Abstract
Inclusion body myositis is a rare, late-onset myopathy. Both inflammatory and myodegenerative features play an important role in their pathogenesis. Overlapping clinicopathological entities are the familial inclusion body myopathies with or without dementia. These myopathies share several clinical and pathological features with the sporadic inflammatory disease. Therefore, better understanding of the genetic basis and pathomechanism of these rare familial cases may advance our knowledge and enable more effective treatment options in sporadic IBM, which is currently considered a relentlessly progressive incurable disease.
Keywords: sporadic inclusion body myositis, genetics, GNE, DES, VCP, IBMPFD, TDP-43
1 Introduction
Inclusion body myositis (IBM), as well as polymyositis, and dermatomyositis are inflammatory myopathies [1]. Sporadic (sIBM) is a late-onset myopathy, representing the most common muscle disease above the age of 50 years [2]. The clinical phenotype of the disease is well-defined, both inflammatory and myodegenerative features can be highlighted during their pathogenesis [1]. However, IBM is a slowly progressive disease with no effective treatment [3]. In inclusion body myositis the typical features are muscle weakness and atrophy in the quadriceps femoris muscle and in the flexors of the wrist and fingers. The most frequent concomitant signs are dysphagia and asymmetric weakness. The serum creatine kinase level is usually under 2000 U/L [4]. The prevalence of the disease is 3.5/100.000, but it moves on a range scale, and varies among different countries, and ethnic groups [5]. This intense diversity may be caused by the combination of different genetic, and environmental factors of the affected racial groups [6]. This disease is most common in males, the male:female ratio is 3:1. The characteristic clinical features of IBM display some overlap with hereditary inclusion body myopathy (hIBM) which makes the diagnosis of sIBM more difficult. However the genetic characteristics of sIBM and hIBM are different [1]. In this review, we briefly introduce the genetic susceptibility factors for sIBM as well as the typical genetic background of hIBM. Furthermore, we describe the main processes associated with the pathogenesis of this disorder. Finally, we introduce the inclusion body myopathy with early-onset Paget disease and frontotemporal dementia, which is a special subtype of IBM.
2 Genetic background of IBM
Inclusion body myopathies can be classified as familial or hereditary inclusion body myopathies (hIBM) or sporadic inclusion body myositis (sIBM) [2]. Hereditary and sporadic IBM share several common pathological features, including rimmed vacuoles, intracytoplasmic, and intranuclear tubulofilamentous inclusions. In spite of sporadic IBM, the muscle biopsies from a patient affected with the hereditary form of IBM, do not show inflammation. This phenomenon explains the ’myopathy’ nomination. Moreover, hIBM can be further characterised by the earlier age of onset and negative MHCI staining of the muscle biopsy. On the other hand, the role of both inflammatory and degenerative processes are important in the pathogenesis of sIBM [7]. In addition to this, there are also differences in the genetic background in these two types of IBM. Moreover, the clinical features of sIBM display some overlap with hereditary inclusion body myopathies [8]. In the next section of our review, we briefly highlight the main genetic differences between sporadic and hereditary IBM.
2.1 Genetic susceptibility factors for sIBM
As we mentioned in the introduction, multiple genetic factors may play an important role during the development and the pathogenesis of sporadic IBM [6]. Similar to other autoimmune diseases, in Caucasians the sIBM is associated with the HLA-DR3 allele and the 8.1 ancestral MHC haplotype: HLA-A*01:01, HLA-B*08:01, HLA-DRB1* 03:01, HLA-DRB3*01:01 and HLA-DQA1*05:01 [9]. Moreover, the HLA-DRB1*03:01 and HLA-DRB1*13:01 alleles may have an influence on the onset of the disease, and severity of muscle weakness [10]. Koffman and colleagues described that sIBM is a distinct disorder with a strong immunogenetic background. They analysed the HLA class II associations in patients with sIBM, and hIBM. The frequency of HLA-DRB1*03:01, HLA-DRB3*01:01 or HLA-DRB3*02:02, as well as HLA-DQB1*02:01 alleles were more than 77% in sIBM, but they did not find a significant association with these alleles in the DR and DQ haplotypes among the 15 hIBM patients [7]. In a recent study, Rojana-Udomsart et al. analysed the HLA-DRB3 alleles and typical MHCII genotypes in sIBM. They found associations with the carriage of the DRB3*01:01 allele which was accounted for by its linkage disequilibrium with the DRB1*03:01 allele. Moreover, the carriage of the HLA-DRB4 allele was found to be protective, in addition it dissolved the risk effect of HLA-DRB1*03:01. Their results indicate that haplotypic combinations of alleles at the HLA-DRB1 and secondary HLA-DRB loci have an important risk modifying effect in sIBM [10]. Cai et al. analysed the three frequent mutated genes of hIBM (DES, GNE, and MYHC2A), as well as two additional genes (VCP, and ZASP) in 21 sIBM patients. They did not find any missense mutations in the DES, VCP or GNE genes. On the other hand, three patients carried a missense mutation c.2542T>C (p.V805A) in the MYHC2A gene, and another patient carried a missense mutation c.1719G>A (p.V566M) in the ZASP gene [8].
2.2 Genetics of hereditary IBM
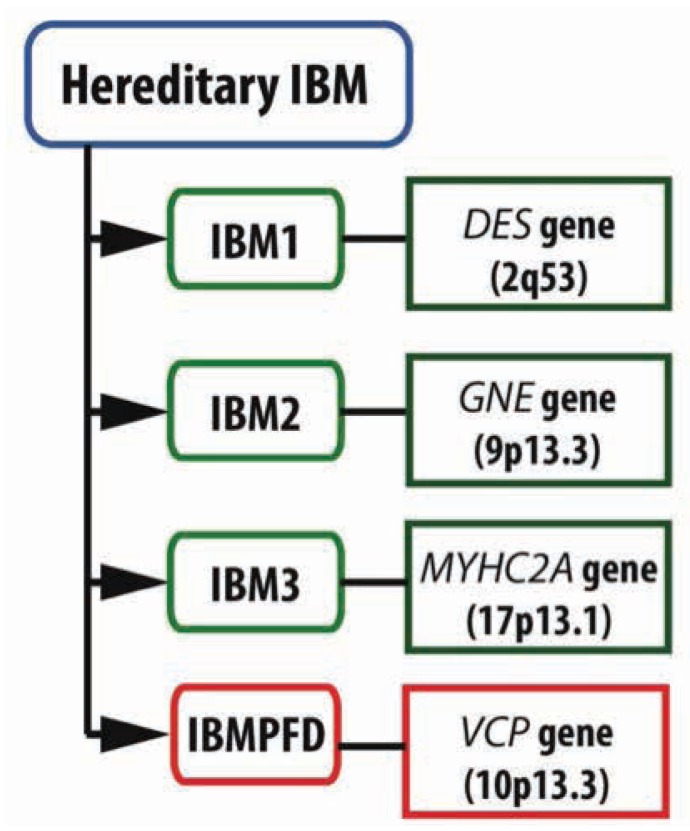
Hereditary IBM is a heterogeneous group of adult-onset muscle disorders with autosomal-dominant (AD) or autosomal-recessive (AR) pattern of inheritance (Figure 1.). According to the affected genes hIBM can be divided into three different types: IBM1, IBM2, and IBM3 [8]. Mutations in the DES (Desmin, 2q35) gene can lead to IBM1, otherwise known as desmin related myopathy. The majority of known mutations of the DES gene show an autosomal dominant inheritance pattern, but some of these alterations are autosomal recessive [6]. The characteristic genetic alteration of the second type of hereditary IBM (IBM2) is the mutated GNE gene (UDP-N-acetylglucosamine-2-epimerase/N-acetylmannosomine kinase). This gene is localised on chromosome 9p13.3, and its mutations are autosomal recessive [8]. At present, we know more than 50 mutations of the GNE gene that are associated with hIBM have been described worldwide [11]. Fischer et al. analysed for mRNA-expression markers of cellular stress, inflammation and β-amyloid of IBM2 patients and compared to non-myopathic controls. The cellular stress molecules αB-crystallin and iNOS are overexpressed in IBM2 muscle and may identify early disease mechanisms. This result helps us to better understand the pathological processes of IBM2 [12]. Sequence analysis of GNE revealed IBM2 individuals were compound heterozygous for a mutation in the 5’ splice donor site of intron 10 (c.1816+5G>A), and a missense mutation (c.2086G>A, p.V696M), confirming the diagnosis as IBM2. The splice site mutation correlated with the exclusion of exon 10 from the transcript, which is predicted to produce an in-frame deletion (p. G545_D605del) of 61 amino acids in the kinase domain of the GNE protein [13]. The third type of IBM (IBM3) follows a pattern of AD inheritance. IBM3 results from mutations in the MYHC2A gene, located on chromosome 17p13.1, and it encodes the myosin heavy chain IIa [8].
Figure 1.
The genetic features of the three types of hereditary inclusion body myopathy (IBM) and the IBM with early-onset Paget disease and frontotemporal dementia (IBMPFD). There are 4 types of familial IBM. Types 1, 3 and IBMPFD are autosomal dominant whereas IBM2 is autosomal recessive disease (for details see Discussion). IBM: Inclusion-body myopathy, DES: Desmin, GNE: UDP-N- acetylglucosamine-2-epimerase/N-acetylmannosomine kinase, IBMPFD: IBM with early-onset Paget disease and frontotemporal dementia, MYHC2A: myosin heavy chain IIa, VCP: valosin-containing protein.
3 Pathogenesis of IBM
The pathomechanism of IBM is a complex process, including many factors. Both the degeneration of the muscle fibres and the presence of mononuclear cells are an important during the pathogenesis; however their relationship is still unknown. Ascanas et al. summarise the typical cellular abnormalities of the disease in their review, and they emphasised that degeneration is the key factor in the muscle weakness. Several proteins can be accumulated as inclusions in the cytoplasm of muscle fibres: amyloid-β42, and its different oligomers, as well as phosphorylated tau protein in double helical filaments. These proteins probably cause the degeneration of the fibres in IBM. In addition, there are other proteins that could play a part in the muscle fibre destruction: prion protein, ubiquitin, cathepsin B and D, α1-antichimotripsin, ApoE, BACE1, BACE2, TGFβ1, cystatin C, superoxide-dismutase, many interleukins, transglutaminase 1 and 2, and γ-tubulin [14]. The suspected factors also include the stress upon rough endoplasmic reticulum and the reduced deacetylase activity of SIRT1. According to Askanas, all these phenomena are provoked by the aging of the intracellular milieu [15]. Weihl et al. described three additional aggregates: TDP-43, p62, and LC3. Among these proteins TDP-43 differentiates IBM from other inflammatory myopathies [16]. In addition, TDP-43 are often associated with optineurin, which are also involved in the pathogenesis of IBM; moreover, optineurin-containing inclusions are observed in neurodegenerative diseases e.g. ALS and FTLD-TDP [17, 18].
The role of the T-cells have been known since the 1980s, and activity of the B-cells have also been described in the affected muscle biopsy [14]. In IBM, the typical non-necrotic fibres are focally surrounded and invaded by CD8+ T-cells and macrophages. Ivanidze et al. classified the muscle fibres of five IBM patients according to muscle fibre invasion by immune cells. Moreover, they isolated the intracellular content of the fibres by laser associated microdissection, then analysed the samples by microarray hybridization and quantitative PCR. They observed HLA upregulation in both invaded and non-invaded fibres, opposite to the controls. Moreover, each chain of the interferon-gamma receptor and other interferon-gamma induced genes were also upregulated [19]. These observations indicate that the increased expression of HLA-1 is not enough for the CD8+ T-cells invading. Salajegheh et al. identified a 43 kDa muscle protein in IBM patients; this protein had never been detected in any other diseases or controls before. Therefore this protein can become a new biomarker in the future [20]. These results suggest examining of the humoral immunity will be the key to understand the exact pathomechanism of disease.
4 Prognosis
As we mentioned, IBM is a slowly progressing disease, and unfortunately there is no effective therapy [3]. Only a subset of patients should respond to steroids temporarily, while others not. In the most cases, using Methotrexate, Cyclosporine A, Azathioprine, or Mycophenolate mofetil therapy are also ineffective [21, 22]. Nevertheless, this agent would be effective only in a subset of the patients, altogether these results are also disappointing. These data emphasise that degenerative process would be more important, than inflammation. Agents stimulating autophagy, under development by cancer and neurodegenerative disease researchers, would be possible candidates for clinical studies in IBM as well [23]. Barca and colleagues collected muscle samples of IBM, PM patients, and controls. They found ANT1 reduction, and NF-κB overexpression in IBM muscle samples, that could explain the lack of apoptosis [24]. A study was carried out with a humanized monoclonal antibody, called alemtuzumab, and though the number of T-cells were decreased in the repeated muscle biopsy, resounding success was not achieved [25, 26]. An Italian study tried to use simvastatin in the therapy of IBM, however it was not successful [27]. Physiotherapy, as in idiopathic inflammatory myopathies, may decelerate the progression of IBM, and improve the quality of life. A key part of this disease is aiding the movements of the patients by wheelchairs and other medical aids that usually become necessary in 5–10 years from the onset [21, 28]. Cox et al. have followed 64 patients for 12 years, and they recognise the importance of long-term clinical follow-up in IBM. Forty-six patients died during the follow-up period, while the others became seriously disabled. In their cases, the lower limbs proved to be weaker, and 47% of the patients had to use a wheelchair. The most frequent cause of death was respiratory disease [29].
5 Inclusion body myopathy with early-onset Paget disease and frontotemporal dementia (IBMPFD) – a special subtype of IBM
In addition to the three types of hIBM, inclusion body myopathy with early-onset Paget disease and frontotemporal dementia (IBMPFD) is another form of the overlapping diseases. The characteristic triad is the following: 1. proximal and distal myopathy, with the morphology of IBM, appearing in the third or fourth decades of life, 2. early onset Paget’s disease of the bone and 3. frontotemporal dementia. This rare autosomal dominant disorder is caused by missense mutations in the VCP (valosin-containing protein) gene, localised chromosome 9p13.3 [30]. The presence of the VCP is not specific for IBMPFD, though. Normally, the VCP protein can be found intramyonuclearly, perinuclearly or in the sarcoplasm, but in case of IBMPFD the VCP forms sarcoplasmic and myonuclear inclusions [31].
Muscle weakness appears only in 30% of the patients. Therefore, it is important to investigate the familial occurrence of dementia, or Paget’s disease [32, 33]. The profound tendon reflexes are often missing. As the disease progresses, patients have to use wheelchairs. The elevated CK level, electromyography, and muscle MRI may help achieve the correct diagnosis. In a subset of the cases, the histological features are similar as in IBM. If the typical basophil-rimmed vacuoles are not present in the majority of patients; they are simply described as non-specific myopathies [34]. On the other hand, varying sizes of the fibres and accumulation of the endomysial connective tissue can be observed. If these signs are present, the vacuoles are localised in subsarcolemmally, as well as in the sarcoplasm. They have an irregular shape and contain basophil debris. Futhermore, eosinophil bodies were described in the muscles of some patients. Frontotemporal dementias (FTDs) are a subsection of neurodegenerative diseases that are characterised by the degeneration of the prefrontal and anterior temporal lobes; therefore should be called frontotemporal lobar degenerations (FTLDs). Approximately half of the patients have a familial occurrence of dementia that suggests a strong genetic background of the disease. A large group of FTLDs belongs to the class of FTLD-TDPs [35]. Inclusion bodies are positive for ubiquitin and TDP-43 are localised in the nuclei and cytoplasm of the affected neurons, and they are typical in FTLD-TDP and suggest a key part in their pathogenesis [36].
6 Conclusions
IBM is a slowly progrediating disease, and unfortunately there is still no effective therapy. The precise pathogenesis of inclusion body myositis remains unknown. In the majority of cases it is diagnosed only a few years after the appearance of the first symptoms. The muscle biopsy typically shows endomysial inflammation in sIBM, with invasion of mononuclear cells into the non-necrotic fibers, as well as rimmed-vacuoles. It appears that both inflammation and degeneration are present at the onset of the disease. The delay of the diagnosis and often inadequate therapy contributes that the disease leads to disability in most of the cases. Furthermore, the importance of the genetic factors could be emphasised during the development and the pathogenesis of IBM. In this review we summarised the main features of this relatively rare, but important entity.
Acknowledgements
The study was supported by the UD Faculty of Medicine Research Found (Bridging Fund 2012) to KD. TH was supported by the Hungarian Brain Research Program (Nemzeti Agykutatási Program) Grant No. KTIA_13_NAP-A-II/7 and the New Széchenyi Plan (Új Széchenyi Terv), Hungary.
Abbreviations
- ALS
Amyotrophic lateral sclerosis
- ApoE
Apolipoprotein E
- BACE
Beta-secretase
- CK
creatine kinase
- FTD
frontotemporal dementia
- FTLDs
frontotemporal lobar degenerations
- GNE
UDP-N-acetylglucosamine-2- epimerase/N-acetylmannosomine kinase
- hIBM
hereditary inclusion body myopathy
- HLA
human leukocyte antigen
- IBM
inclusion body myositis
- IBMPFD
IBM with early-onset Paget disease and frontotemporal dementia
- iNOS
inducible NO synthase
- LC3
1A/1B-light chain 3
- MHC
major histocompatibility complex
- MRI
magnetic resonance imaging
- MYHC2A
myosin heavy chain IIa
- PCR
polymerase chain reaction
- PM
polymyositis
- sIBM
sporadic inclusion body myositis
- SIRT1
NAD-dependent deacetylase sirtuin-1
- TDP-43
TAR DNA-binding protein 43
- TGFβ1
transforming growth factor beta 1
- VCP
valosin-containing protein
- ZASP
Z-band alternatively spliced PDZ-motif protein
Footnotes
Conflict of interest statement: Authors state no conflict of interest
References
- 1.Dalakas MC. Sporadic inclusion body myositis – diagnosis, pathogenesis and therapeutic strategies. Nat Clin Pract Neurol. 2006;2:437–447. doi: 10.1038/ncpneuro0261. [DOI] [PubMed] [Google Scholar]
- 2.Munshi SK, et al. Inclusion body myositis: an underdiagnosed myopathy of older people. Age Ageing. 2006;35:91–94. doi: 10.1093/ageing/afj014. [DOI] [PubMed] [Google Scholar]
- 3.Bodoki L, et al. [Inclusion body myositis]. Ideggy Szle. 2015;68:59–67. [Google Scholar]
- 4.Bodoki L, et al. Inclusion body myositis – a case based clinicopathological update. Cent Eur J Med. 2014;9:80–85. [Google Scholar]
- 5.Greenberg SA. Inclusion body myositis. Curr Opin Rheumatol. 2011;23:574–578. doi: 10.1097/BOR.0b013e32834b53cc. [DOI] [PubMed] [Google Scholar]
- 6.Needham M, et al. Genetics of inclusion-body myositis. Muscle Nerve. 2007;35:549–561. doi: 10.1002/mus.20766. [DOI] [PubMed] [Google Scholar]
- 7.Koffman BM, et al. HLA allele distribution distinguishes sporadic inclusion body myositis from hereditary inclusion body myopathies. J Neuroimmunol. 1998;84:139–142. doi: 10.1016/s0165-5728(97)00245-2. [DOI] [PubMed] [Google Scholar]
- 8.Cai H, et al. Clinical, pathological, and genetic mutation analysis of sporadic inclusion body myositis in Japanese people. J Neurol. 2012;259:1913–1922. doi: 10.1007/s00415-012-6439-0. [DOI] [PubMed] [Google Scholar]
- 9.Mastaglia FL. Sporadic inclusion body myositis: variability in prevalence and phenotype and influence of the MHC. Acta Myol. 2009;28:66–71. [PMC free article] [PubMed] [Google Scholar]
- 10.Rojana-Udomsart A, et al. Analysis of HLA-DRB3 alleles and supertypical genotypes in the MHC Class II region in sporadic inclusion body myositis. J Neuroimmunol. 2013;254:174–177. doi: 10.1016/j.jneuroim.2012.09.003. [DOI] [PubMed] [Google Scholar]
- 11.Broccolini A, et al. Hereditary inclusion-body myopathy with sparing of the quadriceps: the many tiles of an incomplete puzzle. Acta Myol. 2011;30:91–95. [PMC free article] [PubMed] [Google Scholar]
- 12.Fischer C, et al. Cell stress molecules in the skeletal muscle of GNE myopathy. BMC Neurol. 2013;13:24–24. doi: 10.1186/1471-2377-13-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Boyden SE, et al. Molecular diagnosis of hereditary inclusion body myopathy by linkage analysis and identification of a novel splice site mutation in GNE. BMC Medical Genetics. 2011:12. doi: 10.1186/1471-2350-12-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Greenberg SA. Theories of the pathogenesis of inclusion body myositis. Curr Rheumatol Rep. 2010;12:221–228. doi: 10.1007/s11926-010-0102-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Askanas V, et al. Sporadic inclusion-body myositis: conformational multifactorial ageing-related degenerative muscle disease associated with proteasomal and lysosomal inhibition, endoplasmic reticulum stress, and accumulation of amyloid-β42 oligomers and phosphorylated tau. Presse Med. 2011;40:219–235. doi: 10.1016/j.lpm.2010.11.024. [DOI] [PubMed] [Google Scholar]
- 16.Weihl CC, et al. Sporadic inclusion body myositis: possible pathogenesis inferred from biomarkers. Curr Opin Neurol. 2010;23:482–488. doi: 10.1097/WCO.0b013e32833d3897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yamashita S, et al. Optineurin is potentially associated with TDP-43 and involved in the pathogenesis of inclusion body myositis. Neuropathol Appl Neurobiol. 2013;39:406–416. doi: 10.1111/j.1365-2990.2012.01297.x. [DOI] [PubMed] [Google Scholar]
- 18.Hortobagyi T, et al. Optineurin inclusions occur in a minority of TDP-43 positive ALS and FTLD-TDP cases and are rarely observed in other neurodegenerative disorders. Acta Neuropathol. 2011;121:519–527. doi: 10.1007/s00401-011-0813-3. [DOI] [PubMed] [Google Scholar]
- 19.Ivanidze J, et al. Inclusion body myositis: laser microdissection reveals differential up-regulation of IFN-γ signaling cascade in attacked versus nonattacked myofibers. Am J Pathol. 2011;179:1347–1359. doi: 10.1016/j.ajpath.2011.05.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Salajegheh M, et al. Autoantibodies against a 43 KDa muscle protein in inclusion body myositis. PLoS One. 2011:6. doi: 10.1371/journal.pone.0020266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Quick A, et al. Mechanisms of action of intravenous immunoglobulin in inflammatory muscle disease. Curr Rheumatol Rep. 2011;13:192–198. doi: 10.1007/s11926-011-0171-0. [DOI] [PubMed] [Google Scholar]
- 22.Kierdaszuk B, et al. Inclusion body myositis: therapeutic approaches. A case report. Neurol Neurochir Pol. 2011;45:68–73. doi: 10.1016/s0028-3843(14)60062-1. [DOI] [PubMed] [Google Scholar]
- 23.Lloyd TE. Novel therapeutic approaches for inclusion body myositis. Curr Opin Rheumatol. 2010;22:658–664. doi: 10.1097/BOR.0b013e32833f0f4a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Barca E, et al. ANT1 is reduced in sporadic inclusion body myositis. Neurol Sci. 2013;34:217–224. doi: 10.1007/s10072-012-0976-2. [DOI] [PubMed] [Google Scholar]
- 25.Dalakas MC, et al. Effect of Alemtuzumab (CAMPATH 1-H) in patients with inclusion-body myositis. Brain. 2009;132:1536–1544. doi: 10.1093/brain/awp104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Benveniste O, et al. International Workshop on Inclusion Body Myositis held at the Institute of Myology, Paris, on 29 May 2009. Neuromuscular Disorders. 2010;20:414–421. doi: 10.1016/j.nmd.2010.03.014. [DOI] [PubMed] [Google Scholar]
- 27.Sancricca C, et al. Pilot trial of simvastatin in the treatment of sporadic inclusion-body myositis. Neurological Sciences. 2011;32:841–847. doi: 10.1007/s10072-011-0657-6. [DOI] [PubMed] [Google Scholar]
- 28.Alexanderson H. Exercise in inflammatory myopathies, including inclusion body myositis. Curr Rheumatol Rep. 2012;14:244–251. doi: 10.1007/s11926-012-0248-4. [DOI] [PubMed] [Google Scholar]
- 29.Cox FM, et al. A 12-year follow-up in sporadic inclusion body myositis: an end stage with major disabilities. Brain. 2011;134:3167–3175. doi: 10.1093/brain/awr217. [DOI] [PubMed] [Google Scholar]
- 30.Fanganiello RD, et al. A Brazilian family with hereditary inclusion body myopathy associated with Paget disease of bone and frontotemporal dementia. Braz J Med Biol Res. 2011;44:374–380. doi: 10.1590/s0100-879x2011007500028. [DOI] [PubMed] [Google Scholar]
- 31.Greenberg SA, et al. Nuclear localization of valosin-containing protein in normal muscle and muscle affected by inclusion-body myositis. Muscle Nerve. 2007;36:447–454. doi: 10.1002/mus.20823. [DOI] [PubMed] [Google Scholar]
- 32.Kimonis VE, et al. VCP disease associated with myopathy, Paget disease of bone and frontotemporal dementia: review of a unique disorder. Biochim Biophys Acta. 2008;1782:744–748. doi: 10.1016/j.bbadis.2008.09.003. [DOI] [PubMed] [Google Scholar]
- 33.Weihl CC, et al. Valosin-containing protein disease: inclusion body myopathy with Paget’s disease of the bone and frontotemporal dementia. Neuromuscul Disord. 2009;19:308–315. doi: 10.1016/j.nmd.2009.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kimonis VE, et al. Clinical studies in familial VCP myopathy associated with Paget disease of bone and frontotemporal dementia. Am J Med Genet A. 2008;146A:745–757. doi: 10.1002/ajmg.a.31862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mackenzie IRA, et al. Nomenclature and nosology for neuropathologic subtypes of frontotemporal lobar degeneration: an update. Acta Neuropathol. 2010;119:1–4. doi: 10.1007/s00401-009-0612-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Strong M, et al. Amyotrophic lateral sclerosis, Primary lateral sclerosis and Spinal muscular atrophy. In: Dickson DW, et al., editors. Neurodegeneration: The molecular pathology of dementia and movement disorders. Wiley-Blackwell; 2011. [Google Scholar]