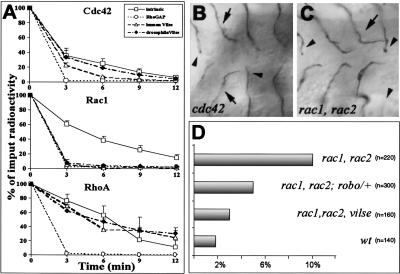
The predicted domain structure of Vilse suggested that it may regulate the activity of RhoGTPases. To examine the ability of Vilse to stimulate GTP hydrolysis, GST-fusion proteins of Drosophila Vilse (amino acids 923–1330) and the corresponding part of the human homolog were incubated with Rac1, Cdc42, or RhoA preloaded with [γ-32P]GTP. At regular intervals, the [γ-32P]GTP that remained bound to the GTPases was measured. The RhoGAP domains of both Drosophila Vilse and human Vilse effectively stimulated GTP hydrolysis of Rac1. In addition, human and Drosophila Vilse also stimulated the GTP hydrolysis of Cdc42, but less efficiently. None of the Vilse RhoGAP domains was able to stimulate the GTPase activity of RhoA. In contrast, p50RhoGAP was effective against all three GTPases and provided a positive control for the reactions (Fig. 6A).
Figure 6.
Vilse RacGAP activity is required at the midline. (A) In vitro GAP activity of fly and human Vilse. GTPase activation of Cdc42, Rac1, and RhoA stimulated by the RhoGAP domains of fly Vilse (black diamonds), human Vilse (empty triangles), and p50RhoGAP (empty circles), and the intrinsic GTPase activity of each GTPase (empty squares); 100% corresponds to the input of [γ-32P]GTP bound protein. Each measurement represents the means of three readings. (B,C) Ventral views of cdc42 and rac1, rac2 zygotic mutants stained to visualize the tracheal lumen. The GB phenotypes of cdc42 and rac1, rac2 are similar; they include mainly arrested branches (arrowheads) and GBs turning prematurely away from the midline (arrows). (D) Graph representing the numbers of GBs with premature turns in mutants of the indicated genotypes. robo acts as a dominant suppressor of rac1, rac2, and removal of the zygotic function of vilse ameliorates the early-turn phenotype.
Vilse down-regulates Rac activity at the midline
To identify the GTPase substrate of Vilse during GB1 pathfinding, we first analyzed the tracheal phenotypes of Drosophila zygotic mutants in cdc42 and rac1 rac2 (Fehon et al. 1997; Ng et al. 2002). Removal of the maternal contribution for cdc42 and rac1 rac2 results in defects in oogenesis and early embryonic phenotypes in epithelial morphogenesis and axonal growth, respectively (Genova et al. 2000; Hakeda-Suzuki et al. 2002). We therefore analyzed embryos lacking the zygotic products of these genes. Those embryos are likely to exhibit reduced levels of cdc42 or rac1 and rac2 at late stages of embryogenesis, resulting in a sensitized background for genetic interactions. GB phenotypes of rac and cdc42 zygotic mutants were heterogeneous, perhaps not surprisingly given the large number of cellular processes mediated by these GTPases. The migration of most GBs was affected in both rac and cdc42, resulting mainly in arrested branches (34% in rac1, rac2, n = 220; 69% in cdc42, n = 180), but no midline crosses were observed. Interestingly, both rac and cdc42 mutants also showed a GB phenotype reminiscent of Robo overexpression: A fraction of GBs turned posteriorly prematurely, before reaching the CNS midline (10% in rac1, rac2, n = 220; 7% in cdc42, n = 180; Fig. 6B,C). This phenotype is also similar to that obtained by overactivation of Robo in GBs either by the ectopic expression of Slit in longitudinal glia or in commissureless mutants (Englund et al. 2002). To test whether reduction of Robo could suppress the premature GB turns, we analyzed rac1, rac2 embryos lacking one copy of functional robo: In these mutants the number of GBs turning early before reaching the midline was reduced to half (5%, n = 300; Fig. 6D), suggesting that reduction of Rac can be overcome by a decrease in Robo signaling.
The GB phenotype of rac1, rac2 and cdc42 mutants is also similar to the one generated by Vilse overexpression in the trachea, which caused 35% of the branches to stall and 8% (n = 168) to turn early before reaching the midline. To test whether the premature-turn phenotype caused by the reduction in Cdc42 or Rac may be ameliorated by the removal of zygotic Vilse, we analyzed rac1, rac2, vilse and cdc42; vilse embryos. Reduction of Vilse in rac mutants effectively suppressed the early-turn phenotype (3%, n = 160; Fig. 6D). This indicates that removal of Vilse, like reduction of Robo, counteracts the effects of reduced Rac activity on migrating GBs. Decrease of Vilse had little or no effect on cdc42 early turns (6%, n = 200). Nevertheless, Vilse reduction in cdc42 mutants resulted in an overall improvement of the fraction of branches that migrated normally (1%–6%, n = 200). This implies that both Rac and Cdc42 are Vilse targets during GB migration, but that Vilse mediates Robo repulsion at the midline mainly through the inactivation of Rac.
Discussion
Vilse and its targets
Vilse promotes the hydrolysis of RacGTP and to a lesser extent that of Cdc42GTP. It is thus expected to antagonize the activity of these GTPases on their known effectors. Increasing experimental evidence indicates that Rac and Cdc42 regulate a multitude of cellular responses ranging from establishment of epithelial polarity and integrity to membrane trafficking and the control of planar polarity, in addition to their well known function in modulating the actin cytoskeleton (Van Aelst and Symons 2002). The tracheal phenotypes of vilse embryos are remarkably specific; all branches form, fuse, and grow toward their targets without any apparent defects on epithelial polarity, integrity, or shape. In addition, the rest of the terminal cells that target other internal organs concurrently with GB1 migrate correctly and associate with their targets. We therefore conclude that the primary function of Vilse in the trachea is in the guidance of GB1 migration.
How then does Vilse fulfill its role in cell navigation? Both of its target GTPases are key regulators of the actin cytoskeleton in several cell types. In fibroblasts, GTP-bound Cdc42 generates actin bundles and characteristic filopodial extensions, possibly through its association with the WASP protein and subsequent stimulation of the actin polymerizing activity of the Arp2/3 complex (Rohatgi et al. 1999; Pollard and Borisy 2003). RacGTP, on the other hand, generates distinct cytoskeletal attributes, membrane ruffling, and lamellipodial protrusions (Ridley et al. 1992). Rac controls actin polymerization through the intermediary protein IRSp53, which associates to the SCAR/WAVE regulator of Arp2/3 activity (Miki et al. 2000). Cdc42 also binds to the IRSp53 adaptor, suggesting that both GTPases regulate actin polymerization through SCAR/WAVE (Govind et al. 2001).
An additional regulatory role for Rac and Cdc42 in cytoskeletal dynamics is exerted through their activating role on PAK (p21 activated kinase; Manser et al. 1994). PAK in turn activates the LIM kinase, which can phosphorylate the actin depolymerization factor (ADF/cofilin; Arber et al. 1998). Cofilin mediates depolymerization of actin filaments and can also function as a filament-severing factor. Its phosphorylation by LIM-kinase down-regulates its activity and inhibits F-actin depolymerization (Yang et al. 1998). Thus, the two GTPases in their active form promote the growth of actin filaments by both enhancing polymerization through the Arp2/3 complex and inhibiting severing and depolymerization at the minus end of the filaments. The phenotypic analysis of vilse and GTPase mutants leads us to propose that Vilse antagonizes the function of Rac in promoting actin polymerization locally at the migrating tip of GB1.
Vilse and Robo signaling
Cell guidance through Slit signaling, apart from its role in axonal pathfinding, has been implicated in a large number of morphogenetic events involving cell migration. It is involved in the migration of leukocytes in vertebrates (Wu et al. 2001), epithelial sheets and muscle cells in Drosophila (Schimmelpfeng et al. 2001), and in the branching of the lung epithelium in mice (Xian et al. 2001), and more recently it was found to control movements of endothelial cells during angiogenesis (Wang et al. 2003). Slit signal interpretation and the cellular responses it elicits depend on the intracellular domains of Robo receptors (Bashaw and Goodman 1999). The best characterized examples of Robo signal transduction derive from studies of migrating neurons (Giger and Kolodkin 2001; Luo 2002; Schmucker 2003). These studies highlight two basic mechanisms for Slit signaling through Robo. In the first paradigm, derived from studies of early-stage-22 Xenopus spinal neurons, Robo silences the attractive Netrin signal in response to Slit through the binding of its CC1 domain to the intracellular part of the DCC receptor. This Netrin-silencing function of Robo is different from the repulsive response to Slit, which is acquired by the same neurons only later, at stage 28 (Stein and Tessier-Lavigne 2001).
Slit-mediated repulsive responses involve the regulation of cytoskeletal organization in the growth cone. In Drosophila, the Abelson kinase (Abl) binds to CC3 and phosphorylates a tyrosine in CC1, thereby modulating Robo activity. On the other hand, the Abl substrate Enabled (Ena), a member of the profilin-binding family of proteins, associates with CC2 and mediates the repulsive role of Robo through an unknown mechanism that may involve control of cytoskeletal organization (Bashaw et al. 2000). More recently, Abl was found to collaborate with the cyclase-associated protein CAP, this time to mediate Robo repulsion (Wills et al. 2002). srGAPs bind to the CC3 domain of Robo in response to Slit and aid Cdc42GTP hydrolysis to directly mediate the repulsive response to Slit in cultured anterior subventricular rat neurons. This Cdc42GTP hydrolysis at the site of Robo activation would then result in actin filament depolymerization and severing, thus promoting the turn of the growth cone to the opposite direction. The functional analysis of Vilse identifies a direct transducer of the Slit signal to the inactivation of Rac. vilse, robo, and slit mutants show qualitatively the same phenotypes of midline crossings of tracheal cells and axons. The effect of vilse overexpression on the robo tracheal phenotypes and the dose-dependent interaction between slit, robo, and vilse, combined with the biochemical analysis indicate that Vilse acts downstream of Robo. Hence, Vilse may play an analogous role to srGAP in locally down-regulating actin polymerization through the hydrolysis of RacGTP and facilitating turning away from the midline.
Paradoxically, both activation and inactivation of Rac appear to interfere with midline crossing and Slit signaling. Expression of constitutively activated Rac causes longitudinal axons to cross the midline, and reduction of Robo signaling enhances this phenotype (Fritz and VanBerkum 2002). On the other hand, rac mutants show strong phenotypes in axonal growth and guidance including midline crosses (Hakeda-Suzuki et al. 2002; Ng et al. 2002), and the latter phenotype becomes more prominent by reduction of Slit (Fan et al. 2003). One possible explanation is that Rac might be involved in multiple cellular processes affecting different aspects of the Slit/Robo pathway. For example Rac might mediate Slit secretion by midline cells or intracellular trafficking of Robo in the axons, in addition to its effect on cytoskeletal dynamics downstream of Robo. The protein adaptor Dock has also been implicated in midline repulsion downstream of Robo. In response to Slit, Dock's binding to the intracellular domain of Robo is enhanced, leading to the recruitment of the Rac effector kinase Pak. This chain of events has been proposed to bring activated Rac to Robo in response to Slit (Fan et al. 2003). Yet, it is not clear how Rac becomes activated in response to Slit, or how the recruitment of active Rac and Pak might translate in the cellular events that lead to repulsion from Slit. The contradicting models of the function of rac downstream of robo may be reconciled by considering a sequential interaction of the effectors with the receptor. For example, Vilse may be required initially for severing of actin filaments at the cell extensions that first encounter Slit. The inducible recruitment of Pak to Robo might occur subsequently, perhaps in response to higher concentrations of Slit, promoting cytoskeletal reorganizations that lead to a sustained turning response. This involves a new function of Rac in the context of repulsion from the signal source (Fan et al. 2003). The genetic analysis of midline repulsion reveals that Slit signaling relies on the dynamic and spatially coordinated control of Rac activity. Vilse provides both the first direct link from Robo to the inactivation of Rac, and a molecular handle to address the complex interactions that control repulsion during cell migration.
Materials and methods
Fly strains
The 2240, vilse enhancer trap line is from the third chromosome collection of lethals from the Department of Genetics, University of Szeged, Hungary (Deak et al. 1997). Excision alleles were generated as described (Robertson et al. 1988). Excision alleles (211) were balanced over TM3UbxlacZ and screened for lethality and embryonic tracheal phenotype. The screen yielded several homozygous viable revertants and the lethal vilse1 (see below). This excision allele failed to complement Df(3R)e-N12 (breakpoints 93B1-2;D6-7). vilse1 mutants die as third instar larvae. Germ-line clones were generated as described (Chou and Perrimon 1996). The tracheal cell marker strain used was trachealess-lacZ (1-eve-1; Perrimon et al. 1991). The following null or strong loss-of-function alleles were used: roboz570 (Kidd et al. 1998), slit,robo (Kidd et al. 1999), cdc42-3 (Fehon et al. 1997), rac1J10rac2 and rac1J11rac2 (Hakeda-Suzuki et al. 2002). For the genetic interaction experiments we constructed the following strains: roboz570/CyOftzlacZ; vilse1//TM3UbxlacZ, slitrobo/CyOftzlacZ; vilse1//TM3UbxlacZ, rac1J11rac2 vilse1//TM3UbxlacZ, cdc42-3/FM7ftzlacZ; vilse1//TM3UbxlacZ, roboz570/CyOftzlacZ; rac1J11rac2//TM3UbxlacZ, cdc42-3/FM7ftzlacZ; roboz570/CyOftzlacZ. These were crossed to wild-type Oregon-R or to vilse1//TM3UbxlacZ. Antibody staining against β-gal allowed the identification of genotypes in embryos.
The following GAL4 and UAS strains were used: SRF-GAL4 (Jarecki et al. 1999), btl-GAL4 (Shiga et al. 1996).
Antibodies, embryo staining, and whole-mount in situ hybridizations
Embryo fixation, antibody staining, and light and confocal fluorescence microscopy were performed as described (Samakovlis et al. 1996). Primary antisera were mAb2A12 against tracheal lumen (diluted 1:3), mAb2-161 against DSRF (1:2000, M. Gilman, Ariad Corp., Boston, MA), anti-β-galactosidase (1:1500, Cappel), mAb1D4 against Fasciclin II (1:5, from C. Goodman; Van Vactor et al. 1993), mAb BP102 that labels all CNS axons (1:50, from B. Dickson, Institute of Molecular Biotechnology of the Austrian Academy of Sciences, Vienna, Austria). Biotin-, Cy2-, and Cy3-conjugated (Jackson Laboratories) and Alexa Fluor-568- and Alexa Fluor-488-conjugated (Molecular Probes) secondary antibodies were used at 1:300, 1:500, and 1:400, respectively. Signal was developed using the Vectastain Elite ABC Kit (Vector Laboratories) or Cy3 Tyramide Signal Amplification (TSA, NEN Life Science Products). Embryos were visualized with a Zeiss Axioplan2 microscope under Nomarski optics or a Leica TCS SP2 confocal microscope.
To prepare the polyclonal Vilse antiserum, a DNA fragment from vilse cDNA LD10379 that encodes amino acids 35–463 was inserted in frame to the 6xHis coding sequence in pRSETB (Invitrogen). This histidine-tagged Vilse-fusion protein was expressed in Escherichia coli, purified on Ni-NTA resin (QIAGEN), and used for immunization of guinea pigs at Med-Probe. The antiserum was used at 1:2000 for immunohistochemistry and Western blotting. Whole-mount in situ hybridization was performed using random-primed, digoxigenin-labeled vilse cDNA (LD10379, Research Genetics) as probe (Lehmann and Tautz 1994). Embryo staging was according to the method of Campos-Ortega and Hartenstein (1985).
Molecular analysis of the vilse1 mutation
The 2240 enhancer trap contains a single P-element, as analyzed by Southern blot. Genomic DNA flanking the P-element was obtained by plasmid rescue in E. coli after cleavage of the genomic DNA with EcoRI or BamHI. This DNA was sequenced and used to search the databases. The search identified several cDNA clones that were obtained from Research Genetics. cDNA LD10379 was selected for complete sequencing. The search also identified a predicted gene CG3421 in GadFly (Celera Genomics; Adams et al. 2000). The homology searches were performed using NCBI BLAST (http://www.ncbi.nlm.nih.gov).
The P-element was inserted at position 16923181 in the assembled 3R chromosome arm sequence (release 3, FlyBase) and 172 bp upstream of the ATG in the 5′-untranslated region of cDNA LD10379. To analyze the Vilse protein, ExPASy (http://ca.expasy.org/cgi-bin/blast), Pfam (http://www.sanger.ac.uk), and InterPro (http://www.ebi.ac.uk) pattern prediction searches were used.
vilse1 genomic DNA was amplified by PCR using primers specific for the region flanking the P-element insertion site (primer A [5′-GCTCTGCTACACAATGCGGGACAAC-3′] and primer B [5′-GCGCAAACAAGTGACCCCGAGTAG-3′] deriving from genomic DNA sequences located 2777 bp and 1859 bp upstream and downstream from the P-element insertion site, respectively). The PCR product was purified and sequenced. Sequence comparisons between PCR-amplified fragment from the vilse1 mutant and from the corresponding region in wild type revealed a 1077-bp deletion in vilse1, upstream of the original P-element insertion site and deleting most of the first exon (corresponding to positions 16923182–16924258 of the 3R chromosome arm sequence, release 3). In situ hybridization revealed that zygotic transcripts were absent from vilse1 embryos. In addition, antibody staining revealed severely reduced Vilse protein levels after stage 10 of embryogenesis.
Generation of UAS-vilse transgenic lines
The UAS-vilse comprises a NotI–ApaI (blunted) fragment from pBluescript-vilse cDNA (LD10379, Research Genetics) subcloned into the NotI–XbaI (blunted) sites of pUAST expression vector (Brand and Perrimon 1993). Germ-line transformation of w1118 embryos was performed as described (Spradling 1986). Independent transformant lines were obtained on the second and third chromosome. For UAS-GAL4 experiments, embryos were collected for 6 h at room temperature, then transferred to 29°C for 10 h to maximize GAL4 activity. Embryos were then fixed and stained as above.
Yeast two-hybrid assays
Yeast two-hybrid assays were done using the Dual-bait hybrid hunter yeast two-hybrid system (Invitrogen). Using PCR, we cloned vilse sequences encoding amino acid residues 1–1330, 105–1330, 923–1330, 1131–1330, 1–922, 1–104, and 105–922 into the ApaI–NotI sites of the pHybLex/Zeo vector to generate fusion proteins with the LexA DNA-binding domain. PCR was also used to clone robo sequences encoding amino acid residues 941–1395, 941–1256, 1257–1395, 941–1033, 941–1097, 1098–1256, and 1098–1122 and a robo2 sequence encoding residues 1013–1463 into the EcoRI–NotI sites of the pYESTrp2 vector to generate fusion proteins with the B42 activation domain. In a similar manner, we cloned human vilse sequences encoding amino acid residues 1–1094 (starting at the first amino acid in the largest open reading frame), 1–135, 139–697, and 701–1094 into the EcoRI–XhoI sites of pHybLex/Zeo. Human robo sequences encoding amino acid residues 921–1651, 921–1154, 1149–1651, and 1180–1213 were cloned into the EcoRI–NotI sites of pYESTrp2. Interactions were detected on plates lacking leucine.
Glutathione S-transferase (GST) pull-down experiments
Vilse sequences encoding amino acid residues 1–1330, 1–104, and 105–1330 were cloned into the SalI–NotI sites of the pGEX-5X-3 vector (Amersham), creating GST-Vilse, GST-WW, and GST-VilseΔWW. Expression of the GST-fusion proteins in E. coli BL21pLys cells was induced with IPTG for 3 h at 30°C and confirmed by Coomassie stainings of the bacterial extracts run on SDS/PAGE gels. Robo sequences encoding amino acid residues 941–1395, 1257–1395, 941–1256, and 941–1097 were cloned into the EcoRI–NotI sites of Bluescript SK generating the entire Robo intracellular domain (Robo IC), cc3 motif only, RoboΔcc3, and RoboΔcc2Δcc3, respectively. These clones were designed so that they contain a Kozak sequence and an ATG in front of the robo sequences. For in vitro translation reactions, the TNT T7-coupled reticulocyte lysate system was used (Promega) in the presence of in vitro translation grade Redivue L-[35S] methionine (Amersham). In vitro translated proteins (2–10 μL) and GST-fusion protein extracts (50 μL) were incubated at4°C for 2 h in MTPBS (150 mM NaCl, 16 mM NaHPO4, 4 mM NaH2PO4) in a total volume of 300 μL. Glutathione sepharose 4B (Amersham Pharmacia) beads were washed five times in MTPBS + 0.5% dry milk before adding them to the mixture. The beads were incubated with the mix for 20 min at 4°C. Beads were washed five times in MTPBS containing 0.75% NP40. Proteins bound to the beads were eluted by addition of SDS-PAGE sample buffer and analyzed by 15% SDS polyacrylamide gel electrophoresis and autoradiography.
GTPase activation assays
GST-fusion proteins of Rac1, Cdc42 (brain isoform), and RhoA were expressed in E. coli, purified on glutathione-Sepharose beads (Amersham Biosciences) and isolated from GST-fusion proteins by thrombin cleavage as described (Richnau and Aspenstrom 2001). GST-fusion proteins of the RhoGAP domains of Drosophila melanogaster Vilse (amino acid residues 923–1330), the corresponding human Vilse residues, and p50RhoGAP (amino acid residues 230–439) were expressed in E. coli as described (Richnau and Aspenstrom 2001). Briefly, the bacteria were lysed in a buffer containing 50 mM Tris-HCl at pH 7.5, 5 mM MgCl2, 50 mM NaCl, 10% glycerol, 0.1% Triton X-100, 1 mM PMSF, 1% aprotinin (Trasylol, Beyer), and 1 mM dithiothreitol (DTT). The proteins were thereafter eluted from the glutathione-Sepharose beads with 5 mM reduced glutathione, desalted on PD10 prepacked chromatography columns (Amersham Pharmacia Biotech), equilibrated in 20 mM Tris-HCl at pH 7.5, 10% glycerol and 1 mM DTT, and thereafter concentrated using Centricon-10 (Millipore). Protein concentrations were determined with the Bradford method. The GTPase assay was as described by Richnau and Aspenstrom (2001). Briefly, 0.1 μg of recombinant wild-type Rac1, RhoA, or Cdc42 was preloaded with 10 μCi [γ-32P]GTP (Amersham Pharmacia Biotech) in 20 μL of 20 mM Tris-HCl at pH 7.5, 25 mM NaCl, 5 mM EDTA, and 0.1 mM DTT. The mixture was incubated for 10 min at 30°C, after which the reaction was terminated by adding 5 μL 0.1 M MgCl2, and the resulting [γ-32P]GTP-loaded GTPase solutions were stored on ice. For the GAP assays, equimolar amounts of the GTPases and the GST-GAP domains were used. Three μL of the [γ-32P]GTP-loaded GTPase was added to a 30-μL mixture of 20 mM Tris-HCl at pH 7.5, 1 mM nonradioactive GTP, 0.87 mg/mL bovine serum albumin, 0.1 mM DTT, with either GST-RhoGAP domains of Drosophila Vilse, human Vilse, or p50RhoGAP. The mixture was incubated at 30°C, and 5-μL aliquots were removed after 0, 3, 6, 9, and 12 min; the reaction was stopped by the addition of 1 mL ice-cold buffer A (50 mM Tris-HCl at pH 7.5, 50 mM NaCl, 5 mM MgCl2). The samples were collected on nitrocellulose filters, then washed with 10 mL ice cold buffer A; the portion of [γ-32P]GTP remaining bound to the GTPases was determined by scintillation counting.
Acknowledgments
We thank R. Fehon and C. Goodman for fly strains. This work was supported by grants from the Swedish Research Council (VR), the Swedish Foundation for strategic research (SSF), and the Wallenberg Consortium North (WCN) to C.S.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.310204.
References
- Adams M.D., Celniker, S.E., Holt, R.A., Evans, C.A., Gocayne, J.D., Amanatides, P.G., Scherer, S.E., Li, P.W., Hoskins, R.A., Galle, R.F., et al. 2000. The genome sequence of Drosophila melanogaster. Science 287: 2185-2195. [DOI] [PubMed] [Google Scholar]
- Arber S., Barbayannis, F.A., Hanser, H., Schneider, C., Stanyon, C.A., Bernard, O., and Caroni, P. 1998. Regulation of actin dynamics through phosphorylation of cofilin by LIM-kinase. Nature 393: 805-809. [DOI] [PubMed] [Google Scholar]
- Bashaw G.J. and Goodman, C.S. 1999. Chimeric axon guidance receptors: The cytoplasmic domains of Slit and Netrin receptors specify attraction versus repulsion. Cell 97: 917-926. [DOI] [PubMed] [Google Scholar]
- Bashaw G.J., Kidd, T., Murray, D., Pawson, T., and Goodman, C.S. 2000. Repulsive axon guidance: Abelson and Enabled play opposing roles downstream of the Roundabout receptor. Cell 101: 703-715. [DOI] [PubMed] [Google Scholar]
- Brand A.H. and Perrimon, N. 1993. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118: 401-415. [DOI] [PubMed] [Google Scholar]
- Campos-Ortega A.J. and Hartenstein, V. 1985. The embryonic development of Drosophila melanogaster, pp. 10-84. Springer-Verlag, New York.
- Chou T.B. and Perrimon, N. 1996. The autosomal FLP-DFS technique for generating germline mosaics in Drosophila melanogaster. Genetics 144: 1673-1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deak P., Omar, M.M., Saunders, R.D.C., Pal, M., Komonyi, O., Sidonya, J., Maroy, P., Zhang, Y., Ashburner, M., Benos, P., et al. 1997. P-element insertion alleles of essential genes on the third chromosome of Drosophila melanogaster: Correlation of physical and cytogenetic maps in chromosomal region 86E-87F. Genetics 147: 1697-1722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickson B.J. 2002. Molecular mechanisms of axon guidance. Science 298: 1959-1564. [DOI] [PubMed] [Google Scholar]
- Englund C., Uv, A.E., Cantera, R., Mathies, L.D., Krasnow, M.A., and Samakovlis, C. 1999. adrift, a novel bnl-induced Drosophila gene, required for tracheal pathfinding into the CNS. Development 126: 1505-1514. [DOI] [PubMed] [Google Scholar]
- Englund C., Steneberg, P., Falileeva, L., Xylourgidis, N., and Samakovlis, C. 2002. Attractive and repulsive functions of Slit are mediated by different receptors in the Drosophila trachea. Development 129: 4941-4951. [DOI] [PubMed] [Google Scholar]
- Fan X., Labrador, J.P., Hing, H., and Bashaw, G.J. 2003. Slit stimulation recruits Dock and Pak to the roundabout receptor and increases Rac activity to regulate axon repulsion at the CNS midline. Neuron 40: 113-127. [DOI] [PubMed] [Google Scholar]
- Fehon R.G., Oren, T., LaJeunesse, D.R., Melby, T.E., and McCartney, B.M. 1997. Isolation of mutations in the Drosophila homologues of the human Neurofibromatosis 2 and yeast CDC42 genes using a simple reverse-genetic method. Genetics 146: 245-252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritz J.L. and VanBerkum, M.F. 2002. Regulation of rho family GTPases is required to prevent axons from crossing the midline. Dev. Biol. 252: 46-58. [DOI] [PubMed] [Google Scholar]
- Genova J.L., Jong, S., Camp, J.T., and Fehon, R.G. 2000. Functional analysis of Cdc42 in actin filament assembly, epithelial morphogenesis, and cell signaling during Drosophila development. Dev. Biol. 221: 181-194. [DOI] [PubMed] [Google Scholar]
- Giger R.J. and Kolodkin, A.L. 2001. Silencing the siren: Guidance cue hierarchies at the CNS midline. Cell 105: 1-4. [DOI] [PubMed] [Google Scholar]
- Govind S., Kozma, R., Monfries, C., Lim, L., and Ahmed, S. 2001. Cdc42Hs facilitates cytoskeletal reorganization and neurite outgrowth by localizing the 58-kD insulin receptor substrate to filamentous actin. J. Cell Biol. 152: 579-594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakeda-Suzuki S., Ng, J., Tzu, J., Dietzl, G., Sun, Y., Harms, M., Nardine, T., Luo, L., and Dickson, B.J. 2002. Rac function and regulation during Drosophila development. Nature 416: 438-442. [DOI] [PubMed] [Google Scholar]
- Jarecki J., Johnson, E., and Krasnow, M.A. 1999. Oxygen regulation of airway branching in Drosophila is mediated by branchless FGF. Cell 99: 211-220. [DOI] [PubMed] [Google Scholar]
- Kidd T., Brose, K., Mitchell, K.J., Fetter, R.D., Tessier-Lavigne, M., Goodman, C.S., and Tear, G. 1998. Roundabout controls axon crossing of the CNS midline and defines a novel subfamily of evolutionarily conserved guidance receptors. Cell 92: 205-215. [DOI] [PubMed] [Google Scholar]
- Kidd T., Bland, K.S., and Goodman, C.S. 1999. Slit is the midline repellent for the Robo receptor in Drosophila. Cell 96: 785-794. [DOI] [PubMed] [Google Scholar]
- Kramer S.G., Kidd, T., Simpson, J.H., and Goodman, C.S. 2001. Switching repulsion to attraction: Changing responses to slit during transition in mesoderm migration. Science 292: 737-740. [DOI] [PubMed] [Google Scholar]
- Lehmann R. and Tautz, D. 1994. In situ hybridization to RNA (ed. L.S.B. Goldstein and E.A. Fyrberg), pp. 576-597. Academic Press, San Diego. [DOI] [PubMed]
- Luo L. 2002. Actin cytoskeleton regulation in neuronal morphogenesis and structural plasticity. Annu. Rev. Cell Dev. Biol. 18: 601-635. [DOI] [PubMed] [Google Scholar]
- Manser E., Leung, T., Salihuddin, H., Zhao, Z.S., and Lim, L. 1994. A brain serine/threonine protein kinase activated by Cdc42 and Rac1. Nature 367: 40-46. [DOI] [PubMed] [Google Scholar]
- Miki H., Yamaguchi, H., Suetsugu, S., and Takenawa, T. 2000. IRSp53 is an essential intermediate between Rac and WAVE in the regulation of membrane ruffling. Nature 408: 732-735. [DOI] [PubMed] [Google Scholar]
- Nagase T., Kikuno, R., Hattori, A., Kondo, Y., Okumura, K., and Ohara, O. 2000. Prediction of the coding sequences of unidentified human genes. XIX. The complete sequences of 100 new cDNA clones from brain which code for large proteins in vitro. DNA Res. 7: 347-355. [DOI] [PubMed] [Google Scholar]
- Ng J., Nardine, T., Harms, M., Tzu, J., Goldstein, A., Sun, Y., Dietzl, G., Dickson, B.J., and Luo, L. 2002. Rac GTPases control axon growth, guidance and branching. Nature 416: 442-447. [DOI] [PubMed] [Google Scholar]
- Nguyen Ba-Charvet K.T., Brose, K., Marillat, V., Kidd, T., Goodman, C.S., Tessier-Lavigne, M., Sotelo, C., and Chedotal, A. 1999. Slit2-mediated chemorepulsion and collapse of developing forebrain axons. Neuron 22: 463-473. [DOI] [PubMed] [Google Scholar]
- Perrimon N., Noll, E., McCall, K., and Brand, A. 1991. Generating lineage-specific markers to study Drosophila development. Dev. Genet. 12: 238-252. [DOI] [PubMed] [Google Scholar]
- Pollard T.D. and Borisy, G.G. 2003. Cellular motility driven by assembly and disassembly of actin filaments. Cell 112: 453-465. [DOI] [PubMed] [Google Scholar]
- Richnau N. and Aspenstrom, P. 2001. Rich, a rho GTPase-activating protein domain-containing protein involved in signaling by Cdc42 and Rac1. J. Biol. Chem. 276: 35060-35070. [DOI] [PubMed] [Google Scholar]
- Ridley A.J., Paterson, H.F., Johnston, C.L., Diekmann, D., and Hall, A. 1992. The small GTP-binding protein rac regulates growth factor-induced membrane ruffling. Cell 70: 401-410. [DOI] [PubMed] [Google Scholar]
- Robertson H.M., Preston, C.R., Phillis, R.W., Johnson-Schlitz, D.M., Benz, W.K., and Engels, W.R. 1988. A stable genomic source of P element transposase in Drosophila melanogaster. Genetics 118: 461-470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohatgi R., Ma, L., Miki, H., Lopez, M., Kirchhausen, T., Takenawa, T., and Kirschner, M.W. 1999. The interaction between N-WASP and the Arp2/3 complex links Cdc42-dependent signals to actin assembly. Cell 97: 221-231. [DOI] [PubMed] [Google Scholar]
- Samakovlis C., Manning, G., Steneberg, P., Hacohen, N., Cantera, R., and Krasnow, M.A. 1996. Genetic control of epithelial tube fusion during Drosophila tracheal development. Development 122: 3531-3536. [DOI] [PubMed] [Google Scholar]
- Schimmelpfeng K., Gogel, S., and Klambt, C. 2001. The function of leak and kuzbanian during growth cone and cell migration. Mech. Dev. 106: 25-36. [DOI] [PubMed] [Google Scholar]
- Schmucker D. 2003. Downstream of guidance receptors: Entering the baroque period of axon guidance signaling. Neuron 40: 4-6. [DOI] [PubMed] [Google Scholar]
- Shiga Y., Tanaka-Matakatsu, M., and Hayashi, S. 1996. A nuclear GFP/β-galactosidase fusion protein as a marker for morphogenesis in living Drosophila. Dev. Growth Diff. 38: 99-106. [Google Scholar]
- Spradling A.C. 1986. P element-mediated transformation (ed. D.B. Roberts), pp. 175-199. IRL Press Limited, Oxford, UK.
- Stein E. and Tessier-Lavigne, M. 2001. Hierarchical organization of guidance receptors: Silencing of netrin attraction by slit through a Robo/DCC receptor complex. Science 291: 1928-1938. [DOI] [PubMed] [Google Scholar]
- Sutherland D., Samakovlis, C., and Krasnow, M.A. 1996. branchless encodes a Drosophila FGF homolog that controls tracheal cell migration and the pattern of branching. Cell 87: 1091-1101. [DOI] [PubMed] [Google Scholar]
- Tessier-Lavigne M. and Goodman, C.S. 1996. The molecular biology of axon guidance. Science 274: 1123-1133. [DOI] [PubMed] [Google Scholar]
- Uv A., Cantera, R., and Samakovlis, C. 2003. Drosophila tracheal morphogenesis: Intricate cellular solutions to basic plumbing problems. Trends Cell Biol. 13: 301-309. [DOI] [PubMed] [Google Scholar]
- Van Aelst L. and Symons, M. 2002. Role of Rho family GTPases in epithelial morphogenesis. Genes & Dev. 16: 1032-1054. [DOI] [PubMed] [Google Scholar]
- Van Vactor D., Sink, H., Fambrough, D., Tsoo, R., and Goodman, C.S. 1993. Genes that control neuromuscular specificity in Drosophila. Cell 73: 1137-1153. [DOI] [PubMed] [Google Scholar]
- Wang B., Xiao, Y., Ding, B.B., Zhang, N., Yuan, X., Gui, L., Qian, K.X., Duan, S., Chen, Z., Rao, Y., et al. 2003. Induction of tumor angiogenesis by Slit–Robo signaling and inhibition of cancer growth by blocking Robo activity. Cancer Cell 4: 19-29. [DOI] [PubMed] [Google Scholar]
- Wills Z., Emerson, M., Rusch, J., Bikoff, J., Baum, B., Perrimon, N., and Van Vactor, D. 2002. A Drosophila homolog of cyclase-associated proteins collaborates with the Abl tyrosine kinase to control midline axon pathfinding. Neuron 36: 611-622. [DOI] [PubMed] [Google Scholar]
- Wong K., Ren, X.-R., Huang, Y.-Z., Xie, Y., Liu, G., Saito, H., Tang, H., Wen, L., Brady-Kalnay, S.M., Mei, L., et al. 2001. Signal transduction in neuronal migration: Roles of GTPase activating proteins and the small GTPase Cdc42 in the Slit–Robo pathway. Cell 107: 209-221. [DOI] [PubMed] [Google Scholar]
- Wu J.Y., Feng, L., Park, H.T., Havlioglu, N., Wen, L., Tang, H., Bacon, K.B., Jiang, Z., Zhang, X., and Rao, Y. 2001. The neuronal repellent Slit inhibits leukocyte chemotaxis induced by chemotactic factors. Nature 410: 948-952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xian J., Clark, K.J., Fordham, R., Pannell, R., Rabbitts, T.H., and Rabbitts, P.H. 2001. Inadequate lung development and bronchial hyperplasia in mice with a targeted deletion in the Dutt1/Robo1 gene. Proc. Natl. Acad. Sci. 98: 15062-15066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang N., Higuchi, O., Ohashi, K., Nagata, K., Wada, A., Kangawa, K., Nishida, E., and Mizuno, K. 1998. Cofilin phosphorylation by LIM-kinase 1 and its role in Rac-mediated actin reorganization. Nature 393: 809-812. [DOI] [PubMed] [Google Scholar]
- Zallen J.A., Yi, B.A., and Bargmann, C.I. 1998. The conserved immunoglobulin superfamily member SAX-3/Robo directs multiple aspects of axon guidance in C. elegans. Cell 92: 217-227. [DOI] [PubMed] [Google Scholar]