Abstract
Background
The association between the hypermethylation of CHFR gene and gastric cancer risk has been investigated by a number of studies. However, the sample size of the majority of these studies was very small. To get a more a convincing conclusion, here we performed a meta-analysis of the previously published studies to assess the association between CHFR methylation and the risk of gastric cancer.
Methods
Eligible studies were identified by searching the MEDLINE/PubMed, Embase, and Web of Science databases before May 2016 without any language restriction. The strength of the association was estimated by odds ratio with its 95% confidence interval (CI).
Results
Totally 1,399 samples, including 758 gastric cancer cases and 641 controls, from 13 studies were included in the present meta-analysis. Compared with non-cancer controls, the pooled OR of CHFR methylation in gastric cancer patients was 9.08 (95% CI: 6.40–12.88, P<0.001), suggesting that the methylation of CHFR was significantly associated with increased risk of gastric cancer. Similar results were observed when subgroup analyses were performed stratified by country, ethnicity, and methylation testing methods.
Conclusion
Our meta-analysis showed a strong positive correlation between CHFR methylation and risk of gastric cancer, suggesting that CHFR methylation might be a promising biomarker for the diagnosis of gastric cancer.
Keywords: CHFR, methylation, tumor suppressor gene, gastric cancer, risk
Introduction
Gastric cancer is one of the most commonly diagnosed human cancers, and it is among the leading causes of cancer-related death worldwide.1 More than 70% of new gastric cancer cases and deaths occur in developing countries. The incidence rate of gastric cancer is high in Eastern Asia, Central and Eastern Europe, and South America, but low in Northern America and Africa.2 It is well established that chronic infection with Helicobacter pylori is the most common risk factor for gastric cancer, since about 90% of new noncardia gastric cancer cases worldwide attributed to this bacteria.3,4 In addition to bacteria infection, genetic and epigenetic changes of some oncogenes and tumor suppressor genes (TSG) have been involved in the initiation and development of gastric cancer.5,6
The CHFR gene is localized to chromosome 12q24.33, and it was identified as a cell-cycle checkpoint gene.7,8 In response to mitotic stress induced by micro-tubule inhibitors, the CHFR protein causes a delay in chromosome condensation and entry into metaphase. However, cancer cells lacking CHFR entered metaphase without delay.8 The CHFR protein possesses an N-terminal forkhead-associated (FHA) domain, a central RING finger (RF) domain, and a C-terminal cysteine-rich (CR) region. The FHA and CR regions are essential for its checkpoint function, and the RF domain is required for the ubiquitin ligase activity of CHFR.9 CHFR is widely expressed in normal tissues, and loss or reduced expression of CHFR has been reported in several primary tumors. Interestingly, in cancer cell lines and several types of primary cancer, the decreased CHFR expression was reported to be caused by the hypermethylation of the CpG island in the promoter region of this gene,10–12 including gastric cancer.13,14
Ever since the initial report of hypermethylation of CHFR in gastric cancer,15 a growing number of studies have investigated the association of CHFR methylation and risk of gastric cancer. However, the sample size of these studies was very small; most of them enrolled less than 100 cancer cases. Based on the notion that the statistical power is low when there is only a small number of cases enrolled in a case-control study, therefore, we conducted a meta-analysis of the previously published studies to assess whether there is an association between CHFR methylation and risk of gastric cancer.
Methods
Literature search strategy
The MEDLINE/PubMed, Embase, and Web of Science databases were used for searching literatures. The search was carried out before May 2016 without any language restriction. The keywords used for paper searching were CHFR, methylation, stomach, gastric, and cancer. To search for additional relevant publications, the reference lists from relevant primary studies and review articles were also checked manually.
Study selection and data extraction
We selected studies if they met all of the following criteria: 1) the study had a case-control design; 2) the study focused on the relationship between CHFR hypermethylation and risk of gastric cancer; 3) the frequency of the CHFR methylation status had been reported or could be calculated; and 4) if several studies had overlapping cancer or control cases, the studies with the largest sample size were selected in the present study.
The following information were extracted, respectively, by two investigators: last name of the first author, year of the publication, country where study conducted, subject ethnicity, testing materials, numbers of cases and controls, and the method for methylation testing in each study. The two investigators reached a consensus on all items.
Statistical analyses
The strength of the association between CHFR methylation and gastric cancer risk was assessed by odds ratio (OR) with its 95% confidence interval (CI). The heterogeneity among the studies was estimated by a chi-square-based Q-test and further quantified by the I2 metric.16 The fixed-effects model was selected to calculate the pooled OR when the between-study heterogeneity was absent.17 Otherwise, the random-effects model was selected.18 Begg’s funnel plots and Egger’s linear regression test were used to examine whether the results were affected by publication bias.19 If publication bias was observed, the nonparametric “trim and fill” method was carried out for estimating the effect of missing studies on the overall outcome.20 Moreover, subgroup analyses were also performed stratified by country, ethnicity, and methylation testing methods, respectively. All of the statistical analyses were carried out by the Stata software (version 10; Stata Corp, College Station, TX, USA). All the P-values were two-sided, and P<0.05 was considered to be statistically significant.
Results
Characteristics of included studies
According to the literature search strategy and selection criteria, 13 independent articles were eventually included in the present meta-analysis.13–15,21–30 The characteristics of all the included studies are summarized in Table 1. The 13 studies were published between 2003 and 2016, and all of them were written in English. Among the 13 studies, eight studies came from investigations involving Japanese populations, three came from China, one came from Korea, and one came from the USA. For all the enrolled studies, the gastric cancer samples were all obtained from gastric cancer tissues, and the controls were all from corresponding non-neoplastic gastric mucosa. Seven of the 13 studies used methylation-specific PCR (MSP) to detect CHFR methylation status in gastric cancer and control samples, two studies used bisulfite treatment and combined bisulfite restriction analysis (COBRA), and four studies used other methylation detection methods. Totally, 1,399 samples, including 758 gastric cancer cases and 641 controls, were involved in the present meta-analysis.
Table 1.
Characteristics of studies included in the present meta-analysis
| Author | Year | Country | Ethnicity | Materials | Cases (n) | Controls (n) | Testing methods |
|---|---|---|---|---|---|---|---|
| Satoh et al15 | 2003 | Japan | Asian | Tissue | 61 | 44 | COBRA |
| Honda et al23 | 2004 | Japan | Asian | Tissue | 71 | 71 | MSP |
| Kang et al24 | 2004 | Korea | Asian | Tissue | 43 | 14 | Bisulfite PCR and sequencing |
| Homma et al22 | 2005 | Japan | Asian | Tissue | 52 | 52 | MSP |
| Koga et al25 | 2006 | Japan | Asian | Tissue | 46 | 46 | MSP |
| Morioka et al26 | 2006 | Japan | Asian | Tissue | 53 | 53 | MSP |
| Yoshida et al30 | 2006 | Japan | Asian | Tissue | 41 | 41 | COBRA |
| Gao et al13 | 2008 | China | Asian | Tissue | 20 | 20 | MSP |
| Oki et al27 | 2009 | Japan | Asian | Tissue | 59 | 59 | MSP |
| Hiraki et al21 | 2010 | Japan | Asian | Tissue | 49 | 49 | qMSP |
| Hu et al14 | 2011 | China | Asian | Tissue | 123 | 123 | MSP |
| Wang et al29 | 2014 | China | Asian | Tissue | 117 | 46 | MethyLight |
| Sepulveda et al28 | 2016 | USA | Caucasian | Tissue | 23 | 23 | Next-generation sequencing |
Abbreviations: COBRA, combined bisulfite restriction analysis; MSP, methylation-specific PCR; qMSP, quantitative methylation-specific PCR; PCR, polymerase chain reaction.
Quantitative data analysis
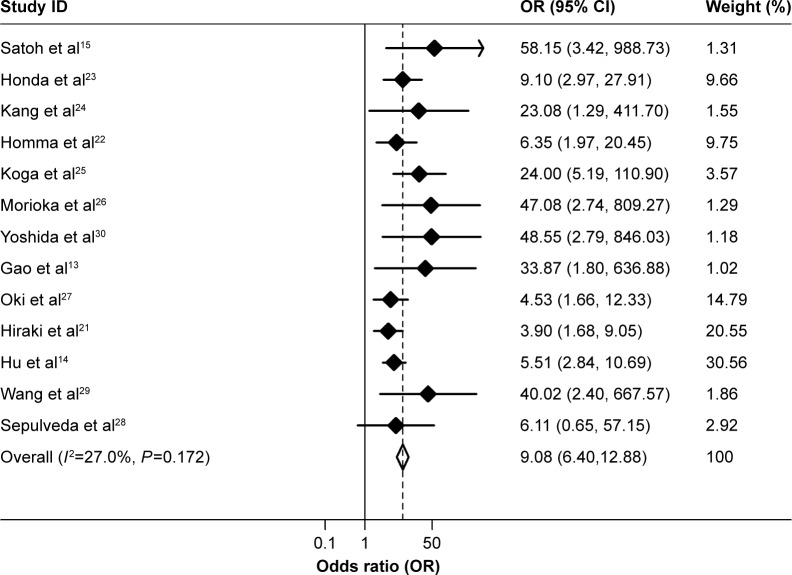
The between-study heterogeneity of all the 13 studies included in the present study was firstly analyzed, and no significant heterogeneity among them was found (P=0.172, I2=27.0%, Figure 1, Table 2). Therefore, the strength of the association between methylation of CHFR and risk of gastric cancer was determined by the fixed-effects model. Overall, compared with non-cancer controls, the pooled OR of CHFR methylation in gastric cancer patients was 9.08 (95% CI: 6.40–12.88, P<0.001, Figure 1, Table 2), suggesting that CHFR methylation was associated with an increased risk of gastric cancer.
Figure 1.
Forest plots of the association between CHFR methylation and gastric cancer risk.
Abbreviations: CI, confidence interval; OR, odds ratio.
Table 2.
Summary of the association between hypermethylation of CHFR and gastric cancer risk
| Variables | Study no | Cases/controls | OR (95% CI) | P-value | PH* | I2 (%) |
|---|---|---|---|---|---|---|
| Total | 13 | 758/641 | 9.08 (6.40–12.88) | <0.001 | 0.172 | 27.0 |
| Country | ||||||
| Japan | 8 | 432/415 | 9.29 (6.00–14.39) | <0.001 | 0.098 | 42.1 |
| China | 3 | 260/189 | 8.30 (4.48–15.40) | <0.001 | 0.170 | 43.7 |
| Ethnicities | ||||||
| Asian | 12 | 735/618 | 9.17 (6.44–13.07) | <0.001 | 0.125 | 33.1 |
| Testing methods | ||||||
| MSP | 7 | 424/424 | 8.02 (5.29–12.16) | <0.001 | 0.315 | 15.1 |
| COBRA | 2 | 102/85 | 53.60 (7.14–402.38) | <0.001 | 0.930 | 0.0 |
| Others | 4 | 232/132 | 7.75 (3.83–15.72) | <0.001 | 0.216 | 32.8 |
Note:
P-value from the Q-test for heterogeneity.
Abbreviations: OR, odds ratio; CI, confidence interval; MSP, methylation-specific PCR; COBRA, combined bisulfite restriction analysis; PCR, polymerase chain reaction.
We next performed subgroup analyses stratified by country, ethnicity, and methylation testing methods, respectively. Country-specific OR showed an increased risk for individuals carrying the methylated CHFR compared with those without CHFR gene methylation in Japan (OR=9.29, 95% CI: 6.00–14.39, P<0.001) and China (OR=8.30, 95% CI: 4.48–15.40, P<0.001, Table 2). When combining the studies regarding Japanese, Chinese and Korean together, a strong association between CHFR methylation and gastric cancer risk was found in Asian populations (OR=9.17, 95% CI: 6.44–13.07, P<0.001, Table 2). In the stratified analysis by testing methods, significantly increased risks were found in MSP (OR=8.02, 95% CI: 5.29–12.16, P<0.001) and COBRA (OR=53.60, 95% CI: 7.14–402.38, P<0.001, Table 2).
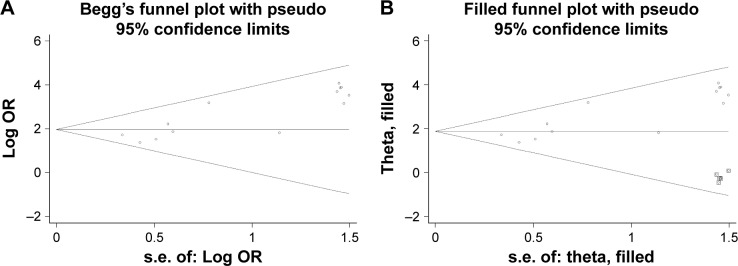
Publication bias
The shape of the funnel plots showed asymmetry in the overall analysis (Figure 2A), meanwhile the results from Egger’s test provided statistical evidence for funnel plot asymmetry (P<0.001), indicating the existence of publication bias. To adjust publication bias, we carried out a nonparametric trim and fill method to estimate potential missing studies and assess the effect that these studies might have had on the outcome. As a result, five missing studies were added to the dataset, and the filled dataset showed no evidence of publication bias (Figure 2B). The new dataset moved the estimated pooled OR from 9.68 (95% CI: 6.40–12.88) to 6.04 (95% CI: 4.26–8.57). The correction for publication bias did not change the overall interpretation of the dataset, indicating that the strong association between CHFR methylation and gastric cancer risk was statistically robust and reliable.
Figure 2.
Begg’s funnel plot of publication biases. Each point represents a separate study. (A) Begg’s funnel plot of publication bias test. (B) Begg’s funnel plot of publication bias test after trim and fill method.
Abbreviations: s.e., standard error; OR, odds ratio.
Discussion
The development of gastric cancer involves genetic or epigenetic alterations that lead to the functional loss of critical genes such as TSG, DNA repair genes, or cell-cycle checkpoint genes.31 Increasing number of cancer-related genes have been reported to be methylated in CpG islands of genes’ promoter regions. Such type of epigenetic change results in the inactivation of TSG and plays a key role in the epigenetically mediated loss-of-gene function.32 Actually, aberrant DNA methylation in the promoter regions of TSG is the most well-defined epigenetic hallmark in gastric cancer.33
In recent years, aberrant methylation of the checkpoint gene CHFR associated with gene silencing has been identified in several cancer types, including gastric cancer.10–12,15 Based on the studies on CHFR methylation and gastric cancer, we focused on the correlation between CHFR hypermethylation and risk of gastric cancer in the present study. To the best of our knowledge, this is the first meta-analysis study to assess the association of CHFR methylation with gastric cancer risk. Our analysis, combining 13 studies with 758 cancer cases and 641 control samples, revealed that the hypermethylation of CHFR increased the risk of gastric cancer. Particularly, the overall OR for CHFR methylation status in gastric cancer vs non-cancer samples was 9.08 (95% CI: 6.40–12.88), suggesting a strong association of CHFR methylation with the risk of gastric cancer.
As gastric cancer has a high prevalent rate in Asia, particularly in Japan and China,2 we summarized the effect of CHFR methylation on gastric cancer risk in these two countries, and as a result we found that CHFR methylation increased the risk of gastric cancer in both of the two countries. When we pooled the data from 12 studies on Asian populations together, a strong positive association was also observed. In addition to studies from Asian populations, one study came from the Caucasian population. The results of this study showed that the CHFR methylation was significantly increased in gastric cancer compared with non-metaplastic mucosa.28 Taken together, these data showed a parallel effect of CHFR methylation on gastric cancer risk among different ethnicities.
The advances in diagnostics and therapeutics in recent years have prolonged survival for gastric cancer patients diagnosed at the early stage. Unfortunately, the overall prognosis is very poor because most of the gastric cancer patients are diagnosed at the advanced stage.34 Therefore, an urgent task for gastric cancer research is the development of efficient diagnostic approaches to enable early detection. Several studies investigated the correlation between CHFR methylation and clinicopathological factors,13,14,21,23,24,26,27 and none of the studies showed an significant association between CHFR methylation and advanced clinicopathological factors, including tumor stage III/IV or lymphnodemetastasis. These results suggest that the CHFR methylation is an early epigenetic event in gastric cancer. To facilitate DNA methylation detection, many powerful molecular techniques have been invented in the past decades. The well-known techniques includes bisulfite sequencing, COBRA, MSP, quantitative methylation-specific PCR, as well as bisulfite pyrosequencing.35 In the present study, the association of CHFR methylation with gastric cancer risk has been stratified by methylation testing methods, and we found significant ORs in subgroups with all testing methods. Moreover, the strong association between CHFR methylation and gastric cancer risk shown in this study is consistent with previous findings that CHFR methylation could be used as an diagnostic factor for several other types of cancer, including lung, colorectal, and esophageal cancers.10–12,36 Herein, CHFR methylation is likely to be a molecular marker for early detection in gastric cancer as well as in certain other types of cancer.
Notably, aberrant methylation of CHFR had been reported to be associated with the sensitivity of microtubule inhibitors in several cancer cells including gastric cancer cells in vitro.15 However, the clinical significance of CHFR methylation and chemosensitivity of microtubule inhibitors (paclitaxel or docetaxel) in gastric cancer patients was investigated only in a few studies,25,29,30 and none of the study showed an significant association between CHFR methylation and the sensitivity of paclitaxel, docetaxel, or a combination of docetaxel and S-1. Since all these studies had very limited number of patients, further research using larger number of patients is necessary to clarify whether CHFR methylation correlates with drug response to microtubule inhibitors.
In summary, this meta-analysis showed a strong association between CHFR methylation and risk of gastric cancer. Although further studies with large number of samples are required to confirm it, the findings in the present study suggest CHFR methylation as a promising molecular marker for early detection in gastric cancer.
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Stadtländer CT, Waterbor JW. Molecular epidemiology, pathogenesis and prevention of gastric cancer. Carcinogenesis. 1999;20(12):2195–2208. doi: 10.1093/carcin/20.12.2195. [DOI] [PubMed] [Google Scholar]
- 2.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65(2):87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 3.Uemura N, Okamoto S, Yamamoto S, et al. Helicobacter pylori infection and the development of gastric cancer. N Engl J Med. 2001;345(11):784–789. doi: 10.1056/NEJMoa001999. [DOI] [PubMed] [Google Scholar]
- 4.Parsonnet J, Friedman GD, Vandersteen DP, et al. Helicobacter pylori infection and the risk of gastric carcinoma. N Engl J Med. 1991;325(16):1127–1131. doi: 10.1056/NEJM199110173251603. [DOI] [PubMed] [Google Scholar]
- 5.González CA, Sala N, Capellá G. Genetic susceptibility and gastric cancer risk. Int J Cancer. 2002;100(3):249–260. doi: 10.1002/ijc.10466. [DOI] [PubMed] [Google Scholar]
- 6.Smith MG, Hold GL, Tahara E, El-Omar EM. Cellular and molecular aspects of gastric cancer. World J Gastroenterol. 2006;12(19):2979–2990. doi: 10.3748/wjg.v12.i19.2979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kang D, Chen J, Wong J, Fang G. The checkpoint protein Chfr is a ligase that ubiquitinates Plk1 and inhibits Cdc2 at the G2 to M transition. J Cell Biol. 2002;156(2):249–259. doi: 10.1083/jcb.200108016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Scolnick DM, Halazonetis TD. Chfr defines a mitotic stress checkpoint that delays entry into metaphase. Nature. 2000;406(6794):430–435. doi: 10.1038/35019108. [DOI] [PubMed] [Google Scholar]
- 9.Chaturvedi P, Sudakin V, Bobiak ML, et al. Chfr regulates a mitotic stress pathway through its RING-finger domain with ubiquitin ligase activity. Cancer Res. 2002;62(6):1797–1801. [PubMed] [Google Scholar]
- 10.Toyota M, Sasaki Y, Satoh A, et al. Epigenetic inactivation of CHFR in human tumors. Proc Natl Acad Sci U S A. 2003;100(13):7818–7823. doi: 10.1073/pnas.1337066100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mizuno K, Osada H, Konishi H, et al. Aberrant hypermethylation of the CHFR prophase checkpoint gene in human lung cancers. Oncogene. 2002;21(15):2328–2333. doi: 10.1038/sj.onc.1205402. [DOI] [PubMed] [Google Scholar]
- 12.Shibata Y, Haruki N, Kuwabara Y, et al. Chfr expression is down-regulated by CpG island hypermethylation in esophageal cancer. Carcinogenesis. 2002;23(10):1695–1699. doi: 10.1093/carcin/23.10.1695. [DOI] [PubMed] [Google Scholar]
- 13.Gao YJ, Xin Y, Zhang JJ, Zhou J. Mechanism and pathobiologic implications of CHFR promoter methylation in gastric carcinoma. World J Gastroenterol. 2008;14(32):5000–5007. doi: 10.3748/wjg.14.5000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hu SL, Huang DB, Sun YB, et al. Pathobiologic implications of methylation and expression status of Runx3 and CHFR genes in gastric cancer. Med Oncol. 2011;28(2):447–454. doi: 10.1007/s12032-010-9467-6. [DOI] [PubMed] [Google Scholar]
- 15.Satoh A, Toyota M, Itoh F, et al. Epigenetic inactivation of CHFR and sensitivity to microtubule inhibitors in gastric cancer. Cancer Res. 2003;63(24):8606–8613. [PubMed] [Google Scholar]
- 16.Higgins JP, Thompson SG, Deeks JJ, Altman DG. Measuring inconsistency in meta-analyses. BMJ. 2003;327(7414):557–560. doi: 10.1136/bmj.327.7414.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mantel N, Haenszel W. Statistical aspects of the analysis of data from retrospective studies of disease. J Natl Cancer Inst. 1959;22(4):719–748. [PubMed] [Google Scholar]
- 18.DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials. 1986;7(3):177–188. doi: 10.1016/0197-2456(86)90046-2. [DOI] [PubMed] [Google Scholar]
- 19.Egger M, Davey Smith G, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997;315(7109):629–634. doi: 10.1136/bmj.315.7109.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Duval S, Tweedie R. Trim and fill: a simple funnel-plot-based method of testing and adjusting for publication bias in meta-analysis. Biometrics. 2000;56(2):455–463. doi: 10.1111/j.0006-341x.2000.00455.x. [DOI] [PubMed] [Google Scholar]
- 21.Hiraki M, Kitajima Y, Sato S, et al. Aberrant gene methylation in the lymph nodes provides a possible marker for diagnosing micrometastasis in gastric cancer. Ann Surg Oncol. 2010;17(4):1177–1186. doi: 10.1245/s10434-009-0815-8. [DOI] [PubMed] [Google Scholar]
- 22.Homma N, Tamura G, Honda T, et al. Hypermethylation of Chfr and hMLH1 in gastric noninvasive and early invasive neoplasias. Virchows Arch. 2005;446(2):120–126. doi: 10.1007/s00428-004-1146-6. [DOI] [PubMed] [Google Scholar]
- 23.Honda T, Tamura G, Waki T, Kawata S, Nishizuka S, Motoyama T. Promoter hypermethylation of the Chfr gene in neoplastic and non-neoplastic gastric epithelia. Br J Cancer. 2004;90(10):2013–2016. doi: 10.1038/sj.bjc.6601849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kang HC, Kim IJ, Park JH, et al. Promoter hypermethylation and silencing of CHFR mitotic stress checkpoint gene in human gastric cancers. Oncol Rep. 2004;12(1):129–133. [PubMed] [Google Scholar]
- 25.Koga Y, Kitajima Y, Miyoshi A, Sato K, Sato S, Miyazaki K. The significance of aberrant CHFR methylation for clinical response to microtubule inhibitors in gastric cancer. J Gastroenterol. 2006;41(2):133–139. doi: 10.1007/s00535-005-1732-7. [DOI] [PubMed] [Google Scholar]
- 26.Morioka Y, Hibi K, Sakai M, et al. Aberrant methylation of the CHFR gene in digestive tract cancer. Anticancer Res. 2006;26(3A):1791–1795. [PubMed] [Google Scholar]
- 27.Oki E, Zhao Y, Yoshida R, et al. Checkpoint with forkhead-associated and ring finger promoter hypermethylation correlates with microsatel-lite instability in gastric cancer. World J Gastroenterol. 2009;15(20):2520–2525. doi: 10.3748/wjg.15.2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sepulveda JL, Gutierrez-Pajares JL, Luna A, et al. High-definition CpG methylation of novel genes in gastric carcinogenesis identified by next-generation sequencing. Mod Pathol. 2016;29(2):182–193. doi: 10.1038/modpathol.2015.144. [DOI] [PubMed] [Google Scholar]
- 29.Wang M, Shen L, Deng D. Association between CHFR methylation and chemosensitivity of paclitaxel in advanced gastric cancer. Med Oncol. 2014;31(4):907. doi: 10.1007/s12032-014-0907-6. [DOI] [PubMed] [Google Scholar]
- 30.Yoshida K, Hamai Y, Suzuki T, Sanada Y, Oue N, Yasui W. DNA methylation of CHFR is not a predictor of the response to docetaxel and paclitaxel in advanced and recurrent gastric cancer. Anticancer Res. 2006;26(1A):49–54. [PubMed] [Google Scholar]
- 31.Tamura G. Alterations of tumor suppressor and tumor-related genes in the development and progression of gastric cancer. World J Gastroenterol. 2006;12(2):192–198. doi: 10.3748/wjg.v12.i2.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Das PM, Singal R. DNA methylation and cancer. J Clin Oncol. 2004;22(22):4632–4642. doi: 10.1200/JCO.2004.07.151. [DOI] [PubMed] [Google Scholar]
- 33.Qu Y, Dang S, Hou P. Gene methylation in gastric cancer. Clin Chim Acta. 2013;424:53–65. doi: 10.1016/j.cca.2013.05.002. [DOI] [PubMed] [Google Scholar]
- 34.Karimi P, Islami F, Anandasabapathy S, Freedman ND, Kamangar F. Gastric cancer: descriptive epidemiology, risk factors, screening, and prevention. Cancer Epidemiol Biomarkers Prev. 2014;23(5):700–713. doi: 10.1158/1055-9965.EPI-13-1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Umer M, Herceg Z. Deciphering the epigenetic code: an overview of DNA methylation analysis methods. Antioxid Redox Signal. 2013;18(15):1972–1986. doi: 10.1089/ars.2012.4923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Derks S, Cleven AH, Melotte V, et al. Emerging evidence for CHFR as a cancer biomarker: from tumor biology to precision medicine. Cancer Metastasis Rev. 2014;33(1):161–171. doi: 10.1007/s10555-013-9462-4. [DOI] [PMC free article] [PubMed] [Google Scholar]