Abstract
In cycling between the mammalian host and the tsetse fly vector, trypanosomes undergo major changes in energy metabolism and surface coat composition. Early procyclic (insect) forms in the tsetse fly midgut are coated by glycoproteins known as EP and GPEET procyclins. EP expression continues in late procyclic forms, whereas GPEET is down-regulated. In culture, expression of GPEET is modulated by glycerol or glucose. Here, we demonstrate that a glycerol-responsive element of 25 nucleotides within the 3′ untranslated region of GPEET mRNA also controls expression by glucose and during development in the fly. In trypanosomes, mitochondrial ATP is produced mainly by the acetate: succinate-CoA transferase/succinyl-CoA synthetase (ASCT) cycle, the citric acid cycle, and the cytochromes. Silencing of the pyruvate dehydrogenase or succinyl-CoA synthetase from the ASCT cycle by RNA interference induces reexpression of GPEET in late procyclic forms, whereas inhibition of the citric acid cycle or the cytochromes has no effect. In contrast, inhibition of the alternative oxidase, the second branch of the electron transport chain, with salicylhydroxamic acid overrides the effect of glucose or glycerol and causes a reduction in the level of GPEET mRNA. Our results reveal a new mechanism by which expression of a surface glycoprotein is controlled by the activity of mitochondrial enzymes.
INTRODUCTION
The protozoan parasite Trypanosoma brucei, which causes human sleeping sickness, encounters different nutrients as it cycles between the mammalian host and the tsetse fly vector. In the glucose-rich environment of the mammalian bloodstream, long slender forms rely solely on glycolysis, which occurs in a specialized organelle, the glycosome, for their energy production (Opperdoes, 1987). At this stage of the life cycle, the single mitochondrion has a rudimentary structure and does not possess a citric acid cycle or a cytochrome-dependent electron transport chain. One of the main functions of this organelle in bloodstream forms is to reoxidize reducing equivalents generated during glycolysis by transferring electrons to the alternative oxidase (AO) (Clarkson et al., 1989). In contrast, procyclic (insect) forms are not dependent on glucose, although they are able to metabolize it. At this stage of the life cycle, the parasite has a highly developed mitochondrion, containing the complete set of proteins involved in the citric acid cycle and the electron transport chain, which enables the parasite to produce ATP by oxidative phosphorylation (Tielens and Van Hellemond, 1998). The mitochondrion also is equipped with an acetate:succinate CoA transferase/succinyl CoA synthetase (ASCT) cycle, which converts acetyl CoA to acetate and results in the formation of extra ATP (Van Hellemond et al., 1998). Surprisingly, procyclic forms can survive as long as either the ASCT cycle or the citric acid cycle is functional, but ablation of both pathways results in growth arrest (Bochud-Allemann and Schneider, 2002; van Weelden et al., 2003).
Differentiation of bloodstream to procyclic forms is also marked by shedding of the variant surface glycoprotein coat and expression of a new coat composed of procyclins (Roditi et al., 1989). There are two major classes of procyclin, GPEET and EP, which differ mainly by their internal pentapeptide (GPEET) or dipeptide (EP) repeat motifs (Roditi and Clayton, 1999). EP can be further subdivided into three isoforms, which are distinguished by the presence or absence of an N-linked glycan. In early procyclic forms, which predominate during the establishment of an infection of the tsetse fly midgut, GPEET is the major surface protein. GPEET is repressed after 4-7 d, whereas glycosylated EP continues to be expressed in late procyclic forms (Vassella et al., 2000; Acosta-Serrano et al., 2001). The same sequence of events is also observed in vitro unless glycerol is present in the medium, in which case GPEET is not repressed (Vassella et al., 2000, 2001). GPEET expression can also be modulated by other factors in the environment. Exposing trypanosomes to hypoxic conditions accelerates GPEET repression and overrides the effect of glycerol. More recently, it was demonstrated that silencing of hexose transporters or glycosomal enzymes involved in glucose metabolism by double-stranded RNA interference (RNAi) induces reexpression of GPEET in late procyclic forms (Morris et al., 2002). Consistent with these results, glucose depletion also induces GPEET in these cells.
Regulation of GPEET by glycerol is posttranscriptional, presumably at the level of RNA stability, and is conferred by the 3′ untranslated region (UTR) of GPEET mRNA (Vassella et al., 2000). In the presence of glycerol, the GPEET 3′ UTR gives rise to twofold higher expression of a reporter gene when compared with the EP1 3′ UTR, whereas in the absence of glycerol it mediates a 50-fold reduction in expression, suggesting a dual function of this sequence in early and late procyclic forms. Analysis of chimeras of the GPEET and EP1 3′ UTR allowed the glycerol-responsive element (GRE) to be mapped to 60 nucleotides (nt) within this region. The GPEET 3′ UTR also regulates expression of a reporter gene in the fly and down-regulates GPEET mRNA under hypoxic conditions (Vassella et al., 2000), but it is not known whether this is controlled by the same element.
Several regulatory elements have been identified in the 3′ UTR of EP1 procyclin. This sequence contains three stem-loop domains (LI-LIII) (Furger et al., 1997; Drozdz and Clayton, 1999) each of which plays a role in procyclin expression. LI and LIII stabilize the mRNA in bloodstream and procyclic forms (Furger et al., 1997; Schürch et al., 1997) and a 16-mer in the LIII domain was shown to function both as a determinant of RNA stability (Furger et al., 1997) and as a translational enhancer (Hehl et al., 1994) by promoting association of the mRNA with polysomes (Roditi et al., 1998). In contrast, the LII domain contains a 26-mer that destabilizes the mRNA (Hotz et al., 1997). However, the reduction in steady-state mRNA is not sufficient to account for the difference in the level of EP procyclin between bloodstream and procyclic forms, suggesting that there is an additional layer of translational control (Furger et al., 1997; Schürch et al., 1997). The 16-mer and the 26-mer are also conserved in the GPEET 3′ UTR, suggesting that they may coordinate expression of EP1 and GPEET procyclins, but this has not been confirmed experimentally.
In trypanosomes, both energy metabolism and surface coat composition can differ considerably between different life cycle stages, but are these processes linked? In this study, we have analyzed the effect of specific inhibitors and conditional knockdowns or knockout mutants of enzymes involved in energy metabolism of glucose or glycerol and demonstrate that GPEET expression is modulated by the activity of mitochondrial enzymes. To investigate whether these signals regulate GPEET expression by the same mechanism, we have defined the glycerol-responsive element in the 3′ UTR of GPEET mRNA more precisely and could show that the same element also regulates expression of a reporter gene in response to glucose or during development in the fly. Our results provide a new mechanism by which expression of a major surface protein is controlled by the activity of mitochondrial pathways.
MATERIALS AND METHODS
Trypanosomes
Procyclic forms of the pleomorphic strain T. b. brucei AnTat1.1 (Le Ray et al., 1977), and procyclic forms of T. brucei stock 427 (Cross and Manning, 1973), clone 29-13 (Wirtz et al., 1999), and RNAi cell lines derived from this clone (Bochud-Allemann and Schneider, 2002) were used in this study. Cells were cultured in SDM-79 (Brun and Schönenberger, 1979) supplemented with 10% fetal bovine serum. Only freshly differentiated AnTat 1.1 cells, which were kept in culture for a maximum of 1 mo, were used. Short stumpy bloodstream forms were triggered to differentiate to the procyclic form by the addition of 6 mM cis-aconitate to the culture medium and lowering the incubation temperature to 27°C as described previously (Brun and Schönenberger, 1981). Normal SDM-79 contains ∼5 mM glucose. Low glucose SDM-79 containing ∼0.3 mM glucose was prepared according to Brun and Schöenenberger (1979) by using the following modifications: the medium was supplemented with glucose-free MEM Earle's balanced salt solution (Invitrogen, Inotech, Dottikon, Switzerland), and additional glucose was omitted. In addition, the concentration of TC199 was reduced fourfold, and 26 mM NaCl, 9 mM KCl, 0.3 mM CaCl2, 111 μM sodium acetate, 0.4 μM hypoxanthine, 0.2 μM l-cysteine, 0.2 mM d/l-glutamate, 0.1 mM glycine, and 0.8 mM d/l-aspartate were added to compensate for the major components of TC199. The medium was supplemented with 10% dialyzed fetal bovine serum (Invitrogen).
Stable and transient transfection of trypanosomes and chloramphenicol acetyltransferase (CAT) assays were performed as described previously (Hehl et al., 1994; Vassella et al., 2000).
Constructs
For in vitro transcription of the entire GPEET 3′ UTR, pBS-GPEET3′UTR was constructed as follows. The GPEET 3′ UTR was amplified from pC-CAT/GPEET (Vassella et al., 2000) by using the primer pair 226 from the chloramphenicol acetyltransferase (CAT) gene and RNA D from the 3′ end of the 3′ UTR (Hehl et al., 1994). The PCR product was digested with Sau3A and inserted into the BamHI site of pBluescript KS(+) (Stratagene, La Jolla, CA). The sequences between the T3 promoter and the start of the GPEET 3′ UTR were excised with KpnI and SmaI, the ends were treated with T4 polymerase and religated. For in vitro transcription, the plasmid was linearized with XbaI.
Mutations were introduced into the GPEET 3′ UTR by site-directed mutagenesis in two sequential amplification steps. For each construct, two complementary primers were designed containing specific mutations as indicated in Figure 1. The 5′ end of the GPEET 3′ UTR and upstream sequences were amplified by PCR by using the antisense primer containing the mutations and the primer SPU (Ruepp et al., 1997) from the procyclin promoter. The 3′ end of the UTR and intergenic sequences were amplified using the corresponding sense primer and primer bleNheas (5′-CCATGATCAAGCTAGCTTGT-3′) from the phleomycin-resistance gene. For construction of CAT/GPEETM1-5, pC-CAT/GPEET was used as a template for both PCR reactions. For construction of pC-CAT/GPEETM24 and M234, pC-CAT/GPEETM4 was used. In a second step, the products of the two PCR reactions were allowed to anneal by hybridization of the complementary sequences, amplified using the primers SPU and bleNheas, and cloned between the BamHI and NheI sites of pCAT/EP1-ble (Vassella et al., 2000), thereby replacing the 3′ UTR and intergenic sequences of the EP1 gene.
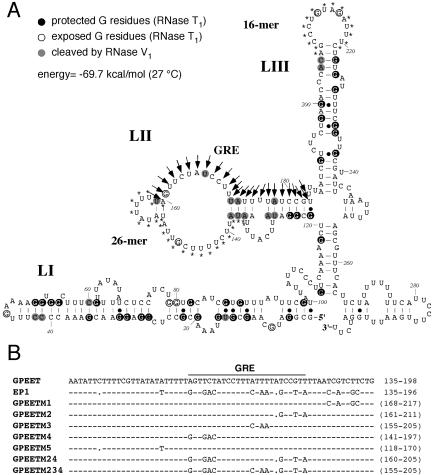
Figure 1.
Analysis of GRE by site-directed mutagenesis. (A) Secondary structure of the GPEET 3′ UTR, modeled by the MFOLD program without constraints (Zucker and Jacobson, 1998), by using the server of the Institute of Medical Computing (Washington University, St. Louis, MO). The temperature was set to 27°C. GRE is indicated by arrows, and the 26-mer (Hotz et al., 1997) and 16-mer (Hehl et al., 1994) by asterisks. (B) Sequence alignment of the GPEET and EP1 3′ UTRs within the region containing the regulatory element, together with the constructs containing mutations introduced into the GPEET 3′ UTR by site-directed mutagenesis. The numbering starts at the beginning of the 3′ UTR. The numbers in brackets indicate the 5′ and 3′ positions of the oligonucleotides used for site-directed mutagenesis.
pC-CAT/GPEETLII also was constructed by two sequential amplification steps. The LII domain of the GPEET 3′ UTR was amplified from pC-CAT/GPEET by using the primer pair GPEETLIIs (5′-CGGGATCCGCGGATATTCATTTAATATT-3′) and GPEETLIIas (5′-CAAAGGAAAACGGATAAAATAAAGGA-3′). The latter is a hybrid primer containing the sequences from nt positions 167-184 of the GPEET 3′ UTR followed by the sequences from nt positions 282-289 of the end of the 3′ UTR. The intergenic region downstream of the 3′ UTR was amplified using the primer GPEETpAS (5′-TTTTATCCGTTTTCCTTTGAATTTGGATC-3′), which overlaps with GPEETLIIas, and bleNheIas. In a second amplification step, the PCR products were allowed to anneal and amplified using the primer pair GPEETLIIs and bleNheIas. The PCR product was cloned between the BamHI and NheI sites of pC-CAT/EP1. All constructs were checked by sequencing.
For construction of GARP cassettes containing the mutated 3′ UTRs, GARP was excised from pGAPRONE (Furger et al., 1997) and cloned between the HindIII and BamHI sites of the CAT constructs, thereby replacing the CAT gene. For stable transformation, they were linearized with SalI and XbaI.
Infection of Tsetse Flies and Immunofluorescence
Infection of tsetse flies and isolation of midgut forms were performed as described previously (Ruepp et al., 1997). For immunofluorescence, cell smears were fixed with acetone (Vassella et al., 2000). EP and the phosphorylated form of GPEET were detected using the monoclonal antibodies (mAbs) TRBP1/247 (Richardson et al., 1988) and 5H3 (Bütikofer et al., 1999), respectively, and the unphosphorylated form of GPEET was detected with the polyclonal rabbit antiserum K1 (Ruepp et al., 1997). A rabbit polyclonal antiserum against GARP was kindly provided by D. Jefferies and J. D. Barry (Wellcome Centre of Molecular Parasitology, Glasgow, Scotland). All antibodies were used at a dilution of 1:500. For double immunofluorescence, Alexa 488-conjugated anti-rabbit (Molecular Probes, Eugene, OR) and tetramethylrhodamine B isothiocyanate-conjugated anti-mouse (Sigma-Aldrich, St. Louis, MO) secondary antibodies were used at dilutions of 1:500-1:1000. Cells were counterstained with 4,6-diamidino-2-phenylindole (DAPI).
Northern and Western Blot Analyses
Northern blot analysis was performed using standard procedures (Sambrook et al., 1989). Megaprime-labeled probes (Amersham Biosciences, Freiburg, Germany) used for hybridization were generated from internal regions of the EP1 and GPEET genes encoding the dipeptide or pentapeptide repeats, respectively. Hybridization and washing were performed at Tm-7°C. An oligonucleotide that hybridizes to the 18S rRNA (Flück et al., 2003) was used as an internal control for sample loading.
For Western blots, total cell protein extracts were separated on 12% polyacrylamide gels and transferred to Immobilon-P (Millipore, Bedford, MA). An mAb directed against the AO (Chaudhuri et al., 1998), obtained from G. C. Hill (Vanderbilt University Medical Center, Nashville, TN), was used at a dilution of 1:100.
In Vitro Transcription
In vitro transcription of the GPEET 3′ UTR was carried out in a total volume of 50 μl, containing 1 μg of linearized plasmid DNA, 20 U of T3 RNA polymerase (Stratagene), 0.6 mM of each rNTP and 20 U RNasin (Roche Diagnostics, Indianapolis, IN). Incubation was overnight at 37°C. The product was purified by phenol extraction and ethanol precipitation and subjected to dephosphorylation, by using 2 U of calf intestinal phosphatase (Roche Diagnostics) and 40 U of RNasin for 1 h at 37°C. The product was separated on a 10% polyacrylamide gel. After electrophoresis, the gel was stained with 3 μg/ml ethidium bromide, the band containing the RNA was excised and eluted overnight with 400 μl of elution butter (0.5 M ammonium acetate, 1 mM EDTA, and 0.1% SDS) in the presence of 50 μl of acidic phenol. The RNA was collected by ethanol precipitation, subjected to 5′ labeling using 10 U of T4 polynucleotide kinase (Roche Diagnostics) and 1 MBq of [γ-32P]ATP (Hartmann Analytic, Braunschweig, Germany) for 45 min at 37°C and gelpurified.
Nuclease Digestion
RNase T1 digestion under denaturing conditions was carried out in a total volume of 20 μl containing 2.5 × 104-2.5 × 105 cpm of radiolabeled RNA, 9 μg of tRNA, and 5 U of RNase T1 (Sigma-Aldrich) in Gc buffer (10 mM citrate, 0.5 mM EDTA, and 2 M urea, pH 5.4) for 10 min at 55°C. RNase T1 digestion under nondenaturing conditions was performed in GE buffer (25 mM Tris-HCl, 100 mM KCl, and 10 mM MgCl2, pH 7.9) at 25°C for 30 min. RNase V1 digestion was carried out in a total volume of 15 μl containing 2.5 × 104 cpm of radiolabeled RNA and 0.036 U RNase V1 (Amersham Biosciences) in 25 mM Tris-HCl, 10 mM MgCl2, and 0.2 M NaCl, pH 7.2. After digestion, the RNA was purified by phenol extraction and ethanol precipitation, and the products were denatured and separated on a 10% polyacrylamide gel. A single nucleotide RNA ladder (end-labeled RNA partially hydrolyzed by boiling for 1 min in 50 mM NaOH) was run on the same gel.
RESULTS
Secondary Structure of the GPEET 3′ UTR
Before embarking on a detailed analysis of GRE, we needed to determine the secondary structure of the GPEET 3′ UTR because this would allow us to design specific mutants without perturbing the conformation of the mRNA. Computer modeling using MFOLD (Zucker and Jacobson, 1998) predicts that the GPEET and EP1 3′ UTRs have very similar secondary structures (compare Figure 1A and Furger et al., 1997; Drozdz and Clayton, 1999), but only the secondary structure of the EP1 3′ UTR has been confirmed experimentally (Drozdz and Clayton, 1999). In both cases, LI and LIII are predicted to form hairpin-like structures, whereas LII is present mainly in a single-stranded conformation. The 3′ UTR of GPEET was transcribed in vitro, radioactively labeled at its 5′ end and subjected to digestion with RNase T1. Under denaturing conditions, all G residues are accessible to cleavage with this enzyme, but under native conditions only those residues which are present in single-stranded conformation are cleaved. According to the pattern of degradation products, protected and exposed G residues of the GPEET 3′ UTR were identified. In addition, the RNA was subjected to RNase V1 cleavage, which is specific for nt in double-stranded regions. The results from this analysis are summarized in Figure 1A and confirm that the computer prediction of the secondary structure and the experimental data are in very good agreement. However, weak cleavage by RNase V1 in the upper part of LII indicates that this region may alternate between single- and double-stranded conformations.
Mapping of the Glycerol-responsive Element
Sequence comparison of the EP1 and GPEET 3′ UTRs revealed only 17 mismatches in the region encompassing GRE (Figure 1B). Our strategy to map GRE was to replace stretches of the GPEET gene by the corresponding sequence of EP1 by site-directed mutagenesis, because the two 3′ UTRs have similar secondary structures. As a basic construct for the introduction of these mutations, we used pC-CAT/GPEET (Vassella et al., 2000), containing the promoter and 5′ UTR of GPEET, followed by the CAT coding region and the GPEET 3′ UTR and downstream intergenic sequences. Five constructs (GPEETM1-5) were generated containing two to four nt changes in the GPEET 3′ UTR as indicated in Figure 1B. The CAT activity of trypanosomes, transiently transfected with these constructs, was expressed relative to that obtained with pC-CAT/EP1 containing the EP1 3′ UTR. We have shown previously that this construct gives rise to similar absolute enzyme activities in trypanosomes cultured in the presence or absence of glycerol (Vassella et al., 2000). Consistent with published results, construct pC-CAT/GPEET produced ∼100-fold less CAT activity in cells cultured without glycerol than in glycerol-treated cells (Figure 2A; Vassella et al., 2000). Mutation M3 resulted in approximately threefold less CAT activity relative to pC-CAT/GPEET in cells cultured in the presence of glycerol, whereas M2 and M4 resulted in approximately threefold more activity in the absence of glycerol and the remaining mutations had no significant effect (Figure 2A). Thus, none of the mutated sequences could prevent down-regulation of GPEET in the absence of glycerol. It is possible, however, that these sequences may collectively contribute to the binding of a factor to this region, and that mutating only a few nt may not be sufficient to abolish binding. Two further constructs were made containing longer stretches of mutated sequences. By combining the mutations from pC-CAT/GPEETM2 and M4 in pC-CAT/GPEETM24, an approximately ninefold increase in activity was obtained relative to pC-CAT/GPEET in trypanosomes cultured in the absence of glycerol. However, only when we combined three mutations, giving rise to construct pC-CAT/GPEETM234, did the element become completely independent of glycerol. The CAT activity produced by this construct was ∼48-fold higher than that of pC-CAT/GPEET in trypanosomes cultured in the absence of glycerol and approximately twofold lower in the presence of glycerol. From these results, we concluded that GRE maps to a region encompassing nt positions 160-184 of the GPEET 3′ UTR and thus is situated immediately downstream of the 26-mer in the LII domain (Figure 1A).
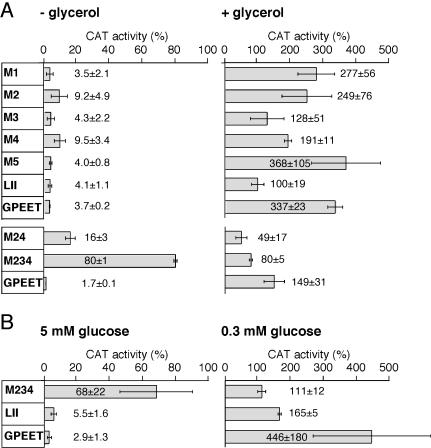
Figure 2.
Transient CAT assays to delineate GRE. (A) Transient transfection of AnTat1.1 procyclic forms cultured in the presence (+) or absence (-) of 20 mM glycerol. (B) Transient transfection of procyclic forms of T. brucei stock 427 cultured in 0.3 or 5 mM glucose, respectively. For direct comparisons, the same clone of trypanosomes was used in this experiment as in Morris et al. (2002) and for the RNAi clones in Table 1 (also see Bochud-Allemann and Schneider, 2002). Values are presented as the means (n = 4) ± SD of CAT activity relative to the activity obtained with the construct pCCAT/EP1. This construct gave rise to similar absolute enzyme activities in early or late procyclic forms (Vassella et al., 2000). Consistent with previous observations, the relative enzyme activity produced by pC-CAT/GPEET in the presence of glycerol may vary slightly depending on the length of time of the procyclic forms in culture (Vassella et al., 2000, 2001; unpublished data). Individual panels show CAT activities from the same series of experiments.
To investigate whether the LII domain could function autonomously, the LI domain (nt positions 1-120) and LIII domain (nt positions 185-279) were deleted giving rise to construct pC-CAT/GPEETLII. As shown in Figure 2A, the truncated UTR of 84 nt was able to mediate complete down-regulation of CAT activity in the absence of glycerol. In contrast, this construct produced threefold less CAT activity than the wild-type construct in the presence of glycerol, indicating the requirement of additional elements outside the LII domain for full activity.
GRE Also Regulates Expression of GPEET by Glucose and during Development in the Fly
Glucose depletion induces reexpression of GPEET in late procyclic forms, but it is not known whether this is controlled by the same mechanism as in early procyclic forms. To establish whether this is controlled by the GPEET 3′ UTR, late procyclic forms were cultured in low concentrations of glucose (0.3 mM), giving rise to ≥90% GPEET-positive cells. When these were subjected to transient transfection with pC-CAT/GPEET, they produced 154-fold more CAT activity than GPEET-negative trypanosomes cultured in 5 mM glucose (Figure 2B). To investigate whether this was due to GRE, cells were transfected with construct pC-CAT/GPEETM234. This resulted in 4 times less activity than the construct with the wild-type (wt) 3′ UTR in cells cultured in 0.3 mM glucose and 23 times more activity in cells cultured in 5 mM glucose. Construct pC-CAT/GPEETLII gave rise to a 30-fold difference in activity between the two cultures, consistent with the results obtained with cells cultured with or without glycerol. In conclusion, expression of GPEET by glucose and glycerol is controlled by the same element.
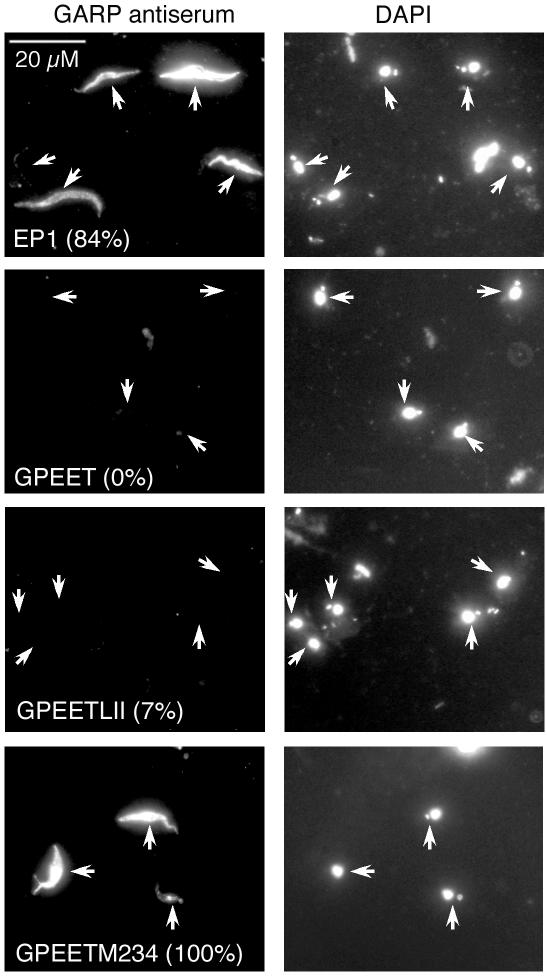
To investigate whether GRE down-regulates GPEET during development in the fly, we generated a series of stable T. brucei transformants expressing the T. congolense surface protein glutamic acid/alanine-rich protein (GARP) (Bayne et al., 1993; Beecroft et al., 1993) as a reporter gene. For these experiments, bicistronic constructs were used to target the GPEET/PAG3 locus (Roditi and Clayton, 1999) and to replace tandemly linked GPEET and EP3 genes by GARP and an antibiotic-resistance gene, respectively (Vassella et al., 2000). In these constructs, GARP is flanked by the different 3′ UTRs. Correct integration of the constructs was confirmed by Southern blot analysis (unpublished data). By Western blot analysis, we were able to show that early procyclic forms of all cell lines expressed GARP at similar levels (unpublished data). These clones were then used for fly infections and analyzed for GARP expression after 14 d. At this time point all cells had completely repressed GPEET, as indicated by immunofluorescence (unpublished data). Consistent with previous results (Vassella et al., 2000), clone GARP/GPEET, in which GARP is flanked by the GPEET 3′ UTR, also had completely repressed GARP, whereas clone GARP/EP1, in which GARP is fused to the EP1 3′ UTR, still expressed it (Figure 3). Expression of GARP from construct pGARP/GPEETLII, in which the reporter gene is fused to the LII domain of GPEET, mirrored that of pGARP/GPEET with only a minority of cells (7%) still expressing GARP at this time point. In the clone GARP/GPEETM234, however, the majority of cells still expressed GARP 14 d postinfection. Thus, the same element also controls expression of GPEET during development in the fly.
Figure 3.
Immunofluorescence of transgenic trypanosomes of T. brucei 427 isolated from the midgut of tsetse flies 14 d after infection. Cells were stained with an antiserum against GARP (left image) and counterstained with DAPI (right image). Trypanosomes from the same fields are highlighted with white arrows. The numbers indicate the percentage of GARP-positive cells. All cells were GPEET-negative at this time point.
Inhibition of the ASCT Cycle, but Not the Citric Acid Cycle, Induces GPEET Expression
Morris et al. (2002) demonstrated that ablation of the activity of glycosomal enzymes by RNAi induces reexpression of GPEET by late procyclic forms. GPEET expression might be regulated by either the energy metabolism in the glycosome, or alternatively, by that in the mitochondrion, because metabolic pathways of both organelles are presumably affected in these conditional knockdowns. To test whether this was due to changes in mitochondrial metabolism, we analyzed an RNAi knockdown of the succinyl-CoA synthetase (SCoAS) (Bochud-Allemann and Schneider, 2002), which is involved in both the ASCT cycle and the citric acid cycle (Figure 4A). Tetracycline-induced ablation of the activity of this enzyme resulted in strong expression of GPEET in 25% of the population after 7 d (Table 1). A similar percentage of GPEET-positive cells also was observed by Morris et al. (2002), suggesting that GPEET reappears with slow kinetics. This was clearly due to RNA interference, however, because only ∼2% GPEET-positive cells were obtained in the uninduced control. To discriminate between the ASCT cycle and the citric acid cycle, we analyzed a series of RNAi cell lines involved in only one of the two pathways. Ablation of the pyruvate dehydrogenase (PDH) in the ASCT cycle also resulted in GPEET expression. In contrast, ablation of α-ketoglutarate dehydrogenase (KDH) or succinate dehydrogenase (SDH) of the citric acid cycle did not induce GPEET, nor did an RNAi clone in which both the KDH and SDH were down-regulated (Table 1). It was shown previously that transcript ablation of these RNAi clones resulted in 85-90% less activity of the respective enzyme relative to the uninduced control (Bochud-Allemann and Schneider, 2002). Because we could not formally exclude that GPEET was not induced because of residual activity of KDH and SDH, we also analyzed an aconitase null mutant (van Weelden et al., 2003). In agreement with the results from the RNAi cell lines, null mutant cells were GPEET-negative (unpublished data). In conclusion, the ASCT cycle, but not the citric acid cycle, is important for GPEET expression.
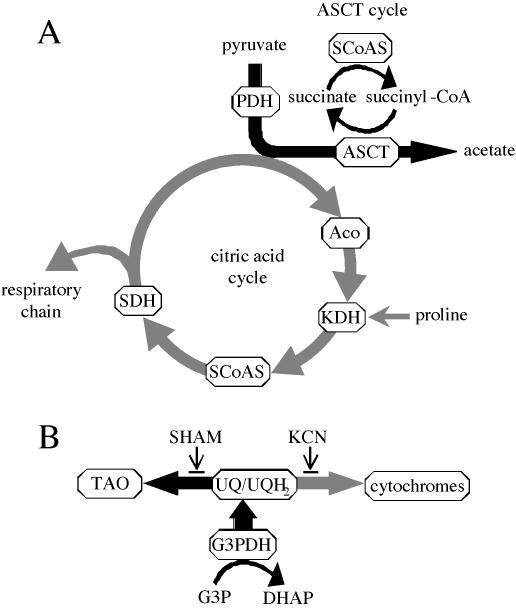
Figure 4.
Major metabolic pathways of energy production in the mitochondrion of T. brucei. (A) Depiction of the three major metabolic pathways for energy production from pyruvate and proline: the ASCT cycle, the citric acid cycle, and the respiratory chain. (B) Pathways of glycerol metabolism. Black arrows indicate pathways modulating GPEET expression. Aco, aconitase; DHAP, dihydroxyacetone phosphate; G3P, glycerol 3-phosphate; G3PDH, glycerol-3-phosphate dehydrogenase; UQ/UQH2, ubiquinone/ubiquinol.
Table 1.
% GPEET-positive cells of the different conditional knockdowns
| Gene ablated by RNAi
|
+ Tetracyclinea
|
− Tetracycline
|
||
|---|---|---|---|---|
| % GPEET +b | nc | % GPEET +b | nc | |
| PDH | 15 ± 4 | 4 | 2 ± 3 | 4 |
| SCoAS | 25 ± 7 | 4 | 2 ± 2 | 3 |
| KDH | 1 ± 1 | 4 | 1 ± 1 | 4 |
| SDH | 1 ± 1 | 4 | 1 ± 1 | 4 |
| KDH/SDH | 1 ± 1 | 3 | 0.3 ± 0.6 | 3 |
Cells were induced with 2 μg/ml tetracycline for 6-8 d.
Percentage of GPEET-positive cells from at least 100 cells analyzed. The mean (±SD) is presented.
Number of independent experiments.
GPEET Expression Is Controlled by Electron Transport to the AO
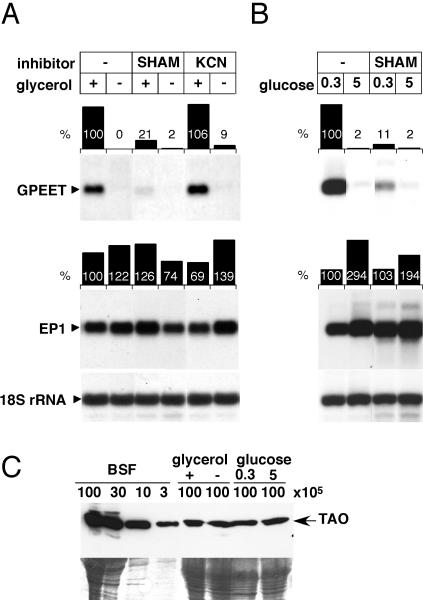
Glycerol is converted to glycerol 3-phosphate by a glycerol kinase in the glycosomes (Krakow and Wang, 1990). The subsequent oxidation to dihydroxyacetone phosphate is catalyzed by a FAD-linked glycerol-3-phosphate dehydrogenase localized in the intermembrane space of the mitochondrion (Opperdoes, 1987) (Figure 4B). This reaction is coupled to a concomitant transfer of electrons to ubiquinone (Opperdoes, 1987). In procyclic forms, cytochrome c oxidase and the AO can both function as terminal oxidases for the electron transport chain, but only the former produces ATP. We first analyzed whether GPEET mRNA stability in early procyclic forms might be affected by oxidative phosphorylation. Cells were cultured in the presence of cyanide, a specific inhibitor of cytochrome c oxidase, but GPEET was not repressed under these conditions. To investigate whether the second branch of the transport chain is important, cells were cultured in the presence of salicylhydroxamic acid (SHAM), an inhibitor of the AO. As shown in Figure 5A, this resulted in almost complete disappearance of GPEET mRNA in the presence of glycerol. Under the same conditions the levels of EP1 mRNA were not significantly changed. These results suggest that expression of GPEET is controlled by the activity of the AO. To investigate whether this was due to alterations in the level of AO expression, Western blot analyses were performed. However, trypanosomes cultured in the presence or absence of glycerol expressed AO at similar levels (Figure 5C). In summary, our results suggest that glycerol-dependent expression of GPEET is linked to electron flow to the AO.
Figure 5.
GPEET expression is controlled by the activity of the AO. (A) Procyclic forms of AnTat 1.1 cultured in SDM-79 in the presence (+) or absence (-) of 20 mM glycerol. (B) Procyclic forms of stock 427 adapted to SDM-79 containing 0.3 mM glucose, or cultured in the same medium supplemented with 5 mM glucose. Subsequently, cells were incubated in the same medium in the presence or absence of 1 mM SHAM or 1 mM KCN for 3 d. Total RNA was hybridized with radioactively labeled probes from the coding region of GPEET and EP1 or from the 18S rRNA gene as indicated under Materials and Methods. Hybridization signals were quantified using a PhosphorImager and normalized to the 18S rRNA. (C) Expression of the AO protein. Top, lysates from equal numbers of procyclic forms were analyzed by SDS-PAGE and immunoblotting by using an mAb against the trypanosomal AO. Serial dilutions of lysates from bloodstream forms (BSF) were loaded on the same gel. Numbers above the figure indicate the cell equivalents loaded per lane. Bottom, after electroblotting, the residual proteins in the polyacrylamide gel were stained with Coomassie Blue as a control for the amount of protein loaded per lane.
To investigate whether expression of GPEET by late procyclic forms is also controlled by the activity of the AO, cells were cultured in the presence of SHAM. Consistent with the results obtained with glycerol, SHAM prevented the accumulation of GPEET mRNA in glucose-depleted medium (Figure 5B). Again, the level of AO protein in cells cultured in 0.3 or 5 mM glucose was unchanged (Figure 5C). These cells did not survive in the presence of cyanide, however, presumably because they were unable to produce sufficient amounts of ATP under these conditions.
DISCUSSION
Exogenous glycerol, glucose and an unidentified factor in the tsetse fly midgut are all capable of modulating expression of GPEET by procyclic form trypanosomes. Glycerol prevents the differentiation of early procyclic forms to late procyclic forms and concomitant GPEET repression (Vassella et al., 2000), whereas glucose depletion below a certain threshold induces GPEET expression in late procyclic forms (Morris et al., 2002). In this study, we have shown that all three external signals converge on a discrete element that maps between positions 160 and 184 in the LII domain. GRE is positioned immediately downstream of another regulatory element, the 26-mer (Hotz et al., 1997), but the two elements function in different life cycle stages. The 26-mer exerts its main effects in bloodstream forms, whereas GRE is most active in late procyclic forms.
Converting GRE to the corresponding sequence in the 3′ UTR of EP1 resulted in high level expression that was independent of the amount of exogenous glycerol or glucose in the culture medium and prevented the down-regulation of a reporter gene during fly infections. In transient assays, destruction of GRE resulted in a ∼48-fold increase in CAT activity in late procyclic forms, consistent with the removal of a destabilizing element. Two possible interpretations of these results are that GRE recruits factors to the 3′ UTR, causing specific degradation of GPEET mRNA in late procyclic forms, or that it interferes with RNA processing at this stage of the life cycle. In contrast, in early procyclic forms, destruction of GRE resulted in approximately twofold reduction of CAT activity, indicating that it can function as a weakly positive element. Under these conditions, factors binding to this element might protect the mRNA from nucleases or enhance processing. Additional elements in the 3′ UTR must also contribute to high level expression of GPEET in early procyclic forms, however, because the LII domain alone was not sufficient to confer full activity.
Culturing late procyclic forms in low concentrations of glucose or ablating components of the ASCT cycle by RNAi both resulted in expression of GPEET. These processes are probably linked because pyruvate, the major intermediate of glycolysis, is degraded in the mitochondrion by the ASCT cycle (Van Hellemond et al., 1998). These results indicate that the activity of the mitochondrial enzymes PDH and SCoAS can influence GPEET mRNA stability. In contrast, ablation of the activity of enzymes of the citric acid cycle did not induce GPEET, in agreement with data from van Weelden et al. (2003) showing that glucose is not degraded via this pathway. However, another mitochondrial enzyme, AO, was implicated in GPEET regulation because addition of the inhibitor SHAM to the culture medium resulted in ≥5-fold decrease the steady state level of GPEET mRNA. This effect was seen in early procyclic forms cultured in the presence of glycerol or late procyclic forms subjected to glucose depletion, suggesting that the expression of GPEET is controlled by the same AO-dependent mechanism in both life cycle stages. Our previous finding that hypoxia overrides the effect of glycerol and accelerates the disappearance of GPEET mRNA (Vassella et al., 2000) can now be explained as this is likely to inhibit AO activity. Although GPEET is regulated by electron transport to the AO, we can exclude that this is purely due to alterations in the level of ATP because this is mainly produced by electron transport to the cytochromes and, furthermore, treatment with cyanide did not influence the steady state level of GPEET mRNA.
In early procyclic forms glycerol is used as a substrate for energy metabolism in preference to glucose and results in electron transport to both AO and the cytochromes (Ryley, 1962). AO is also required for GPEET expression in late procyclic forms, but at present it is not clear whether its activation is a consequence of suppressing the ASCT cycle. It is possible, however, that alterations in metabolites or in redox balance might lead to AO activation, as is the case in other organisms. In plants, AO is a homodimeric protein whose activity can be regulated by a redox-sensitive intersubunit sulfhydryl/disulfide system that depends on the electron transport chain (Umbach et al., 1994). The activity of this enzyme also can be regulated by a redox-independent pathway which is induced by pyruvate, succinate or malate (Rhoads et al., 1998). It is possible that the activity of the AO is controlled by either of these two mechanisms in trypanosomes. It can be excluded, however, that this is controlled solely by the amount of AO protein (Figure 5C).
Because the electron transport chain to the AO is not coupled to ATP production, it produces heat. In plants, thermogenic respiration is a mechanism to volatilize aromatic compounds to attract pollinators. AO also may play an important role in maintaining the balance of redox equivalents or as a defense against reactive oxygen species (Popov et al., 1997; Vanlerberghe and McIntosh, 1997; Maxwell et al., 1999). In the case of trypanosomes, it has recently been shown that inhibition of the AO in procyclic forms leads to an increase in superoxide production and protein oxidation (Fang and Beattie, 2003). Because GPEET is controlled by the activity of the AO, this might conceivably provide a mechanism by which energy metabolism can translate into gene expression.
Although carbon sources are among the best-documented factors known to affect growth and development of lower eukaryotic cells, the pathways transducing the extracellular signals are largely obscure. Mainly from work in yeast, it is known that nutrient transporters or G protein-coupled receptors can function as extracellular sensors for carbon sources, whereas hexose kinases may function as intracellular sensors [for reviews, see Ronne (1995); Forsberg and Ljungdahl, 2001)]. In most of these cases, the gene products controlled by changes in carbon sources are involved in metabolic functions such as gluconeogenesis, mitochondrial energy metabolism, or catabolism of other nutrient sources. This is in contrast to what is found in trypanosomes, in which metabolic pathways are linked to expression of a major surface protein.
Acknowledgments
We thank Dave Barry (Wellcome Centre of Molecular Parasitology, Glasgow, Scotland), Terry Pearson (University of Victoria, Victoria, Canada), and George C. Hill (Vanderbilt University Medical Center, Nashville, TN) for antibodies and Michael Boshart (Ludwig-Maximilian University, Munich, Germany) for the aconitase null mutant clone. This research was supported by a grant from the Swiss National Science Foundation (31-63987.00) to I.R. and by grants from the Swiss National Science Foundation (31-64900.01) and Hans Sigrist Foundation to E.V.
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E04-04-0341. Article and publication date are available at www.molbiolcell.org/cgi/doi/10.1091/mbc.E04-04-0341.
Abbreviations used: AO, alternative oxidase; ASCT, acetate:succinate CoA transferase/succinyl CoA synthetase; CAT, chloramphenicol acetyltransferase; GRE, glycerol-responsive element; KDH, α-ketoglutarate dehydrogenase; nt, nucleotide(s); PDH, pyruvate dehydrogenase; SCoAS, succinyl-CoA synthetase; SDH, succinate dehydrogenase; SHAM, salicylhydroxamic acid; UTR, untranslated region.
References
- Acosta-Serrano, A., Vassella, E., Liniger, M., Kunz Renggli, C., Brun, R., Roditi, I., and Englund, P.T. (2001). The surface coat of procyclic Trypanosoma brucei: programmed expression and proteolytic cleavage of procyclin in the tsetse fly. Proc. Natl. Acad. Sci. USA 98, 1513-1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayne, R.A., Kilbride, E.A., Lainson, F.A., Tetley, L., and Barry, J.D. (1993). A major surface antigen of procyclic stage Trypanosoma congolense. Mol. Biochem. Parasitol. 61, 295-310. [DOI] [PubMed] [Google Scholar]
- Beecroft, R.P., Roditi, I., and Pearson, T.W. (1993). Identification and characterization of an acidic major surface glycoprotein from procyclic stage Trypanosoma congolense. Mol. Biochem. Parasitol. 61, 285-294. [DOI] [PubMed] [Google Scholar]
- Bochud-Allemann, N., and Schneider, A. (2002). Mitochondrial substrate level phosphorylation is essential for growth of procyclic Trypanosoma brucei. J. Biol. Chem. 277, 32849-32854. [DOI] [PubMed] [Google Scholar]
- Brun, R., and Schönenberger, M. (1979). Cultivation and in vitro cloning of procyclic culture forms of Trypanosoma brucei in a semi-defined medium. Acta Trop. 36, 289-292. [PubMed] [Google Scholar]
- Brun, R., and Schönenberger, M. (1981). Stimulating effect of citrate and cis-aconitate on the transformation of Trypanosoma brucei bloodstream forms to procyclic forms in vitro. Z. Parasitenkd. 66, 17-24. [DOI] [PubMed] [Google Scholar]
- Bütikofer, P., Vassella, E., Ruepp, S., Boschung, M., Civenni, G., Seebeck, T., Hemphill, A., Mookherjee, N., Pearson, T.W., and Roditi, I. (1999). Phosphorylation of a major GPI-anchored surface protein of Trypanosoma brucei during transport to the plasma membrane. J. Cell Sci. 112, 1785-1795. [DOI] [PubMed] [Google Scholar]
- Chaudhuri, M., Ajayi, W., and Hill, G. (1998). Biochemical and molecular properties of the Trypanosoma brucei alternative oxidase. Mol. Biochem. Parasitol. 95, 53-68. [DOI] [PubMed] [Google Scholar]
- Clarkson, A.B., Bienen, E.J., Pollakis, G., and Grady, R.W. (1989). Respiration of bloodstream forms of the parasite Trypanosoma brucei brucei is dependent on a plant-like alternative oxidase. J. Biol. Chem. 264, 17770-17776. [PubMed] [Google Scholar]
- Cross, G.A.M., and Manning, J.C. (1973). Cultivation of Trypanosoma brucei spp. in semi-defined and defined media. Parasitology 67, 315-331. [DOI] [PubMed] [Google Scholar]
- Drozdz, M., and Clayton, C.E. (1999). Structure of a regulatory 3′ untranslated region from Trypanosoma brucei. RNA 5, 1632-1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang, J., and Beattie, D.S. (2003). Alternative oxidase present in procyclic Trypanosoma brucei may act to lower the mitochondrial production of superoxide. Arch. Biochem. Biophys. 14, 294-302. [DOI] [PubMed] [Google Scholar]
- Flück, C., Salomone, J.Y., Kurath, U., and Roditi, I. (2003). Cycloheximide-mediated accumulation of transcripts from a procyclin expression site depends on the intergenic region. Mol. Biochem. Parasitol. 127, 93-97. [DOI] [PubMed] [Google Scholar]
- Forsberg, H., and Ljungdahl, P.O. (2001). Sensors of extracellular nutrients in Saccharomyces cerevisiae. Curr. Genet. 40, 91-109. [DOI] [PubMed] [Google Scholar]
- Furger, A., Schürch, N., Kurath, U., and Roditi, I. (1997). Elements in the 3′ untranslated region of procyclin mRNA regulate expression in insect forms of Trypanosoma brucei by modulating RNA stability and translation. Mol. Cell. Biol. 17, 4372-4380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hehl, A., Vassella, E., Braun, R., and Roditi, I. (1994). A conserved stem-loop structure in the 3′ untranslated region of procyclin mRNAs regulates expression in Trypanosoma brucei. Proc. Natl. Acad. Sci. USA 91, 370-374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hotz, H.R., Hartmann, C., Huober, K., Hug, M., and Clayton, C. (1997). Mechanisms of developmental regulation in Trypanosoma brucei: a polypyrimidine tract in the 3′ untranslated region of a surface protein mRNA affects RNA abundance and translation. Nucleic Acids Res. 25, 3017-3026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krakow, J.L., and Wang, C.C. (1990). Purification and characterization of glycerol kinase from Trypanosoma brucei. Mol. Biochem. Parasitol. 43, 17-25. [DOI] [PubMed] [Google Scholar]
- Le Ray, D., Barry, J.D., Easton, C., and Vickerman, K. (1977). First tsetse fly transmission of the “AnTat” serodeme of Trypanosoma brucei. Ann. Soc. Belge Méd. Trop. 57, 369-381. [PubMed] [Google Scholar]
- Maxwell, D.P., Wang, Y., and McIntosh, L. (1999). The alternative oxidase lowers mitochondrial reactive oxygen production in plant. Proc. Natl. Acad. Sci. USA 96, 8271-8276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris, J.C., Wang, Z., Drew, M.E., and Englund, P.T. (2002). Glycolysis modulates trypanosome glycoprotein expression as revealed by an RNAi library. EMBO J. 21, 4429-4438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opperdoes, F.R. (1987). Compartmentation of carbohydrate metabolism in trypanosomes. Annu. Rev. Microbiol. 41, 127-151. [DOI] [PubMed] [Google Scholar]
- Popov, V.N., Simonian, R.A., Skulachev, V.P., and Starkov, A.A. (1997). Inhibition of the alternative oxidase stimulates H2O2 production in plant mitochondria. FEBS Lett. 415, 87-90. [DOI] [PubMed] [Google Scholar]
- Rhoads, D.M., Umbach, A.L., Sweet, C.R., Lennon, A.M., Rauch, G.S., and Siedow, J.N. (1998). Regulation of the cyanide-resistant alternative oxidase of plant mitochondria. Identification of the cysteine residue involved in alpha-keto acid stimulation and intersubunit disulfide bond formation. J. Biol. Chem. 273, 30750-30756. [DOI] [PubMed] [Google Scholar]
- Richardson, J.P., Beecroft, R.P., Tolson, D.L., Liu, M.K., and Pearson, T.W. (1988). Procyclin: an unusual immunodominant glycoprotein surface antigen from the procyclic stage of African trypanosomes. Mol. Biochem. Parasitol. 31, 203-216. [DOI] [PubMed] [Google Scholar]
- Roditi, I., and Clayton, C. (1999). An unambiguous nomenclature for the major surface glycoproteins of the procyclic form of Trypanosoma brucei. Mol. Biochem. Parasitol. 103, 99-100. [DOI] [PubMed] [Google Scholar]
- Roditi, I., Furger, A., Ruepp, S., Schürch, N., and Bütikofer, P. (1998). Unravelling the procyclin coat of Trypanosoma brucei. Mol. Biochem. Parasitol. 91, 117-130. [DOI] [PubMed] [Google Scholar]
- Roditi, I., et al. (1989). Procyclin gene expression and loss of the variant surface glycoprotein during differentiation of Trypanosoma brucei. J. Cell Biol. 108, 737-746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronne, H. (1995). Glucose repression in fungi. Trends Genet. 11, 12-17. [DOI] [PubMed] [Google Scholar]
- Ruepp, S., Furger, A., Kurath, U., Renggli Kunz, C., Hemphill, A., Brun, R., and Roditi, I. (1997). Survival of Trypanosoma brucei in the tsetse fly is enhanced by the expression of specific forms of procyclin. J. Cell Biol. 137, 1369-1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryley, J.F. (1962). Studies on the metabolism of the protozoa. Biochem. J. 85, 211-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook, J., Fritsch, E.F., and Maniatis, T. (eds.) (1989). Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press.
- Schürch, N., Furger, A., Kurath, U., and Roditi, I. (1997). Contributions of the procyclin 3′ untranslated region and coding region to the regulation of expression in bloodstream forms of Trypanosoma brucei. Mol. Biochem. Parasitol. 89, 109-121. [DOI] [PubMed] [Google Scholar]
- Tielens, A.G.M., and Van Hellemond, J.J. (1998). Differences in energy metabolism between Trypanosomatidae. Parasitol. Today 14, 265-271. [DOI] [PubMed] [Google Scholar]
- Umbach, A.L., Wiskich, J.T., and Siedow, J.N. (1994). Regulation of alternative oxidase kinetics by pyruvate and intermolecular disulfide bond redox status in soybean seedling mitochondria. FEBS Lett. 348, 181-184. [DOI] [PubMed] [Google Scholar]
- Van Hellemond, J.J., Opperdoes, F.R., and Tielens, A.G. (1998). Trypanosomatidae produce acetate via a mitochondrial acetate:succinate CoA transferase. Proc. Natl. Acad. Sci. USA 95, 3036-3041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Weelden, S., Fast, B., Vogt, A., van der Meer, P., Saas, J., van Hellemond, J., Tielens, A., and Boshart, M. (2003). Procyclic Trypanosoma brucei do not use Krebs cycle activity for energy generation. J. Biol. Chem. 278, 12854-12863. [DOI] [PubMed] [Google Scholar]
- Vanlerberghe, G.C., and McIntosh, L. (1997). Alternative oxidase: from gene to function. Annu. Rev. Plant. Mol. Biol. 48, 703-734. [DOI] [PubMed] [Google Scholar]
- Vassella, E., Acosta-Serrano, A., Studer, E., Lee, S.H., Englund, P.T., and Roditi, I. (2001). Multiple procyclin isoforms are expressed differentially during the development of insect forms of Trypanosoma brucei. J. Mol. Biol. 312, 597-607. [DOI] [PubMed] [Google Scholar]
- Vassella, E., van Den Abbeele, J., Bütikofer, P., Renggli Kunz, C., Furger, A., Brun, R., and Roditi, I. (2000). A major surface glycoprotein of Trypanosoma brucei is expressed transiently during development and can be regulated post-transcriptionally by glycerol or hypoxia. Genes Dev. 14, 615-626. [PMC free article] [PubMed] [Google Scholar]
- Wirtz, E., Leal, S., Ochatt, C., and Cross, G.A. (1999). A tightly regulated inducible expression system for conditional gene knock-outs and dominant-negative genetics in Trypanosoma brucei. Mol. Biochem. Parasitol. 99, 89-101. [DOI] [PubMed] [Google Scholar]
- Zucker, M., and Jacobson, A.B. (1998). Using reliability information to annotate RNA secondary structures. RNA 4, 669-679. [DOI] [PMC free article] [PubMed] [Google Scholar]