Abstract
Using comparative FISH and genomics, we have studied and compared the evolution of chromosome 3 in primates and two human neocentromere cases on the long arm of this chromosome. Our results show that one of the human neocentromere cases maps to the same 3q26 chromosomal region where a new centromere emerged in a common ancestor of the Old World monkeys ∼25-40 million years ago. Similarly, the locus in which a new centromere was seeded in the great apes' ancestor was orthologous to the site in which a new centromere emerged in the New World monkeys' ancestor. These data suggest the recurrent use of longstanding latent centromeres and that there is an inherent potential of these regions to form centromeres. The second human neocentromere case (3q24) revealed unprecedented features. The neocentromere emergence was not accompanied by any chromosomal rearrangement that usually triggers these events. Instead, it involved the functional inactivation of the normal centromere, and was present in an otherwise phenotypically normal individual who transmitted this unusual chromosome to the next generation. We propose that the formation of neocentromeres in humans and the emergence of new centromeres during the course of evolution share a common mechanism.
Evolutionary centromere repositioning, a move of the centromere along the chromosome not accompanied by a pericentric inversion, is a phenomenon that we first described while studying primate evolution of chromosome 9. In some species, the centromere position exhibited an evolutionary history that appeared to be independent from the flanking chromosomal markers (Montefalcone et al. 1999). Since then, additional examples of evolutionary centromere repositioning in primates have been reported, for example, on chromosome X (Ventura et al. 2001), on chromosome 10 (Carbone et al. 2002), on chromosome 6 (Eder et al. 2003), and on chromosome 14 and 15 (Ventura et al. 2003). This phenomenon also appears to be present in nonprimate mammals. Band et al. (2000) have compared cattle radiation hybrid maps with humans. According to these authors, “41 translocation events, a minimum of 54 internal rearrangements, and repositioning of all but one centromere can account for the observed organizations of the cattle and human genomes”. Horse and donkey are two Equidae species that diverged about 1.9-2.3 million years ago (Mya) (Oakenfull et al. 2000). Comparative FISH studies have suggested that at least four different centromere repositioning events occurred during this very short evolutionary time (Yang et al. 2004). An example of centromere repositioning has been reported in birds (Kasai et al. 2003). Nagaki et al. (2004) have invoked a new centromere emergence event to explain the unusual intermediate state of heterochromatization of chromosome 8 centromere of rice. Altogether, these data suggest that the centromere repositioning phenomenon may be a widespread property of genome evolution.
Hence, the phenomenon of centromere repositioning should not be regarded purely as an oddity. Further, the examples found in chromosomes 6 and 15 show that such events deeply affect both the old and new centromeric regions (Eder et al. 2003; Ventura et al. 2003). Inactivated ancestral centromeres at 6p22.1 and 15q25 outline a common scenario accompanying their silencing. The strong constraint against recombination acting on normal centromeres progressively weakens following inactivation. We speculate that nonallelic homologous exchanges, favored by repetitive DNA and segmental duplications, trigger an accelerated elimination of satellite DNA, and at the same time, the dispersal of pericentromeric duplications over a range as large as 10 Mb. This scenario is supported by the well-known example of centromere inactivation at 2q21, which followed the telomere-telomere fusion that generated human chromosome 2 (Ijdo et al. 1991). This human lineage-specific inactivation occurred ∼4-6 Mya, and the corresponding centromere regions seem to be in the early stages of restructuring, as can be seen for 6p22.1 and 15q25. The biological implications of the centromere inactivation processes have been recently discussed by Jackson (2003).
The evolutionary emergence and progression of new centromeres is more obscure. The only example from which specific information can be derived is that of Old World Monkeys' (OWM) homolog to HSA6 in a region corresponding to the human 6q24.3, inside of the sequence defined by the BAC RP11-474A9 (Eder et al. 2003). This evolutionary new centromere appears to have progressed toward the normal complex organization typical of a mammalian centromere without affecting the displaced flanking sequences.
In recent years, clinical cytogenetics has identified a substantial number of human analphoid neocentromere cases. In the vast majority of these cases, the seeding of the neocentromere was concomitant to a rearrangement that generated an acentric fragment whose mitotic survival was rescued by neocentromere activation. An analysis of the distribution of neocentromeres suggests clustering in hot spot regions 15q25, 3q, and 13q (Amor and Choo 2002). The precise sequence underlying some neocentromeres has been identified (Lo et al. 2001; Satinover et al. 2001; Alonso et al. 2003), but analysis and comparison of these sequences did not reveal a clear association. The only unambiguous conclusion is that the neocentromerization event is an epigenetic phenomenon, whose nature, however, remains to be elucidated.
The existence of latent centromeres has been proposed as a potential primer for neocentromere emergence (Choo 1997; du Sart et al. 1997). With this hypothesis in mind, we have recently investigated the evolutionary history of the human chromosome 15, where one of the neocentromere hot spots is located (15q25). We have discovered that an ancestral centromere at 15q25 was inactivated following the noncentromeric fission of an ancestral chromosome that gave rise to the present-day chromosomes 14 and 15 (Ventura et al. 2003). We also found that duplicons that flanked the ancestral centromere dispersed in a wide area (about 10 Mb), and that two human neocentromeres mapped to duplicons that flanked the ancestral centromere. These findings have established a still unclear, but intriguing connection between duplicon remains in a dispersed ancestral centromeric area and neocentromere emergence in clinical cases, as if the region retained a dormant centromeric functionality that was able to emerge opportunistically in the absence of the normal centromere.
The association of human neocentromeres and the location of ancestral centromeres prompted us to undertake a detailed study of the evolutionary history of chromosome 3, which harbors, at 3q24-3q28, another neocentromere hot spot (Amor and Choo 2002). We searched for relevant human cases showing neocentromere occurrence in this region and identified two independent cases. In the first case, the neocentromere spanned the region in which our evolutionary studies identified the appearance of a centromere in the ancestor of OWM. The second case revealed unprecedented features. The centromere moved along the chromosome from its normal position to 3q24 in the absence of any other chromosomal rearrangement, and was found in an otherwise phenotypically normal individual. This exceptional repositioning event can be regarded as a present-day episode equivalent to new centromere appearance that occurred during evolution, and which was subsequently fixed in the population.
RESULTS
Chromosome 3 evolution was studied by cohybridization FISH experiments using a panel of 14 single-copy human BAC probes distributed along chromosome 3 (Table 1, clones in regular style; Fig. 1,8 letters in black). The probes were hybridized on metaphase spreads of the following primate species. Great apes: common chimpanzee (Pan troglodytes, PTR), gorilla (Gorilla gorilla, GGO), Borneo (Pongo pygmaeus pygmaeus, PPY-B), and Sumatra (Pongo pygmaeus abelii PPY-S) orangutan; Old World Monkeys (OWM): rhesus monkey (Macaca mulatta, MMU, Cercopithecinae), sacred baboon (Papio hamadryas, PHA, Cercopithecinae), African green monkey (Cercopithecus aethiops, CAE, Cercopithecinae), silvered leaf-monkey (Presbytis cristata, PCR, Colobinae); New World Monkeys (NWM): wooly monkey (Lagothrix lagothricha, LLA, Atelinae), common marmoset (Callithrix jacchus, CJA, Callitrichinae), dusky titi (Callicebus moloch, CMO, Callicebinae), squirrel monkey (Saimiri boliviensis, SBO); prosimians: ring-tailed lemur (Lemur catta, LCA). The domestic cat (Felix Catus, FCA) was used as an outgroup because its karyotype is highly conserved and closely resembles the ancestral karyotype of mammals (Murphy et al. 2000; Yang et al. 2000). The sequence encompassed by each human BAC reported in Table 1 (BACs in regular style) was searched for conservation against mouse genome. “Overgo” probes (see Methods) were designed on the most conserved region of each BAC (or very close to it) and were then used to screen the cat library RPCI-86. This approach was aimed at assembling a panel of BACs mapping to chromosomal loci orthologous to the loci encompassed by the human BACs, thus facilitating mapping comparison (Table 2).
Table 1.
Bacterial Artificial Chromosome Probes (BAC) Used in This Study
| Code | BAC | Acc. N. | map | UCSC July 2003 |
|---|---|---|---|---|
| A | RP11-183N22 | AL512885 (BLAT) | 3p26.1 | ch3:4,346,716-4,508,442 |
| A1 | RP11-732C9 | BES | 3p25.2 | ch3:12,441,757-12,649,037 |
| A2 | RP11-616M11 | AC090954 | 3p25.1 | ch3:15,045,781-15,213,797 |
| B | RP11-109D5 | AC074287 | 3p24.2 | ch3:25,469,450-25,670,575 |
| C | RP11-491D6 | AC006583 | 3p22.3 | ch3:37,019,771-37,122,141 |
| C1 | RP11-603D9 | BES | 3p22.1 | ch3:42,572,315-42,803,228 |
| D | RP11-395P16 | AC130472 | 3p21.31 | ch3:47,570,335-47,764,494 |
| E | RP11-380J21 | AC092419 | 3p14.1 | ch3:64,035,784-64,195,049 |
| E1 | RP11-634L22 | BES | 3p12.3 | ch3:75,290,471-75,466,812 |
| E2 | RP11-601J5 | BES | 3p12.3 | ch3:75,715,213-75,874,660 |
| F | RP11-536K4 | AC016942 | 3p12.3 | ch3:76,521,107-76,673,120 |
| G | RP11-655A17 | AC107204 | 3p11.1 | ch3:86,937,903-87,108,701 |
| cen | ||||
| H | RP11-454H13 | AC084198 | 3q12.3 | ch3:102,636,899-102,831,853 |
| I | RP11-305I9 | AC092981 | 3q13.33 | ch3:120,303,388-120,463,387 |
| I1 | RP11-924M2 | BES | 3q22.1 | ch3:130,965,112-131,192,514 |
| I2 | RP11-687B8 | AC112646 | 3q22.1 | ch3:131,258,851-131,297,451 |
| J | RP11-21N8 | AC023438 | 3q22.1 | ch3:131,589,362-131,747,328 |
| K | RP11-505J9 | AC024897 | 3q24 | ch3:149,683,332-149,888,125 |
| nc1 | RP11-498P15 | AC112906 | 3q26.1 | ch3:163,368,542-163,486,312 |
| K1 | RP11-355I21 | AC025826 | 3q26.1 | ch3:163,755,042-163,941,067 |
| d1 | RP11-449O23 | AC139338 | 3q26.1 | ch3:163,893,071-164,059,219 |
| d2 | RP11-535I24 | AC128685 | 3q26.1 | ch3:164,057,229-164,178,429 |
| d3 | RP11-486A08 | BES | 3q26.1 | ch3:164,138,756-164,304,389 |
| K2 | RP11-418B12 | AC079910 | 3q26.1 | ch3:164,377,941-164,545,374 |
| L | RP11-526M23 | AC048332 | 3q26.1 | ch3:166,693,588-166,880,817 |
| M | RP11-114M1 | AC026355 | 3q26.32 | ch3:178,593,777-178,751,215 |
| M1 | RP11-35G16 | AC016926 | 3q27.3 | ch3:187,493,799-187,646,665 |
| N | RP11-313F11 | AC016953 | 3q29 | ch3:196,921,579-197,093,785 |
| Tel | ch3:199,344,050 |
Probes in regular style were used to characterize all primate species. Probes in Italics were used to define specific rearrangements. (BES) BAC ends; (nc1) the BAC spanning the neocentromere in case 1.
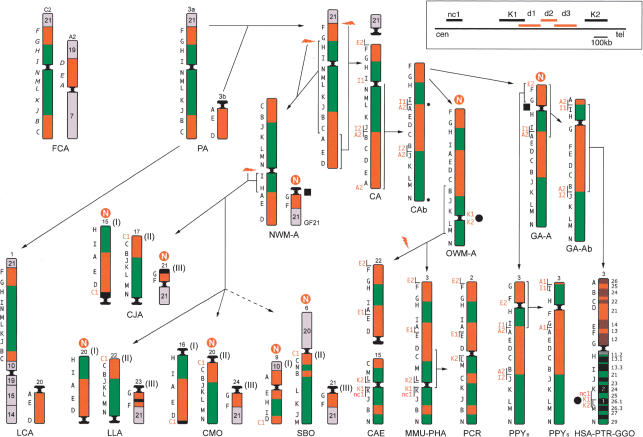
Figure 1.
Reconstruction of the chromosome 3 phylogeny in primates. Some chromosomes are upside down to facilitate comparison. (PA) Primate Ancestor; (CA) Catarrhini Ancestor. N in a red circle stands for new centromere. The number that identifies the chromosome in each species is reported on top of the chromosome. The black letters on the left of each primate chromosome refer to the panel of BAC probes reported in Table 1 (human BACs); letters on cat chromosomes refer to the corresponding probes reported in Table 2. Letters in red are the additional probes used to delimit breakpoints, centromeres, and telomeres, reported in italics in Table I. Arrows with hatched lines point to species for which an intermediate ancestor has not been drawn. A white N in a red circle indicates the sites where new centromeres appeared during evolution. Probes used on the cat (in italics) are cat BACs identified using Overgo probes. For details see text. The black circles and the black squares are positioned at the loci where an evolutionary new centromere or a neocentromere appeared. In the rectangle on the top, right corner is reported the map of the region 3q26, in which the nc1, K1, K2, and the deleted marker d1, d2, and d3 markers are located. For more details see text.
Table 2.
Overgo Probes Used to Screen the Cat RPCI-86 Library
| Probe | cat BAC | Overgo sequence | Location—UCSC July 2003 | Corresponding human probes (as reported in Table 1) |
|---|---|---|---|---|
| A | 231P20 | AGCTATGTGTTTTGCATTTGAAGACCATCATGCTATCAGGG | ch3:4,726,374-4,726,414 | ch3:4,346,716-4,508,442 |
| B | 300M8 | TGGTATCCAGACCGCCAGGACCTTGAGGAACCGACAAAAG | ch3:25,610,965-25,611,005 | ch3:25,469,450-25,670,575 |
| C | 325L14 | AGATGCCACGAACTAGGTGGCAATGCCTTAACCGTATGTG | ch3:36,970,320-36,970,360 | ch3:37,019,771-37,122,141 |
| D | 330H13 | TTTGCTGATTGGCACTGGGACAGTCCACCAAGAGAGAAAGC | ch3:50,369,515-50,369,555 | ch3:47,570,335-47,764,494 |
| E | 355B19 | CCGCATGCTAATGAAGCTGTGATGTCAGCAAGTCAGTCAC | ch3:64,168,491-64,168,531 | ch3:64,035,784-64,195,049 |
| F | 366O21 | CCTATGCCACAGTTGAATTTAAAGCTGTGACCTGTGATCT | ch3:76,675,869-76,675,909 | ch3:76,521,107-76,673,120 |
| G | 212B15 | CAGCAAGGCTATGACAGATGACACTTTGAGAGTGTTGGGG | ch3:89,002,905-89,002,945 | ch3:86,937,903-87,108,701 |
| H | 529L18 | TTCCAGATCCGACGTATCCTGAAGGGAAAGTCCATTCAGC | ch3:102,899,056-102,899,096 | ch3:102,636,899-102,831,853 |
| I | 449M23 | AGTTCACAGTGGTTGAGGTAGGTCTTGCCATTACTGCCACA | ch3:121,451,664-121,451,704 | ch3:120,303,388-120,463,387 |
| J | 230E22 | CACAGGATTCTTAGTCATCTGTAATTCTGACCAGCTGTTC | ch3:131,528,185-131,528,225 | ch3:131,589,362-131,747,328 |
| K | 352L13 | GCAGAGGCATGGTAAAAGCATTGTTTGCACCAAAAGGAAC | ch3:150,113,000-150,113,040 | ch3:149,683,332-149,888,125 |
| L | 230K14 | AACCAGTTTGGCACCTGCTGTGATCATGAGTCTACTGCTG | ch3:166,571,710-16,657,1750 | ch3:166,693,588-166,880,817 |
| M | 292I22 | ACCAAATATTGAGGAGTTACCAGCCCTGGTCAGTGAGTGT | ch3:180,739,803-180,739,842 | ch3:178,593,777-178,751,215 |
| N | 222D17 | CTTGAGTTTGGTCCCTCTCTCAGCACAGTGAGCTGATGTCT | ch3:197,401,159-197,401,199 | ch3:196,921,579-197,093,785 |
About 150 additional BACs were used to delimit regions of interest, for example, centromeres, telomeres, and chromosomal rearrangements. The most informative probes are reported in italics in Table 1 (red in Fig. 1). Further experiments were performed to clarify specific inconsistencies with the recent results on the evolution of chromosome 3 reported by Tsend-Ayush et al. (2004). Some probes gave multiple signals because of segmental duplications. BAC RP11-418B12 (K2 in Table 1) gave, as expected, a signal at 3q26.1, and unexpectedly, a second large signal at the 1q12 heterochromatic block (Fig. 2d; see below). The overall FISH results are summarized in Figure 1. The figure also shows the putative ancestral chromosome organization and the most parsimonious rearrangements necessary to reconcile the chromosome 3 organization in the different extant primate species. These reconstructions took advantage of the GRIMM software package, designed to outline the most parsimonious scenario of evolutionary marker order changes (Bourque and Pevzner 2002; http://www.cs.ucsd.edu/groups/bioinformatics/GRIMM/).
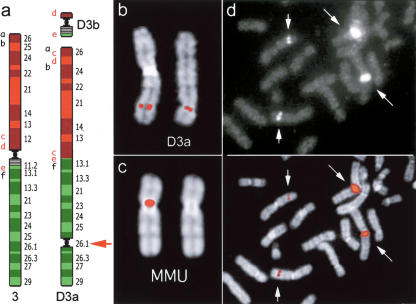
Figure 2.
Neocentromere case 1. (a) Diagram of the rearrangement that generated the derivative chromosomes D3a and D3b of neocentromere case 1. Several BAC probes were used to define the rearrangement. The most informative BACs, reported in a as letters in lower case, are reported in the Supplemental Table 3. Letters in red refer to probes yielding splitting signals. These data suggest that the segment defined by BACs c-e was excised from its original position and inserted between a and b; then a second excision extracted the fragment d-e, containing the centromere, to form the minichromosome D3b. The red arrow indicates the position of the neocentromere. (b) Partial metaphase from case 1 showing the normal chromosome 3 (left) and the derivative D3a (right) with FISH signals of the BAC RP11-498P15 (nc1 in Fig. 1), mapping at the neocentromere (see text). (c) (Left) FISH signal of BAC RP11-498P15 on macaque chromosome 3. (Right) The chromosome is shown without the signal for a better identification of the centromere. Note that MMU chromosome 3 is upside down in Figure 1. (d) Partial metaphase from a normal individual showing FISH signals of probe RP11-418B12 (K2 in Fig. 1 and Table 1), mapping on the opposite side of the MMU centromere with respect to BAC RP11-498P15, showing a strong signal on the heterochromatic block of chromosome 1. The image with signals only (top) and the merged image (bottom) are separately reported to better show how the signals appear on the microscope. Note the normal signals on chromosome 3 (short arrows) and the large signals on the heterochromatic block of chromosome 1 (long arrows).
Conserved marker order in cat and in ring-tailed lemur strongly suggests that their marker order occurred within the primate ancestor (PA in Fig. 1). The 3/21 association is well established in LCA (Cardone et al. 2002) and in mammals (Muller et al. 2000; Murphy et al. 2001). In NWMs, the association of markers G, F, and chromosome 21 sequences form a separate chromosome (GF21 in Fig. 1), which appears derivative in this family of primates. Contrary to previously published reports, our results indicated that chromosome 3 was not a unique chromosome in PA, markers A, B, and D defining a small chromosome (3b in Fig. 1), as in LCA. This hypothesis is further supported by the finding that this association is part of a distinct, larger chromosome in the cat. The 3a/3b fusion took place after prosimians, but before Catarrhini/Platyrrhini divergence.
The location of the centromere of chromosome 3a in Primate Ancestor (PA), between markers N and I, was assumed taking into account the centromere position in the cat and in NWMs. In Catarrhini Ancestor (CA) marker, A was brought close to I by an inversion encompassing the entire long arm of the PA chromosome 3 (A-N inversion). The A-I association was conserved unchanged in all OWM and the Bornean orangutan. BAC probes from the human chromosomal segment encompassed by markers I-J were used in reiterative FISH experiments on chromosomal metaphase spreads of the Bornean orangutan to identify the first breakpoint of the A-N inversion. The rearrangement was localized between BACs I1 and I2 (see Fig. 1), which are 66 Kb apart (see Table 1). This region is part of a large multicopy Olfactory Receptor (OR) gene family, which prevented a more precise location of the breakpoint. We similarly mapped the second breakpoint using BAC probes centromeric to A in humans. A2 BAC probe generated a split signal. In the interpretation of these results, it should be kept in mind that an inversion in HSA-PTRGGO ancestor (GA-Ab in Fig. 1) broke the A2-I1 association and led to the present-day marker organization of HSA, PTR, and GGO. A pericentric inversion differentiated the Bornean orangutan from the derivative subspecies, the Sumatran orangutan. FISH experiments performed revealed that the breakpoints of the inversion are located in a duplication-reach region (OR clusters), as described by Tsend-Ayush et al. (2004).
The chromosome 3 organization of Catarrhini Ancestor (CA) following the A-N inversion is indicated as CA-b in Figure 1. It could not be unequivocally determined, however, whether the inversion was paracentric or pericentric in nature, that is, whether the centromere (small black circle in Fig. 1) mapped to the N telomeric region or between the A2-I1 markers. The centromere position in OWM and great apes, in any case, is unequivocally distinct, and located in regions where a centromere was not present in either of their ancestors. The most parsimonious explanation is that a centromere-repositioning event occurred in both the OWM ancestor (OWM-A) and in the great apes ancestor (GA-A), and that the ancestral CA centromere was inactivated. The inactivation took place after Catarrhini-Platyrrhini divergence, which is estimated to have occurred ∼40 Mya (Goodman 1999). We searched for sequence features of pericentromeric regions near the ancestral CA centromere (A2, I1, and N regions). The segmental duplication survey reported by Bailey et al. (2002), as most of the similar surveys within the human genome, has been performed using percent sequence identity thresholds set at >90%, which aim to detect duplication events that emerged over the last 35 million years of evolution. Analyses targeting more divergent genomic segmental duplications, and therefore, more ancient duplication events, have only recently been introduced (X. She, J.E. Horvath, Z. Jian, G. Liu, T.S. Furey, L. Christ, R. Clark, T. Graves, C.L. Gulden, C. Alkan, et al., in prep.). These analyses identify duplications with sequence identity as low as 75% and provide, at least in theory, a more comprehensive view of historical segmental duplications during primate evolution. Using this new set of data (www://humanparalogy.gene.cwru.edu), we examined segmental duplications within 2 Mb of markers A2, I1, and N. Each of these three regions was characterized by an abundance of segmental duplications when compared with the current human chromosome 3 pericentromeric region (Supplemental Table 1). Many of the interchromosomal alignments corresponding to regions I1 and A2 mapped to similar locations within the human genome. In the case of region N, most of the interchromosomal alignments corresponded to subtelomeric portions of other human chromosomes, consistent with the hypothesis that this region represents an ancestral subtelomeric region.
Human Neocentromere Cases
Case 1
A neocentromere at 3q26 was observed in a father, mildly mentally retarded, and his daughter, on an abnormal chromosome 3 lacking the centromeric region that appeared excised to form a supernumerary minichromosome, as already described (Wandall et al. 1998). His parents were normal. No α-satellite DNA was detected, by FISH, at the 3q26 neocentromere. CREST immunotyping, reported by Wandall et al. (1998), detected centromeric proteins at the neocentromere site (3q26). Reiterated FISH experiments using BAC clones as a probe were performed to precisely define the rearrangement that generated the two derivative chromosomes (D3a and D3b in the Fig. 2a) and to identify the location of the neocentromere. D3a and D3b revealed a complex rearrangement. Marker order of the segment 3q12-3qter remained unchanged (data not shown). The BAC RP11-498P15 (nc1 in Fig. 1; Fig. 2b) was found to be the most (neo)centromeric one among the several overlapping BACs tested by FISH. This conclusion was based mainly on the observation that in FISH experiments using successive probes along the finished genome sequence, the two FISH signals on the two chromatids appear ever closer as the (neo)centromere is approached. The complete list of the BACs used in these experiments is available as Supplemental Table 2. The location of RP11-498P15 on macaque chromosome 3 is reported in Figure 2c. Interestingly, the BAC RP11-418B12 (K2 in Fig. 1 and Table 1), mapping 892 kb telomeric to RP11-98P15 and flanking the OWM centromere on the opposite side with respect to RP11-498P15, gave, in addition to the signal at 3q26.1, a strong signal on the heterochromatic block at 1q12 (Fig. 2d). Sequence analysis of RP11-498P15 did not reveal any repeat that would account for the signal on 1q12, indicating an occasional inadequacy of sequence coverage.
Case 2
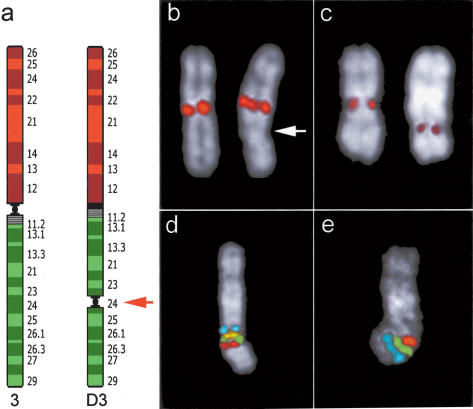
A prenatal cytogenetic analysis due to increased maternal age showed a male fetus with trisomy 21. An abnormal chromosome 3 was also evident, with an abnormal centromere location. FISH analysis with a chromosome 3-specific alphoid probe indicated that the inactivated centromere was unchanged (Fig. 3b). The same abnormal chromosome 3 was found in the father, whereas the father's parents and his son were normal. Immunotyping using CREST antibodies gave specific signals on normal centromeres and on the neocentromere at 3q24, whereas no reaction was detected at the chromosome 3-inactivated centromere (Fig. 3c). The signal on neocentromere was always fainter. FISH experiments narrowed the precise location of the neocentromere, and indicated that it emerged in the region encompassed by BAC RP11-505J9 (K in Fig. 1; red in Fig. 3d,e), located about 14 Mb centromeric to the neocentromere of case 1. The hypothesis of a pericentric inversion was not supported by marker order analysis around the neocentromere that was found unchanged (Fig. 3d,e). The list of BACs used to restrict the location of the neocentromere is available as Supplemental Table 2.
Figure 3.
Neocentromere case 2. (a) Diagram of chromosome 3 showing the position of the neocentromere in the derivative chromosome (D3). (b) FISH signals on normal (left) and abnormal chromosome 3 (right) using an alphoid probe specific for chromosome 3. Arrow points to the neocentromere. (c) Immunotyping experiment using CREST antibodies. Signals are present at the centromere of normal chromosome 3 (left) and on the neocentromere (right). (d,e) Cohybridization experiments performed to show normal marker arrangement around the neocentromeric region. (d) FISH signals of probes RP11-21N8 (blue); RP11-220J13 (yellow); RP11-383G6 (green); RP11-505J9 (red). (e) Signals of probes RP11-505J9 (red); RP11-426N12 (green); RP11-203L15 (blue). RP11-505J9 (red, K in Fig. 1), present in both experiments, is the most (neo) centromeric probe (see text). Both experiments indicate that marker order around the neocentromere is perfectly conserved. The position of each of these BACs in UCSC database is reported in Supplemental Table 2.
DISCUSSION
On the basis of our analysis of FISH marker order in 13 primate species, we have reconstructed the evolutionary history of this chromosome in primates. Marker arrangement in the cat, used as outgroup, was crucial to pinpoint the position of the centromere in Primate Ancestor, encompassed by markers I and N (Fig. 1). The position of the centromere in both OWM and great apes is distinct, and could not be reconciled with the PA centromere position. Our data, therefore, suggest in both cases the emergence of a new centromere. Intra- and interchromosomal segmental duplication clustering in the pericentromeric, as well as subtelomeric regions of primate chromosomes, humans in particular, is well established (Jackson et al. 1999; Bailey et al. 2002). The pericentromeric region of chromosome 3 is one of the few exceptions (X. She, J.E. Horvath, Z. Jian, G. Liu, T.S. Furey, L. Christ, R. Clark, T. Graves, C.L. Gulden, C. Alkan, et al., in prep.), showing little evidence of pericentromeric duplication near the mapped centromere. Although we cannot exclude the possibility that pericentromeric duplications remain to be identified for this region of the genome, the apparent absence of duplication is consistent with our finding that the centromere was seeded there quite recently during evolution. Several other evolutionary new centromeres have been documented in NWMs (Fig. 1). It is worth noting that the fission that generated the GF21 chromosome in Platyrrhini also triggered the appearance of an evolutionary new centromere in the vicinity of marker G. This same region has been used again later in evolution (see below).
Riethman et al. (2004) have reported a detailed characterization of human subtelomeric regions. The human 3p telomere, in contrast to 3q, is virtually devoid of segmental duplications. Once again, the evolutionary history of this region might provide an adequate explanation for this disparity. Our data show that the 3p telomeric sequence reached its present day position only recently, after orangutan, but before gorilla divergence, ∼7-14 Mya (Goodman 1999).
The OWM centromere is encompassed by human K1 and K2 markers (BACs RP11-355I21 and RP11-418B12, respectively), located ∼430 kb apart (Fig. 4a,b; see also Fig. 1, top, right); BACs mapping inside this gap (d1, d2, and d3 in Table 1 and Fig. 1) failed to give detectable signals in MMU, CAE, and PCR, while giving normal FISH signals in all great apes (data not shown). These findings suggested that the loss of the region in OWM could be specifically attributed to the emergence of the new centromere. The OWM chromosome 6 centromere also emerged as a new centromere (Eder et al. 2003). In contrast, in this case, a single human BAC generates split signals around this centromere as if the new centromere was seeded, and then progressed without affecting the displaced flanking sequences. The chromosome 3 region encompassed by K1-K2 markers was investigated for gene content. Interestingly, this region appears as a gene desert, harboring only two transcribed, but not translated spliced RNAs (UCSC, http://genome.ucsc.edu, July 2003 release). It is very likely that the 430-kb loss was due to the absence of selective pressure opposing the erosion resulting from the heterochromatization process. Saffery et al. (2003) have reported a normal transcription activity within the constitutively modified, nonheterochromatic chromatin of a human neocentromere. These data and data on the evolutionary new centromere seeding on chromosome 6 (see above) suggest that the centromere appearance has no adverse consequences on transcription per se. Rather, the heterochromatization process, which results in the assembling of large blocks of satellite DNA, creates nonpermissive conditions for gene expression.
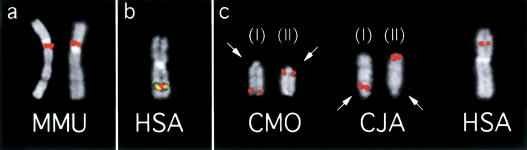
Figure 4.
(a) (Left) FISH experiments on macaque (MMU) using BAC RP11-355I21 (K1 in Fig. 1 and Table 1); (right) using BAC and RP11-418B12 (K2 in Fig. 1 and Table 1). (b) K1 (red) and K2 (yellow) probes cohybridized on human chromosomes. The signals, due to the packaging of metaphase chromosomes, are almost completely overlapping. (c) BAC C1 (RP11-603D9) giving splitting signal in dusky titi (CMO) chromosomes 16 (left) and 20 (right) (arranged and annotated as in Fig. 1) and in common marmoset (CJA) chromosomes 15 (left) and 17 (right) (arranged and annotated as in CMO). Arrows indicate the centromere. The single signal in humans is also shown (right).
The evolution of chromosome 3 has been investigated by Muller et al. (2000), who have already noted centromere position inconsistencies between OWM and great apes. Their assumption that the Bornean orangutan chromosome 3 arrangement was shared by a Catarrhini ancestor is not supported by the present analysis. The remaining inconsistencies directly descend from this discrepancy. More recently, Tsend-Ayush et al. (2004) have also investigated the chromosome 3 evolution in primates. Some of their conclusions do not agree with the conclusions of the present work, and again, these inconsistencies can be attributed to the different reconstruction of the ancestral organization of chromosome 3. Toward this reconstruction, the authors used a single species in NWM (Callithrix Geoffrey) and in OWM (Presbytis cristata). These species appear to not be representative of the Platyrrhini and Cercopithecidae, respectively. Our data, for instance, strongly suggest that chromosome 3 organization of Presbytis cristata (PCR, Colobinae) is derivative in comparison with Cercopithecinae (CAE, MMU, and PAN), as already reported by Bigoni et al. (1997). Furthermore, the order of markers in close proximity is difficult to resolve without using appropriate cohybridization experiments.
Centromeres and Telomeres
In the four NWM species examined (CJA, LLA, CMO, and SBO), HSA chromosome 3 is split into three chromosomes annotated as (I), (II), and (III) in Figure 1. Chromosomes (I) and (II) have an identical marker order in CJA, LLA, and CMO, but show a peculiar position of the centromere which is located, in CMO, at the opposite telomere in both (I) and (II) chromosomes in CMO with respect to the ancestral form of CJA and LLA (see examples in Fig. 4c). Additionally, the A region, centromeric in FCA and LCA, is a telomeric region in great apes. Noteworthy, the telomerically located centromere of the present-day human chromosome 15 emerged in the telomeric region of the short arm of the submetacentric chromosome composed by the ancestral 15/14 association (Ventura et al. 2003). Duplicon exchanges between pericentromeric and subtelomeric duplications are relatively frequent (Bailey et al. 2002). It could be theorized that the spread of pericentromeric duplications might play a major role in evolutionary new centromere appearance in telomeric regions.
Human Neocentromeres
Two human neocentromere cases on 3q were investigated. They have distinct, but important implications toward a better understanding of new centromere emergence phenomenon both in evolution and in clinical cases. In case 1, the neocentromere emergence process arose as part of a complex rearrangement. The functional centromere was excised, and a neocentromere seeded in the acentric derivative chromosome. The striking interest of this case is based on the fact that the neocentromere location is extremely close to the position of the chromosome 3 centromere in Old World monkeys, which also arose as a new centromere in the OWM ancestor (black circles in Fig. 1). This coincidence suggests that the same region maintained a longstanding latent centromere competence. It is noteworthy, in addition, that K2 probe, defining one side of the OWM (new) centromere, gave strong signals on the heterochromatic nonalphoid satellite block of human chromosome 1 (Fig. 2d), providing an indirect link of this locus to a centromere.
Furthermore, a new centromere appeared very close to marker G before the great apes divergence and a new centromere was also seeded close to marker G in the NWM ancestor in the chromosome reported as GF21 (black squares in Fig. 1). These findings strongly suggest that the marker identifies a latent centromeric region that was activated in both great apes and NWM ancestors. The reuse of breakpoints in evolution has been recently suggested by Pevzner and Tesler (2003) on the basis of computational analysis of mouse-human synteny. These two examples suggest a similar concept—the existence of recurrent sites for both evolutionary new centromere seedings and neocentromere appearance in humans. In this context, it would be interesting to define the precise location of 3q28-3q29 neocentromere cases to test whether the hypothesized ancestral centromeric region located at 3q29 is also a latent neocentromere emergence site.
Case 2 also sheds light on the evolutionary new centromere emergence process. It represents an unprecedented example of repositioning events that took place in evolution. It occurred as the sole karyotype variation, in absence of any rearrangement that usually accompanies human neocentromere cases. The father transmitted this chromosome to the fetus, supporting a potential normal behavior at meiosis of the abnormal chromosome. Most of human neocentromere cases arose as a consequence of a rearrangement that generated an acentric fragment. As a consequence, the patient is usually trisomic or tetrasomic for a chromosomal segment and shows an abnormal phenotype, indirectly preventing the transmission of the neocentromere to next generations. Neocentromere appearance not accompanied by the loss of the normal centromere has been reported on chromosome Y (Bukvic et al. 1996; Rivera et al. 1996; Tyler-Smith et al. 1999). In one case, however, the chromosome was unstable (Rivera et al. 1996), and in the other two cases, the neocentromere was embedded in the heterochromatic block of Yq. Very recently, Amor et al. (2004) have reported a two-generation family, in which the active centromere of one chromosome 4 has been relocated to 4q21.3 in otherwise cytogenetically normal and mitotically stable karyotypes. Our case 2, and the case reported by Amor et al. (2004) may be regarded as a present-day episode equivalent to new centromere appearance that occurred during the course of evolution.
The evolutionary history of the chromosome 15 disclosed a clear link between two human neocentromere cases at 15q24-15q26 and an ancestral centromere at 15q25 (Ventura et al. 2003). This was not the case for the two neocentromeres we have studied in the present study. On the other hand, the first case clearly identifies a region prone to neocentromere emergence that may represent a vestige of a very ancient centromere inactivation event. The search for ancestral centromeres, however, becomes very difficult when the investigation extends beyond primates. We think that this topic will warrant further attention as soon as additional data on genome evolution of mammals becomes available.
As stated, segmental duplications are biased to accumulate near pericentromeric regions. Large nonpericentromeric olfactory receptor duplicon clusters, however, are present in the human genome (Niimura and Nei 2003). Two nonpericentromeric olfactory receptor clusters lying on chromosome 3 are precisely located at domains identified by A2 and I1 markers. The evolutionary history of chromosome 3 linked together these two markers, with a pericentromeric location in the primate ancestor. This observation may suggest that segmental duplication clusters now positioned within euchromatin may be pericentromeric in origin. The significance of such euchromatization events of former pericentromeric sequence awaits further characterization.
METHODS
Primate metaphases used to track the evolutionary history of chromosome 3 were obtained from lymphoblastoid or fibroblast cell lines as previously described (Montefalcone et al. 1999). Human metaphase spreads were obtained from PHA-stimulated peripheral lymphocytes of normal donors by standard procedures. All human BACs used in this study are from the RP11 library (P. de Jong; http://www.chori.org/bacpac/). Chromosome preparations were cohybridized in situ with probes directly labeled, by nick-translation, with Cy3 (Perkin-Elmer), FluorX-dCTP (Amersham), DEAC (Molecular Probes), Cy5 (Amersham) essentially as already described (Ventura et al. 2003). Chromosome identification was obtained by simultaneous DAPI staining, producing a Q-banding pattern. Digital images were obtained using a Leica DMRXA2 epifluorescence microscope equipped with a cooled CCD camera (Princeton Instruments). DAPI, DEAC, FluorX, Cy3, and Cy5 fluorescence signals, detected with specific filters, were recorded separately as gray scale images. Pseudocoloring and merging of images were performed using Adobe Photoshop software.
All of the probes used in the present study were first validated in human chromosomes.
Fourteen overgo probes of 40 bp each were designed on sequences conserved between the human and mouse genomes according to the HomoloGene database (http://www.ncbi.nlm.nih.gov/HomoloGene/) as described by McPherson et al. (2001). The probes were hybridized to high-density filters and the images were analyzed with ArrayVision Ver6.0 (Imaging Research, Inc.). The sequence and location of Overgo probes are reported in Table 2.
Acknowledgments
Telethon, CEGBA (Centro di Eccellenza Geni in campo Biosanitario e Agroalimentare), MIUR (Ministero Italiano della Universita' e della Ricerca; Cluster C03, Prog. L.488/92), European Commission (INPRIMAT, QLRI-CT-2002-01325) are gratefully acknowledged for financial support. This work was supported, in part, by NIH grants GM58815 and HG002385 to E.E.E. Critical reading by Dr. R. Stanyon is also acknowledged.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
[Supplemental material is available online at www.genome.org.]
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.2608804.
Orthologous chromosomes can have a different chromosome number in different species because, in each species, they are ordered on the basis of chromosome size only. To avoid confusion, the human nomenclature will be used. The actual chromosome number in the different species is reported on top of each chromosome in Figure 1.
References
- Alonso, A., Mahmood, R., Li, S., Cheung, F., Yoda, K., and Warburton, P.E. 2003. Genomic microarray analysis reveals distinct locations for the CENP-A binding domains in three human chromosome 13q32 neocentromeres. Hum. Mol. Genet. 12: 2711-2721. [DOI] [PubMed] [Google Scholar]
- Amor, D.J. and Choo, K.H. 2002. Neocentromeres: Role in human disease, evolution, and centromere study. Am. J. Hum. Genet. 71: 695-714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amor, D.J., Bentley, K., Ryan, J., Perry, J., Wong, L., Slater, H., and Choo, K.H. 2004. Human centromere repositioning “in progress”. Proc. Natl. Acad. Sci. 101: 6542-6547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey, J.A., Gu, Z., Clark, R.A., Reinert, K., Samonte, R.V., Schwartz, S., Adams, M.D., Myers, E.W., Li, P.W., and Eichler, E.E. 2002. Recent segmental duplications in the human genome. Science 297: 1003-1007. [DOI] [PubMed] [Google Scholar]
- Band, M.R., Larson, J.H., Rebeiz, M., Green, C.A., Heyen, D.W., Donovan, J., Windish, R., Steining, C., Mahyuddin, P., Womack, J.E., et al. 2000. An ordered comparative map of the cattle and human genomes. Genome Res. 10: 1359-1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bigoni, F., Stanyon, R., Koehler, U., Morescalchi, A.M., and Wienberg, J. 1997. Mapping homology between human and black and white colobine monkey chromosomes by fluorescent in situ hybridization. Am. J. Primatol. 42: 289-298. [DOI] [PubMed] [Google Scholar]
- Bourque, G. and Pevzner, P.A. 2002. Genome-scale evolution: Reconstructing gene orders in the ancestral species. Genome Res. 12: 26-36. [PMC free article] [PubMed] [Google Scholar]
- Bukvic, N., Susca, F., Gentile, M., Tangari, E., Ianniruberto, A., and Guanti, G. 1996. An unusual dicentric Y chromosome with a functional centromere with no detectable α-satellite. Hum. Genet. 97: 453-456. [DOI] [PubMed] [Google Scholar]
- Carbone, L., Ventura, M., Tempesta, S., Rocchi, M., and Archidiacono, N. 2002. Evolutionary history of chromosome 10 in primates. Chromosoma 111: 267-272. [DOI] [PubMed] [Google Scholar]
- Cardone, M.F., Ventura, M., Tempesta, S., Rocchi, M., and Archidiacono, N. 2002. Analysis of chromosome conservation in Lemur catta studied by chromosome paints and BAC/PAC probes. Chromosoma 111: 348-356. [DOI] [PubMed] [Google Scholar]
- Choo, K.H.A. 1997. Centromere DNA dynamics: Latent centromeres and neocentromere formation. Am. J. Hum. Genet. 61: 1225-1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- du Sart, D., Cancilla, M.R., Earle, E., Mao, J., Saffery, R., Tainton, K.M., Kalitsis, P., Martin, J., Barry, A.E., and Choo, K.H.A. 1997. A functional neo centromere formed through activation of a latent human centromere and consisting of non-α-satellite DNA. Nat. Genet. 16: 144-153. [DOI] [PubMed] [Google Scholar]
- Eder, V., Ventura, M., Ianigro, M., Teti, M., Rocchi, M., and Archidiacono, N. 2003. Chromosome 6 phylogeny in primates and centromere repositioning. Mol. Biol. Evol. 20: 1506-1512. [DOI] [PubMed] [Google Scholar]
- Goodman, M. 1999. The genomic record of Humankind's evolutionary roots. Am. J. Hum. Genet. 64: 31-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ijdo, J.W., Baldini, A., Ward, D.C., Reeders, S.T., and Wells, R.A. 1991. Origin of human chromosome 2: An ancestral telomere-telomere fusion. Proc. Natl. Acad. Sci. 88: 9051-9055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson, M. 2003. Duplicate, decouple, disperse: The evolutionary transience of human centromeric regions. Curr. Opin. Genet. Dev. 13: 629-635. [DOI] [PubMed] [Google Scholar]
- Jackson, M.S., Rocchi, M., Thompson, G., Hearn, T., Crosier, M., Guy, J., Kirk, D., Mulligan, L., Ricco, A., Piccininni, S., et al. 1999. Sequences flanking the centromere of human chromosome 10 are a complex patchwork of arm-specific sequences, stable duplications and unstable sequences with homologies to telomeric and other centromeric locations. Hum. Mol. Genet. 8: 205-215. [DOI] [PubMed] [Google Scholar]
- Kasai, F., Garcia, C., Arruga, M.V., and Ferguson-Smith, M.A. 2003. Chromosome homology between chicken (Gallus gallus domesticus) and the red-legged partridge (Alectoris rufa); evidence of the occurrence of a neocentromere during evolution. Cytogenet. Genome Res. 102: 326-330. [DOI] [PubMed] [Google Scholar]
- Lo, A.W., Magliano, D.J., Sibson, M.C., Kalitsis, P., Craig, J.M., and Choo, K.H. 2001. A novel chromatin immunoprecipitation and array (cia) analysis identifies a 460-kb cenp-a-binding neocentromere dna. Genome Res. 11: 448-457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McPherson, J.D., Marra, M., Hillier, L., Waterston, R.H., Chinwalla, A., Wallis, J., Sekhon, M., Wylie, K., Mardis, E.R., Wilson, R.K., et al. 2001. A physical map of the human genome. Nature 409: 934-941. [DOI] [PubMed] [Google Scholar]
- Montefalcone, G., Tempesta, S., Rocchi, M., and Archidiacono, N. 1999. Centromere Repositioning. Genome Res. 9: 1184-1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller, S., Stanyon, R., Finelli, P., Archidiacono, N., and Wienberg, J. 2000. Molecular cytogenetic dissection of human chromosomes 3 and 21 evolution. Proc. Natl. Acad. Sci. 97: 206-211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy, W.J., Sun, S., Chen, Z.q., Yuhki, N., Hirschmann, D., Menotti-Raymond, M., and O'Brien, S.J. 2000. A Radiation hybrid map of the cat genome: Implications for comparative mapping. Genome Res. 10: 691-702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy, W.J., Stanyon, R., and O'Brien, S.J. 2001. Evolution of mammalian genome organization inferred from comparative gene mapping. Genome Biol. (www) 2: REVIEWS0005. [DOI] [PMC free article] [PubMed]
- Nagaki, K., Cheng, Z., Ouyang, S., Talbert, P.B., Kim, M., Jones, K.M., Henikoff, S., Buell, C.R., and Jiang, J. 2004. Sequencing of a rice centromere uncovers active genes. Nat. Genet. 36: 138-145. [DOI] [PubMed] [Google Scholar]
- Niimura, Y. and Nei, M. 2003. Evolution of olfactory receptor genes in the human genome. Proc. Natl. Acad. Sci. 100: 12235-12240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oakenfull, E., Lim, H., and Ryder, O. 2000. A survey of equid mitochondrial DNA: Implications for the evolution, genetic diversity and conservation of Equus. Conservation Genet. 1: 341-355. [Google Scholar]
- Pevzner, P. and Tesler, G. 2003. Human and mouse genomic sequences reveal extensive breakpoint reuse in mammalian evolution. Proc. Natl. Acad. Sci. 100: 7672-7677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riethman, H., Ambrosini, A., Castaneda, C., Finklestein, J., Hu, X.-L., Mudunuri, U., Paul, S., and Wei, J. 2004. Mapping and initial analysis of human subtelomeric sequence assemblies. Genome Res. 14: 18-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivera, H., Vassquez, A.I., Ayala-Madrigal, M.L., Ramirez-Duenas, M.L., and Davalos, I.P. 1996. Alphoidless centromere of a familial unstable inverted Y chromosome. Ann. Genet. 39: 236-239. [PubMed] [Google Scholar]
- Saffery, R., Sumer, H., Hassan, S., Wong, L.H., Craig, J.M., Todokoro, K., Anderson, M., Stafford, A., and Choo, K.H. 2003. Transcription within a functional human centromere. Mol. Cell 12: 509-516. [DOI] [PubMed] [Google Scholar]
- Satinover, D.L., Vance, G.H., Van Dyke, D.L., and Schwartz, S. 2001. Cytogenetic analysis and construction of a BAC contig across a common neocentromeric region from 9p. Chromosoma 110: 275-283. [DOI] [PubMed] [Google Scholar]
- Tsend-Ayush, E., Grutzner, F., Yue, Y., Grossmann, B., Hansel, U., Sudbrak, R., and Haaf, T. 2004. Plasticity of human chromosome 3 during primate evolution. Genomics 83: 193-202. [DOI] [PubMed] [Google Scholar]
- Tyler-Smith, C., Gimelli, G., Giglio, S., Floridia, G., Pandya, A., Terzoli, G., Warburton, P.E., Earnshaw, W.C., and Zuffardi, O. 1999. Transmission of a fully functional human neocentromere through three generations. Am. J. Hum. Genet. 64: 1440-1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ventura, M., Archidiacono, N., and Rocchi, M. 2001. Centromere emergence in evolution. Genome Res. 11: 595-599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ventura, M., Mudge, J.M., Palumbo, V., Burn, S., Blennow, E., Pierluigi, M., Giorda, R., Zuffardi, O., Archidiacono, N., Jackson, M.S., et al. 2003. Neocentromeres in 15q24-26 map to duplicons which flanked an ancestral centromere in 15q25. Genome Res. 13: 2059-2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wandall, A., Tranebjaerg, L., and Tommerup, N. 1998. A neocentromere on human chromosome 3 without detectable α-satellite DNA forms morphologically normal kinetochores. Chromosoma 107: 359-365. [DOI] [PubMed] [Google Scholar]
- Yang, F., Graphodatsky, A.S., O'Brien, P.C., Colabella, A., Solanky, N., Squire, M., Sargan, D.R., and Ferguson-Smith, M.A. 2000. Reciprocal chromosome painting illuminates the history of genome evolution of the domestic cat, dog and human. Chromosome Res. 8: 393-404. [DOI] [PubMed] [Google Scholar]
- Yang, F., Fu, B., O'Brien, P.C.M., Nie, W., Ryder, O.A., and Ferguson-Smith, M.A. 2004. Refined genome-wide comparative map of the domestic horse, donkey and human based on cross-species chromosome painting: Insight into the occasional fertility of mules. Chromosome Res. 12: 65-76. [DOI] [PubMed] [Google Scholar]
WEB SITE REFERENCES
- http://www.cs.ucsd.edu/groups/bioinformatics/GRIMM/; GRIMM software home page.
- www://humanparalogy.gene.cwru.edu; Segmental Duplication database.
- http://www.chori.org/bacpac/; Pieter de Jong libraries home page.
- http://www.ncbi.nlm.nih.gov/HomoloGene/; HomoloGene database.
- http://genome.ucsc.edu; University California Santa Cruz, Human Genome Browser Gateway.