Abstract
COP10 is a ubiquitin-conjugating enzyme variant (UEV), which is thought to act together with COP1, DET1, and the COP9 signalosome (CSN) in Arabidopsis to repress photomorphogenesis. Here, we demonstrate that COP10 interacts with ubiquitin-conjugating enzymes (E2s) in vivo, and can enhance their activity in vitro, an activity distinct from previous characterized UEVs such as MMS2 and UEV1. Furthermore, we show that COP10 forms a complex with UV-damaged DNA-binding protein 1a (DDB1a) and de-etiolated 1 (DET1), and physically interacts with COP1 and the CSN. Purified CDD (COP10, DDB1, DET1) complex also shows enhancement of E2 activity (UEA) similar to that observed with COP10 itself. Our data suggests that COP10, along with COP1 and the CSN, promotes the degradation of positive regulators of photomorphogenesis, such as the transcription factor HY5, via the ubiquitin/26S proteasome system. Thus, the CDD complex may act as a ubiquitylation-promoting factor to regulate photomorphogenesis.
Keywords: CDD complex, COP10, E2 enhancer, photomorphogenesis, ubiquitylation, UEV
Light is an important regulator of plant development. One of the most dramatic effects of light in plant development is seen in the development of the Arabidopsis seedling. Arabidopsis seedlings show different development patterns depending upon the presence or absence of light. In the light seedlings show photomorphogenic development with short hypocotyls and open and expanded cotyledons. In contrast, in the dark seedlings show skotomorphogenic development characterized by long hypocotyls with closed unexpanded cotyledons protected by an apical hook (von Arnim and Deng 1996). The repression of photomorphogenic development in dark-grown Arabidopsis seedlings requires the action of a group of essential constitutive photomorphogenesis/de-etiolated/fusca (COP/DET/FUS) genes, mutations in which all show photomorphogenic development in darkness (Serino and Deng 2003; Sullivan and Deng 2003). It has been shown that photomorphogenesis-promoting factors such as HY5 and LAF1 are degraded in darkness, with the COP/DET/FUS proteins promoting their degradation (Osterlund et al. 2000; Saijo et al. 2003; Seo et al. 2003).
One of the most widely studied protein degradation mechanism is the ubiquitin–proteasome (Ub/26S) system. The Ub/26S system degrades short-lived or misfolded and is highly conserved in all eukaryotes. Target proteins first are polyubiquitylated, then recognized, and subsequently degraded by the 26S proteasome (Vierstra 2003). The process of ubiquitylation involves the sequential action of a ubiquitin-activating enzyme (E1), a ubiquitin-conjugating enzyme (E2), followed by a ubiquitin-ligase enzyme (E3; Vierstra 2003). In this cascade the ubiquitin moiety is transferred from the E2 to a lysine residue in the target protein in a process facilitated by the E3, which by binding both the E2 and the target provides substrate specificity (Vierstra 2003). The 26S proteasome is composed of a core 20S proteasome and two 19S/22S/PA700 regulatory particles (Tanaka 1998; DeMartino and Slaughter 1999; Ferrell et al. 2000). The 19S regulatory particle can also be further divided into base and lid subcomplexes (Glickman et al. 1998).
One of the COP/DET/FUS genes, COP1, encodes a ring-finger-type ubiquitin E3 ligase, which is responsible for the ubiquitylation of photomorphogenesis-promoting factors such as HY5 and LAF1 in darkness (Osterlund et al. 2000; Saijo et al. 2003; Seo et al. 2003). In vivo, COP1 acts as a large protein complex of 700 kDa (Saijo et al. 2003). A further dozen of the COP/DET/FUS genes define the COP9 signalosome (CSN), which is a nuclear enriched protein complex showing homology to the lid subcomplex of the 26S proteasome and acts to deconjugate NEDD8/Rub1 from the cullin subunit of SCF (SKP1/Cullin/F-box)-type E3 ligases (Lyapina et al. 2001; Schwechheimer et al. 2001; Serino and Deng 2003; Wei and Deng 2003). It has been shown that the CSN interacts with 26S proteasome (Kwok et al. 1999; Peng et al. 2003). DET1 is also a negative regulator of photomorphogenesis (Pepper et al. 1994), and its mammalian homolog has recently been reported to regulate the ubiquitylation of c-Jun together with COP1 (Wertz et al. 2004).
COP10 was originally identified as a negative regulator of photomorphogenesis essential for COP1-mediated degradation of HY5 (Wei et al. 1994; Osterlund et al. 2000). COP10 belongs to a family of ubiquitin E2 variant (UEV) proteins that contain the ubiquitin-conjugating motif (Ubc) but lack a critical cysteine residue required for conjugation (Sanch et al. 1998; Schwechheimer et al. 2001). UEVs have been shown to function in numerous cellular processes, including postreplicative DNA repair and control of the cell cycle (Hofmann and Pickart 1999; Li et al. 2001). To date, the best-characterized UEVs belong to the MMS2/UEV1 group of proteins, which function in the RAD6 DNA repair system (Hofmann and Pickart 1999). Both the yeast MMS2 and the human UEV1 proteins work together with an active E2 enzyme, Ubc13, to produce noncanonical ubiquitin chains linked through a K63 residue, which, unlike K48-linked ubiquitin chains, do not target a protein for proteasome-mediated degradation (Hofmann and Pickart 1999). Interestingly, COP10 shows higher homology to active E2s than other UEVs such as MMS2 and UEV1 and exists in a nuclear-localized complex of ∼300 kDa in vivo (Suzuki et al. 2002). Genetic studies underscore the role of the COP10-containing complex in photomorphogenesis and in ubiquitin-mediated protein degradation. However, the biochemical function of the COP10 and the subunit composition of the COP10-containing complex remain unclear.
Here we show that COP10 forms a complex together with DDB1a and DET1 and physically interacts with COP1, CSN, and proteasome subunits. We also demonstrate that COP10 physically interacts with E2s and has the ability to enhance E2 activity in promoting both K48- and K63-linked polyubiquitin chain formation. These observations suggest that the COP10-containing complex acts in a novel fashion to promote ubiquitylation and thus provides a new insight into the regulation of the photomorphogenesis by the COP/DET/FUS proteins.
Results
Biochemical purification of the COP10-containing complex
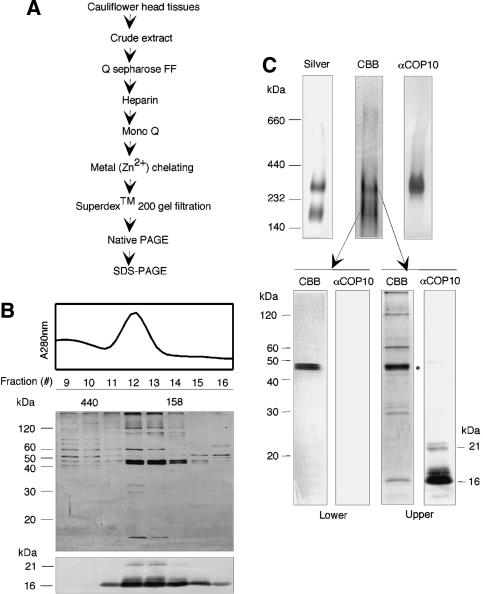
To identify the subunit components of the COP10-containing complex, a biochemical purification procedure for the complex from cauliflower, a Brassica species closely related to Arabidopsis, was developed. Following several chromatographic steps (for details, see Fig. 1A; Materials and Methods), the COP10-enriched fraction was subjected to gel filtration chromatography. The majority of COP10 was found in fractions corresponding to ∼300 kDa, as predicted from the size of the COP10-containing complex (Fig. 1B). Several protein bands were found in two fractions (no. 12 and no. 13) that were enriched for COP10 (Fig. 1B).
Figure 1.
Purification of COP10-containing complex. (A) The COP10-containing complex was purified from cauliflower extract following Q Sepharose fast flow, heparin, Mono Q, metal (Zn2+) chelating, and Superdex 200 gel filtration column chromatographs, and native PAGE. The fractions containing the COP10-containing complex were determined by using immunoblotting with a COP10 antibody. (B) The peak fractions of Superdex 200 gel filtration chromatography were identified by using absorbance values 280 nm (top) and protein bands visualized by silver staining (middle) or immunoblotting with COP10 antibody (bottom). (C) Native PAGE of protein complexes in peak fractions 12 and 13 (B) visualized using silver staining (top left) and immunoblotting with COP10 antibodies (top right). Lower and upper complexes of native gel stained with Coomassie brilliant blue (CBB, top middle) were eluted and then applied onto SDS-PAGE followed CBB staining or immunoblotting with COP10 antibodies (bottom). Note that ∼100-fold amount of sample was used for the native gel stained with CBB than for the native gel stained with silver.
To further purify the complex to homogeneity, the peak fractions were combined and subjected to native PAGE in an attempt to separate the COP10-containing complex from other copurifying complexes. As shown in Figure 1C, two major protein complexes with approximate sizes of 300 and 200 kDa were resolved in native PAGE (Fig. 1C, upper left and middle panels). Immunoblotting with COP10 antibodies showed that only the upper 300-kDa complex contained COP10, whereas the lower 200-kDa band can be separated away from COP10 (Fig. 1C, upper right panel). The upper complex was excised from the preparative scale native gel (Fig. 1C, upper middle panel) and applied onto an SDS-PAGE in which four major proteins of 120, 62, 29, and 16 kDa were identified (Fig. 1C, lower right panels, CBB). In addition, four bands of between 44 and 46 kDa corresponding to the same four proteins in the smaller 200-kDa complex were also observed (Fig. 1C, lower left panels, CBB), probably due to incomplete separation of the two complexes in the preparative native gel. It is possible that these proteins represent contaminants or proteins associated with the COP10-containing core complex.
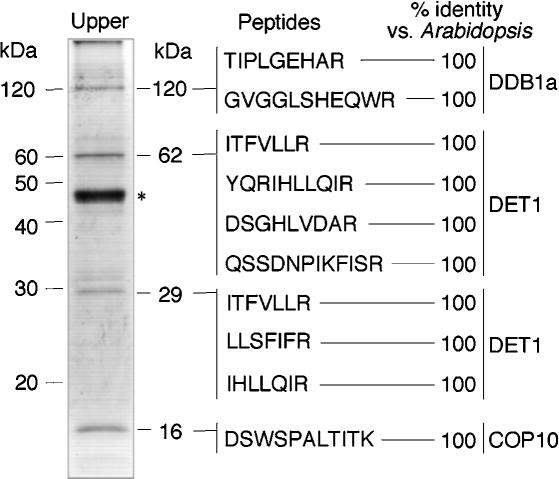
Mass spectroscopy analysis and/or peptide sequencing (Shen et al. 2002) of two and four peptides obtained from the 62- and 120-kDa proteins, respectively, showed a 100% match to Arabidopsis DET1 (62 kDa) and one of the two highly similar (91% identity) Arabidopsis 120 kDa DDB1 proteins, DDB1a (Fig. 2; Schroeder et al. 2002). Three peptide sequences were also obtained for the 29-kDa protein, which also showed 100% identity to an N-terminal portion of Arabidopsis DET1. It should be noted that the predicted size for COP10 is 21 kDa, and although a protein of 21 kDa was not present in sufficient quantity to be detected by Coomassie brilliant blue (CBB) staining, proteins of 21 and 16 kDa were identified following immunoblotting with COP10 antibodies (Fig. 1C, lower right panels). One peptide sequence was obtained for the 16-kDa protein (Fig. 2), which, as expected, showed 100% identity to Arabidopsis COP10. Because the purified COP10-containing complex still fractionated as a ∼300-kDa complex, the 16-kDa COP10 in the complex was likely a partial degradation product produced during chromatography (data not shown). Despite this degradation, the 16-kDa stable COP10 degradation product still remained associated with the complex throughout chromatography, gel filtration, and native PAGE. The identities of the four proteins of the 200-kDa copurifying complex could not be determined as the peptide sequences obtained failed to identify any homologous proteins in the Arabidopsis proteome database. Although we cannot completely rule out the possibility that the 200-kDa complex may associate with the COP10-containing complex, it is clear from these results that the “core COP10 complex” consists of COP10, DDB1, and DET1. The presence of both DET1 and DDB1 in the COP10-containing complex is consistent with the copurification of DDB1 with an epitope-tagged DET1 protein using a tobacco cell culture system (Schroeder et al. 2002). Therefore, we designate this complex as CDD (COP10, DDB1, and DET1) complex from this point onward.
Figure 2.
Component identification of the COP10-containing complex. Amino acid sequences were obtained following mass spectroscopy and/or peptide sequencing.
COP10, DDB1, and DET1 cofractionate, and the CDD complex is undetectable in the det1-1 mutant
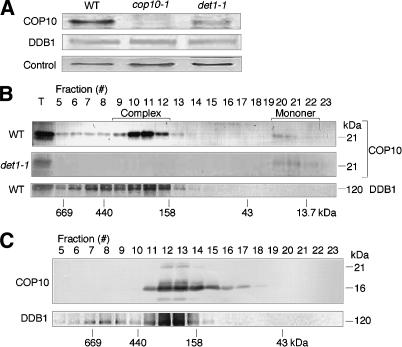
To substantiate the finding that COP10, DDB1, and DET1 form a stable complex in Arabidopsis, we next investigated the accumulation of CDD complex components in Arabidopsis det1 and cop10 mutants. Immunoblotting with COP10 antibodies revealed that COP10 is absent from cop10-1 and is significantly reduced in det1-1 seedlings (Fig. 3A). In contrast, DDB1 abundance is unchanged in either cop10-1 or det1-1 (Fig. 3A). Because DDB1 has two highly identical family members in Arabidopsis, DDB1a and DDB1b (Schroeder et al. 2002), it is likely that the anti-DDB1 antibody, which was raised against rice DDB1 (Ishibashi et al. 2003), detects both isoforms of Arabidopsis DDB1. In addition, DDB1 is known to exist in multiple complexes such as the DDB2- and CSA-containing complexes (Groisman et al. 2003), and it is possible that these other DDB1 complexes are not affected by loss of COP10.
Figure 3.
DET1 and DDB1 are components of the COP10-containing complex. (A) Immunoblot analysis of light-grown wild-type, cop10-1, and det1-1 seedlings using COP10 and DDB1 antibodies; a nonspecific cross-reacting band was used as a loading control. (B) Immunoblot analysis using COP10 and/or DDB1 antibodies following gel filtration of crude extracts from 6-day-old wild-type and det1-1 seedlings. (C) Immunoblot analysis using COP10 and DDB1 antibodies on COP10-containing complex purified from cauliflower following gel filtration. Samples are same as Figure 1B, and the fraction numbers correspond to Figure 1B.
To further investigate the formation of the CDD complex in det1-1 seedlings, gel filtration analysis was performed. As reported previously (Suzuki et al. 2002) in wild-type seedlings, COP10 fractionated predominantly as a complex of ∼300 kDa with a small amount of monomeric proteins (Fig. 3B). In the det1-1 mutant, however, the CDD complex was undetectable, and essentially all COP10 was found in monomeric form (Fig. 3B). DDB1 also cofractionated with the CDD complex in wild-type seedlings (Fig. 3B), although no clear difference in the DDB1 gel filtration profile was observed between wild-type and cop10 or det1 mutants, possibly due to its involvement in multiple protein complexes and broad fractionation profile (data not shown). To confirm if DDB1 copurified with the CDD complex, immunoblotting using rice DDB1 antibodies was performed against the gel filtration samples of purified cauliflower CDD complex. As expected, DDB1 was strongly detected in the fractions corresponding to the CDD complex (Fig. 3C).
Flag-tagged COP10 is functional and associates with DDB1 and DET1 in vivo
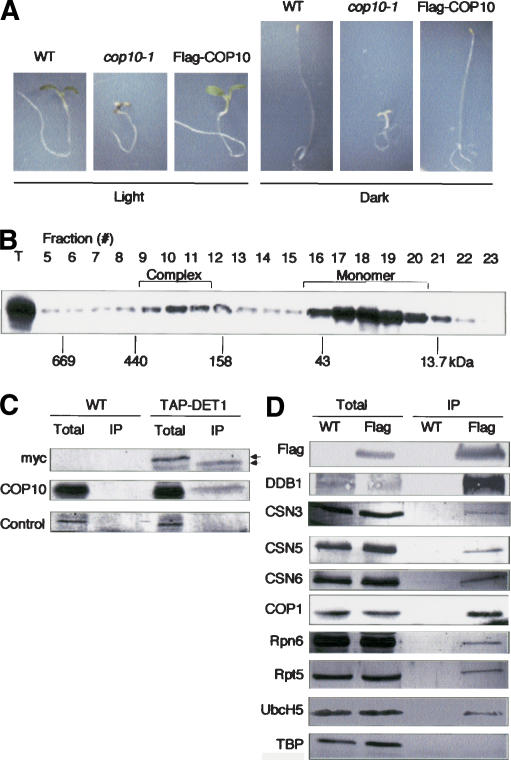
To further confirm the physical association of the CDD complex components in Arabidopsis, a 3xFlag epitope-tagged COP10 cDNA (FlagCOP10) driven by the constitutive cauliflower mosaic virus (CaMV) 35S promoter was introduced into cop10-1 plants. As shown in Figure 4A, the FlagCOP10 transgene completely rescued the light- and dark-grown phenotype of cop10-1 seedlings, verifying the functionality of FlagCOP10. As predicted, FlagCOP10 protein fractionated corresponding to a ∼300-kDa complex as well as monomeric form (Fig. 4B). It should be noted that a much larger proportion of Flag-COP10 was found in monomeric form compared with endogenous COP10, which is probably due to the overexpression of FlagCOP10. We then performed immunoprecipitation with Flag antibodies in extracts from Flag-COP10 seedlings. As shown in Figure 4D, DDB1 was strongly coprecipitated with Flag antibodies in Flag-COP10 extracts, but not wild-type seedlings. To confirm the association between COP10 and DET1, we used transgenic Arabidopsis lines that contained a CaMV 35S-driven fusion between DET1 and a modified tandem affinity purification (TAP) tag (Saijo et al. 2003). As expected, COP10 coprecipitated with DET1 in extracts from TAP-DET1 seedlings but not from wild type (Fig. 4C).
Figure 4.
Analysis of Flag-tagged COP10 transgenic Arabidopsis lines. (A) Morphological phenotype of wild-type (WT), cop10-1, and Flag-COP10 in a cop10-1 background following growth in light or dark. (B) Immunoblot analysis using Flag antibodies on crude extracts from Flag-COP10 transgenic seedlings (in cop10-1 background) following gel filtration chromatography. (C) Coimmunoprecipitation of proteins using IgG Sepharose in extracts from wild-type (WT) and TAP-DET1 transgenic seedlings. DET1 was detected by using anti-myc antibodies; a nonspecific cross-reacting band from the COP10 antibody is shown as a negative control. DET1 was eluted from the beads by cleavage with 3C protease. Upper and lower arrows indicate the DET1 bands before and after cleavage, respectively. (D) Coimmunoprecipitation of proteins using Flag antibodies in extracts from wild-type (WT) and Flag-COP10 transgenic seedlings (Flag). FlagCOP10 immunoprecipitates DDB1, CSN3, CSN5, CSN6, COP1, Rpn6, Rpt5, and E2. Note there are 10 UbcH5 homologs in Arabidopsis (Bachmair et al. 2001).
The CDD complex interacts with COP1, CSN, proteasome subunits, and E2 in vivo
It has been shown previously that the CDD complex was unstable in csn mutants and that several CSN subunits and COP1 interact with COP10 in a yeast two-hybrid assay (Suzuki et al. 2002). To investigate the interaction between the CDD complex and the CSN in vivo, we performed immunoblotting with CSN3, CSN5, and CSN6 antibodies following immunoprecipitation of FlagCOP10. As shown in Figure 4D, all three CSN subunits coprecipitated with FlagCOP10. In addition, we also observed coprecipitation of COP1, Rpn6, and Rpn5 with FlagCOP10 (Fig. 4D). Furthermore, we performed immunoblotting with antibodies raised against the E2 UbcH5 following immunoprecipitation of FlagCOP10. As shown in Figure 4D, FlagCOP10 coprecipitated with a UbcH5 cross-reacting E2, but not with an antibody raised against the rice E2 Rad6 (data not shown). However, it is unclear from these results which of the 10 UbcH5 homologs found in Arabidopsis interacts with COP10. As COP1 is present in vivo as a large protein complex (Saijo et al. 2003), together this coimmunoprecipitation study indicated that the CDD complex is associated with the CSN, the COP1 complex, the 26S proteasome, and an UbcH5-related E2 (or E2s).
COP10 promotes both K48- and K63-linked ubiquitin chain formation
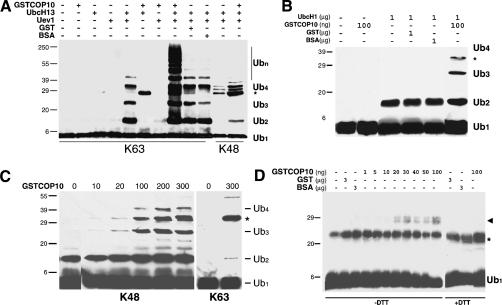
To investigate the biochemical activity of COP10, we performed ubiquitylation assays in vitro. Because both MMS2 and UEV1 are functionally equivalent in ubiquitylation assays in vitro (Ashley et al. 2002), we performed an in vitro ubiquitylation assay to see if recombinant COP10 could function with human Ubc13 to produce K63-linked ubiquitin chains. To facilitate the characterization of UEV activity, we used single lysine ubiquitin, in which all the lysine resides except one have been mutated to arginine and are no longer available for chain formation (Hodgins et al. 1996). K63 chain formation was only observed with a combination of Ubc13 and Uev1; addition of recombinant COP10 to Ubc13 did not result in the formation of K63-linked chains (Fig. 5A). However, we observed a large enhancement in K63-chain formation by Ubc13/Uev1 following the addition of GSTCOP10 but not GST alone, BSA (Fig. 5A), or GSTHY5 (data not shown). It should also be noted that E1 activity was unaffected by the addition of COP10 and that COP10 itself was not ubiquitylated in the reaction (data not shown). COP10-mediated enhancement did not seem to affect the specificity of Ubc13/Uev1 activity, as very little chain formation was observed using K48 ubiquitin (Fig. 5A).
Figure 5.
In vitro ubiquitylation assays with recombinant COP10. (A) GSTCOP10, but not GST or BSA, enhances K63 but not K48 single lysine ubiquitin chain formation mediated by Ubc13/Uev1. Asterisk indicates a reductant stable nonspecific ubiquitylated protein(s) associated with the addition of UbcH13. (B) GSTCOP10 enhances UbcH1-mediated ubiquitin chain formation. Note longer chain formation mediated by UbcH1 alone is visible upon extended exposure. (C) GSTCOP10 enhances UbcH1-mediated ubiquitin chain formation in a concentration-dependent manner with K48 single lysine ubiquitin but has little effect on K63 single lysine ubiquitin. (B,C) Asterisk indicates a reductant stable nonspecific ubiquitylated protein associated with the addition of UbcH1 (amount of GSTCOP10 used shown in nanograms). (D) GSTCOP10 enhances ubiquitin-thiolester formation mediated by AtUbc9 in a concentration-dependent manner. Reactions were terminated with nonreducing sample buffer (-DTT) or reducing sample buffer (+DTT). Arrowhead indicates E2-ubiquitin thiolester conjugate, asterisk indicates nonspecific biotinylated protein contaminate of ubiquitin (data not shown). Ubiquitin conjugates were detected by immunoblotting with monoclonal ubiquitin antibodies for single lysine ubiquitin, or streptavidin-conjugated horseradish peroxidase for biotinylated-ubiquitin, followed by chemiluminescence visualization.
To determine if the enhancement of E2 activity by COP10 was specific to K63-linked chains, we used the E2 UbcH1 that produces K48-linked ubiquitin chains in the absence of an E3 (Hodgins et al. 1996). As shown in Figure 5B, significant enhancement in UbcH1 activity was also observed following the addition of COP10, but not GST-alone, BSA, or GSTHY5 (data not shown). This enhancement was dependent upon the concentration of COP10 (Fig. 5C) and did not seem to affect the specificity of chain formation by UbcH1 (Fig. 5C). It should be noted, as observed with UbcH13/Uev1 and UbcH1, that a small amount of chain formation by UbcH1 was observed with K63 ubiquitin following the addition of COP10. It is unclear if this represents some residual affinity for K63 by UbcH1 (or K48 by UbcH13/Uev1) or the misincorporation of lysine at arginine codons that sometimes occurs with recombinant proteins produced in bacteria (Hodgins et al. 1996). However, it is clear from these results that the formation of both K63-linked ubiquitin chains by Uev1/Ubc13 and K48-linked chains by UbcH1 can be greatly enhanced by the addition of COP10.
COP10 enhances thiolester formation
Because most E2s do not form stable ubiquitin chains but instead form a thiolester linkage with ubiquitin to allow transfer to a target protein via the action of an E3 (Vierstra 2003), we investigated the effect of recombinant COP10 on thiolester formation mediated by Arabidopsis Ubc9 (AtUbc9). AtUbc9 shows homology to the yeast E2s Ubc4 and Ubc5 and has been shown to function with COP1 in the ubiquitylation of HY5 (Saijo et al. 2003). COP10 enhanced AtUbc9-mediated thiolester formation in a concentration-dependent manner (Fig. 5D). A similar enhancement was also observed with several other E2s, including UbcH2, UbcH5, OsRad6, and AtUbc8 (data not shown), suggesting that COP10 enhancement of E2 activity in in vitro assays affects a broad range of E2 types. Although the exact mechanism for COP10 E2 enhancement activity remains to be determined, it seems possible that increased thiolester formation as observed with AtUbc9 may be at least partially responsible. This enhancement in thiolester formation could ultimately lead to the increased polyubiquitin chain formation mediated by UbcH1 and UbcH13/Uev1.
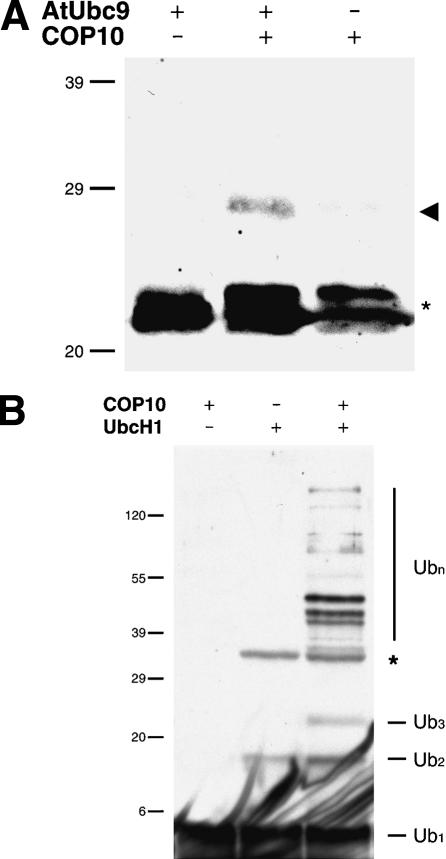
The CDD complex also has the ability to enhance E2 activity
To determine if the CDD complex also possesses E2 enhancement ability, we used the CDD complex purified from cauliflower in our in vitro reactions. It was evident that despite the partial degradation of COP10 within the complex, the purified CDD complex enhanced the ubiquitin thiolester formation mediated by AtUbc9 (Fig. 6A). It is also clear that the purified CDD complex can enhance ubiquitin chain formation mediated by UbcH1 (Fig. 6B), although we can not rule out the possibility that some high-molecular ubiquitin-containing bands may be mono-ubiquitylated proteins that copurify with the CDD complex rather than free ubiquitin chains. However, the lack of an E3 in the in vitro reactions, which would allow ubiquitylation of target proteins, suggests this is unlikely.
Figure 6.
In vitro ubiquitin E2 activity assays with purified CDD complex. (A) Purified cauliflower CDD complex enhances ubiquitin-thiolester formation mediated by AtUbc9. Note that to facilitate detection of weak signal, free ubiquitin was allowed to run off the bottom of the gel. Reactions were terminated with nonreducing sample buffer; arrowhead indicates E2-thiolester conjugate and asterisk indicates biotinylated contaminate described in Figure 5D. (B) Purified cauliflower CDD complex enhances ubiquitin chain formation mediated by UbcH1. Asterisk indicates a reductant stable nonspecific ubiquitylated protein associated with the addition of UbcH1. Ubiquitin conjugates were detected by immunoblotting with monoclonal ubiquitin antibodies for single lysine ubiquitin, or streptavidin-conjugated horseradish peroxidase for biotinylated-ubiquitin, followed by chemiluminescence visualization.
Discussion
COP10 forms a CDD complex with DET1 and DDB1
From our results it is clear that COP10 forms a stable complex with DET1 and DDB1 in plants. Both COP10 and DET1 have been previously identified through genetic screens as repressors of photomorphogenesis (Pepper et al. 1994; Wei et al. 1994). Null mutations in Arabidopsis DDB1a only show a phenotype in the context of a det1 mutation, whereas null mutations of DDB1b show pre-embryonic lethality, which may explain why DDB1 was not identified as one of the COP/DET/FUS proteins (Schroeder et al. 2002). DET1 has been shown to bind Histone 2B in a nucleosome context and has been implicated in the regulation of gene expression through chromatin remodeling (Benvenuto et al. 2002). Because the ubiquitin E2 enhancement ability (UEA) of the CDD complex can be explained by the activity of COP10 alone, it seems likely that DET1 and DDB1 may specify other aspects of the CDD complex function. For example, they may act to link the CDD complex to functional targets or other cellular pathways or define target specificity of its UEA activity. COP10 has no nuclear localization signal (NLS), but the CDD complex localizes in the nucleus (Suzuki et al. 2002). Previously, we showed that the fusion protein GUS–COP10 does not enter the nucleus in onion cells, presumably because the GUS domain of the fusion protein prevents assembly of the complex (Suzuki et al. 2002). DDB1 also has no NLS and is known to have capability to enter the nucleus with DDB2, which contains a NLS (Hwang et al. 1999; Nichols et al. 2000; Nag et al. 2001) and is prevented from entering the nucleus in ddb2 mutants (Shiyanov et al. 1999). Thus, for complex formation DET1, which has a NLS, (Robbins et al. 1991) is probably necessary for nuclear localization. Because COP10 is dissociated from the CDD complex in det1-1 mutant, DET1 must also act to stabilize the CDD complex.
Recently, DDB1 and DET1 were reported to regulate c-Jun ubiquitylation together with COP1 in mammals, and it was suggested that COP1 and DET1 exist as part of an E3 complex (Wertz et al. 2004). From the results presented here and elsewhere (Saijo et al. 2003), we propose that DET1, but not COP1, is a stoichiometric component of the CDD core complex, which interacts with the COP1 complex, although the identity of the plant COP1 complex components remains to be determined. Given the size of the CDD complex in gel filtration, it is likely that multiple subunits of COP10, DET1, and/or DDB1 are present in the complex. Alternatively, it is possible that other proteins are associated with the CDD core complex, which were lost during the purification procedure. However, because the size of the purified CDD complex was ∼300 kDa in native PAGE, the association of a large number of accessory proteins seems unlikely.
The CDD complex has UEA
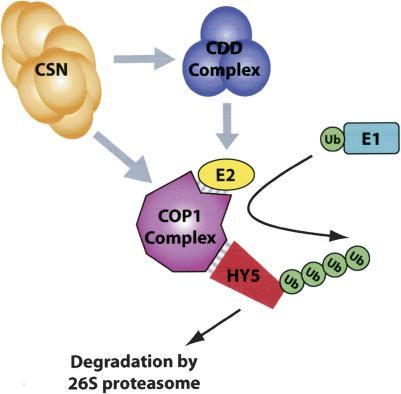
Previous characterized UEV proteins such as MMS2 and UEV1 act to form noncanonical ubiquitin chains together with an active E2 Ubc13 (Hofmann and Pickart 1999). Clearly the activity of COP10 is distinct from these UEV proteins and appears not to be directly involved in formation of noncanonical ubiquitin chains. FlagCOP10 physically interacts with E2s, implying that the COP10 might act to change conformation of E2s and/or enhance their affinity for ubiquitin. The results presented here demonstrate that Arabidopsis COP10 enhances the activity of several different E2s from both plants and animals in vitro. However, the observation that FlagCOP10 can coimmunoprecipitate a Ubc5-type E2 but not Rad6 (the activity of which was enhanced by COP10 in vitro) suggests that in planta the UEA activity of COP10 may be more discriminating. By sequence analysis, no homologous gene to COP10 has so far been found in mammals (Suzuki et al. 2002), implying that COP10 is a plant-specific E2 regulator that acts as part of the CDD complex. However, because COP10 enhances the activity of E2s from animals, mammalian cells might contain a protein with similar function but that lacks close homology to Arabidopsis COP10. In plants, COP1 acts as an E3 ligase for photomorphogenesis-promoting factors (Oaterlund et al. 2000; Saijo et al. 2003; Seo et al. 2003), and because the CDD complex interacts with COP1 and E2s, it is probable that the CDD complex supports COP1 to ubiquitylate photomorphogenesis-promoting factors, possibly by acting as an E2 enhancer. Because the CSN stabilizes the CDD complex (Suzuki et al. 2002), the CDD complex together with the COP1 complex and the CSN are three protein complexes defined by the COP/DET/FUS group of genes that act together to regulate ubiquitin–proteasome-mediated degradation of photomorphogenesis-promoting transcription factors in darkness (Fig. 7).
Figure 7.
Working model for the functional relationships of the COP/DET/FUS proteins. In the dark the COP9 signalosome (CSN) directly interacts with the CDD complex and possibly the COP1 complex to regulate their assembly and/or activity. The COP1 complex contains ubiquitin E3 ligase activity and, by binding both the E2 and HY5, facilitates the transfer of ubiquitin from the E2 to HY5. The activity of the E2 enzyme is also enhanced by the CDD complex, which also directly interact with the COP1 complex. Subsequent rounds of ubiquitylation lead to the polyubiquitylation of HY5 and degradation via the 26S proteasome.
Materials and methods
Plant materials and growth condition
All Arabidopsis growth conditions have been described previously (Wei et al. 1994). The cop10-1 (Wei et al. 1994) and det1-1 (Pepper et al. 1994) mutants were isolated from Wassilewskija (WS) and Columbia, respectively.
Purification of CDD complex
All procedures were performed at 4°C. The fractions containing the CDD complex were determined by immunoblot with COP10 antibody. One kilogram of fresh cauliflower head tissue was homogenized with equal volume of extraction buffer (25 mM Tris-HCl at pH 7.5, 0.2 M NaCl, 0.5 mM phenylmethylsulfonyl fluoride [PMSF], 5 mM β-mercaptoethanol, complete protease inhibitor [Roche], 10% v/v glycerol). The extract was centrifuged at 10,000 rpm for 15 min and then centrifuged again at 13,000 rpm for 30 min. The supernatant was applied to a 250 mL Q Sepharose fast-flow ion-exchange column (Amersham Biosciences) equilibrated with buffer A (25 mM Tris-HCl at pH 7.5, 5 mM EDTA, 0.5 mM PMSF, 10 μM leupeptin, 5 mM β-mercaptoethanol, 10% v/v glycerol) containing 0.2 M NaCl. The CDD complex was eluted with buffer A containing 0.3 M NaCl. The salt concentration of sample was adjusted to 50 mM NaCl with buffer A and then applied to a 20 mL heparin affinity column (Amersham Biosciences) equilibrated with buffer A, including 50 mM NaCl. The complex was eluted with buffer A containing 0.2 M NaCl and applied to a 1 mL Mono Q ionexchange column (Amersham Biosciences) equilibrated with buffer A containing 0.2 M NaCl. The complex was eluted at buffer A containing 0.2–0.4 M NaCl gradient. The complexcontaining fractions were applied to a desalting column (Amersham Biosciences) equilibrated with buffer B (25 mM NaPi at pH 7.5, 0.2 M NaCl, 0.5 mM PMSF, 5 mM β-mercaptoethanol, 10 μM leupeptin, 10% v/v glycerol). The eluate was applied to a 1 mL zinc chelating affinity column, and the complex was eluted with 50 mM imidazol in buffer B. The eluate was concentrated to 500 μL with Microsep 50 (Amicon), and applied onto a Superdex 200 gel filtration chromatography column (Amersham Biosciences) equilibrated with buffer C (25 mM Tris-HCl at pH 7.5, 0.15 M NaCl, 5 mM β-mercaptoethanol, 10% v/v glycerol). Samples were pooled and were subjected to native gel electrophoresis. For protein MS/sequence analysis, the COP10 protein complex was eluted from preparative scale native PAGE gel slices with buffer D (125 mM Tris-HCl at pH 6.8, 20% w/v glycine, 2% w/v SDS) and then applied onto SDS-PAGE.
Immunoblot analysis and gel filtration chromatography
For immunoblot analysis and gel filtration chromatography, 6-day-old seedlings were homogenized with buffer A with 0.15 M NaCl. Gel filtration chromatography was performed as previously described (Suzuki et al. 2002). The polyclonal antibodies against COP10, DDB1, CSN3, CSN5, CSN6, COP1, Rpt5, and Rpn6 were prepared previously (McNellis et al. 1994; Kwok et al. 1998, 1999; Peng et al. 2001a,b; Suzuki et al. 2002; Ishibashi et al. 2003).
Construction of FlagCOP10 and TAP-DET1 transgenic plants and immunoprecipitation
The COP10 cDNA was amplified by PCR from pETCOP10 (Suzuki et al. 2002) with the primers 5′-GGTACCATGATGAC ACCTGGCGGAAG-3′ with KpnI site at the 5′ terminus and 5′-GAGCTCTCACTTGGCAAATCGCAATG-3′ with SacI site at the 3′ terminus. The COP10 fragment was cloned into pF3PZPY122, which besides three copies of Flag tag also carries a gentamicin-resistance marker and a 35S promoter from CaMV to drive the expression of the transgene (Feng et al. 2003). The construct was transformed into cop10-1 heterozygous plants via vacuum infiltration and homozygous lines were selected in the T3 generation.
A DET1 cDNA was amplified by PCR with the primers 5′-CACCACAAAATGTTCACAAGCGGTAACGTCACC-3′ at the 5′ terminus and 5′-TCGCCTAAAATGGATATTGACGA CAG-3′ at the 3′ terminus. This fragment was cloned into a pENTR/D-TOPO vector (Invitrogen) and transferred, by using the BP reaction (Gateway Cloning Technology; GIBCO-BRL), into a modified N-terminal TAP tag containing vector (Saijo et al. 2003).
For the FlagCOP10 protein immunoprecipitation, total protein extracts from Arabidopsis seedlings were used according to our previous procedure (Schwechheimer et al. 2001). Immunoprecipitation of IgG Sepharose (Amersham Biosciences) was performed as described previously (Saijo et al. 2003), and then proteins were eluted by cleavage of the IgG-binding domain with 3C protease (Amersham Biosciences). A monoclonal antibody against Flag was purchased from Sigma. UbcH5 antibody was purchased from Boston Biochem.
In vitro ubiquitylation assays
Recombinant glutathione-S-transferase (GST)-tagged COP10 was expressed in BL21 codon plus (Stratagene) Escherichia coli cells and purified as described previously (Suzuki et al. 2002). Ubiquitylation reactions were performed in a total volume of 30 μL, consisting of 50 mM Tris-HCl (pH 7.5), 25 mM ZnCl2, 10 mM MgCl2, 10 mM ATP, 125 ng rabbit E1 (Boston Biochem), 200 ng GST-COP10 (unless otherwise stated), and 10 μg single lysine (K48, K63) ubiquitin (Boston Biochem) or 1 μg biotinylated ubiquitin (Boston Biochem) supplemented with 4 μg unlabeled ubiquitin (Sigma). In addition, 1 μg UbcH1 (Sigma), 1 μg UbcH13 (Boston Biochem), 1 μg Uev1 (Boston Biochem), 3 μg of GST (unless otherwise stated), 3 μg (unless otherwise stated) bovine serum albumin (BSA, Sigma), 250 ng AtUbc9 (Saijo et al. 2003), or 2 μg (∼50–100 ng of COP10) of purified CDD complex were added to the reactions. Reactions were incubated for 2 h or for 30 min at 37°C for thiolester assays, before being terminated by the addition of SDS-sample buffer containing DTT or 8 M urea sample buffer (without DTT) in the case of thiolester assays.
Acknowledgments
We thank T. Yamamoto and K. Sakaguchi for the OsRad6 expression construct and antibody and C. Pickard for advice with the single lysine ubiquitin experiments. We also thank Drs. N. Wei, L. Li, T. Nelson, and V. Irish for critical reading of this manuscript. This research is supported by a grant from National Institutes of Health (GM-47850 to X.W.D.). J.A.S. and V.R. are supported by human frontiers science program long-term fellowships. Y.Y. and Y.S. are supported by postdoctoral researcher abroad fellowships from the Japanese Society for the Promotion of Science (JSPS).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.1229504.
References
- Ashley C., Pastushok, L., McKenna, S., Ellison, M.J., and Xiao, W. 2002. Roles of mouse UBC13 in DNA postreplicative repair and Lys63-linked ubiquitination. Gene 285: 183-191. [DOI] [PubMed] [Google Scholar]
- Bachmair A., Novatchkova, M., Potuschak, T., and Eisenhaber, F. 2001. Ubiquitylation in plants: A post-genomic look at a post-translational modification. Trends Plant Sci. 6: 463. [DOI] [PubMed] [Google Scholar]
- Benvenuto G., Formiggini, F., Laflamme, P., Malakhov, M., and Bowler, C. 2002. The photomorphogenesis regulator DET1 binds the amino-terminal tail of histone H2B in a (space) nucleosome context. Curr. Biol. 12: 1529-1534. [DOI] [PubMed] [Google Scholar]
- DeMartino G.N., and Slaughter, C.A. 1999. The proteasome, a novel protease regulated by multiple mechanisms. J. Biol. Chem. 274: 22123-22126. [DOI] [PubMed] [Google Scholar]
- Feng S., Ma, L., Wang, X., Xie, D., Dinesh-Kumar, S.P., Wei, N., and Deng, X.W. 2003. The COP9 signalosome interacts physically with SCFCOI1 and modulates jasmonate responses. Plant Cell 15: 1083-1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrell K., Wilkinson, C.R.M., Dubiel, W., and Gordon, C. 2000. Regulatory subunit interactions of the 26S proteasome, a complex problem. Trends Biochem. Sci. 25: 83-88. [DOI] [PubMed] [Google Scholar]
- Glickman M.H., Rubio, D.M., Coux, O., Wefes, I., Pfeifer, G., Cjeka, Z., Baumeister, W., Fried, V.A., and Finley, D. 1998. A subcomplex of the proteasome regulatory particle required for ubiquitin-conjugate degradation and related the COP9-signalosome and eIF3. Cell 94: 615-623. [DOI] [PubMed] [Google Scholar]
- Groisman R., Polanowska, J., Kuraoka, I., Sawada, J., Saijo, M., Drapkin, R., Kisselev, A.F., Tanaka, K., and Nakatani, Y. 2003. The ubiquitin ligase activity in the DDB2 and CSA complexes is differentially regulated by the COP9 signalosome in response to DNA damage. Cell 113: 357-367. [DOI] [PubMed] [Google Scholar]
- Hodgins R., Gwozd, C., Arnason, T., Cummings, M., and Ellison, M.J. 1996. The tail of a ubiquitin-conjugating enzyme redirects multi-ubiquitin chain synthesis from the lysine 48-linked configuration to a novel nonlysine-linked form. J. Biol. Chem. 271: 28766-28771. [DOI] [PubMed] [Google Scholar]
- Hofmann R.M. and Pickart, C.M. 1999. Noncanonical MMS2-encoded ubiquitin-conjugating enzyme functions in assembly of novel polyubiquitin chain for DNA repair. Cell 96: 645-653. [DOI] [PubMed] [Google Scholar]
- Hwang B.J., Ford, J.M., Hanawalt, P.C., and Chu, G. 1999. Expression of the p48 xeroderma pigmentosum gene is p53-dependent and is involved in global genomic repair. Proc. Natl. Acad. Sci. 96: 424-428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishibashi T., Kimura, S., Yamamoto, T., Furukawa, T., Takata, K., Uchiyama, Y., Hashimoto, J., and Sakaguchi, K. 2003. Rice UV-damaged DNA binding protein homologues are most abundant in proliferating tissues. Gene 308: 79-87. [DOI] [PubMed] [Google Scholar]
- Kwok S.F., Solano, R., Tsuge, T., Chamovitz, D., Ecker, J.R., Matsui, M., and Deng, X.W. 1998. Arabidopsis homologs of a c-Jun coactivator are present both in monomeric form and in the COP9 complex, and their abundance is differentially affected by the pleiotropic cop/det/fus mutations. Plant Cell 10: 1779-1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwok S.F., Staub, J.M., and Deng, X.W. 1999. Characterization of two subunits of Arabidopsis 19S proteasome regulatory complex and its possible interaction with the COP9 complex. J. Mol. Biol. 285: 85-95. [DOI] [PubMed] [Google Scholar]
- Li L., Kiao, J., Ruland, J., Mak, T.W., and Cohen, S.N. 2001. A TSG101/MDM2 regulatory loop modulates MDM2 degradation and MDM2/p53 feedback control. Proc. Natl. Acad. Sci. 98: 1619-1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyapina S., Cope, G., Shevchenko, A., Serino, G., Tsuge, T., Zhou, C., Wolf, D.A., Wei, N., Shvchenko, A., and Deshaies, R.J. 2001. Promotion of NEDD8-CUL1 cunjugate cleavage by COP9 signalosome. Science 292: 1382-1385. [DOI] [PubMed] [Google Scholar]
- McNelis T.W., von Arnim, A.G., Araki, T., Komeda, Y., Misera, S., and Deng, X.W. 1994. Genetic and molecular analysis of an allelic series of cop1 mutants suggests functional roles for the multiple protein domains. Plant Cell 6: 487-500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nag A., Bondar, T., Shiw, S., and Raychaudhuri, P. 2001. The xeroderma pigmentosum group E gene product DDB2 is a specific target of Cullin4A in mammalian cells. Mol. Cell. Biol. 21: 6738-6747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols A.F., Itoh, T., Graham, J.A., Liu, W., Yamazaki, M., and Linn, S. 2000. Human damage-specific DNA-binding protein p48. J. Biol. Chem. 275: 21422-21428. [DOI] [PubMed] [Google Scholar]
- Osterlund M.T., Hardtke, C.S., Wei, N., and Deng, X.W. 2000. Targeted destabilization of HY5 during light-regulated development of Arabidopsis. Nature 405: 462-466. [DOI] [PubMed] [Google Scholar]
- Peng Z., Serino, G., and Deng, X.W. 2001a. A role of Arabidopsis COP9 signalosome in multifaceted developmental processes revealed by the characterization of its subunit 3. Development 128: 4277-4288. [DOI] [PubMed] [Google Scholar]
- ____. 2001b. Molecular characterization of subunit 6 of the COP9 signalosome and its role in multifaceted developmental processes in Arabidopsis. Plant Cell 13: 2393-2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng Z., Shen, Y., Feng, S., Wang, X., Chitteti, B.N., Vierstra, R.D., and Deng, X.W. 2003. Evidence for a physical association of the COP9 signalosome, the proteasome, and specific SCF E3 ligases in vivo. Curr. Biol. 13: R504-R505. [DOI] [PubMed] [Google Scholar]
- Pepper A., Delaney, T., Washburn, T., Poole, D., and Chory, J. 1994. DET1, a negative regulator of light-mediated development and gene expression in Arabidopsis, encodes a novel nuclear-localized protein. Cell 78: 109-116. [DOI] [PubMed] [Google Scholar]
- Robbins J., Dilworth, S., Laskey, R., and Dingwall, C. 1991. Two interdependent basic domains in nucleoplasmin nuclear targeting sequence. Cell 64: 615-623. [DOI] [PubMed] [Google Scholar]
- Saijo Y., Sullivan, J.A., Haiyang, W., Yang, J., Shen, Y., Rubio, V., Ma, L., Hoecker, U., and Deng, X.W. 2003. The COP1–SPA1 interaction defines a critical step in phytochrome A-mediated regulation of HY5 activity. Genes & Dev. 17: 2642-2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanch E., Vila, M.R., Sanchez-Pulido, L., Lozano, J.J., Paciucci, R., Nadal, M., Fox, M., Harvery, C., Bercovich, B., Loukili, N., et al. 1998. Role of UEV-1, an inactive variant of the E2 ubiquitin-conjugating enzymes, in in vitro differentiation and cell cycle behavior of HT-29-M6 intestinal mucosecretory cells. Mol. Cell. Biol. 18: 576-589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder D.F., Gahrtz, M., Maxwell, B.B., Cook, R.K., Kan, J.M., Alonso, J.M., Ecker, J.R., and Chory, J. 2002. De-etiolated 1 and damaged DNA binding protein 1 interact to regulate Arabidopsis photomorphogenesis. Curr. Biol. 12: 1462-1472. [DOI] [PubMed] [Google Scholar]
- Schwechheimer C., Serino, G., Callis, J., Crosby, W.L., Lyapina, S., Deshaies, R.J., Gray, W.M., Estelle, M., and Deng, X.W. 2001. Interactions of the COP9 signalosome with the E3 ubiquitin ligase SCFTIRI in mediating auxin response. Science 292: 1379-1382. [DOI] [PubMed] [Google Scholar]
- Seo H.S., Yang, J-Y., Ishikawa, M., Bolle, C., Ballesteros, M.L., and Chua, N-H. 2003. LAF1 ubiquitination by COP1 controls photomorphogenesis and is stimulated by SPA1. Nature 423: 995-999. [DOI] [PubMed] [Google Scholar]
- Serino G. and Deng, X.W. 2003. The COP9 signalosome: Regulating plant development through the control of proteolysis. Ann. Rev. Plant Biol. 54: 165-182. [DOI] [PubMed] [Google Scholar]
- Shen S., Matsubae, M., Takao, T., Tanaka, N., and Komatsu, S. 2002. A proteomic analysis of leaf sheaths from rice. J. Biochem. 132: 613-620. [DOI] [PubMed] [Google Scholar]
- Shiyanov P., Hayes, S.A., Donepudi, M., Nichols, A.F., Linn, S., Slagle, B.L., and Raychaudhuri, P. 1999. The naturally occurring mutants of DDB are impaired in stimulating nuclear import of the p125 subunit and E2F1-activated transcription. Mol. Cell. Biol. 19: 4935-4943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan J.A. and Deng, X.W. 2003. From seed to seed: The role of photoreceptors in Arabidopsis development. Dev. Biol. 260: 289-297. [DOI] [PubMed] [Google Scholar]
- Suzuki G., Yanagawa, Y., Kwok, S.F., Matsui, M., and Deng, X.W. 2002. Arabidopsis COP10 is an ubiquitin-conjugating enzyme variant that acts together with COP1 and the COP9 signalosome in repressing photomorphogenesis. Genes & Dev. 16: 554-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka K. 1998. Proteasome: Structure and biology. J. Biochem. 123: 195-204. [DOI] [PubMed] [Google Scholar]
- Vierstra R.D. 2003. The ubiquitin/26S proteasome pathway, the complex last chapter in the life of many plant proteins. Trends Plant Sci. 8: 135-142. [DOI] [PubMed] [Google Scholar]
- von Arnim A.G. and Deng, X.W. 1996. Light control of seedling development. Annu. Rev. Plant Physiol. Plant Mol. Biol. 47: 215-243. [DOI] [PubMed] [Google Scholar]
- Wei N. and Deng, X.W. 2003. The COP9 signalosome. Annu. Rev. Cell Dev. Biol. 19: 261-286. [DOI] [PubMed] [Google Scholar]
- Wei N., Kwok, S.F., von Arnim, A.G., Lee, A., McNellis, T.W., Piekos, B., and Deng, X.W. 1994. Arabidopsis COP8, COP10, and COP11 genes are involved in repression of photomorphogenic development in darkness. Plant Cell 6: 629-643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wertz I.E., O'Rourke, K.M., Zhang, Z., Dornan, D., Arnott, D., Deshaies, R.J., and Dixit, V.M. 2004. Human de-etiolated-1 regulates c-Jun by assembling a CUL4A ubiquitin ligase. Science 303: 1371-1374. [DOI] [PubMed] [Google Scholar]