Abstract
Many CpG islands have tissue-dependent and differentially methylated regions (T-DMRs) in normal cells and tissues. To elucidate how DNA methyltransferases (Dnmts) participate in methylation of the genomic components, we investigated the genome-wide DNA methylation pattern of the T-DMRs with Dnmt1-, Dnmt3a-, and/or Dnmt3b-deficient ES cells by restriction landmark genomic scanning (RLGS). Approximately 1300 spots were detected in wild-type ES cells. In Dnmt1-/- ES cells, additional 236 spots emerged, indicating that the corresponding loci are methylated by Dnmt1 in wild-type ES cells. Intriguingly, in Dnmt3a-/-Dnmt3b-/- ES cells, the same 236 spots also emerged, and no additional spots appeared differentially. Therefore, Dnmt1 and Dnmt3a/3b share targets in CpG islands. Cloning and virtual image RLGS revealed that 81% of the RLGS spots were associated with genes, and 62% of the loci were in CpG islands. By contrast to the previous reports that demethylation at repeated sequences was severe in Dnmt1-/- cells compared with Dnmt3a-/-Dnmt3b-/- cells, a complete loss of methylation was observed at RLGS loci in Dnmt3a-/-Dnmt3b-/- cells, whereas methylation levels only decreased to 16% to 48% in the Dnmt1-/- cells. We concluded that there are CpG islands with T-DMR as targets shared by Dnmt1 and Dnmt3a/3b and that each Dnmt has target preferences depending on the genomic components.
It is estimated that the human and mouse genomes consist of ∼3 × 109 bp/haploid genome but contain only 30,000 to 40,000 genes. A large amount of the genome is composed of nongenic repetitive elements (∼41% and 48% in human and mouse, respectively), including interspersed repeats and satellites (Lander et al. 2001; Venter et al. 2001; Waterston et al. 2002). Most CpGs are methylated in the mammalian genome; the value varies between 60% and 90% depending on the report (Gruenbaum et al. 1981; Razin et al. 1984; Cross and Bird 1995). It is likely that overall levels of DNA methylation reflect the hypermethylation status of nongenic repeated sequences rather than gene encoding areas.
CpG islands are genomic regions with high GC content and frequent CpG appearance compared with the entire genome (Gardiner-Garden and Frommer 1987) and are often located in promoter regions in the 5′ flanking region of housekeeping genes and many tissue-specific genes (Bird 1987; Gardiner-Garden and Frommer 1987; Larsen et al. 1992). The numbers of housekeeping genes and tissue-specific genes in the human and mouse were calculated by Antequera and Bird (1993). The human and mouse genome projects identified ∼15,500 and 29,000 CpG islands in the mouse and human genomes, respectively (Lander et al. 2001; Venter et al. 2001; Waterston et al. 2002), and a recent database analysis suggested that approximately half of the tissue-specific promoters are linked with CpG islands (Suzuki et al. 2001). Although most CpG dinucleotides in the mammalian genome have been found to be methylated at the cytosine residue (Cooper and Krawczak 1989), CpG islands were once considered to be unmethylated regions in normal tissue. However, there are numerous tissue-dependent and differentially methylated regions (TDMRs) in CpG islands in normal cells or tissues, and the methylation profile of the CpG islands is unique in each tissue or cell type (Shiota et al. 2002).
Information is still limited concerning DNA methylation in gene coding regions, although aberrant DNA methylation in these regions is known to occur in cancers (Jones and Baylin 2002). It is still obscure how DNA methylation, which is regulated by DNA methyltransferases (Dnmts), occurs at T-DMRs of CpG islands. To date, three classes of Dnmts—Dnmt1, Dnmt2, and Dnmt3—have been identified in mammals. Methyltransferases Dnmt1 and Dnmt3a/3b are found to be functional both in vitro and in vivo. In vitro studies indicated that Dnmt1 prefers hemimethylated DNA compared to unmethylated DNA (Bestor 1992; Yoder et al. 1997; Pradhan et al. 1999), whereas Dnmt3a/3b methylate CpG dinucleotides without preference for hemimethylated or unmethylated DNA (Okano et al. 1998). Collectively, the fact that Dnmt1 is localized to DNA replication foci (Leonhardt et al. 1992) and is associated with MeCP2, which directs DNA methyltransferase activity to hemimethylated DNA (Kimura and Shiota 2003), implies that Dnmt1 is involved in maintenance methylation in vivo to preserve methylation patterns in genomic DNA, and that Dnmt3a/3b function as de novo methyltransferases. Studies using Dnmt1-/- and Dnmt3a-/-Dnmt3b-/- ES cells demonstrated that demethylation occurred at repeated sequences, such as endogenous C-type retroviruses, more extensively in Dnmt1-/- cells than in Dnmt3a-/-Dnmt3b-/- cells, and that some imprinted genes such as Igf2r and H19 became demethylated only in Dnmt1-/- cells but not in Dnmt3a-/-Dnmt3b-/- cells (Okano et al. 1999). Recently, a direct association between DNMTs was reported (Kim et al. 2002), and it was demonstrated that DNMT1 and DNMT3B function cooperatively for maintenance methylation in the human cancer cell line (Rhee et al. 2002). Therefore, categorizing Dnmts into maintenance and de novo DNA methylation may not be appropriate when attempting to characterize in vivo mechanisms involved in determining or establishing DNA methylation profiles in the genome.
Dnmts are of critical importance for development (Li et al. 1992; Okano et al. 1999). Formation of DNA methylation profiles underlies mammalian development (Shiota and Yanagimachi 2002), and abnormal methylation status induces various abnormal phenotypes (Ohgane et al. 2001). Systematic survey of DNA methylation patterns regarding CpG islands is one of the most interesting ways to study DNA methylation from the aspect of epigenetic participation in the differentiation and development of mammals. To address the question how DNA methylation in the T-DMR of CpG islands is regulated by Dnmts in vivo, the genome-wide DNA methylation in Dnmt1-, Dnmt3a-, and/or Dnmt3b-deficient ES cells was assessed by restriction landmark genomic scanning (RLGS). We used NotI as a landmark enzyme to focus on CpG islands, because >90% of NotI recognition sites are located in CpG islands (Lindsay and Bird 1987).
RESULTS
Genome-Wide Demethylation of Gene Areas Associated With CpG Islands in Dnmt-Deficient ES Cells
In the RLGS profile, the spot is visible if the corresponding NotI site in the genome is unmethylated or hypomethylated, whereas it is invisible if the site is hypermethylated. We have observed that the methylation status of the NotI site reflects those of the surrounding CpGs in several cases (Imamura et al. 2001; Ohgane et al. 2001). There were ∼1300 spots when wild-type ES cells were assessed (data not shown), suggesting that these 1300 genomic sites are hypomethylated in ES cells despite the expression of all types of Dnmts (Chen et al. 2003). Considering our previous observation that there are both methylated and unmethylated loci in cells and tissues and that the methylation pattern at such loci is unique depending on the cell type (Shiota et al. 2002), there should be methylated and undetectable loci in addition to the unmethylated 1300 loci in wild-type ES cells. If so, Dnmts responsible for DNA methylation at NotI sites, probably in CpG islands, can be identified by comparing the RLGS profile of wild-type cells with those of Dnmt-deficient cells.
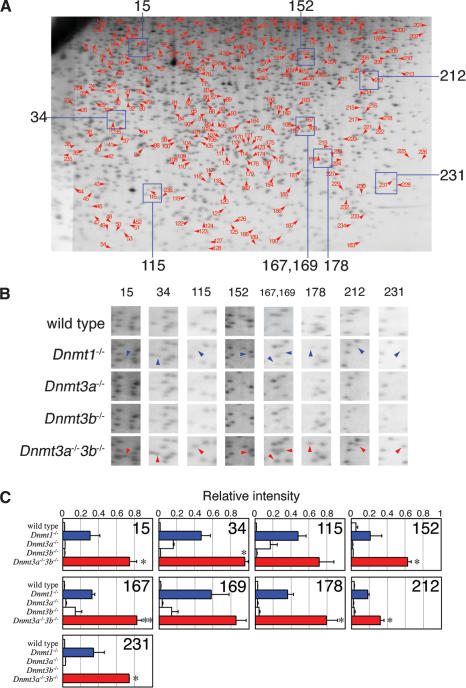
Two hundred thirty-six spots emerged in the RLGS profile of Dnmt1-/- ES cells in addition to the basal 1300 spots in wild-type cells (Fig. 1A). This implies that there are many loci that are methylated by Dnmt1 in ES cells, considering the fact that ≥15,000 CpG islands exist in the mouse genome (Waterston et al. 2002). The intensities of these emerging spots in Dnmt1-/- cells were considerably less than those of the unchanged authentic spots (Fig. 1B,C). This suggests that demethylation of T-DMR caused by Dnmt1 deficiency was not complete. Intriguingly, the identical 236 spots found in Dnmt1-/- ES cells also emerged in the RLGS profile of the double-mutant (Dnmt3a-/-Dnmt3b-/-) ES cells, suggesting that all spots emerging in the RLGS profiles of Dnmt1-/- and Dnmt3a-/-Dnmt3b-/- ES cells should be common targets of both Dnmt1 and Dnmt3a/3b. There is no RLGS spot that differentially appeared or disappeared between Dnmt1-/- and Dnmt3a-/-Dnmt3b-/- cells. Therefore, DNA methylation targets were completely overlapped and shared by Dnmt1 and Dnmt3a/3b. However, there might be some spots that differentially increased their intensities in Dnmt1-/- and Dnmt3a-/-Dnmt3b-/- cells. It is also likely that some spots were visible in the wild-type ES cells but increased their spot intensities in Dnmt1-/- and Dnmt3a-/-Dnmt3b-/- cells. More importantly, the intensities of the 236 spots were greater in Dnmt3a-/-Dnmt3b-/- ES cells than Dnmt1-/- cells (Fig. 1C). These results suggest that the levels of demethylation are more extensive in Dnmt3a-/-Dnmt3b-/- cells than in Dnmt1-/- cells.
Figure 1.
DNA methylation status of NotI sites spread throughout the genome in wild-type and Dnmt-deficient ES cells. (A) RLGS profile of Dnmt3a-/-Dnmt3b-/- ES cells is shown as a representative; 236 spots indicated by arrowhead with number were invisible in the wild-type, Dnmt3a-/-, and Dnmt3b-/- single-mutant ES cells but emerged in Dnmt1-/- and Dnmt3a-/-Dnmt3b-/- ES cells. Corresponding genomic loci of these spots were hypomethylated by the loss of Dnmt1 or Dnmt3a/3b. (B) Enlarged RLGS spots of wild-type and Dnmt-deficient ES cells. Areas with indicated numbers are represented in A. Spots with arrowhead exist in the profiles in Dnmt1-/- and Dnmt3a-/-Dnmt3b-/- ES cells but are invisible in those of wild-type, Dnmt3a-/-, and Dnmt3b-/- ES cells. Note that intensity of all spots in Dnmt3a-/-Dnmt3b-/- ES cells (red arrowheads) is greater than those in Dnmt1-/- (blue arrowheads). (C) Relative intensities of the representative RLGS spots. The intensity of each spot was digitized and averaged from three independent RLGS profiles and normalized by the average intensity of the surrounding invariant spots. Differences between Dnmt1-/- and Dnmt3a-/-Dnmt3b-/- ES cells were compared by t test. *P < 0.1 **P < 0.01 (n = 3)
Each single mutant (Dnmt3a-/- or Dnmt3b-/-) ES cell line showed a similar RLGS profile to that of wild-type cells; thus the deficiency of Dnmt3a or Dnmt3b alone seemed to result in no change in DNA methylation at CpG islands (Fig. 1B,C). Therefore, the roles of Dnmt3a and Dnmt3b appear to be redundant, and a compensatory mechanism between Dnmt3a and Dnmt3b exists regarding DNA methylation at least at the genomic sites corresponding to these 236 spots.
Loss of Methylation at the Flanking CpGs of NotI Sites Was Severe in the Double Mutant of Dnmt3a and Dnmt3b Compared With That in Dnmt1 Mutant Cells
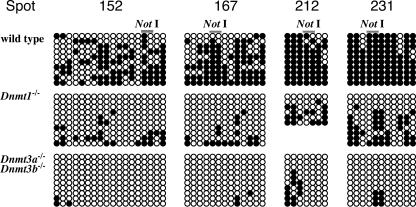
We further investigated the methylation status of the flanking regions up to -600 bp, including the NotI sites by sodium bisulfite genomic sequencing at four genomic loci (Fig. 2). There are 18, 13, seven, and 11 CpGs within 588-, 504-, 490-, and 486-bp genomic regions corresponding to spots 152, 167, 212, and 231, respectively. In the wild-type ES cells, the NotI sites in all regions were almost completely methylated as seen in the RLGS experiment, and overall, the regions were hypermethylated. Methylation levels at the NotI sites vary 30% to 60% in Dnmt1-/- ES cells, whereas they are 0% to 30% in Dnmt3a-/-Dnmt3b-/- cells. Furthermore, demethylation in Dnmt3a-/-Dnmt3b-/- ES cells was more extensive than Dnmt1-/- cells in the flanking CpGs of the NotI site. These results support the notion that DNA methylation in the regions, including the NotI site, depends on Dnmt3a/3b more significantly than Dnmt1. In the present study, however, we used Dnmt1-/- ES cells collected after 36 passages in culture, whereas the mutant cell lines of Dnmt3a-/-, Dnmt3b-/-, and Dnmt3a-/-Dnmt3b-/- were collected after 16, 19, and 19 passages, respectively. Taking the fact that culturing time could affect DNA methylation levels, it is also possible that the lower demethylation levels observed in Dnmt1-/- ES cells might be due to remethylation activity of Dnmt3a and/or Dnmt3b, which are still present and active in Dnmt1-/- ES cells.
Figure 2.
DNA methylation status at CpG sites around the NotI sites. Methylation status around ∼600-bp genomic region, including the NotI site, was assessed by sodium bisulfite sequencing method. Open circles and closed circles represent unmethylated and methylated cytosine residues, respectively. In wild-type ES cells, all genomic regions are hypermethylated, including the NotI sites, indicating that DNA methylation is maintained at these regions in the wild-type ES cells. In contrast, CpG sites in the same regions are demethylated in Dnmt1-/- and Dnmt3a-/-Dnmt3b-/- ES cells. Although the demethylation is moderate in Dnmt1-/- ES cells, almost all CpG sites are totally demethylated in Dnmt3a-/-Dnmt3b-/- ES cells, indicating that RLGS analysis reflects DNA methylation status not only at the NotI site but surrounding CpG dinucleotides of the NotI site. These data suggest that Dnmt3a/3b are more significant for DNA methylation in CpG islands than Dnmt1.
Identification of the Target for DNA Methylation in CpG Island
To identify the sequence information for the methylation target in CpG islands, we applied virtual image RLGS (Vi-RLGS), which is the recently developed in silico method for analyzing database sequences (Matsuyama et al. 2003). By matching the RLGS profile with the Vi-RLGS image, we selected the candidate loci for the RLGS spots. Of the total 236 candidate spots, ∼54.2% of the candidate spots are in CpG islands, and ∼89.4% of the spots are in the gene coding region (Table 1).
Table 1.
Summary of 236 Loci Predicted by Vi-RLGS
| Genes
|
Nongenic sequences
|
|||||||
|---|---|---|---|---|---|---|---|---|
| Function knowna | Function unknownb | Predictedc | Repeat sequencesd | Nonrepeat sequences | Total | NDe | ||
| CpG island | 69 (29.2%) | 46 (19.5%) | 8 (3.4%) | 2 (0.8%) | 3 (1.3%) | 128 (54.2%) | ||
| Non-CpG island | 37 (15.7%) | 24 (10.2%) | 27 (11.4%) | 11 (4.7%) | 6 (2.5%) | 105 (44.5%) | ||
| Total | 211 (89.4%) | 22 (9.3%) | 3 (1.3%) | |||||
Genes that encode proteins with known functions.
Including predicted genes supported by mRNA expression and/or ESTs.
Genes predicted by computer software such as Genescan.
Sequences including LINEs, SINEs, LTR elements or DNA elements.
Not determined.
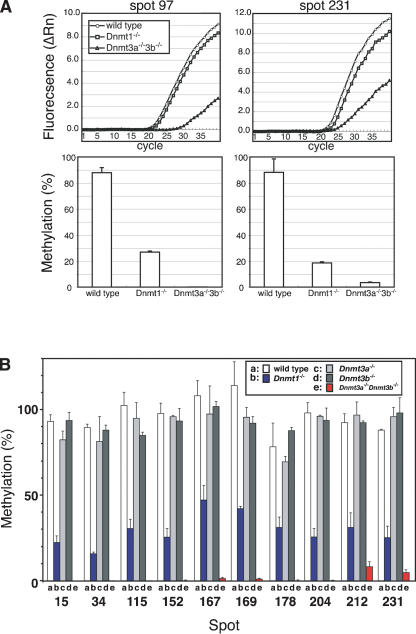
We initially confirmed four spots (spot 12, 15, 18, and 235), which are demethylated in Dnmt1 - / - and Dnmt3a-/-Dnmt3b-/- ES cells by spot cloning with the electroelution method. To confirm the methylation status of more candidate genomic loci in Vi-RLGS image, we used the method based on the methylation-sensitive restriction digestion and quantitative PCR. PCR amplification of genomic DNA following NotI digestion was dependent on the methylation levels revealed by RLGS (Fig. 3A). Methylation levels at selected loci in Dnmt1-/- ES cells were estimated as 16.1% to 47.5% (average, 41.4 ± 3.2%) and those in Dnmt3a-/-Dnmt3b-/- ES cells were evaluated as 0% to 8.5% (average, 5.3 ± 1.0%), whereas the methylation levels in the wild-type, Dnmt3a-/-, and Dnmt3b-/- ES cells were ∼100% (Fig. 3B). These data indicate that the genomic locus of each spot can be identified with the combination of Vi-RLGS and the quantitative PCR. We identified 40 genomic loci, including the above four that are methylated in the wild-type ES cells but demethylated in Dnmt1-/- or Dnmt3a-/-Dnmt3b-/- ES cells (Table 2). Among these identified loci, 59.0% (23/39) are located in CpG islands, and more importantly, 84.6% (33/39) are in the gene coding regions. Thus, we confirmed that most of the NotI sites reside in the gene coding regions. As far as data obtained in the present study, no imprinted gene was found in Dnmt-deficient ES cells, probably because we focused on spots that did not exist in the wild-type ES cells but emerged in Dnmt-deficient cells.
Figure 3.
Evaluation of DNA methylation by methylation-sensitive quantitative PCR. (A) The amount of undigested genomic DNA after NotI treatment was estimated by real-time PCR using genomic DNA of wild-type, Dnmt1-/-, and Dnmt3a-/-Dnmt3b-/- ES cells. Spots 97 and 231 are chosen as examples. (Top panels) Amplification plots of PCR products from each NotI-treated genomic DNA sample. (Bottom panels) Methylation levels at each corresponding genomic site, which are calculated from the ratio of amounts of NotI treated/untreated genomic DNA. For details, see Methods. (B) Evaluation of methylation levels at the corresponding genomic loci represented in Fig. 1B in the wild-type, Dnmt1-/-, Dnmt3a-/-, Dnmt3b-/-, and Dnmt3a-/-Dnmt3b-/- ES cells. In the wild-type, Dnmt3a-/-, and Dnmt3b-/- ES cells, all NotI sites were hypermethylated and amplified by real-time PCR at the same levels as NotI-untreated genomic DNA. In contrast, NotI sites were only methylated from 16.1% to 47.5% in Dnmt1-/- ES cells and from 0% to 8.5% in Dnmt3a-/-Dnmt3b-/- ES cells.
Table 2.
Target Loci of Dnmts Detected in RLGS Confirmed by Vi-RLGS and the Methylation-Sensitive Real-Time PCR Method
| Spot no.a | Locus | Gene symbolb | Gene name | CpG island |
|---|---|---|---|---|
| 12 | 15B2 | repeat (LTR element) | — | yes |
| 15 | 5G2 | D130017N08Rik | RIKEN cDNA D130017N08 gene | yes |
| 17 | 11E2 | Cbx4 | chromobox homolog 4 | no |
| 18 | 8B1.1 | — | — | no |
| 21 | 12D1 | LOC238316 | predicted gene | yes |
| 25 | 10B4 | 4732470K04 | predicted gene | yes |
| 30 | 13A5 | LOC238623 | predicted gene | yes |
| 31 | 7B3 | Dbx1 | developing brain homeobox 1 | yes |
| 32 | 9A5.1 | D130038B21 Rik | RIKEN cDNA D130038B21 gene | yes |
| 34 | 18C | AA987150 | predicted gene | yes |
| 43 | 2H3 | LOC329560 | predicted gene | yes |
| 47 | 19C3 | Trim8 | tripartite motif protein 8 | yes |
| 53 | 5B1 | BC037112 | gene with protein product | no |
| 73 | 9A4 | D730048106Rik | RIKEN cDNA D730048106 gene | yes |
| 76 | 2H3 | — | — | no |
| 77 | 11A4 | AL732390.10.1.195612.21298.172306 | predicted gene | no |
| 79 | 10C1 | Agpat3 | 1-acylglycerol-3-phosphate O-acyltransferase 3 | yes |
| 87 | 14D1 | Baiap1 | BAI1-associated protein 1 | no |
| 88 | 10B4 | Mm.338763 (unigene) | — | no |
| 93 | 7D1 | Bnc | basonuclin | yes |
| 95 | 14C1 | MGC38922 | predicted gene | yes |
| 97 | 7A3 | Rps19 | ribosomal protein S19 | yes |
| 99 | 17E3 | Cox7a21 | cytochrome c oxidase subunit VIIa polypeptide 2-like | yes |
| 109 | 13A5 | — | — | yes |
| 111 | 7F3 | hmm37743 | predicted gene | no |
| 115 | 7A2 | LOC381956 | predicted gene | yes |
| 144 | 13B1 | Tgfbi | transforming growth factor, beta induced | no |
| 152 | 19C3 | Pik3ap1 | phosphoinositide-3-kinase adaptor protein 1 | yes |
| 161 | 5B2 | Man2b2 | mannosidase 2, alpha B2 | yes |
| 166 | 5F | Cit | citron | no |
| 167 | 4D3 | LOC230868 | predicted gene | no |
| 173 | 11A4 | Odz2 | odd Oz/ten-m homolog 2 | no |
| 178 | 2H1 | Bfzb-pending | basic FGF repressed, zinc binding protein | no |
| 212 | 14D2 | repeat (DNA element) | — | yes |
| 225 | 11B1.3 | A830006N08 | predicted gene | yes |
| 231 | 16C2 | — | — | yes |
| 234 | 11B5 | — | — | no |
| 235 | 8E2 | — | — | no |
| 236 | 10C1 | Pcnt2 | pericentrin 2 | no |
Spot numbers indicate the position in RLGS profile shown in Fig. 1A.
Symbols are shown if the Notl site of the fragment is located within 10 kb of the gene.
DISCUSSION
Repetitive elements, including interspersed repeats such as endogenous retroviruses, occupy a large part of the mammalian genome, whereas the area for protein coding genes is very limited (Lander et al. 2001; Venter et al. 2001; Waterston et al. 2002). CpG islands were considered to be clusters of unmethylated CpG dinucleotides except for those in imprinted genes, X-chromosome inactivation, and aberrant methylation in cancer (Bird and Wolffe 1999; Jones and Takai 2001; Reik 2001). There are, however, several CpG island-associated genes that are regulated by DNA methylation in normal tissues (Imamura et al. 2001; Newell-Price et al. 2001; Pao et al. 2001; Futscher et al. 2002; Strichman-Almashanu et al. 2002). In the present study, there were 236 emerging RLGS spots in Dnmt1- or Dnmt3a/3b-deficient cells in addition to the basal 1300 unmethylated or hypomethylated spots. Cloning of the 236 spots confirmed that they are primarily in the gene encoding regions, and not repeated sequences. In addition to the authentic 247 T-DMRs that are differentially methylated in tissues and cells (Shiota et al. 2002), there are at least 236 T-DMRs that are methylated by Dnmts in ES cells as described below. These T-DMRs are methylated in ES cells but may be unmethylated in other types of cells that we have not explored. The number of T-DMRs in normal cells would be 483, if duplication were not taken into account.
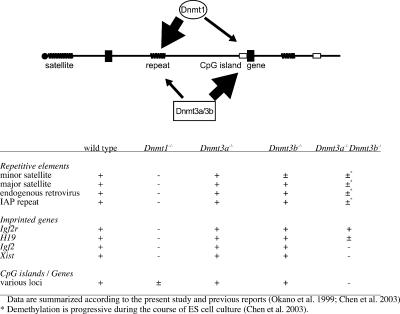
It has been widely accepted that Dnmt1 functions solely as the maintenance DNA methyltransferase, whereas Dnmt3a/3b are involved in de novo methylation. So far, however, maintenance activity by Dnmt1 has been investigated predominantly in repetitive elements such as centromeric/pericentromeric regions as well as broad intergenic regions containing various repetitive elements (Li et al. 1992; Okano et al. 1999). We demonstrated here that Dnmt3a or Dnmt3b is required for DNA methylation in T-DMR of CpG islands. This indicates that Dnmt3a/3b have importance both in maintenance and de novo methylation in CpG islands, at least in ES cells. Dnmt1 deficiency caused total loss of methylation at the endogenous repeated sequences (Li et al. 1992; Okano et al. 1999). The apparent contribution of Dnmt1 was incomplete and appeared to be less than that of Dnmt3a/Dnmt3b, although Dnmt1 was also involved in DNA methylation in the T-DMRs of CpG islands. Alternatively, both Dnmt1 and Dnmt3a/3b might be necessary and equally contribute to the maintenance of DNA methylation in CpG islands, whereas Dnmt3a and Dnmt3b also had the function of de novo DNA methyltransferases in the same regions. In comparing the overall DNA methylation functions of Dnmt1 and Dnmt3a/3b, Dnmt1 functions as a maintenance methyltransferase both in repeated sequences and CpG islands, whereas Dnmt3 functions more importantly in CpG islands than in other genomic regions (Fig. 4).
Figure 4.
Summary of DNA methylation at CpG islands and genes, or repetitive elements such as interspersed repeats and centromeric satellites. Although Dnmt1 and Dnmt3a/3b are involved in DNA methylation at both repeated sequences (zigzag lines) and genes (closed boxes) with/without CpG islands (open boxes), Dnmt1 and Dnmt3a/3b act in a different fashion; Dnmt1 functions as a maintenance methyltransferase both in repeated sequences and CpG islands, whereas Dnmt3 primarily functions in CpG islands. Lower table represents the methylation statuses at various genomic areas in the wild-type and Dnmt-deficient ES cells reported here or elsewhere (Okano et al. 1999; Chen et al. 2003).
Chen et al. (2003) reported that Dnmt3a-/-Dnmt3b-/- ES cells showed loss of DNA methylation in various repeated sequences and some single nonimprinted and imprinted genes. Kinetic analysis indicated that demethylation at repeated sequences in Dnmt1-/- ES cells was more rapid than that in Dnmt3a-/-Dnmt3b-/- ES cells, and that demethylation in Dnmt3a-/-Dnmt3b-/- ES cells was progressive and thus exhibited extensive demethylation at late stages of cell culture. They proposed a model for the distinctive roles of Dnmt1 and Dnmt3a/3b, in which Dnmt3 functions as a proofreader to complement the maintenance activity of Dnmt1. In contrast, in the component of the present study focusing on T-DMR of CpG islands, the complete loss of DNA methylation in Dnmt3a-/-Dnmt3b-/- ES cells was observed even at early stages of cell passage, a time when demethylation in the repeated sequences did not progress extensively. This result indicates that Dnmt3a-/-Dnmt3b-/- ES cells lost both activities of de novo and maintenance DNA methylation in CpG islands. The proofreading functions of Dnmt3a/3b may also be different depending on genomic area.
In contrast to Dnmt3a-/-Dnmt3b-/- ES cells, Dnmt1-/- ES cells had partially demethylated T-DMRs of CpG islands, although the repeated sequences were fully demethylated in Dnmt1-/- ES cells. DNA methylation in CpG islands and gene encoding regions in ES cells may depend predominantly on the activities of Dnmt3a/3b (Fig. 4). Alternatively, the maintenance activities of Dnmt3a/3b may be achieved by the remethylation mechanism of de novo activity in the absence of Dnmt1. The remethylation mechanisms or the proofreading functions (Chen et al. 2003) by Dnmt3a/3b may involve other epigenetic functions such as chromatin remodeling during the course of maintenance of DNA methylation in CpG islands or gene encoding regions. Chromatin configuration affects DNA methylation status and vice versa (Lachner and Jenuwein 2002; Li 2002). To determine the mechanism for maintaining DNA methylation patterns by Dnmt3a/3b, studies on the molecular link between histone modification and DNA methylation will be needed in the future.
The present study demonstrated that Dnmt3a/3b are involved in the maintenance of DNA methylation patterns at CpG islands. Interestingly, T-DMRs with loss of methylation were completely overlapped between Dnmt1- and Dnmt3a/Dnmt3b-deficient ES cells. Collaboration of Dnmts has been reported in some genomic regions. Cooperation of DNMT1 with DNMT3B for maintenance methylation in a human cancer cell line has been reported (Rhee et al. 2002). Although disruption of either DNMT1 or DNMT3B resulted in a minimal effect on DNA methylation in those cells, double knockout of these genes resulted in marked decrease of DNA methylation in Igf2, p16INK4a, and repeated sequences (Rhee et al. 2002). In this case, DNMT1 and DNMT3B complement each other in (or share) the maintenance activity of DNA methylation. Involvement of Dnmt3a/3b in maintenance methylation is suggested also by the genetic study at DMR2 region of Igf2, the 5′ region of Xist, and some repeated sequences (Okano et al. 1999). Dnmt1 and either Dnmt3a or Dnmt3b may cooperate with each other for maintenance of DNA methylation in CpG island in wild-type ES cells, based on the present data that demethylation occurred in CpG islands in both of Dnmt1-/- and Dnmt3a-/-Dnmt3b-/- ES cells. It was reported that DNMT1, DNMT3A, and DNMT3B made direct association with each other, and an enhancement of DNA methylation activity was observed when DNMT1 and DNMT3s interacted (Kim et al. 2002). Collectively, Dnmt1 and Dnmt3a/3b may associate directly and function as a complex for DNA methylation in CpG islands. Interaction among the Dnmts and the methyl CpG-binding domain proteins (MBDs), including MeCP2, should also be considered for the mechanism of maintaining DNA methylation patterns by using the proofreading or remethylation function. Furthermore, the mutual dependency on epigenetic memory systems such as DNA methylation and chromatin remodeling should be considered as well.
The present study suggests that Dnmts have preferences for specific genomic regions; Dnmt3a/3b prefer the T-DMR of CpG islands and genes compared with repeated sequences, whereas the nongenic regions including repeated sequences are less dependent on Dnmt3a/3b for maintenance. It is clear that there is some redundancy between the roles of Dnmt3a/3b and Dnmt1 regarding the methylation of gene encoding regions (Fig. 4). No mammalian Dnmts exhibited any sequence specificities in vitro, although there is a report of preferred nucleotide composition around the CpG site for Dnmt3a (Lin et al. 2002). It was demonstrated previously that disruption of Dnmt3a and Dnmt3b resulted in partial demethylation of endogenous C-type retroviruses, L1-like repeats, and major and minor satellites, whereas the lack of Dnmt1 activity caused almost complete demethylation in these sequences (Okano et al. 1999; Liang et al. 2002; Chen et al. 2003). In addition, it was demonstrated that the DMR2 of Igf2 and 5′ region of Xist gene were unmethylated both in Dnmt1-/- and Dnmt3a-/-Dnmt3b-/- ES cells, whereas region 2 of Igf2r and 5′ upstream region of H19 were unmethylated in Dnmt1-/- ES cells but not or partially demethylated in Dnmt3a-/-Dnmt3b-/- ES cells (Okano et al. 1999). The preference of Dnmts is different depending on the genomic region, providing novel insight into the mechanism of DNA methylation.
METHODS
ES Cells and Genomic DNA Preparation
Mutant ES cell lines deficient of Dnmt1-/- (c/c), Dnmt3a-/- (6aa), Dnmt3b-/- (8bb), and Dnmt3a-/-Dnmt3b-/- (7aabb) were generated in vitro by several rounds of homologous recombination (Li et al. 1992; Lei et al. 1996; Okano et al. 1999), and all ES cells were cultured as previously described (Lei et al. 1996; Okano et al. 1999). Lack of Dnmt protein expression in each mutant ES cell line was previously examined using immunoblotting analyses (Okano et al. 1999; Chen et al. 2003).
Proliferating and undifferentiated ES cells were harvested to isolate genomic DNA as reported previously (Ohgane et al. 1998; Imamura et al. 2001; Shiota et al. 2002). Genomic DNA was extracted when passage numbers of J1, c/c, 6aa, 8bb, and 7aabb were at 11, 36, 16, 19, and 19, respectively.
RLGS and Spot Identification by Cloning and Vi-RLGS
RLGS, which enables us to assess the methylation status of cells or tissues reproducibly (Shiota et al. 2002) was performed according to the previous report (Ohgane et al. 1998; Imamura et al. 2001) using the combination of restriction enzymes, NotI-PvuII-PstI. To block nonspecific labeling, genomic DNA was treated with Klenow fragment (TaKaRa) in the presence of dGTPαS, dCTPαS (Amersham Pharmacia), ddATP, and ddTTP (TaKaRa). DNA was digested with NotI as a landmark enzyme (Nippongene), and the resulting cohesive ends were labeled with Sequenase version 2.0 (USB) in the presence of [α-32P]dCTP and [α-32P]dGTP (Amersham Pharmacia), digested with Pvu II (Nippongene), and then subjected to first dimension electrophoresis in a 0.9% agarose disc gel for 23 h at 230 V. After DNA fragments were treated with PstI (Nippongene) in the disk gel, the resulting DNA fragments were separated in second dimensional 5% polyacrylamide gel for 20 h at 150 V. The gel was dried onto chromatography paper (Whatman) and exposed to an Imaging Screen for scanning with Molecular Imager FX (BioRad) or to X-ray film (Kodak XAR5, Eastman Kodak) at -80°C. The spot profiles were analyzed by a 2D/E analyzing system (PDquest, BioRad). For each cell preparation, three to 10 RLGS experiments were performed to evaluate whether RLGS profiles are reproducible.
To clone each RLGS spot, the mixture of radiolabeled and unlabeled samples was applied to RLGS as described above. DNA fragments were electroeluted from the resulting spots, which were punched out in advance, for 20 min at 200 V. Extracted DNA fragments were connected with a NotI adaptor (5′-ACGCCAGGGTTTTCCCAGTCACGACGC-3′ and 5′-p-GGCCGCGTCGTGACTGGGAAAACCCTGGCGT-3′) and PstI adaptor (5′-p-GTGTACTGCACCAGCAAATCC-3′ and 5′-GGATTTGCTGGTGCAGTACACTGCA-3′) followed by removal of excess adaptors with MicroSpin S-400 column (Amersham Pharmacia Biotech) and amplified in two rounds of PCR (30 cycles each) with primers for adaptors (for NotI adaptor: 5′-AGGGTTTTCCCAGTCACGACGCGG-3′, for PstI adaptor: 5′-TTGCTGGTGCAGTACACTGCAG-3′). Amplified fragments were digested with NotI and PstI, ligated into pBluescript II SK- (Stratagene), and then subjected to sequence analysis.
Vi-RLGS software, which processes any sequence data in GenBank or FASTA format and simulates two-dimensional electrophoresis using the resulting fragments, was developed and described elsewhere (Matsuyama et al. 2003). Mouse draft genome sequence (MGCSv3_release3) was downloaded in masked FASTA format from the GenBank ftp site (ftp://ftp.ncbi.nih.gov/genomes/M_musculus/) and processed with the combination of NotI-PvuII-PstI recognition sequences. By matching the Vi-RLGS and “real” RLGS profiles to identify candidate spots, the corresponding sequences were retrieved by clicking the spot on the virtual image and were used as queries for sequence analysis in BLAST (http://www.ncbi.nlm.nih.gov/genome/seq/MmBlast.html) and Ensemble (http://www.ensembl.org/Mus_musculus/) to obtain the surrounding sequence information, chromosomal position, and gene names. By using the sequence information, the primer and probes were designed for methylation sensitive quantitative real-time PCR described below. Identification of repetitive elements in spot data was performed using RepeatMasker (http://ftp.genome.washington.edu/cgi-bin/RepeatMasker).
Sodium Bisulfite Genomic Sequencing
Sodium bisulfite genomic sequencing was performed as previously described (Hattori et al. 2004). Briefly, genomic DNA was digested with PstI and denatured by adding 0.3 M NaOH and incubating for 15 min at 37°C. After the incubation, sodium metabisulfite (pH 5.0) and hydroquinone were added to final concentrations of 2.0 M and 0.5 mM, respectively, and the mixture was further incubated in the dark for 16 h at 55°C. The modified DNA was purified through the Wizard DNA Clean-Up system (Promega), and the bisulfite reaction was terminated with NaOH at final concentration of 0.3 M for 15 min at 37°C. The solution was then neutralized by adding NH4OAc (pH7.0) to a final concentration of 3 M. The ethanol-precipitated DNA was resuspended in water and then amplified by PCR using primers were designed as follows: spot 152, 5′-GGTGAGTTTTTTTGGATT TAATAAT-3′ and 5′-AACACTAAACACCTAATTTATATCTA-3′; spot 167, 5′-GGTGGTTTTAGAGATTGATATTT-3′ and 5′-CCCTTCATCTACTTCTCTACAA-3′; spot 212, 5′-TTTATAG GAGTTTAGTTGGTTTGTT-3′ and 5′-AAAACAATAATTCTCAAC CCATA-3′; and spot 231, 5′-GTTTGATTTTATATTAAAGTATTGG-3′ and 5′-TCACATCAATTTTAACCTCTAAATA-3′. The amplified PCR fragments were cloned into pGEM T-easy vector (Promega) and sequenced for each sample.
Methylation Analysis Based on the Real-Time PCR
Methylation status at specific loci detected by RLGS was evaluated by using the combination of the methylation-sensitive restriction digestion and quantitative real-time PCR (Heid et al. 1996). Genomic DNA was digested by PstI, and the aliquot was treated subsequently with NotI. Each primer set for PCR was designed to amplify the region that included the NotI site detected in RLGS analysis. Forty nanograms of genomic DNA treated with or without NotI were analyzed by real-time PCR with the primers. The amount of undigested DNA both in NotI-treated and -untreated genomic DNA was estimated by real-time PCR with TaqMan Universal PCR Master Mix by using ABI Prism 7000 Sequence Detection System (Applied Biosystems) according to the manufacturer's protocol. The methylation ratio at each RLGS locus was defined as the proportion of the amount of undigested DNA in NotI-treated genome to that in the NotI-untreated one. Initial DNA amount in the reaction mix was normalized with TaqMan Rodent GAPDH Control Reagents VIC Probe (Applied Biosystems). For all samples, at least three independent PCRs performed in duplicate were repeated, Primer and probe sets for quantitative real-time PCR used in this study are as follows: spot 15, 5′-AACAGCTAGGGCAATCCTCTTC-3′, 5′-ACAGTTACAG GTGAGTGTGATCTG-3′, and 5′-Fam-CCGCCTGCTGATGCT CATGGCTCT-Tamra-3′; spot 34, 5′-CAGGTACACCACATGAG GTCTC-3′, 5′-CTCCCAAGGTCGTTCTAAAGAGTA-3′, and 5′-Fam-ACACAGCCATCTGGTGCTTTGCGT-Tamra-3′; spot 115, 5′-GATGTACCTGGCCCTCAACTG-3′, 5′-AGTCTGCCGAAGTCCT GTGA-3′ and 5′-Fam-CCAGCCCACCTTACCCCAAGCCCATamra-3′; spot 152, 5′-GTCTTAGCTCCGTTTCTTCTTTCG-3′, 5′-GGGCACATAGCAATAACTGGGT-3′, and 5′-Fam-CCAG CAGCTCGCAACGTGAACGGT-Tamra-3′; spot 167, 5′-GATC TGGAGTCAGTCTCTTCACT-3 ′, 5 ′-GTGAACATCTT TACTCCATGACCA-3′, and 5′-Fam-CAAACCCGAGTGTCTCCT GCCTGG-Tamra-3′; spot 169, 5′-CCTCCGGCAGATGAATGC TAA-3′, 5′-CACACTGGCTGACCTGGATC-3′, and 5′-Fam-ACCCGCCTCTCCTTAGTGTGCCCTTamra-3′; spot 178, 5′-GCCCTCCAGTAAAAGTGAAGC-3′, 5′-TGATCTCTGACC TCTGCACATATA-3′, and 5′-Fam-AGCTGCACAATGAC CCCTCCTGAC-Tamra-3′; spot 212, 5′-GTAATTCTGGCTTGGGC TATAAGA-3 ′, 5 ′-ACAGCCGCAAAATTACAATTAACA-3 ′, and 5 ′-Fam-TTCCCTGTTGCTGCTACTGCTGGTTamra-3′; and spot 231, 5′-GCTTTCACATCTAAGAGACT GACC-3′, 5′-CGTTCTTGGATGGGCTGTGT-3′, and 5′-Fam-CCAGCAGCGTGTGCGCGGCC-Tamra-3′.
Acknowledgments
We thank Maddy Roberts for proofreading the original manuscript and Yasushi Kogo, Chiaki Maeda, and Daisuke Sato for technical assistances. This work was supported by the Program for Promotion of Basic Research Activities for Innovative Biosciences and the Grant-in-aid for Scientific Research, Ministry of Education, Culture, Sports, Science and Technology, Japan (15208027, 15080202, 16014300) to K.S.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.2431504. Article published online before print in August 2004.
References
- Antequera, F. and Bird, A. 1993. Number of CpG islands and genes in human and mouse. Proc. Natl. Acad. Sci. 90: 11995-11999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bestor, T.H. 1992. Activation of mammalian DNA methyltransferase by cleavage of a Zn binding regulatory domain. EMBO J. 11: 2611-2617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird, A.P. 1987. CpG island as gene markers in the vertebrate nucleus. Trends Genet. 3: 342-347. [Google Scholar]
- Bird, A.P. and Wolffe, A.P. 1999. Methylation-induced repression: Belts, braces, and chromatin. Cell 99: 451-454. [DOI] [PubMed] [Google Scholar]
- Chen, T., Ueda, Y., Dodge, J.E., Wang, Z., and Li, E. 2003. Establishment and maintenance of genomic methylation patterns in mouse embryonic stem cells by Dnmt3a and Dnmt3b. Mol. Cell. Biol. 23: 5594-5605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper, D.N. and Krawczak, M. 1989. Cytosine methylation and the fate of CpG dinucleotides in vertebrate genomes. Hum. Genet. 83: 181-188. [DOI] [PubMed] [Google Scholar]
- Cross, S.H. and Bird, A.P. 1995. CpG islands and genes. Curr. Opin. Genet. Dev. 5: 309-314. [DOI] [PubMed] [Google Scholar]
- Futscher, B.W., Oshiro, M.M., Wozniak, R.J., Holtan, N., Hanigan, C.L., Duan, H., and Domann, F.E. 2002. Role for DNA methylation in the control of cell type specific maspin expression. Nat. Genet. 31: 175-179. [DOI] [PubMed] [Google Scholar]
- Gardiner-Garden, M. and Frommer, M. 1987. CpG islands in vertebrate genomes. J. Mol. Biol. 196: 261-282. [DOI] [PubMed] [Google Scholar]
- Gruenbaum, Y., Stein, R., Cedar, H., and Razin, A. 1981. Methylation of CpG sequences in eukaryotic DNA. FEBS Lett. 124: 67-71. [DOI] [PubMed] [Google Scholar]
- Hattori, N., Nishino, K., Ko, Y.G., Hattori, N., Ohgane, J., Tanaka, S., and Shiota, K. 2004. Epigenetic control of mouse Oct-4 gene expression in embryonic stem cells and trophoblast stem cells. J. Biol. Chem. 279: 17063-17069. [DOI] [PubMed] [Google Scholar]
- Heid, C.A., Stevens, J., Livak, K.J., and Williams, P.M. 1996. Real time quantitative PCR. Genome Res. 6: 986-994. [DOI] [PubMed] [Google Scholar]
- Imamura, T., Ohgane, J., Ito, S., Ogawa, T., Hattori, N., Tanaka, S., and Shiota, K. 2001. CpG island of rat sphingosine kinase-1 gene: tissue-dependent DNA methylation status and multiple alternative first exons. Genomics 76: 117-125. [DOI] [PubMed] [Google Scholar]
- Jones, P.A. and Baylin, S.B. 2002. The fundamental role of epigenetic events in cancer. Nat. Rev. Genet. 3: 415-428. [DOI] [PubMed] [Google Scholar]
- Jones, P.A. and Takai, D. 2001. The role of DNA methylation in mammalian epigenetics. Science 293: 1068-1070. [DOI] [PubMed] [Google Scholar]
- Kim, G.D., Ni, J., Kelesoglu, N., Roberts, R.J., and Pradhan, S. 2002. Co-operation and communication between the human maintenance and de novo DNA (cytosine-5) methyltransferases. EMBO J. 21: 4183-4195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura, H. and Shiota, K. 2003. Methyl-CpG-binding protein, MeCP2, is a target molecule for maintenance DNA methyltransferase, Dnmt1. J. Biol. Chem. 278: 4806-4812. [DOI] [PubMed] [Google Scholar]
- Lachner, M. and Jenuwein, T. 2002. The many faces of histone lysine methylation. Curr. Opin. Cell. Biol. 14: 286-298. [DOI] [PubMed] [Google Scholar]
- Lander, E.S., Linton, L.M., Birren, B., Nusbaum, C., Zody, M.C., Baldwin, J., Devon, K., Dewar, K., Doyle, M., FitzHugh, W., et al. 2001. Initial sequencing and analysis of the human genome. Nature 409: 860-921. [DOI] [PubMed] [Google Scholar]
- Larsen, F., Gundersen, G., Lopez, R., and Prydz, H. 1992. CpG islands as gene markers in the human genome. Genomics 13: 1095-1107. [DOI] [PubMed] [Google Scholar]
- Lei, H., Oh, S.P., Okano, M., Juttermann, R., Goss, K.A., Jaenisch, R., and Li, E. 1996. De novo DNA cytosine methyltransferase activities in mouse embryonic stem cells. Development 122: 3195-3205. [DOI] [PubMed] [Google Scholar]
- Leonhardt, H., Page, A.W., Weier, H.U., and Bestor, T.H. 1992. A targeting sequence directs DNA methyltransferase to sites of DNA replication in mammalian nuclei. Cell 71: 865-873. [DOI] [PubMed] [Google Scholar]
- Li, E. 2002. Chromatin modification and epigenetic reprogramming in mammalian development. Nat. Rev. Genet. 3: 662-673. [DOI] [PubMed] [Google Scholar]
- Li, E., Bestor, T.H., and Jaenisch, R. 1992. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell 69: 915-926. [DOI] [PubMed] [Google Scholar]
- Liang, G., Chan, M.F., Tomigahara, Y., Tsai, Y.C., Gonzales, F.A., Li, E., Laird, P.W., and Jones, P.A. 2002. Cooperativity between DNA methyltransferases in the maintenance methylation of repetitive elements. Mol. Cell. Biol. 22: 480-491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, I.G., Han, L., Taghva, A., O'Brien, L.E., and Hsieh, C.L. 2002. Murine de novo methyltransferase Dnmt3a demonstrates strand asymmetry and site preference in the methylation of DNA in vitro. Mol. Cell. Biol. 22: 704-723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsay, S. and Bird, A.P. 1987. Use of restriction enzymes to detect potential gene sequences in mammalian DNA. Nature 327: 336-338. [DOI] [PubMed] [Google Scholar]
- Matsuyama, T., Kimura, M.T., Koike, K., Abe, T., Nakano, T., Asami, T., Ebisuzaki, T., Held, W.A., Yoshida, S., and Nagase, H. 2003. Global methylation screening in the Arabidopsis thaliana and Mus musculus genome: Applications of virtual image restriction landmark genomic scanning (Vi-RLGS). Nucleic Acids Res. 31: 4490-4496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newell-Price, J., King, P., and Clark, A.J. 2001. The CpG island promoter of the human proopiomelanocortin gene is methylated in nonexpressing normal tissue and tumors and represses expression. Mol. Endocrinol. 15: 338-348. [DOI] [PubMed] [Google Scholar]
- Ohgane, J., Aikawa, J., Ogura, A., Hattori, N., Ogawa, T., and Shiota, K. 1998. Analysis of CpG islands of trophoblast giant cells by restriction landmark genomic scanning. Dev. Genet. 22: 132-140. [DOI] [PubMed] [Google Scholar]
- Ohgane, J., Wakayama, T., Kogo, Y., Senda, S., Hattori, N., Tanaka, S., Yanagimachi, R., and Shiota, K. 2001. DNA methylation variation in cloned mice. Genesis 30: 45-50. [DOI] [PubMed] [Google Scholar]
- Okano, M., Xie, S., and Li, E. 1998. Cloning and characterization of a family of novel mammalian DNA (cytosine-5) methyltransferases. Nat. Genet. 19: 219-220. [DOI] [PubMed] [Google Scholar]
- Okano, M., Bell, D.W., Haber, D.A., and Li, E. 1999. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell 99: 247-257. [DOI] [PubMed] [Google Scholar]
- Pao, M.M., Tsutsumi, M., Liang, G., Uzvolgyi, E., Gonzales, F.A., and Jones, P.A. 2001. The endothelin receptor B (EDNRB) promoter displays heterogeneous, site specific methylation patterns in normal and tumor cells. Hum. Mol. Genet. 10: 903-910. [DOI] [PubMed] [Google Scholar]
- Pradhan, S., Bacolla, A., Wells, R.D., and Roberts, R.J. 1999. Recombinant human DNA (cytosine-5) methyltransferase, I: Expression, purification, and comparison of de novo and maintenance methylation. J. Biol. Chem. 274: 33002-33010. [DOI] [PubMed] [Google Scholar]
- Razin, A., Webb, C., Szyf, M., Yisraeli, J., Rosenthal, A., Naveh-Many, T., Sciaky-Gallili, N., and Cedar, H. 1984. Variations in DNA methylation during mouse cell differentiation in vivo and in vitro. Proc. Natl. Acad. Sci. 81: 2275-2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reik, W., Dean, W., and Walter, J. 2001. Epigenetic reprogramming in mammalian development. Science 293: 1089-1093. [DOI] [PubMed] [Google Scholar]
- Rhee, I., Bachman, K.E., Park, B.H., Jair, K.W., Yen, R.W., Schuebel, K.E., Cui, H., Feinberg, A.P., Lengauer, C., Kinzler, K.W., et al. 2002. DNMT1 and DNMT3b cooperate to silence genes in human cancer cells. Nature 416: 552-556. [DOI] [PubMed] [Google Scholar]
- Shiota, K. and Yanagimachi, R. 2002. Epigenetics by DNA methylation for development of normal and cloned animals. Differentiation 69: 162-166. [DOI] [PubMed] [Google Scholar]
- Shiota, K., Kogo, Y., Ohgane, J., Imamura, T., Urano, A., Nishino, K., Tanaka, S., and Hattori, N. 2002. Epigenetic marks by DNA methylation specific to stem, germ and somatic cells in mice. Genes Cells 7: 961-969. [DOI] [PubMed] [Google Scholar]
- Strichman-Almashanu, L.Z., Lee, R.S., Onyango, P.O., Perlman, E., Flam, F., Frieman, M.B., and Feinberg, A.P. 2002. A genome-wide screen for normally methylated human CpG islands that can identify novel imprinted genes. Genome Res. 12: 543-554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki, Y., Tsunoda, T., Sese, J., Taira, H., Mizushima-Sugano, J., Hata, H., Ota, T., Isogai, T., Tanaka, T., Nakamura, Y., et al. 2001. Identification and characterization of the potential promoter regions of 1031 kinds of human genes. Genome Res. 11: 677-684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venter, J.C., Adams, M.D., Myers, E.W., Li, P.W., Mural, R.J., Sutton, G.G., Smith, H.O., Yandell, M., Evans, C.A., Holt, R.A., et al. 2001. The sequence of the human genome. Science 291: 1304-1351. [DOI] [PubMed] [Google Scholar]
- Waterston, R.H., Lindblad-Toh, K., Birney, E., Rogers, J., Abril, J.F., Agarwal, P., Agarwala, R., Ainscough, R., Alexandersson, M., An, P., et al. 2002. Initial sequencing and comparative analysis of the mouse genome. Nature 420: 520-562. [DOI] [PubMed] [Google Scholar]
- Yoder, J.A., Soman, N.S., Verdine, G.L., and Bestor, T.H. 1997. DNA (cytosine-5)-methyltransferases in mouse cells and tissues. Studies with a mechanism-based probe. J. Mol. Biol. 270: 385-395. [DOI] [PubMed] [Google Scholar]
WEB SITE REFERENCES
- http://www.ncbi.nlm.nih.gov/genome/seq/MmBlast.html; BLAST.
- http://www.ensembl.org/Mus_musculus/; Ensemble.
- ftp://ftp.ncbi.nih.gov/genomes/M_musculus/; GenBank ftp site.
- http://ftp.genome.washington.edu/cgi-bin/RepeatMasker; RepeatMasker.