Abstract
Short blocks of telomeric-like DNA (Interstitial Telomeric Sequences, ITSs) are found far from chromosome ends. We addressed the question as to how such sequences arise by comparing the loci of 10 human ITSs with their genomic orthologs in 12 primate species. The ITSs did not derive from expansion of pre-existing TTAGGG units, as described for other microsatellites, but appeared suddenly during evolution. Nine insertion events were dated along the primate evolutionary tree, the dates ranging between 40 and 6 million years ago. Sequence comparisons suggest that in each case the block of (TTAGGG)n DNA arose as a result of double-strand break repair. In fact, ancestral sequences were either interrupted precisely by the tract of telomeric-like repeats or showed the typical modifications observed at double-strand break repair sites such as short deletions, addition of random sequences, or duplications. Similar conclusions were drawn from the analysis of a chimpanzee-specific ITS. We propose that telomeric sequences were inserted by the capture of a telomeric DNA fragment at the break site or by the telomerase enzyme. Our conclusions indicate that human ITSs are relics of ancient breakage rather than fragile sites themselves, as previously suggested.
The telomeres of vertebrate chromosomes are composed of extended arrays of the TTAGGG hexamer (McEachern et al. 2000). The specialized enzyme telomerase performs the synthesis of telomeres using as a template a complementary oligoribonucleotide, which is associated to the catalytic protein moiety (Greider 1996; Harrington 2003; Cech 2004). The presence of the telomeric repeats at the chromosome ends is essential for ensuring a correct functioning of telomeres and for the maintenance of chromosome integrity and stability (Blackburn 2001). In several species, including humans and other primates, repetitions of the TTAGGG unit, called interstitial telomeric sequences (ITSs), have been observed also inside the chromosomes away from the termini (Meyne et al. 1990; Azzalin et al. 1997; Ruiz-Herrera et al. 2002, 2004).
Whereas the relationship between the sequence organization of telomeres and their function is clear, the presence of telomere-like sequences inside chromosomes is far from being understood. We have initially approached this problem by analyzing the sequence organization of the interstitial telomeres present in the human genome with the purpose of understanding the basis for their creation. In a previous study (Azzalin et al. 2001) we reported that, in the human genome, more than a hundred intrachromosomal telomeric arrays composed of more than four TTAGGG repeats are present; we then grouped them in three classes: (1) subtelomeric ITSs, composed of extended tandemly oriented arrays containing many degenerate units; (2) fusion ITSs in which two stretches of telomeric repeats are oriented head-to-head; (3) short ITSs composed of few tandemly oriented TTAGGG repeats.
The three classes derive from different mechanisms. It is likely that the degenerate repeats of subtelomeric ITSs (class 1) arose from recombination events involving chromosome ends.
Fusion ITSs (class 2) originate from end fusion of ancestral chromosomes and appear to be very rare in the human genome because only the previously described fusion ITS from Chromosome 2q13 (Ijdo et al. 1991) was found in our search.
More intriguing results were obtained from the analysis of 50 short ITSs (class 3), which allowed us to propose that they probably arose from the repair of DNA double-strand breaks occurring in the germ line during evolution (Azzalin et al. 2001). In fact, several short ITSs interrupt precisely the consensus sequence of known dispersed repetitive elements such as SINEs or LINEs; moreover, some short ITSs were flanked by direct repeats of 7 to 43 nt, suggesting that the ITSs with this particular configuration were inserted during the repair of staggered breaks. Similar conclusions were reached after the analysis of ITSs isolated from the Chinese hamster genome (Faravelli et al. 2002).
In mammals, one of the main mechanisms for the repair of DNA double-strand breaks is the nonhomologous end-joining (NHEJ) pathway, which is carried out by a multiprotein apparatus comprising, among others, the Ku heterodimer, the DNA-PK catalytic subunit, the homolog of the MRE11-RAD50-NBS1 Saccharomyces cerevisiae complex, the DNA ligase IV, and the XRCC4 and Artemis proteins (Valerie and Povirk 2003). The primary function of this apparatus is the recognition of the free ends arising from the break; because double-strand breaks often produce frayed ends, these have to be processed before joining, leading to deletions or insertions (Smith et al. 2001). Although the DNA ends produced by breakage are structurally and functionally different from the telomeric ends, some of the mentioned proteins can bind and act on both structures (Chan and Blackburn 2002; de Lange 2002).
In the work presented here, we used an evolutionary approach to test our hypothesis that short ITSs derive from the repair of double-strand breaks; we analyzed the sequence organization of 22 loci orthologous to human short ITS in 12 primate species. One chimpanzee ITS was also analyzed using the same approach. The comparison of the sequences flanking each ITS allowed us to confirm our starting hypothesis as well as to provide an evolutionary dating of the events leading to the insertion of the telomeric repeats.
RESULTS
We selected 22 sequences out of the >100 short ITSs present in the human genome. We also included in our study one chimpanzee ITS that is absent in the human genome.
Primer pairs for the sequences flanking the 22 selected human ITSs were used to amplify genomic DNA from great apes (Pan troglodytes, PTR, common chimpanzee; Pan paniscus, PPA, pygmy chimpanzee; Gorilla gorilla, GGO, gorilla; Pongo pygmaeus, PPY, Bornean orangutan); gibbons (Hylobates lar, HLA, white-handed gibbon; Hylobates klossii, HKL, Kloss' gibbon); Old World monkeys (Papio anubis, PAN, olive baboon; Presbytis cristata, PCR, leaf-monkey; Macaca mulatta, MMU, rhesus macaque; Cercopithecus aethiops, CAE, vervet monkey); and New World monkeys (Callithrix jacchus, CJA, common marmoset; Callicebus moloch, CMO, dusky titi). A similar analysis was carried out with the chimpanzee ITS. Of the 22 human loci, 16 were considered informative because, for each of them, specific PCR products could be obtained in at least three nonhuman primates. The chimpanzee locus was also informative. The PCR products were sequenced and the orthologous sequences were aligned.
Six human ITS loci were found in all the examined species; therefore, their insertion dates before Catarrhini/Platyrrhini divergence (data not shown). For nine human ITS loci and for the chimpanzee locus, comparison of the sequences in the different primates allowed us both to date the insertion of the telomeric array during evolution and to propose a molecular mechanism underlying the phenomenon (Fig. 1A,B; Fig. 2A,B,C,D,F,G; Fig. 3A,B). For another locus, the sequence comparison allowed us to propose a mechanism for ITS insertion but not to date the insertion event (Fig. 2E). A short portion from the sequence of these 10 human loci and of the chimpanzee locus is reported in the Figures 1, 2, 3. The inserted telomeric arrays are shown in red. Occasional mismatches in the telomeric array (less than 1 mismatch every 12 bp) were observed but are not reported in the figures. The identity values of the sequences flanking the ITSs in all the primates (data not shown) are very high, comprised between 88% and 99%, and are substantially in agreement with their phylogenetic distance.
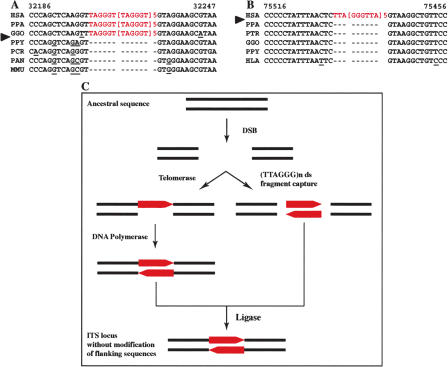
Figure 1.
Interstitial telomeric-like repeat insertion without modification of the flanking sequences. (A,B) Two examples of this type of insertion. A short portion of the DNA sequence of the ITS loci in different primate species is shown. (HSA) Homo sapiens; (PPA) Pan paniscus; (GGO) Gorilla gorilla; (PPY) Pongo pygmaeus; (HLA) Hylobates lar; (PCR) Presbytis cristata; (PAN) Papio anubis; (MMU) Macaca mulatta. The inserted nucleotides are shown in red. In the locus presented in A, the ITS is present in human, chimpanzee, and gorilla and absent in the more distant primates; the ITS presented in B is human-specific. (C) A model for this type of insertion. The telomeric repeats (red boxes) are inserted at a blunt double-strand break site either by the telomerase enzyme or via the capture of a double-stranded telomeric fragment. The human ITS locus in A is the AL033381 sequence from Chromosome 6p24; in B the AC039056 sequence from 15q14.
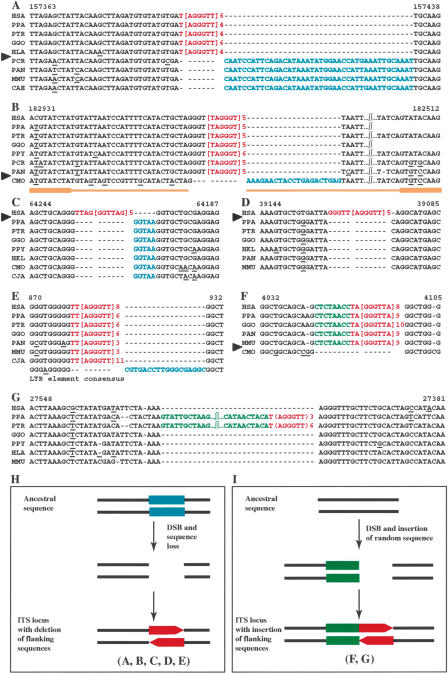
Figure 2.
Interstitial telomeric-like repeat insertion with flanking sequence deletion or insertion. (A,B,C,D,E) Five examples of ITS insertions accompanied by deletions of flanking sequences. The dates of insertion of these ITS vary between 6 and 25-40 Mya. (F,G) The interstitial telomeric repeats appear together with a random sequence. The repeat in G is very recent, being present only in chimpanzees. The inserted telomeric repeats are shown in red, the deleted nucleotides in blue, and the inserted random nucleotides in green. The ITS shown in B is inserted within an LTR/THE1D element (thin yellow lines), which is inserted inside an LTR/MLT1C element (thick yellow lines). (H,I) Models for these types of insertion (deleted sequences: blue boxes; telomeric repeats: red boxes; inserted random nucleotides: green boxes). (HSA) Homo sapiens; (PTR) Pan troglodytes; (PPA) Pan paniscus; (GGO) Gorilla gorilla; (PPY) Pongo pygmaeus; (HLA) Hylobates lar; (HKL) Hylobates klossii; (PAN) Papio anubis; (PCR) Presbytis cristata; (MMU) Macaca mulatta; (CAE) Cercopithecus aethiops; (CMO) Callicebus moloch; (CJA) Callithrix jacchus. (A) The human ITS locus is the AL354723 sequence from Chromosome 9p22; (B) AF216808, Chromosome 8p12; (C) AC009054, Chromosome 16q22; (D) AC025982, Chromosome 3q25; (E) AF236882, Chromosome 7q36; (F) AC005032, Chromosome 7q36. (G) The chimpanzee ITS locus is the BS000043 sequence from chimpanzee Chromosome 22.
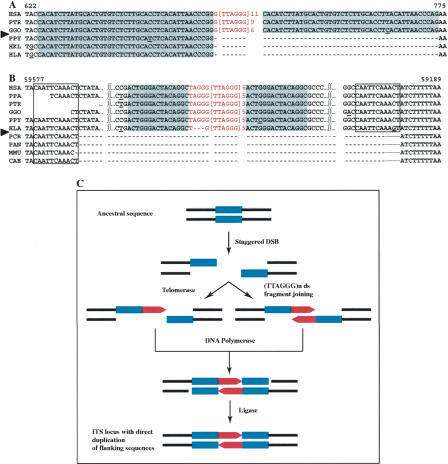
Figure 3.
Interstitial telomeric-like repeat insertion at staggered double-strand break sites. (A,B) Two examples of telomeric repeat insertions associated with flanking sequence duplications. The inserted telomeric nucleotides are shown in red and the direct repeats flanking the insertion are shaded in blue; the direct repeats flanking the AluY transposon are boxed. (C) A staggered double-strand break can give rise to a duplication by a gap-filling mechanism (telomeric arrays: red boxes; direct repeats: light blue boxes). (HSA) Homo sapiens; (PTR) Pan troglodytes; (PPA) Pan paniscus; (GGO) Gorilla gorilla; (PPY) Pongo pygmaeus; (HLA) Hylobates lar; (HKL) Hylobates klossii; (PAN) Papio anubis; (PCR) Presbytis cristata; (MMU) Macaca mulatta; (CAE) Cercopithecus aethiops. (A) The human ITS locus is the AF237881 sequence from Chromosome 2q31; (B) AC026696, Chromosome 5q14.
The sequences of the 10 human loci and of the chimpanzee locus were grouped in four sets according to the proposed models for the insertion of the telomeric repeats. In all four models, the proposed initiating event is a DNA double-strand break. A description of the analyzed sequences and the basis for their grouping is presented below.
IT Insertion Without Modification of the Flanking Sequences
The human AL033381 sequence (Fig. 1A), located on Chromosome 6p24, contains a 39-bp TTAGGG array (from nucleotide 32196 to nucleotide 32234) and does not show any peculiar feature in the region flanking the ITS. The telomeric array was present in HSA (Homo sapiens), PPA, and GGO, while absent in PPY, PCR, PAN, and MMU. A GT dinucleotide, which is part of the telomeric hexamer, was contained in the ancestral sequence at the 5′-end of the insertion. The flanking sequences are highly conserved in all primates analyzed. These data strongly suggest that the ITS was inserted after the divergence of PPY from the Homo/Pan/Gorilla group and before GGO divergence, which is ∼7-14 million years ago (Mya; Fig. 4; Goodman 1999).
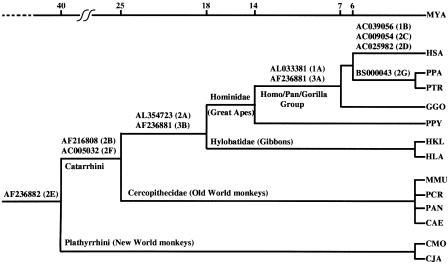
Figure 4.
Phylogenetic tree of primates. The time of insertion of each one of the 11 telomeric repeats is marked on the phylogenetic tree. Each ITS locus is indicated by its accession number. The figure number relative to each locus is indicated in parentheses.
The ITS in the human AC039056 sequence (Fig. 1B), located on Chromosome 15q14, is composed of a 34-bp telomeric-like array (from nucleotide 75468 to nucleotide 75501). Also, this sequence does not show any remarkable feature in the ITS flanking regions. This ITS appears human-specific, because it was not found in any nonhuman primate. Consequently, the insertion took place after the chimpanzee/human divergence, which occurred ∼6 Mya (Fig. 4; Goodman 1999).
In both cases, a similar event has probably occurred, as outlined in Figure 1C; the sudden appearence of a stretch of telomeric repeats at a given point in evolution suggests a mechanism whereby DNA ends are produced in the germ line by a double-strand break; during joining of the broken ends, TTAGGG repeats are inserted either directly by telomerase or via the capture of a double-stranded telomeric repeat fragment. No modification of the flanking sequences was observed in the human locus containing the ITS compared with the ancestral locus, suggesting that the repair process occurred either at a blunt double-strand break or at staggered break that was filled in on one side and blunted on the other side.
Deletion and ITS Insertion at Break Site
The human AL354723 sequence (Fig. 2A), located on Chromosome 9p22, contains a 37-bp telomeric array (nucleotides 157,396-157,432); the flanking regions do not show any particular feature. Specific amplification fragments from PPA, PTR, GGO, HLA, PCR, PAN, MMU, and CAE were sequenced. Sequence comparison showed that the human ITS was conserved in great apes (PPA, PTR, and GGO) and in the gibbon HLA but absent in Old World monkeys (PCR, PAN, MMU, and CAE). A 42-bp sequence following the 3′-end of the telomeric array is present in Old World monkeys but missing in the human and ape genomes. Very likely a unique event generated the ITS insertion and the 42-bp deletion, and occurred in the great apes/gibbons ancestor, ∼18-25 Mya (Fig. 4; Goodman 1999). The number of telomeric repeats appeared to vary in the different species. In this respect, however, it is worth noticing that variations in ITS repeat number among different human individuals has been reported for several ITS loci (Mondello et al. 2000). Variability in ITS repeat number among different species (see also below) is therefore not unexpected.
The human AF216808 sequence (Fig. 2B), located on Chromosome 8p12, contains a 36-bp TTAGGG repeat array (nucleotides 182,863-182,898) inserted within an LTR/THE1D element (thin yellow lines at the bottom of Fig. 2B); the LTR/THE1D sequence is inserted, in turn, inside an LTR/MLT1C element (thick yellow lines). Using a primer pair external to the MLT1C element, we amplified and sequenced the genomic DNA from HSA, PPA, PTR, GGO, PPY, PCR, PAN, and CMO. Figure 2B shows the alignment of the relevant portion of the sequenced fragments; the human sequence organization is conserved in all species except the most distant one, the New World monkey CMO, which contains only the first three nucleotides (TAG) of the array. Conversely, in all the other species, a 23-bp fragment, belonging to the LTR/THE1D element and present in CMO, is missing. Hence, the insertion of the telomeric array must have occurred simultaneously with the loss of the 23-bp fragment. Most likely, the insertion of the THE1D transposon inside a preexisting MLT1C element is ancestral. Subsequently, after Catarrhini and Platyrrhini divergence (22-40 Mya; Fig. 4), the telomeric repeat was inserted and, at the same time, the 23-bp fragment was deleted.
The human AC009054 sequence (Fig. 2C) is located on Chromosome 16q22 and contains an exact 41-bp telomeric array (nucleotides 64,198-64,238); the flanking sequences do not show any particular feature. The orthologous loci from PPA, PTR, GGO, PPY, HKL, CMO, and CJA lack the telomeric array and contain five additional nucleotides at the 3′-end of the ITS. An AGGG tetranucleotide, in frame with the telomeric array, is present in the ancestral sequence at the 5′-end of the insertion. Most likely, also in this case, the insertion event occurred concurrently with a deletion, after the chimpanzee/human divergence (Fig. 4).
The human AC025982 sequence (Fig. 2D), which maps on Chromosome 3q25, contains a 41-bp telomeric repeat (nucleotides 39,092-39,132) inserted within an Alu element. Analysis of the orthologous sequences in PPA, PTR, GGO, HKL, PAN, and MMU showed that the ITS is human-specific and that a TTA trinucleotide, in frame with the telomeric insert, is present in the ancestral sequence. In all the nonhuman primates, however, an extra C nucleotide at the 3′-end of the telomeric array was found. Thus, in this case, the insertion event occurred within a repetitive element, interrupting exactly its consensus sequence except for the 1-bp deletion.
The last example of this group (Fig. 2E; sequence AF236882) shows an ITS, located by FISH on 7q36 (Azzalin et al. 2001) consisting of a 53-bp telomeric array (nucleotides 876-928). The ITS is present, although with different numbers of repeats, in all the analyzed primates: PPA, PTR, GGO, PAN, MMU, and CJA. This ITS interrupts an LTR element that is very conserved in all species and contains a GGG trinucleotide at the site of the insertion. However, sequence alignement with the LTR consensus showed an 18-bp deletion at the 3′-end of the telomeric array. These data suggest that the insertion-deletion was already present in the ancestor of Catarrhini and Platyrrhini, that is, >40 Mya (Fig. 4; Goodman 1999).
All the ITS insertion events described in Figure 2, A to E, probably arose through a mechanism like the one shown in Figure 2G: a double-strand break accompanied by the loss of adjacent sequences is repaired through the insertion of a telomeric array either by the intervention of telomerase or by the capture of a telomeric fragment as described in Figure 1C.
Random Sequence Addition and ITS Insertion at Break Site
The human sequence AC005032 is located on Chromosome 7q36.3 and contains a 50-bp ITS (Azzalin et al. 2001) extending from nucleotides 4051 to 4100 (Fig. 2F). The orthologous sequences from PPA, GGO, PAN, and MMU showed the presence of the telomeric array. In the New World monkey CMO, the ITS and an unrelated 8-bp sequence 5′ to the ITS are absent. These results suggest that in the Catarrhini ancestor, after divergence from Platyrrhini (25-40 Mya; Goodman 1999), a double-strand break was repaired with the mechanism illustrated in Figure 2H: a random sequence is added to one end of the break site and telomeric repeats are then inserted.
After a search of ITS loci in the P. troglodytes genome (PTR), we identified the BS000043 sequence, which contains a recent insertion (Fig. 2G); in fact, the telomeric repeat is present only in the two chimpanzee species PTR and PPA. Similarly to the human AC005032, a 76-bp DNA fragment was introduced together with the ITS in the ancestral sequence (Fig. 2I).
IT Insertion at Staggered Double-Strand Break Site
The human sequence AF236881 isolated from Chromosome 2q31 (Azzalin et al. 2001) is characterized by a peculiar organization (Fig. 3A). A 67-bp telomeric array interrupts a 397-bp LTR element and is flanked on each side by a 43-bp direct repeat (shaded in blue in Fig. 3A) containing only two mismatches; the 43-bp sequence is present only once in the LTR consensus. The ITS and the 43-bp repeat are conserved in PTR and GGO. Conversely, in PPY, HKL, and HLA, the LTR consensus sequence was not interrupted: no trace of telomeric array and only one copy of the 43-bp sequence are found. On the basis of these observations, we propose that a single hit accounts for the insertion of the telomeric sequence and for the duplication of the 43 nucleotides. The insertion-duplication event occurred after the split of the human/chimpanzee/gorilla ancestor from PPY, that is, between 7 and 14 Mya (Fig. 4; Goodman 1999).
A similar organization was found in the human sequence AC026696, located on Chromosome 5q14. This locus contains a 41-bp telomeric array extending from nucleotides 59,378 to 59,418. The ITS interrupts a 303-bp AluY element and is flanked on each side by a perfect 15-bp direct repeat (shaded in blue in Fig. 3B). The AluY consensus contains only one copy of the 15-bp sequence. The AluY element at this locus (nucleotides 59,222-59,591), in its turn, is flanked on each side by an 11-bp direct repeat (boxed in Fig. 3B), which marks the transposition event. The orthologous sequences of great apes (PPA, PTR, GGO, PPY) and of the gibbon HLA showed the same overall organization. Conversely, in PCR, PAN, MMU, and CAE, the AluY element, including the inserted telomeric array, is missing and only one copy of the 11 bp is present. Therefore, ∼18-25 Mya (Fig. 4; Goodman 1999), the AluY sequence was inserted creating the 11-bp target duplication; at the same time or “soon” after, the telomeric array was inserted inside the AluY element, generating the 15-bp direct flanking repeat.
The ITSs shown in Figure 3, A and B, were probably generated by the same mechanism, as outlined in Figure 3C. A staggered double-strand break occurs in the germ line, and, during repair, a telomeric array is inserted; the ITS, similarly to what is depicted in Figure 1C, can be introduced either through the synthesis of telomeric repeats by telomerase, or by the insertion of a double-stranded telomeric fragment. According to this model, the direct repeats are generated by filling of the single strand gaps deriving from the separation of the staggered ends.
DISCUSSION
The relationship between the sequence organization of telomeres and their function is well understood; on the contrary, the presence of intrachromosomal telomeric repeats demands a definition of their possible function and of the molecular events leading to their occurrence. One proposed reason for the existence of ITSs is the telomeric fusion of ancestral chromosomes; however, in the human species, ITSs generated from this mechanism appear to be very rare (Azzalin et al. 2001). In the human genome, more than a hundred intrachromosomal telomeric arrays composed of more than four TTAGGG repeats are present, whereas tandem repeats of other hexameric units are rare (Azzalin et al. 2001). Is the relative abundance of telomeric repeats inside the chromosomes, that is, in a position totally unrelated to the function for which they have evolved, of any significance?
In the present paper, we have reported a detailed characterization of 10 ITSs found in the human genome and of one ITS found in the chimpanzee genome. We have determined the approximate date of their evolutionary occurrence and, from the analysis of their flanking sequences, we have derived four model mechanisms that explain the origins of these particular sequence elements. These models imply a double-strand break as the primary causal event and can be considered as modifications, involving the intervention of telomerase or telomerase products, of the canonical nonhomologous end-joining (NHEJ) process.
According to these models, the telomeric hexamers were introduced either directly by the telomerase enzyme or by the insertion of a blunt-ended, double-stranded telomeric sequence. In the first case, the enzyme uses a 3′-end of the break as a primer to synthesize the G-rich strand of a stretch of telomeric hexamers; a DNA polymerase synthesizes the second strand, and a ligase restores the integrity of the double helix. In the second case, one may suppose that a preexisting (TTAGGG)n fragment is transposed at the site of breakage in a way resembling the capture of filler DNA during NHEJ (Roth et al. 1991; Liang et al. 1998; Lin and Waldman 2001). We can suppose that all these transactions take place with the contribution of several proteins that are deputed to the recognition of the break and maintenance of a close contact between the broken ends.
The ITSs described in Figure 1 were inserted without any modification of the flanking sequences, whereas the ITSs described in Figure 2, A to E, share a common pattern of insertion, that is, the addition of telomeric repeats coupled with the loss of flanking sequences. The size of the deletions varies from 1 to 42 nt. The loss of sequences at the break site has been described as a frequent outcome of NHEJ events (Smith et al. 2001), and is likely due to the action of nucleases involved in end processing. The sequence depicted in Figure 2B has a peculiar organization: the ITS is inserted inside a transposable element, which, on its turn, is inserted into another transposable element. This may point to this site as a possible hot spot for breakage. The ITSs described in Figure 2, F and G, are interpretable as arising from the insertion of a random DNA fragment coupled with the addition of telomeric repeats. Alternatively, these ITSs could have arisen from the direct insertion of a preformed double-stranded fragment composed of the random sequence and the telomeric array.
The sequence organization of the ITSs reported in Figure 3 is particularly intriguing: the telomeric array are precisely inserted between two copies of a sequence present as a single copy in the ancestral genome. The most likely explanation for this observation is that the telomeric repeats were inserted at the sites of staggered double-strand breaks (43 bp in the sequence shown in Fig. 3A and 15 bp in that shown in Fig. 3B), and the duplications were then generated upon the intervention of a DNA polymerase (Fig. 3C). The organization of these two ITSs is surprisingly analogous with those of transposable elements, whose insertion leads to direct duplication of the target sequence.
Thus, all the insertion events described in the present work point to the possibility that telomerase is recruited for the repair of DNA double-strand breaks via a variation of the canonical NHEJ. Whereas the ability of telomerase to add telomeric repeats to free 3′-ends derived from double-strand breaks in a process known as “telomere healing” is well demonstrated (Flint et al. 1994; Sprung et al. 1999), we provide evidence that telomeric sequences can be inserted during joining of two broken ends. In telomere healing, the recognition of the 3′-end by the enzyme is enhanced by the presence of a 1-5-nt homology to the telomeric sequence. Indeed, this appears to be the case in several of the sequences shown here: in the sequences reported in Figures 1A, 2B-E, and 3A, the oligonucleotides GT, TAG, AGGG, TTA, GGG, and GG, respectively, are present at the break sites, in frame with the telomeric inserts. Because the sequence in Figure 2F is not informative in this respect, we can conclude that, in six out of the 10 informative sequences, the telomeric arrays were inserted in frame with telomeric oligonucleotides present at the break site in the ancestral sequences. This observation supports the direct involvement of telomerase, rather than the addition of a preformed telomeric array, in the repair process.
The proposed occasional involvement of telomerase in the repair of double-strand breaks is not surprising; in fact, several reports have shown that factors are shared between the telomere maintenance and double-strand break repair machineries (Bailey et al. 1999; Martin et al. 1999; Wei et al. 2002). It is, therefore, conceivable that telomerase, which plays an essential role in telomere maintenance and is present in germ-line cells where these events must have occurred, may be recruited at some repair sites. As recalled above, the double-strand break repair process via NHEJ is carried out by a multiplicity of proteins, which are geared for recognizing the break, maintaining the ends in close contact, processing, and finally joining them. One might surmise that telomerase molecules are occasionally recruited and contribute to the repair.
Because the telomeric arrays consist of tandem repetitions of short oligonucleotides, they can be classified as microsatellites; it has been shown that microsatellites composed of di-, tri-, or tetranucleotide units appeared during evolution through the creation of a minimum number of repeat units by mutation followed by expansion of the repeat number, probably via replication errors caused by DNA polymerase slippage. This process appears to be very slow on the evolutionary timescale (Messier et al. 1996). As the evolutionary evidence presented here shows, this mechanism does not apply to ITSs: the telomeric arrays appear in one step, inserted in a preexisting and well-conserved unrelated sequence. Hence, the mechanism for the creation of ITSs is clearly distinct from the classical mechanism of origin of the other microsatellites. Although we cannot exclude that some ITSs were generated by DNA polymerase slippage, the relatively low frequency of non-TTAGGG hexameric arrays we have observed in the human genome (Azzalin et al. 2001) suggests that the generation of hexanucleotide repeats by the classical mechanism is a rare event. On the other hand, once the TTAGGG arrays are inserted, their repeat number may be expanded, probably by the DNA polymerase slippage mechanism; in fact, in a previous study on three human ITS loci, we have shown that the number of telomeric repeats is variable in the human population (Mondello et al. 2000). In the present study, because only a single sample of DNA from each species was available to us, we do not know if intraspecific variation in repeat number is present at the studied loci; however, it is likely that the observed interspecific variability of repeat number at several loci is caused by DNA polymerase slippage.
The results here presented allow an evolutionary dating of several ITS insertion events (Fig. 4). Some insertions are very recent (<6 Mya), the ITS being present only in the human species (Figs. 1B and 2C,D) or in the two chimpanzee species (Fig. 2G); two insertions occurred after the divergence of PPY from the Homo/Pan/Gorilla group (7-14 Mya; Figs. 1A and 3A); two insertions occurred during the evolution of Catarrhini and took place in the great ape/gibbon ancestor (18-25 Mya; Figs. 2A and 3B); two ITSs are present in all the Catarrhini species analyzed and therefore were inserted 25-40 Mya (Fig. 2B,F). The ITS shown in Figure 2E is present in all the primates tested and is therefore very ancient; the same conclusion applies to six other ITSs that are not presented here because the sequence results do not provide information on the mechanism generating the insertion. In addition, the data presented in Figure 3B allowed us to date the insertion of an AluY transposable element in the primate lineage ∼18-25 Mya, before the Hominidae radiation.
From the results shown here, we cannot infer any particular role of ITS insertion in the evolutionary process. Most ITSs may simply represent junk DNA without any specific function; however, some of them could have been inserted near or within transcribed regions modifying the gene expression profile. In addition, as suggested for other microsatellites, the presence of many identical repeats could promote illegitimate recombination events leading to genome rearrangements.
Finally, it is worth reminding that the insertions of telomeric repeats here described necessarily occurred in the germ line in the course of primate evolution. ITS insertion at the repair junctions in experimentally induced double-strand breaks in several telomerase positive somatic cell lines has never been reported. Thus, either this type of event cannot occur for unknown reasons in somatic cells, or it is too rare to be detected in the experimental systems so far used.
In conclusion, the data presented here confirm our starting hypothesis that several human ITSs derive from the repair of double-strand breaks during evolution; the evidence presented does not allow us to ascribe any particular function to the ITSs present in the primate genomes; rather, the intrachromosomal telomeric arrays could simply represent “scars” marking sites of DNA double-strand break repair events that occurred in the germ line during evolution.
It has been suggested that ITSs are preferential sites of breakage (Bertoni et al. 1994, 1996; Desmaze et al. 1999); the conclusions derived from the present work support instead the hypothesis that the short primate ITSs are relics of ancient breakage within fragile sites rather than fragile sites themselves.
METHODS
Source of DNA
Human genomic DNA was extracted from blood samples of healthy individuals. Nonhuman primate genomic DNA was extracted from lymphoblastoid cell lines established from the following species: PPA, PTR, GGO, PPY, HLA, HKL, PAN, PCR, MMU, CAE, CJA, and CMO.
Polymerase Chain Reaction Amplification
Genomic DNA (50-100 ng) was amplified by polymerase chain reaction in a 25-μL total volume as previously described (Azzalin et al. 2001). Sequences of oligonucleotide primers are reported in Supplemental material. Reaction profiles are given in comments to appropriate sequences deposited to GenBank. Reaction products were analyzed by electrophoresis on 1%-3% agarose gel, and specific fragments were gel extracted.
Sequence Generation and Analysis
In most cases, gel-purified amplification products were directly sequenced with both the forward and the reverse amplification primers. Alternatively, gel-purified fragments were cloned in the pGEM-T-easy vector (Promega) and sequenced with the M13 forward and reverse primers. Sequencing was carried out using the ABI PRISM BigDye Terminator Cycle Sequencing Kit and an ABI PRISM 310 Genetic Analyzer automated sequencer (Perkin Elmer) according to the manufacturer's instructions.
To prove specificity of amplified fragments from nonhuman primates, their DNA sequences were compared with the corresponding human sequences. The RepeatMasker software at EMBL (http://woody.embl-heidelberg.de/repeatmask/) was used to identify known repetitive elements (such as SINEs, LINEs, microsatellites, etc.). Sequences were aligned using the Multiple sequence Alignment software, MultAlin (http://prodes.toulouse.inra.fr/multalin/multalin.html).
Acknowledgments
We thank Livia Bertoni and Alberto Salzano for helpful suggestions during the preparation of the manuscript. This work was funded by grants from Ministero dell'Università e della Ricerca Scientifica e Tecnologica (Cofinanziamento 2002 and Cluster C03, Prog. L.488/92), from the European Commission (TELOSENS, FIGH-CT 2002-00217 and INPRIMAT, QLRI-CT-2002-01325), and from CEGBA (Centro di Eccellenza Geni in campo Biosanitario e Agroalimentare).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
[Supplemental material is available online at www.genome.org. The nucleotide sequences reported in this work have been submitted to GenBank under accession nos. AY512975-AY513061 and AY585870-AY5858575.]
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.2778904. Article published online ahead of print in August 2004.
References
- Azzalin, C.M., Mucciolo, E., Bertoni, L., and Giulotto, E. 1997. Fluorescence in situ hybridization with a synthetic (T2AG3)n polynucleotide detects several intrachromosomal telomere-like repeats on human chromosomes. Cytogenet. Cell Genet. 78: 112-115. [DOI] [PubMed] [Google Scholar]
- Azzalin, C., Nergadze, S., and Giulotto, E. 2001. Human intrachromosomal telomeric-like repeats: Sequence organization and mechanisms of origin. Chromosoma 110: 75-82. [DOI] [PubMed] [Google Scholar]
- Bailey, S.M., Meyne, J., Chen, D.J., Kurimasa, A., Li, G.C., Lehnert, B.E., and Goodwin, E.H. 1999. DNA double-strand break repair proteins are required to cap the ends of mammalian chromosomes. Proc. Natl. Acad. Sci. 96: 14899-14904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertoni, L., Attolini, C., Tessera, L., Mucciolo, E., and Giulotto, E. 1994. Telomeric and non-telomeric (TTAGGG)n sequences in gene amplification and chromosome stability. Genomics 24: 53-62. [DOI] [PubMed] [Google Scholar]
- Bertoni, L., Attolini, C., Faravelli, M., Simi, S., and Giulotto, E. 1996. Intrachromosomal telomere-like DNA sequences in Chinese hamster. Mamm. Genome 7: 853-855. [DOI] [PubMed] [Google Scholar]
- Blackburn, E.H. 2001. Switching and signaling at the telomere. Cell 106: 661-673. [DOI] [PubMed] [Google Scholar]
- Cech, T.R. 2004. Beginning to understand the end of the chromosome. Cell 116: 273-279. [DOI] [PubMed] [Google Scholar]
- Chan, S.W. and Blackburn, E.H. 2002. New ways not to make ends meet: Telomerase, DNA damage proteins and heterochromatin. Oncogene 21: 553-563. [DOI] [PubMed] [Google Scholar]
- de Lange, T. 2002. Protection of mammalian telomeres. Oncogene 21: 532-540. [DOI] [PubMed] [Google Scholar]
- Desmaze, C., Alberti, C., Martins, L., Pottier, G., Sprung, C.N., Murnane, J.P., and Sabatier, L. 1999. The influence of intertitial telomeric sequences on chromosome instability in human cells. Cytogenet. Cell Genet. 86: 288-295. [DOI] [PubMed] [Google Scholar]
- Faravelli, M., Azzalin, C.M., Bertoni, L., Chernova, O., Attolini, C., Mondello, C., and Giulotto E. 2002. Molecular organization of internal telomeric sequences in Chinese hamster chromosomes. Gene 283: 11-16. [DOI] [PubMed] [Google Scholar]
- Flint, J., Craddock, C.F., Villegas, A., Bentley, D.P., Williams, H.J., Galanello, R., Cao, A., Wood, W.G., Ayyub, H., and Higgs, D.R. 1994. Healing of broken human chromosomes by the addition of telomeric repeats. Am. J. Hum. Genet. 55: 505-512. [PMC free article] [PubMed] [Google Scholar]
- Goodman, M. 1999. The genomic record of humankind's evolutionary roots. Am. J. Hum. Genet. 64: 31-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greider, C.W. 1996. Telomere length regulation. Annu. Rev. Biochem. 65: 337-365. [DOI] [PubMed] [Google Scholar]
- Harrington, L. 2003. Biochemical aspects of telomerase function. Cancer Lett. 194: 139-154. [DOI] [PubMed] [Google Scholar]
- Ijdo, J.W., Baldini, A., Ward, D.C., Reeders, S.T., and Wells, R.A. 1991. Origin of human chromosome 2: An ancestral telomere-telomere fusion. Proc. Natl. Acad. Sci. 88: 9051-9055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang, F., Han, M., Romanienko, P.J., and Jasin, M., 1998. Homology-directed repair is a major double-strand break repair pathway in mammalian cells. Proc. Natl. Acad. Sci. 28: 5172-5177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, Y. and Waldman, A.S. 2001. Promiscuous patching of broken chromosomes in mammalian cells with extrachromosomal DNA. Nucleic Acids Res. 29: 3975-3981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin, S.G., Laroche, N.S., Grunstein, M., and Gasser, S.M. 1999. Relocalization of telomeric Ku and SIR proteins in response to DNA strand breaks in yeast. Cell 97: 621-633. [DOI] [PubMed] [Google Scholar]
- McEachern, M.J., Krauskopf, A., and Blackburn, E.H. 2000. Telomeres and their control. Annu. Rev. Genet. 34: 331-358. [DOI] [PubMed] [Google Scholar]
- Messier, W., Li, S.H., and Steward C.B. 1996. The birth of microsatellites. Nature 381: 483. [DOI] [PubMed] [Google Scholar]
- Meyne, J., Baker, R.J., Hobart, H.H., Hsu, T.C., Ryder, O.A., Ward, O.G., Wiley, J.E., Wurster-Hill, D.H., Yates, T.L., and Moyzis, R.K. 1990. Distribution of non-telomeric sites of the (TTAGGG)n telomeric sequence in vertebrate chromosomes. Chromosoma 99: 3-10. [DOI] [PubMed] [Google Scholar]
- Mondello, C., Pirzio, L., Azzalin, C.M., and Giulotto, E. 2000. Instability of interstitial telomeric sequences in the human genome. Genomics 68: 111-117. [DOI] [PubMed] [Google Scholar]
- Roth, D.B., Proctor, G.N., Stewart, L.K., and Wilson, J.H. 1991. Oligonucleotide capture during end joining in mammalian cells. Nucleic Acids Res. 19: 7201-7205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Herrera, A., Garcia, F., Azzalin, C., Giulotto, E., Egozcue, J., Ponsa, M., and Garcia, M. 2002. Distribution of intrachromosomal telomeric sequences (ITS) on Macaca fascicularis (Primates) chromosomes and their implication for chromosome evolution. Hum. Genet. 110: 578-586. [DOI] [PubMed] [Google Scholar]
- Ruiz-Herrera, A., García, F., Giulotto, E., Attolini, C., Egozcue, J., Ponsa, M., and Garcia, M. 2004. Evolutionary breakpoints are co-localized with fragile sites and intrachromosomal telomeric sequences in Primates. Cytogenet. Genome Res. (in press). [DOI] [PubMed]
- Smith, J., Baldeyron, C., De Oliveira, I., Sala-Trepat, M., and Papadopoulo, D. 2001. The influence of DNA double-strand break structure on end-joining in human cells. Nucleic Acids Res. 29: 4783-4792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprung, C.N., Reynolds, G.E., Jasin, M., and Murnane, J. 1999. Chromosome healing in mouse embryonic stem cells. Proc. Natl. Acad. Sci. 96: 6781-6786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valerie, K. and Povirk, L.F. 2003. Regulation and mechanisms of mammalian double-strand break repair. Oncogene 22: 5792-5812. [DOI] [PubMed] [Google Scholar]
- Wei, C., Skopp, R., Takata, M., Takeda, S., and Price, C.M. 2002. Effects of double-strand break repair proteins on vertebrate telomere structure. Nucleic Acids Res. 30: 2862-2870. [DOI] [PMC free article] [PubMed] [Google Scholar]
WEB SITE REFERENCES
- http://prodes.toulouse.inra.fr/multalin/multalin.html; MultAlin, the Multiple sequence Alignment tool.
- http://woody.embl-heidelberg.de/repeatmask/; RepeatMasker.