Abstract
A set of 1638 informative SNP markers easily assayed by the Amplifluor genotyping system were tested in 102 mouse strains, including the majority of the common and wild-derived inbred strains available from The Jackson Laboratory. Selected from publicly available databases, the markers are on average ∼1.5 Mb apart and, whenever possible, represent the rare allele in at least two strains. Amplifluor assays were developed for each marker and performed on two independent DNA samples from each strain. The mean number of polymorphisms between strains was 608±136 SD. Several tests indicate that the markers provide an effective system for performing genome scans and quantitative trait loci analyses in all but the most closely related strains. Additionally, the markers revealed several subtle differences between closely related mouse strains, including the groups of several 129, BALB, C3H, C57, and DBA strains, and a group of wild-derived inbred strains representing several Mus musculus subspecies. Applying a neighbor-joining method to the data, we constructed a mouse strain family tree, which in most cases confirmed existing genealogies.
For almost a century, mouse models have played an important role in helping us understand the genetics and pathophysiology of human diseases and other traits determined by either single genes or sets of multiple loci. Two recent and substantial advancements have substantially improved these capabilities. First, maps constructed with a high density of simple sequence length polymorphism (SSLP) markers and expressed sequence tag (EST) loci (Dietrich et al. 1996; Rowe et al. 2003) greatly facilitated the process of identifying candidate genes for genetic traits. Second, the comparison of the completely sequenced genome of strain C57BL/6J (Waterston et al. 2002) with sequences from other mouse strains (Grupe et al. 2001; Wade et al. 2002; Wiltshire et al. 2003) revealed an extremely abundant type of genetic variants, single nucleotide polymorphisms (SNPs). The greater abundance and cheaper costs of assaying SNPs promise substantial advantages over the use of SSLPs in genetic mapping. Several groups have described tens of thousands of SNPs each in up to 15 (Grupe et al. 2001), four (Wade et al. 2002), and nine (Wiltshire et al. 2003) mouse strains. In doing so, these groups have laid the foundation for SNP genotyping in the mouse. To further genetic mapping in mice, we have developed a robust set of SNP markers sufficiently polymorphic to perform quantitative trait locus (QTL) analyses between nearly any two mouse strains, including virtually all of the inbred and wild-derived inbred strains available from The Jackson Laboratory. A recent paper from our laboratory described ∼240 SNP markers in 48 strains and showed that markers from only a few strains can successfully be used to type a wide variety of strains, including those that are wild-derived (Petkov et al. 2004). By consulting public databases, we selected an additional set of 2158 evenly spaced markers. After testing them in a variety of mouse strains for their utility and information content, we settled on a panel of 1638, slightly more than one per 1.5 Mb. On average, 608 ± 136 SD of the 1638 were polymorphic between any two mouse strains, providing a powerful, standardized marker system for genetic analyses with an average interstrain resolution of 4.2 Mb.
Using these 1638 markers and the neighbor-joining method (Saitou and Nei 1987), we also constructed the most comprehensive mouse family tree to date, recognizing that it may exaggerate the differences between the strains originally used to select the SNPs.
RESULTS AND DISCUSSION
Selecting SNP Markers and Testing Their Assays
Our two major objectives were to choose markers that would: (1) facilitate genome-wide scans in all possible crosses between the 102 strains we analyzed and (2) be successfully and reliably assayed by Amplifluor technology (Assay Architect software, https://apps.serologicals.com/aaa/login.aspx). From publicly available databases (Wade et al. 2002; Wiltshire et al. 2003) we selected 2158 candidate SNPs using the following four criteria: (1) they were preferably 0.8-1.2 Mb apart (at most 0.6-1.4 Mb apart in SNP-poor regions); (2) their flanking sequences mapped to a single and unequivocal position in the ENSEMBL mouse genomic sequences; (3) available information indicated polymorphism in more than two mouse strains; and (4) they were preferably located in coding regions (in the end, about 3% were cSNPs).
Using Amplifluor assays, we tested the 2158 markers on a panel of six laboratory and two wild-derived strains which had data in publicly available databases, namely 129X1/SvJ, A/J, AKR/J, BALB/cByJ, C3H/HeJ, C57BL/6J, DBA2/J, CAST/EiJ, and SPRET/EiJ. In all, 1654 of the 2158 assays (76.8%) successfully amplified their target SNPs. Of the remainder, 223 (10.3%) did not amplify the corresponding sequence from any sample, 107 (4.9%) amplified both alleles poorly, 61 (2.8%) amplified the same allele from all samples and were considered monomorphic, 53 (2.5%) did not distinguish heterozygotes, 41 (2%) amplified one allele well but not the other, and 18 (0.8%) amplified their target SNPs inconsistently. We made no attempts to optimize any failed assays.
After comparing 1654 of the successfully amplified SNPs to Build 30 of the mouse sequence database (www.ensembl.org), we excluded 16 more: 14 mapped to two regions and obviously represented duplications; one was not found in Build 30, and one was mapped on a different chromosome, making its status uncertain. Our final marker set consisted of 1638 successful SNP assays, which we used to genotype 102 mouse strains (Supplemental Table 1). All assays were identified by their physical position in the mouse genome and by their refSNP ID in the dbSNP (http://www.ncbi.nlm.nih.gov/SNP).
The average distance between them was 1.56 Mb, or about 0.8 cM (Table 1), with a minimum resolution power of 5 Mb for most chromosomal regions. Exceptions were the middle of Chromosome 10, a large part of Chromosome X, both of which have been reported as “SNP deserts” (Wade et al. 2002; Wiltshire et al. 2003), and the distal end of Chromosome 16 (70-90 Mb), where very few of the available SNPs converted to useful assays. The SNP panel described here will be included in the Mouse Phenome Project web site (http://aretha.jax.org/pub-cgi/phenome/mpdcgi?rtn=docs/home). All assays are available from The Jackson Laboratory, Bar Harbor, ME.
Table 1.
Marker Disbribution Along the Chromosomes
| Chromosome | Chromosome length [Mb] (Ensembl) | Number of markers | Minimal distance [Mb] | Maximal distance [Mb] | Average distance [Mb] |
|---|---|---|---|---|---|
| 1 | 196 | 169 | 0.002 | 5.924 | 1.135 |
| 2 | 181 | 99 | 0.001 | 8.492 | 1.806 |
| 3 | 161 | 100 | 0.1 | 6.647 | 1.564 |
| 4 | 153 | 102 | 0.039 | 6.477 | 1.478 |
| 5 | 150 | 96 | 0.016 | 4.925 | 1.534 |
| 6 | 150 | 91 | 0.007 | 8.326 | 1.617 |
| 7 | 135 | 89 | 0.001 | 5.6 | 1.474 |
| 8 | 129 | 84 | 0.021 | 4.877 | 1.477 |
| 9 | 125 | 79 | 0.041 | 6.346 | 1.539 |
| 10 | 131 | 57 | 0.243 | 14.221 | 2.237 |
| 11 | 123 | 78 | 0.094 | 7.537 | 1.529 |
| 12 | 114 | 77 | 0.003 | 8.157 | 1.447 |
| 13 | 116 | 75 | 0.023 | 6.824 | 1.517 |
| 14 | 116 | 86 | 0.037 | 5.303 | 1.308 |
| 15 | 104 | 73 | 0.034 | 6.934 | 1.383 |
| 16 | 99 | 58 | 0.019 | 14.432 | 1.649 |
| 17 | 94 | 60 | 0.001 | 7.737 | 1.517 |
| 18 | 91 | 72 | 0.076 | 4.439 | 1.193 |
| 19 | 61 | 43 | 0.033 | 4.899 | 1.356 |
| X | 150 | 50 | 0.001 | 27.082 | 2.802 |
| Total | 2579 | 1638 | |||
| Average | 0.039 | 8.259 | 1.578 |
Information Content of the Marker Set
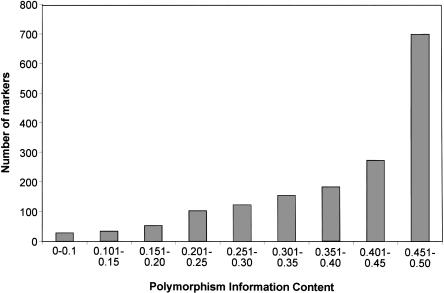
Previous information regarding the distribution of SNPs among inbred mouse strains was limited to a modest number of strains; about two-thirds of the reported polymorphisms were described in two or three strains (Wade et al. 2002), and about one-third were reported in up to nine strains (Wiltshire et al. 2003). To determine whether the SNPs we chose would be informative for genome-wide scans in a variety of crosses, we calculated the polymorphism information content (PIC) for all 102 strains, where PIC = 2pq, p and q being the allelic frequencies of an SNP, and PIC can range from 0.0 to 0.5 (Botstein et al. 1980; Anderson et al. 1993). The SNPs we selected had an average PIC of 0.39, and 96.3% were either highly informative (86.7% with PIC > 0.25) or informative (11.6% with 0.1 < PIC < 0.25; Fig. 1). Although the remaining 3.7% were only slightly informative across the whole set of 102 strains, they are highly valuable for selected strain pairs.
Figure 1.
Distribution of polymorphism information content (PIC) of the SNPs among 102 strains. The PIC was calculated as 2pq, where p and q are the frequencies of the corresponding alleles.
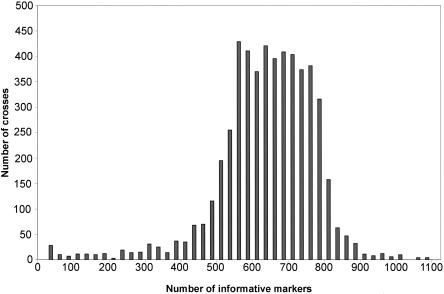
Our SNP set appears to be adequate for performing genome scans in almost all possible crosses. The number of polymorphisms between all possible pairs of strains is shown in Figure 2 and Supplemental Table 2. The average was 608 ± 136 SD, with 97% having at least 300. The average number for all possible crosses between 17 commonly used laboratory strains and five wild-derived strains (the four Mus musculus subspecies and Mus spretus) was 616 ± 136 SD, which corresponds to an average marker density of 4.1 Mb or 2.3 cM, with 98% having at least 300 for a marker density of 8.5 Mb or 4.67 cM (Table 2).
Figure 2.
Distribution of informative markers in all possible crosses of 102 mouse strains.
Table 2.
Distribution of Informative Markers in All Possible Crosses of 22 Mouse Strains
| Strain | 129S1/SvlmJ | A/J | AKR/J | BALB/cByJ | BTBR T+tflJ | BUB BnJ | C3H/HeJ | C57BL/6J | CAST/EiJ | CBA/J | DBA/2J | FVB/NJ | JF1/Ms | KK/HIJ | MRL/MpJ | NOD/LtJ | NZW/LacJ | PL/J | PWK/PhJ | SJL/J | SPRET/EiJ | WSB/EiJ |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 129S1/SvlmJ | ||||||||||||||||||||||
| A/J | 785 | |||||||||||||||||||||
| AKR/J | 778 | 469 | ||||||||||||||||||||
| BALB/cByJ | 816 | 363 | 526 | |||||||||||||||||||
| BTBR T+tflJ | 534 | 623 | 622 | 694 | ||||||||||||||||||
| BUB/BnJ | 730 | 513 | 425 | 562 | 646 | |||||||||||||||||
| C3H/HeJ | 818 | 400 | 519 | 525 | 706 | 530 | ||||||||||||||||
| C57BL/6J | 1047 | 882 | 786 | 880 | 747 | 747 | 981 | |||||||||||||||
| CAST/EiJ | 747 | 710 | 674 | 713 | 707 | 687 | 749 | 769 | ||||||||||||||
| CBA/J | 798 | 455 | 518 | 533 | 675 | 522 | 205 | 902 | 696 | |||||||||||||
| DBA/2J | 744 | 576 | 569 | 631 | 660 | 570 | 479 | 825 | 702 | 456 | ||||||||||||
| FVB/NJ | 729 | 495 | 480 | 567 | 604 | 430 | 558 | 760 | 678 | 526 | 574 | |||||||||||
| JF1/Ms | 774 | 774 | 775 | 790 | 778 | 775 | 808 | 772 | 286 | 765 | 744 | 791 | ||||||||||
| KK/HIJ | 639 | 624 | 590 | 676 | 601 | 560 | 680 | 711 | 684 | 637 | 579 | 539 | 703 | |||||||||
| MRL/MpJ | 754 | 434 | 352 | 530 | 681 | 472 | 432 | 833 | 716 | 463 | 550 | 532 | 784 | 613 | ||||||||
| NOD/LtJ | 731 | 493 | 471 | 552 | 612 | 405 | 590 | 736 | 697 | 582 | 551 | 412 | 777 | 564 | 520 | |||||||
| NZW/LacJ | 582 | 606 | 567 | 649 | 584 | 529 | 658 | 688 | 670 | 609 | 554 | 525 | 715 | 468 | 575 | 540 | ||||||
| PL/J | 712 | 475 | 396 | 511 | 626 | 470 | 518 | 810 | 697 | 527 | 575 | 514 | 770 | 577 | 427 | 502 | 557 | |||||
| PWK/PhJ | 762 | 734 | 742 | 730 | 739 | 736 | 764 | 782 | 300 | 720 | 710 | 748 | 142 | 697 | 755 | 754 | 691 | 735 | ||||
| SJL/J | 660 | 487 | 472 | 510 | 592 | 387 | 525 | 734 | 648 | 501 | 547 | 328 | 738 | 523 | 467 | 414 | 477 | 452 | 699 | |||
| SPRET/EiJ | 627 | 630 | 598 | 649 | 630 | 605 | 649 | 703 | 249 | 617 | 616 | 594 | 300 | 595 | 602 | 609 | 567 | 586 | 291 | 571 | ||
| WSB/EiJ | 722 | 634 | 610 | 653 | 656 | 610 | 671 | 730 | 552 | 630 | 661 | 584 | 662 | 619 | 624 | 614 | 588 | 621 | 633 | 550 | 486 |
Wild-Derived Strains
Because they are becoming increasingly important in QTL analyses and evolutionary studies, we tested our marker set in an extensive group of wild-derived inbred strains. Most of the common laboratory strains are predicted to have a mixed ancestry of M. m. musculus and M. m. domesticus, possibly with a small contribution from M. m. castaneus (Ferris et al. 1982; Bishop et al. 1985; Yonekawa et al. 1994; Beck et al. 2000). In contrast, most of the wild-derived strains are largely pure subspecies of wild mice trapped in various locations, including M. m. domesticus strains WSB/EiJ (MD, USA), LEWES/EiJ (DE, USA), CALB/RkJ (CA, USA), WMP/PasDnJ (Tunisia), TIRANO/EiJ (Italy), ZALENDE/EiJ (Switzerland), PERA/EiJ and PERC/EiJ (Peru); M. m. musculus strains SKIVE (Denmark), CZECHI/EiJ, CZECHII/EiJ and PWK/PhJ (Czech Republic). M. m. molossinus subspecies strains MOLC/RkJ, MOLD/RkJ, MOLF/RkJ, and MSM/Ms were derived from mice trapped in Japan, M. m. castaneus CASA/RkJ and CAST/EiJ originated in Thailand. The musculus, domesticus, and castaneus subspecies are thought to have diverged from a common ancestor about one million years ago (Silver 1995). M. m. molossinus is considered a more recent hybrid between M. m. musculus and M. m. castaneus, although the data supporting these estimates are anecdotal. There are indications that M. m. musculus and M. m. domesticus may represent different species rather than subspecies (Sage et al. 1986; Tucker et al. 1992). Among the other mouse species represented in this study, M. hortulanus (PANCEVO/EiJ) was separated from the common ancestor between it and M. musculus ∼two million years ago, and M. spretus (SPRET/EiJ) ∼three million years ago, according to the same estimate (Silver 1995). Although the wild-derived strains have been separated for one to three million years, our SNP panel successfully genotyped all of them, including the most genetically distant ones, with a marker density comparable to that used for genotyping laboratory strains: 86% of the assays worked well with SPRET/EiJ, over 90% with PANCEVO/EiJ, and over 96% with CAST/EiJ and CASA/RkJ. Our panel of robust SNP assays can provide further studies of wild mouse populations with the necessary tool for establishing the evolutionary relationships between species on firm factual basis.
Evolutionary Origin of SNPs
The distribution of SNPs in wild-derived inbred strains suggested that many of them originated before the progenitors of these strains diverged in evolution. In 42% of our SNP set, both alleles were found in strains belonging to each of the two subspecies M. m. musculus and M. m. domesticus, suggesting that the mutations causing the polymorphisms originated before the two subspecies separated. It is unlikely that these SNPs arose after evolutionary divergence by independent mutation in each lineage, either at the population level or during the process of inbreeding, given the estimated mutation rate in mouse of 1.8×10-10 mutations per nucleotide per generation (Drake et al. 1998) and the separation time of M. musculus subspecies as one million years (∼3×106 generations).
Persistence of Residual Heterozygosity During Inbreeding
An analysis of SNPs in closely related strains suggests that residual heterozygosity has persisted during the process of inbreeding strains long after theoretical calculations suggest it should be gone. For example, C57BL/10J and C57BL/6J were separated from the original C57BL strain in the mid-1930s, after about 40 generations of inbreeding (Festing 1996). Forty-nine (3%) of the 1638 SNPs we tested were polymorphic between these two strains. Thirty-three of these 49 were also polymorphic in a variety of other laboratory and wild-derived strains, indicating that they almost certainly represent old SNPs still segregating at the time the two strains separated. Two percent segregating loci after about 40 generations of inbreeding (the C57BL strain was developed around 1921) is considerably more than would be expected by chance. This suggests that there is selection for residual heterozygosity at some loci during the process of inbreeding.
Further evidence for the persistence of heterozygosity at selected loci during inbreeding came from a comparison of C57BL/6 substrains. The low-frequency allele of seven SNPs from our marker set was found only in C57BL/6J and strains derived from it after it separated from C57BL/10J, possibly representing the loss of this allele in C57BL/10J or new mutations fixed in C57BL/6J after its separation from the BL/10 lineage. Five of the seven SNPs were polymorphic between C57BL/6J and C57BL/6ByJ, the rare allele in all of them being found only in C57BL/6J and strains partially derived from C57BL/6J after the 1960s, including BPL/1J, BPH/2J, BPN/3J, and MOR/RkJ. C57BL/6ByJ was separated in the 1950s from C57BL/6NCr, a substrain maintained at the National Cancer Institute (NCI) since 1951. We tested a group of six more C57BL/6 strains separated at different times and found that: (1) those originating from the substrain maintained at the NCI shared the same genotype as C57BL/6ByJ, (2) two strains separated from The Jackson Laboratory stock after the 1970s shared the five alleles with C57BL/6J, and (3) C57BL/6JEiJ, separated in 1975, shared two alleles with the NCr substrain and three with the J substrain. Although it is possible that new mutations were randomly fixed, it is more likely that this allelic distribution resulted from residual heterozygosity at the time the J and NCr substrains were separated.
Recapitulating Strain Histories
We evaluated the usefulness of our marker set for genome scanning by testing how well it could either verify or detect events known to have happened during the origin of mouse strains. For example, we know that 129S1/SvImJ was created by crossing 129/Sv with C3HeB/FeJ, backcrossing to the parental 129/Sv, and selectively breeding the offspring with the highest teratoma incidence. The C3HeB/FeJ contribution has been located on Chromosome 7 and includes the wild-type alleles of the Tyr and p loci (Simpson et al. 1997; Threadgill et al. 1997). Our SNP set confirmed that the entire 129S1/SvImJ genome has but one substantial C3HeB/FeJ segment, a single DNA block between markers at 22.3 Mb and 82.7 Mb on Chromosome 7. As a second example, we know that another strain of this group, 129X1/SvJ, was contaminated with unknown genetic material around 1987 (Simpson et al. 1997). An analysis with our SNP marker set revealed that the strain has contributions by C57BL/6J on Chromosomes 5, 7, 14, 18, and 19, and by BALB/cJ on Chromosomes 7, 8, 10, 18, 19, and X, suggesting an F1 hybrid between these strains as the most possible contaminant. As a third example, C57BLKS/J had been determined to be derived from C57BL/6J contaminated with DBA/2J (Naggert et al. 1995). In fact, an analysis with our SNP marker set revealed that not only did the C57BLKS/J genome have different size DBA/2J segments on every chromosome except Chromosomes 2 and 13, but it had regions on Chromosomes 4 (27-42 Mb and 96-106 Mb), 9 (18-30 Mb), 11 (23-26 Mb and 80-91 Mb), and 15 (96-103 Mb) that could not be explained solely by DBA/2J contributions on a C57BL/6J background. Although the different markers on Chromosomes 9 and 15 were present in other strains of the C57 group and may have been randomly fixed after C57BLKS/J separated in the 1940s, those on Chromosomes 4 and 11 suggest that C57BLKS/J had genetic segments from another strain, and BTBR T+ tf/J best fits the pattern observed among the strains we tested. In yet another example, NOR/LtJ, a diabetes-free strain, was derived from the diabetic NOD/LtJ through an accidental outcross with C57BLKS/J. An analysis with our SNP marker set not only confirmed the presence of C57BLKS/J sequences on Chromosomes 1, 2, 4, 5, 7, 11, 12, and 18, as originally reported (Prochazka et al. 1992; Serreze et al. 1994), it also revealed additional C57BLKS/J regions on Chromosomes 10 (21-28 Mb) and 14 (77-88 Mb), and it precisely showed the regions of C57BLKS/J ancestry that were ultimately derived from either C57BL/6J or DBA/2J genetic backgrounds.
Family Tree of the Mouse Strains
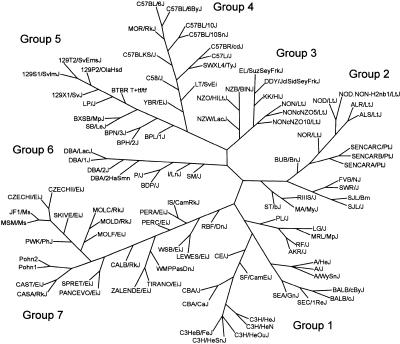
A better understanding of the genetic relationships among inbred strains of mice will improve experimental designs for mapping quantitative trait loci and choosing strategies for developing new genetic resources. The genealogy of inbred mouse strains is well documented (Simpson et al. 1997; Beck et al. 2000): some share a common origin, some were either deliberately or accidentally crossed with pre-existing strains, and others are of entirely independent origin. Several genotypically based mouse family trees involving different sets of strains have been constructed using classical genotypes or SSLP markers (Atchley and Fitch 1993; Schalkwyk et al. 1999; Witmer et al. 2003). To further explore these relationships, we sought to reconstruct the phylogenetic relationships among the 102 inbred and wild-derived inbred strains tested in this study by using our SNP marker set (Fig. 3).
Figure 3.
Mouse family tree. The 102 strains tested are organized into seven groups. Group 1, Bagg albino derivatives; Group 2, Swiss mice; Group 3, Japanese and New Zealand inbred strains; Group 4, C57/58 strains; Group 5, Castle's mice; Group 6, C.C. Little's DBA and related strains; Group 7, wild-derived strains. A detailed description of each group is included in the text. The length and angle of the branches have been optimized for printing and do not reflect the actual evolutionary distances between strains.
As discussed above, most of the SNPs we tested were either informative or highly informative. Moreover, their origins appear to predate the evolutionary divergence of Mus species and subspecies, suggesting that they were present long before the development of inbred mouse strains. However, any tree we construct will be biased, because our markers were originally selected for being polymorphic among a small subset of strains, and thus emphasize divergent relationships among these strains. It is not surprising then that our marker set assigned C57BL/6J (completely sequenced) to one branch, 129S1/SvImJ (partially sequenced) to a second, and BALB/cByJ and C3H/HeJ (both partially sequenced) to a third branch of the tree, because a substantial number of SNPs in the set originated from and were thus polymorphic among these four strains. Accepting this caveat, our markers remain useful for assigning other strains to either existing or new tree branches, and for revealing the relationships among the strains within a branch. The large proportion (42%) of ancient SNPs in the set, being widely distributed among the 102 strains, may actually have resolved many of the tree's relationships quite well despite the “enhanced-resolution” bias for the four strains mentioned above.
Our phylogenetic tree distinguished seven groups. Group 1 has two branches anchored, respectively, by two of the sequenced strains, BALB/c and C3H. The BALB/c branch includes A/J, SEA/GnJ, and SEC/1ReJ (both derived from crosses of BALB/cBy with other strains; Festing 1996) and almost all albino strains and strains derived from Bagg albino mice. The C3H branch includes CBA, the wild-derived SF/CamEiJ, CE/J (a cross with a wild unknown). Group 2 comprises strains derived from Swiss mice and includes BUB/BnJ, NOD/LtJ, and its congenic NOD.NON-H2nb1/LtJ, and derivatives NOR/LtJ, ALR/LtJ, and ALS/LtJ. Group 3 includes strains derived in Japan and the New Zealand strains NZW/LacJ, NZO/HlLtJ, and NZB/BlNJ (all three were developed from Imperial Cancer Research mice). Interestingly, it also includes NON/LtJ, a strain sharing a common predecessor with NOD/LtJ, but closer to the Japanese strains DDY/JclSidSeyFrkJ and EL/SuzSeyFrkJ through our marker set. Group 4 is anchored by C57BL/6J, another of the sequenced strains, and comprises C57, C58, and their derivatives LT/SvEiJ (derived from a cross between C58 and BALB/c), SWXL4/TyJ (recombinant inbred of SWR/J and C57L/J), and MOR/RkJ, partially congenic to C57BL/6J. Group 5 is anchored by the 129 strain, the final sequenced strain, and consists of Castle mice derivatives, including the 129 group and related strains LP/J and SB/LeJ. Groups 4 and 5 seem to be parts of a larger and probably related branch, as previously noted (Witmer et al. 2003). Group 6 is anchored by DBA/2J and consists of C.C. Little's DBA strains, including SM/J, BDP/J, and P/J (all related to DBA). Group 7 consists entirely of wild-derived inbred strains. It is heterogeneous and includes representatives of M. m. musculus, M. m. domesticus, M. m. castaneus, M. m. molossinus, M. hortulanus (PANCEVO/EiJ), and M. spretus (SPRET/EiJ). The clustering of all wild-derived strains in one group should be interpreted with discretion given the known diverse species and subspecies origins of the more common inbred strains. Despite whatever biases are present in its construction, our family tree conforms well to the known history of mouse strains (Beck et al. 2000) and should have utility in experimental planning.
METHODS
Selection of Polymorphisms for Assay Development
The SNPs used in this study were selected from publicly available databases (Wade et al. 2002; Wiltshire et al. 2003). Their physical positions were verified using the ENSEMBL assembly of the mouse genome (www.ensembl.org) and confirmed with Build 30 of the database after the tests were completed.
DNA Isolation
Using a DNEasy kit (QIAGEN), DNA was isolated from livers of two pedigreed mice of each strain from the foundation stocks of The Jackson Laboratory. For some strains with poor breeding performance, either spleen or brain DNA (prepared by phenol-chloroform extraction) was obtained from The Jackson Laboratory DNA Resource service. Before being tested, DNA was diluted to 10 ng/μL working concentration in 10 mM Tris, pH 8.0. Strain identity was confirmed using the 28-marker panel developed for genetic quality control (Petkov et al. 2004).
Assay Development and Scoring
Amplifluor assays (Myakishev et al. 2001) were designed and performed as recommended by the manufacturer (Serologicals). Each PCR reaction mixture contains five primers: two allele-specific primers, each one containing one of two different sequences at its 5′-end, and a 3′-part corresponding to the target sequence with alternating nucleotides at the 3′-end, common reverse primer, and two common primers matching the 5′-portion of the allele-specific primers. The two common primers include either a green or red fluorescence label and a quencher that form a loop in the free primer but become separated when the primer is incorporated in a double-stranded PCR product. A fluorescent signal is emitted only if the label and the quencher are beyond a set distance from each other. In the course of the PCR reaction, only the primers that anneal perfectly to the target DNA will amplify a DNA fragment. The results were visualized with SNPViewer program (www.kbioscience.co.uk). Only data points with unequivocal scores for the two samples from each strain were included in the final table.
Parsimony Analysis
The clustering analysis for constructing the parsimony tree of mouse strains was performed by the neighbor-joining method (Saitou and Nei 1987) using Phylip3.6 software developed by Dr. Joe Felsenstein (http://evolution.genetics.washington.edu/phylip.html).
Acknowledgments
We thank Drs. Kenneth Paigen, Edward Leiter, Muriel Davisson, Eva M. Eicher, and Ms. Moyha Lennon-Pierce for critical reading and discussion of the manuscript, Dr. Gary Churchill for advising mouse family tree preparation, and Mr. Raymond Lambert for helping in the manuscript preparation. This study was partially supported by a Howard Hughes Medical Institute grant and by an NCI grant to The Jackson Laboratory.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
[Supplemental material is available online at www.genome.org.]
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.2825804.
References
- Anderson, J.A., Churchill, G.A., Autrique, J.E., Tanksley, S.D., and Sorrells, M.E. 1993. Optimizing parental selection for genetic linkage maps. Genome Res. 36: 181-186. [DOI] [PubMed] [Google Scholar]
- Atchley, W.R. and Fitch, W. 1993. Genetic affinities of inbred mouse strains of uncertain origin. Mol. Biol. Evol. 10: 1150-1169. [DOI] [PubMed] [Google Scholar]
- Beck, J.A., Lloyd, S., Hafezparast, M., Lennon-Pierce, M., Eppig, J.T., Festing, M.F., and Fisher, E.M. 2000. Genealogies of mouse inbred strains. Nat. Genet. 24: 23-25. [DOI] [PubMed] [Google Scholar]
- Bishop, C.E., Boursot, P., Baron, B., Bonhomme, F., and Hatat, D. 1985. Most classical Mus musculus domesticus laboratory mouse strains carry a Mus musculus musculus Y chromosome. Nature 315: 70-72. [DOI] [PubMed] [Google Scholar]
- Botstein, D., White, R.L., Skolnick M., and David, R. 1980. Construction of a genetic linkage map in man using restriction fragment length polymorphisms. Am. J. Hum. Genet. 32: 314-331. [PMC free article] [PubMed] [Google Scholar]
- Dietrich, W.F., Miller, J., Steen, R., Merchant, M.A., Damron-Boles, D., Husain, Z., Dredge, R., Daly, M.J., Ingalls, K.A., and O'Connor, T.J. 1996. A comprehensive genetic map of the mouse genome. Nature 380: 149-152. [DOI] [PubMed] [Google Scholar]
- Drake, J.W., Charlesworth, B., Charlesworth, D., and Crow, J.F. 1998. Rates of spontaneous mutation. Genetics 148: 1667-1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferris, S.D., Sage, R.D., and Wilson, A.C. 1982. Evidence from mtDNA sequences that common laboratory strains of inbred mice are descended from a single female. Nature 295: 163-165. [DOI] [PubMed] [Google Scholar]
- Festing, M.F.W. 1996. Origins and characteristics of inbred strains of mice. Oxford University Press, Oxford, UK.
- Grupe, A., Germer, S., Usuka, J., Aud, D., Belknap, J.K., Klein, R.F., Ahluwalia, M.K., Higuchi R., and Peltz, G. 2001. In silico mapping of complex disease-related traits in mice. Science 292: 1915-1918. [DOI] [PubMed] [Google Scholar]
- Myakishev, M.V., Khripin, Y., Hu, S., and Hamer, D.H. 2001. High-throughput SNP genotyping by allele-specific PCR with universal energy-transfer-labeled primers. Genome Res. 11: 163-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naggert, J.K., Mu, J.L., Frankel, W., Bailey, D.W., and Paigen, B. 1995. Genomic analysis of the C57BL/Ks mouse strain. Mamm. Genome 6: 131-133. [DOI] [PubMed] [Google Scholar]
- Petkov, P.M., Cassell, M.A., Sargent, E.E., Donnelly, C.J., Robinson, P., Crew, V., Asquith, S., Haar, R.V., and Wiles, M.V. 2004. Development of a SNP genotyping panel for genetic monitoring of the laboratory mouse*1. Genomics 83: 902-911. [DOI] [PubMed] [Google Scholar]
- Prochazka, M., Serreze, D.V., Frankel, W.N., and Leiter, E.H. 1992. NOR/Lt mice: MHC-matched diabetes-resistant control strain for NOD mice. Diabetes 41: 98-106. [DOI] [PubMed] [Google Scholar]
- Rowe, L.B., Barter, M.E., Kelmenson, J.A., and Eppig, J.T. 2003. The comprehensive mouse radiation hybrid map densely cross-referenced to the recombination map: A tool to support the sequence assemblies. Genome Res. 13: 122-133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sage, R.D., Heyneman D., Lim, K.C., and Wilson, A.C. 1986. Wormy mice in a hybrid zone. Nature 324: 60-63. [DOI] [PubMed] [Google Scholar]
- Saitou, N. and Nei, M. 1987. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4: 406-425. [DOI] [PubMed] [Google Scholar]
- Schalkwyk, L.C., Jung, M., Daser, A., Weiher, M., Walter, J., Himmelbauer, H., and Lehrach, H. 1999. Panel of microsatellite markers for whole-genome scans and radiation hybrid mapping and a mouse family tree. Genome Res. 9: 878-887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serreze, D.V., Prochazka, M., Reifsnyder, P.C., Bridgett, M.M., and Eleiter, E.H. 1994. Use of recombinant congenic and congenic strains of NOD mice to identify a new insulin-dependent diabetes resistance gene. J. Exp. Med. 180: 1553-1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silver, L.M. 1995. Mouse Genetics Oxford University Press, Oxford, UK.
- Simpson, E.M., Linder, C.C., Sargent, E.E., Davisson, M.T., Mobraaten, L.E., and Sharp, J.J. 1997. Genetic variation among 129 substrains and its importance for targeted mutagenesis in mice. Nat. Genet. 16: 19-27. [DOI] [PubMed] [Google Scholar]
- Threadgill, D.W., Yee, D., Matin, A., Nadeau, J.H., and Magnuson, T. 1997. Genealogy of the 129 inbred strains: 129/SvJ. is a contaminated inbred strain. Mamm. Genome 8: 390-393. [DOI] [PubMed] [Google Scholar]
- Tucker, P.K., Sage, R.D., Warner, J., Wilson, A.C., and Eicher, E.M. 1992. Abrupt cline for sex chromosomes in a hybrid zone between two species of mice. Evolution 46: 1146-1163. [DOI] [PubMed] [Google Scholar]
- Wade, C.M., Kulbokas III, E.J., Kirby, A.W., Zody, M.C., Mullikin, J.C., Lander, E.S., Lindblad-Toh, K., and Daly, M.J. 2002. The mosaic structure of variation in the laboratory mouse genome. Nature 420: 574-578. [DOI] [PubMed] [Google Scholar]
- Waterston, R.H., Lindblad-Toh, K., Birney, E., Rogers, J., Abril, J.F., Agarwal, P., Agarwala, R., Ainscough, R., Alexandersson, M., An, P., et al. 2002. Initial sequencing and comparative analysis of the mouse genome. Nature 420: 520-562. [DOI] [PubMed] [Google Scholar]
- Wiltshire, T., Pletcher, M.T., Batalov, S., Barnes, S.W., Tarantino, L.M., Cooke, M.P., Wu, H., Smylie, K., Santrosyan, A., Copeland, N.G., et al. 2003. Genome-wide single-nucleotide polymorphism analysis defines haplotype patterns in mouse. Proc. Natl. Acad. Sci. 100: 3380-3385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witmer, P.D., Doheny, K.F., Adams, M.K., Boehm, C.D., Dizon, J.S., Goldstein, J.L., Templeton, T.M., Wheaton, A.M., Dong, P.N., Pugh, E.W., et al. 2003. The development of a highly informative mouse Simple Sequence Length Polymorphism (SSLP) marker set and construction of a mouse family tree using parsimony analysis. Genome Res. 13: 485-491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yonekawa, H., Takahama, S., Gotoh, O., Miyashita, N., and Moriwaki, K. 1994. Genetic diversity and geographic distribution of Mus musculus subspecies based on the polymorphism of mitochondrial DNA. Japan Sci. Soc. Press, Tokyo.
WEB SITE REFERENCES
- https://apps.serologicals.com/aaa/login.aspx; Serologicals Corporation page.
- www.ensembl.org; The Wellcome Trust Sanger Institute site. [DOI] [PubMed]
- http://www.ncbi.nlm.nih.gov/SNP; NCBI dbSNP database page.
- http://aretha.jax.org/pub-cgi/phenome/mpdcgi?rtn=docs/home; Mouse Phenome Project Web site.
- www.kbioscience.co.uk; KBiosciences home page.
- http://evolution.genetics.washington.edu/phylip.html; Web pages of Joe Felsenstein of the Dept. of Genome Sciences at the University of Washington.