Abstract
The CHIP ubiquitin ligase turns molecular chaperones into protein degradation factors. CHIP associates with the chaperones Hsc70 and Hsp90 during the regulation of signaling pathways and during protein quality control, and directs chaperone-bound clients to the proteasome for degradation. Obviously, this destructive activity should be carefully controlled. Here, we identify the cochaperone HspBP1 as an inhibitor of CHIP. HspBP1 attenuates the ubiquitin ligase activity of CHIP when complexed with Hsc70. As a consequence, HspBP1 interferes with the CHIP-induced degradation of immature forms of the cystic fibrosis transmembrane conductance regulator (CFTR) and stimulates CFTR maturation. Our data reveal a novel regulatory mechanism that determines folding and degradation activities of molecular chaperones.
INTRODUCTION
Molecular chaperones are well known to assist intracellular protein folding (Frydman, 2001; Hartl and Hayer-Hartl, 2002). However, the cochaperone CHIP is able to switch chaperone activity from protein folding to protein degradation by virtue of its ubiquitin ligase activity (Connell et al., 2001; Demand et al., 2001; Höhfeld et al., 2001; Meacham et al., 2001; Murata et al., 2001). CHIP interacts with the carboxy termini of the cytoplasmic and nuclear chaperones Hsc70 and Hsp90, and mediates the covalent attachment of the degradation marker ubiquitin to chaperone-bound client proteins. In this way, CHIP triggers the turnover of chaperone clients such as the glucocorticoid hormone receptor and the oncogenic receptor tyrosine kinase ErbB2 by the 26S proteasome (Connell et al., 2001; Demand et al., 2001; Xu et al., 2002). CHIP apparently alters the balance between folding and degradation during the Hsc70- and Hsp90-mediated regulation of signaling pathways. In addition, CHIP plays a prominent role in protein quality control, as is illustrated by its involvement in the degradation of immature forms of cystic fibrosis transmembrane conductance regulator (CFTR) (Meacham et al., 2001). CFTR is an ion channel in the plasma membrane of epithelial cells and loss-of-function mutations in the gene encoding CFTR cause cystic fibrosis (Kopito, 1999; Riordan, 1999). The most common disease-causing allele, ΔF508, drastically interferes with the protein's ability to fold, essentially barring it from functional expression in the plasma membrane. However, also wild-type CFTR folds very inefficiently, and <30% of the protein reaches the plasma membrane (Kopito, 1999). While trafficking through the endoplasmic reticulum (ER), immaturely glycosylated and aberrantly folded forms of CFTR are recognized by quality control systems and are eventually directed to the proteasome for degradation (Jensen et al., 1995; Ward et al., 1995; Kostova and Wolf, 2003). Hsc70 and Hsp90 participate in CFTR quality control, because the membrane protein exposes large domains into the cytoplasm. The chaperones assist the folding of these cytoplasmic domains (Yang et al., 1993; Loo et al., 1998; Meacham et al., 1999; Farinha et al., 2002). However, through binding to Hsc70, CHIP is able to gain access to immature CFTR and induces the ubiquitylation and proteasomal degradation of the membrane protein (Meacham et al., 2001).
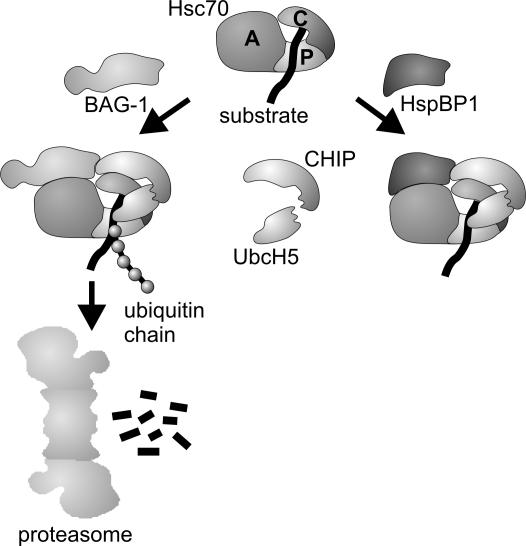
During the sorting of chaperone clients to the proteasome, CHIP can cooperate with the Hsc70 cochaperone BAG-1. The two cochaperones interact simultaneously with Hsc70, as BAG-1 binds to the amino terminal ATPase domain of the chaperone and leaves the carboxy terminus of Hsc70 available for an association with CHIP (Demand et al., 2001; Alberti et al., 2002). BAG-1 is intimately involved in the regulation of the ATP-dependent peptide binding and release cycle of Hsc70 by stimulating nucleotide exchange (Höhfeld and Jentsch, 1997; Sondermann et al., 2001). Furthermore, BAG-1 associates with the proteasome through an integrated ubiquitin-like domain and thereby recruits Hsc70 chaperone complexes to the proteasome (Lüders et al., 2000). As a consequence BAG-1 stimulates the CHIP-induced degradation of the glucocorticoid hormone receptor (Demand et al., 2001). A cooperation of diverse cochaperones apparently provides additional levels of regulation to alter chaperone-assisted folding and degradation pathways. Conceivably, such a cooperation of cochaperones also may allow to confine the destructive activity of CHIP. We therefore investigated how cochaperones other than BAG-1 influence CHIP-mediated degradation. We were particularly interested in the cochaperone HspBP1, which is a nucleotide release factor of Hsc70 of unknown cellular function (Raynes and Guerriero, 1998; Kabani et al., 2002). HspBP1 is expressed at a similar level as CHIP in human HeLa cells (0.05% of cellular protein; Alberti, unpublished observation) and displays a similar tissue distribution (Raynes and Guerriero, 1998; Ballinger et al., 1999). Furthermore, HspBP1 interacts with the ATPase domain of Hsc70, like BAG-1, and may therefore modulate the function of the Hsc70/CHIP complex. In fact, here we identify HspBP1 as an inhibitor of the CHIP ubiquitin ligase and as an essential component of the cellular surveillance system that monitors the folded state of CFTR.
MATERIALS AND METHODS
Antibodies and Proteins
The following antibodies were used for immunoblotting: anti-BAG-1 (C-16; Santa Cruz Biotechnology, Santa Cruz, CA), anti-CFTR (M3A7; Upstate Biotechnology, Lake Placid, NY), anti-CHIP (Ballinger et al., 1999), anti-FLAG (M2; Sigma-Aldrich, St. Louis, MO), anti-Hsc/Hsp70 (SPA-820; StressGen Biotechnologies, San Diego, CA), anti-HspBP1 (FL4; Delta Biolabs, Campbell, CA), anti-luciferase (Promega, Madison, WI), and anti-raf-1 (Santa Cruz).
Rat Hsc70, wheat E1, and BAG-1S, Hsp40, UbcH5b, and CHIP (all of human origin) were purified after recombinant expression as described previously (Höhfeld and Jentsch, 1997; Lüders et al., 2000; Demand et al., 2001; Alberti et al., 2002). Purified bovine ubiquitin was purchased from Sigma-Aldrich. For recombinant expression of His-tagged HspBP1 Escherichia coli BL21(DE3) cells were transformed with pET28a-hspBP1 (Raynes and Guerriero, 1998). Bacterial lysates were applied to Ni2+-NTA agarose chromatography. Affinity purified HspBP1 was further purified through anion exchange chromatography on a DEAE Sepharose column. For purification of untagged HspBP1, the hspBP1 coding region was cloned into pTYB12 and purified via a self-cutting intein/chitin binding domain as described by the manufacturer (New England BioLabs, Beverly, MA).
Immunoprecipitation and Binding Experiments
To isolate HspBP1 and CHIP immunocomplexes, HeLa cells were transfected with the plasmids pCMVTag2-hspBP1 and pCMVTag2-CHIP, respectively, by using the CalPhos transfection system (BD Biosciences Clontech, Palo Alto, CA). When effects of HspBP1 on the association of CHIP with Hsc70 were investigated, cells were cotransfected with pCMVTag2-CHIP and pcDNA3.1-hspBP1 or an equal amount of empty pcDNA3.1. Two days after transfection, HeLa cells were lysed in 25 mM 3-(N-morpholino)propanesulfonic acid (MOPS), pH 7.2, 100 mM KCl, and 0.5% Tween 20 (buffer A) containing 5 mM EDTA and Complete protease inhibitors (Roche Diagnostics, Indianapolis, IN). The lysate was centrifuged at 20,000 × g for 20 min at 4°C, and the soluble supernatant fraction was used for immunoprecipitation. Anti-FLAG antibody (M2; Sigma-Aldrich) was added at a concentration of 10 μg/ml, and samples were incubated for 1 h at 4°C. After addition of protein G-Sepharose, samples were incubated for another hour. Alternatively, M2-agarose (Sigma-Aldrich) was used at a concentration of 30 μl of agarose per milliliter of lysate, and samples were incubated for 3 h at 4°C. The Sepharose was collected by centrifugation and washed five times with buffer A and once with buffer A lacking detergent. Isolated immunocomplexes were incubated for 20 min at 30°C in detergent-free buffer A containing 2 mM MgCl2, 1 mM ATP, and 1 mM phenylmethylsulfonyl fluoride, followed by the precipitation of ATP-eluted proteins with trichloroacetic acid. The Sepharose beads were washed once more with buffer A, followed by elution of associated proteins by boiling in SDS-PAGE loading buffer. When M2-agarose was used, agarose-bound proteins were eluted with 0.1 M glycine, pH 3.5.
To test for interactions of HspBP1 with Hsc70, in vitro binding assays with immobilized His-tagged HspBP1 were performed. For immobilization of His-tagged HspBP1 10 μg of purified protein was incubated with 50 μl of Ni2+-NTA agarose in buffer B (20 mM Tris-HCl, pH 7.6, 100 mM KCl, 20 mM imidazole) for 2 h with gentle agitation, resulting in ∼20% binding. Samples were washed once with buffer B, followed by addition of Hsc70, BAG-1S, and CHIP at one-fifth of the initial HspBP1 concentration. Samples were incubated for additional 2 h at 4°C, followed by four washing steps with buffer B. Proteins were eluted in 20 mM Tris, pH 7.6, 100 mM KCl, 200 mM imidazole for 15 min at room temperature and precipitated by the addition of trichloroacetic acid.
The competition of BAG-1 and HspBP1 in Hsc70 binding was analyzed with purified BAG-1 immobilized to protein G-Sepharose beads with a specific anti-BAG-1 antibody. The binding reaction was performed with 1.33 μg of BAG-1S and equimolar amounts of Hsc70 for 1 h at 4°C in 200 μl of buffer A containing 5 mM EDTA. When indicated HspBP1 was added in equimolar amounts or in a two- or fivefold molar excess over BAG-1. Samples were mixed with 4 μg of anti-BAG-1 antibody or control IgG and incubated for another hour at 4°C. After addition of protein G-Sepharose, samples were further incubated for 1 h at 4°C. The Sepharose was collected by centrifugation and was washed five times with buffer A. Sepharose-associated proteins were eluted by addition of SDS-PAGE loading buffer.
In Vitro Ubiquitylation
Ubiquitylation of Raf-1 in vitro was performed as described previously in the presence of 0.3 μM Hsp40, 3 μM Hsc70, 4 μM UbcH5, and 3 μM CHIP (Demand et al., 2001). When indicated, His-tagged HspBP1 was added. Samples were analyzed by SDS-PAGE and immunoblotting with anti-raf-1, anti-Hsc/Hsp70, and anti-HspBP1 antibodies.
To investigate the ubiquitylation of heat-denatured firefly luciferase, the protein (0.26 μM) was incubated at 43°C for 10 min in buffer C (20 mM MOPS, pH 7.2, 50 mM KCl, 4 mM MgCl2, 4 mM ATP, and 2 mM DTT) in the presence of Hsp40 (3.2 μM) and Hsc70 (4.7 μM); or in the presence of Hsp90 (11 μM) under conditions described previously (Murata et al., 2001). After heating, samples were chilled on ice, and subsequent ubiquitylation was performed under the same conditions as described above for Raf-1 without further addition of Hsc70 and Hsp40.
For reconstitution of CFTR ubiquitylation, in vitro transcription of CFTR was performed with linearized pcDNA3.1-CFTR as template by using the T7 RiboMax system according to the protocol of the manufacturer (Promega). Using nuclease-treated rabbit reticulocyte lysate (Promega), radiolabeled CFTR was synthesized in the presence of porcine pancreas microsomes (0.1 eq/μl), which were isolated according to Walter and Blobel (1983). After incubation at 30°C for 90 min, translation reactions were stopped by addition of 2 mM puromycin, 0.5 mM cycloheximide, 2 mM unlabeled methionine, and 10 μM MG-132 (Calbiochem, San Diego, CA). For ubiquitylation of CFTR, 20 μl of the translation reactions received 1 μM UbcH5, 1 μM CHIP, and increasing amounts of HspBP1 (1, 2, and 5 μM) as indicated. The total volume of the samples was adjusted to 25 μl with 20 mM MOPS, pH 7.2, 100 mM KCl containing 10 μM MG-132 and EDTA-free Complete protease inhibitor. Samples were incubated for 1 h at 16°C and were subsequently subjected to immunoprecipitation to isolate ubiquitylated CFTR. To this end, samples were split into two equal aliquots, diluted 50-fold in buffer D (25 mM Tris-HCl, pH 8.0, 150 mM NaCl, 0.1% SDS, 0.5% sodium deoxycholate, 1% Nonidet P-40, 10% glycerol, 2 mM EDTA, Complete protease inhibitor) and mixed with either 5 μg/ml control IgG or 5 μg/ml anti-ubiquitin antibody (FK2; Affiniti Research Products, Biomol International, Plymouth Meeting, PA). The samples were incubated for 2 h at 4°C with gentle agitation, followed by addition of protein G-Sepharose and an additional incubation for 1 h at 4°C. Sepharose beads were collected by centrifugation and washed three times with buffer D. Sepharose-bound proteins were eluted by addition of SDS-PAGE sample buffer.
Luciferase Aggregation Assay
To assay luciferase aggregation, 3 μM Hsc70, 2 μM Hsp40, 3 μM His-HspBP1, and 3 μM CHIP were preincubated in indicated combinations for 5 min at room temperature in 20 mM MOPS, pH 7.2, 50 mM KCl, 2 mM MgCl2, 1 mM DTT, and 2 mM ATP. Subsequently, luciferase (0.6 μM) was added, and samples were incubated at 42°C for 15 min. The reaction was stopped by addition EDTA to a concentration of 5 mM, and samples were centrifuged at 120,000 × g for 20 min at 4°C. Supernatant and pellet fractions were separated and analyzed by SDS-PAGE and immunoblotting with an anti-luciferase antibody, followed by densitometric quantification.
CFTR Turnover and Maturation
To determine the steady-state levels of CFTR in the presence of overexpressed HspBP1 and CHIP, human embryonic kidney (HEK)293 cells were seeded in six-well plates and transfected with 0.38 μg of pcDNA3.1-CFTR or pcDNA3-CFTRΔ F508 by using Effectene transfection reagent (QIAGEN, Valencia, CA). When indicated, 0.38 μg of pcDNA3.1-chip and 0.38 or 0.75 μg of pcDNA3.1-hspBP1 was cotransfected. The total amount of added DNA was kept constant at 1.5 μg by addition of pcDNA3.1. Twenty-four hours after transfection the cells were washed once with phosphate-buffered saline (PBS) and lysed in lysis buffer (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 1% Nonidet P-40, 0.5% deoxycholate, and 0.2% sodium dodecyl sulfate) containing Complete protease inhibitor. The lysate was centrifuged at 20,000 × g for 20 min at 4°C, and the supernatant was used as a soluble extract. For immunoblotting, 60 μg of total protein was loaded onto an SDS-PAGE gel, and extracts were analyzed for CFTR, Hsc70, and cochaperone expression.
To analyze the degradation kinetics of CFTR, HEK293 cells were seeded in six-well plates and were transfected with 0.25 μg of pcDNA3.1-CFTR and 0.375 μg of pcDNA3.1-CHIP or the same amount of pcDNA3.1-hspBP1 as indicated. The total amount of added DNA was kept constant at 1 μg/well by addition of pcDNA3.1. After 24 h, cells were shifted to methionine-free medium for 1 h, followed by metabolic labeling for 30 min in the presence of [35S]methionine at a concentration of 150 μCi/ml. Cells were washed once with PBS containing 0.1 mM CaCl2 and 1 mM MgCl2 and lysed immediately in lysis buffer containing Complete protease inhibitor (time point 0) or incubated in medium supplemented with 1 mM methionine and 0.5 mM cycloheximide for the indicated chase periods. The lysates were centrifuged at 20,000 × g for 20 min at 4°C, and the resultant supernatants were precleared by addition of protein G-Sepharose and incubation for 30 min at 4°C. After removal of the Sepharose by centrifugation, supernatant fractions were mixed with 0.4 μg of anti-CFTR antibody (24-1; R&D Systems, Minneapolis, MN). Samples were incubated for 1 h at 4°C, protein G-Sepharose was added, and samples were further incubated for 1 h. Immunocomplexes were washed three times with lysis buffer. Isolated CFTR was deglycosylated for 2 h at 25°C with 250 U of PNGase F (New England BioLabs) in digestion buffer (50 mM sodium phosphate, pH 7.5, 1% Nonidet P-40, 10% glycerol, 0.5% SDS, and 100 mM β-mercaptoethanol). Deglycosylation was stopped by addition of SDS-PAGE loading buffer, and samples were analyzed by SDS-PAGE. Quantification of CFTR was carried out by densitometric analysis using a PhosphorImager (PerkinElmer Life and Analytical Sciences, Boston, MA).
Biotinylation of glycosylated CFTR exposed on the surface of intact cells was performed as described by Lisanti et al. (1989). Twenty-four hours after transfection, cells were washed twice with ice-cold PBS containing 0.1 mM CaCl2 and 1 mM MgCl2 (PBS-c/m) and were incubated with 10 mM NaIO4 in PBS-c/m for 30 min at 4°C in the dark to oxidize the carbohydrate moieties of surface glycoproteins. Cells were washed twice with PBS-c/m and once with acetate buffer (100 mM sodium acetate, pH 5.5, 0.1 mM CaCl2, and 1 mM MgCl2) and were biotinylated using 2 mM biotin-LC-hydrazide (Pierce Chemical, Rockford, IL) in acetate buffer for 30 min at 4°C. Unbound biotin was removed by washing three times with ice-cold PBS-c/m followed by cell lysis in lysis buffer containing Complete protease inhibitor. Lysates were centrifuged for 20 min at 20,000 × g, and the supernatant (1 mg of protein in 1 ml of lysis buffer) was incubated with 100 μl of Avidin beads (UltraLink NeutrAvidin; Pierce Chemical) at 4°C for 1 h. Beads were collected by centrifugation and washed three times with lysis buffer followed by elution of Avidin-bound proteins in SDS-PAGE sample buffer.
HspBP1 levels of HEK293 cells were reduced using a small hairpin RNA (shRNA) expression plasmid, which was generated according to the pSUPER RNAi system (Oligoengine, Seattle, WA). The siRNA produced in pSUPERHspBP1-transfected cells targets the sequence CAT GGA CAA TGC CGC AGA C of the human HspBP1 gene. HEK293 cells were seeded in six-well plates and were transfected with 0.38 μg of pcDNA3.1-CFTR by using the Effectene transfection reagent (QIAGEN). When indicated 0.56 μg of pSUPERHspBP1, the same amount of pSUPER, 0.56 μg of pcDNA3.1 and pcDNA3.1-hspBP1 were transfected. Cells were harvested 72 h after transfection and processed as described above.
RESULTS
HspBP1 and CHIP Simultaneously Associate with Hsc70
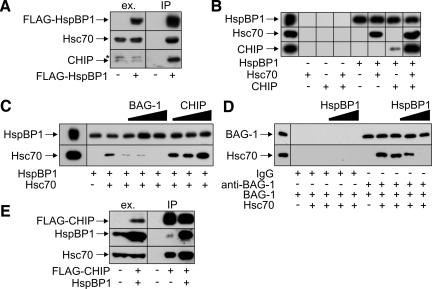
A potential interaction between HspBP1 and CHIP was analyzed by immunoprecipitation of HspBP1-containing protein complexes. For this purpose, a FLAG epitope tagged form of HspBP1 was expressed in human HeLa cells and was immunoprecipitated with an anti-FLAG antibody. In addition to Hsc70, CHIP was readily detectable in HspBP1-containing complexes (Figure 1A). In contrast, Hsp90, the chaperonin TRiC, and the Hsc70 cochaperones BAG-1, Hip, and Hop could not be detected in such complexes (unpublished data). HspBP1 thus seems to cooperate specifically with CHIP to regulate Hsc70 function.
Figure 1.
HspBP1 is associated with Hsc70 and CHIP. (A) HeLa cells were transfected with pCMVTag2-hspBP1, which encodes a FLAG-tagged form of HspBP1 (+ FLAG-HspBP1), or with an equal amount of a control plasmid (-). HspBP1-containing protein complexes were immunoprecipitated with an anti-FLAG antibody (IP) and analyzed for the presence of Hsc70 and CHIP by immunoblotting. The chaperone and CHIP were detected in ATP-eluates of the isolated complexes. To monitor expression levels, 40 μg of protein extracts were loaded (ex.). The star denotes a polypeptide that cross-reacts with the anti-CHIP antibody in cell extracts. (B) Purified His-tagged HspBP1 was immobilized and incubated with Hsc70 and CHIP as indicated. Proteins retained on the affinity resin were detected using specific antibodies. As negative control, proteins were incubated with the affinity resin in the absence of HspBP1. The panel on the very left shows 33% of the input. (C) Binding of Hsc70 to immobilized HspBP1 was analyzed in the presence of different concentrations of BAG-1 and CHIP, respectively, ranging from an equimolar amount to a two- and fivefold molar excess over HspBP1. The left panel represents 66% of the input. (D) Hsc70 binding to BAG-1 is blocked in the presence of increasing amounts of HspBP1. All reactions received purified BAG-1 protein. When indicated equimolar amounts of Hsc70 were added. The HspBP1 concentration was increased from equimolar amounts to a two- and fivefold molar excess over BAG-1. BAG-1 and associated Hsc70 were isolated using a specific anti-BAG-1 antibody. When indicated rabbit IgG was used instead of anti-BAG-1 antibody to monitor unspecific binding. The left panel represents 20% of the input. (E) Overexpression of HspBP1 increases the amount of Hsc70 detectable in CHIP-containing protein complexes. HeLa cells were transfected with pCMVTag2-CHIP (+ FLAG-CHIP) and pcDNA3.1-hspBP1 (+ HspBP1) as indicated. The total amount of added DNA was kept constant by addition of pcDNA3.1. CHIP-containing protein complexes were isolated using an immobilized anti-FLAG antibody (IP). Immunoprecipitated CHIP was detected in glycine eluates using a specific antibody. HspBP1 and Hsc70 were detected in ATP eluates of the isolated CHIP complexes by immunoblotting. Extracts (40 μg) were loaded to monitor expression levels (ex.).
Because HspBP1 and CHIP bind to different domains of Hsc70, it was conceivable that the chaperone provides a physical link between the two cochaperones. Therefore, His-tagged HspBP1 was immobilized on an affinity resin and incubated with purified Hsc70 and CHIP in different combinations. An interaction between immobilized HspBP1 and CHIP was indeed largely dependent on the presence of Hsc70 (Figure 1B). However, a low amount of CHIP also interacted with HspBP1 in the absence of Hsc70, suggesting that a low-affinity direct interaction between the two cochaperones may contribute to the formation of the ternary complex (Figure 1B). The formed complex resembles the previously described BAG-1/Hsc70/CHIP complex (Demand et al., 2001). We therefore investigated whether HspBP1 and BAG-1 compete in binding to Hsc70. Addition of purified BAG-1 to binding reactions indeed strongly reduced the amount of Hsc70 retained on immobilized HspBP1 (Figure 1C). Similarly, HspBP1 interfered with the binding of Hsc70 to BAG-1, when the latter cochaperone was immobilized on an anti-BAG-1 antibody resin (Figure 1D). The data identify HspBP1 as a novel component of Hsc70/CHIP complexes, which competes with the degradation-stimulating cochaperone BAG-1 in complex binding. In contrast to the findings for BAG-1, addition of CHIP to binding reactions stimulated the interaction between HspBP1 and Hsc70 (Figure 1C). In this regard, it is also noteworthy that overexpression of HspBP1 in HeLa cells increased the amount of Hsc70 detectable in CHIP complexes (Figure 1E). HspBP1 and CHIP apparently interact with Hsc70 in a cooperative manner.
HspBP1 Inhibits the Ubiquitin Ligase Activity of CHIP When Complexed with Hsc70
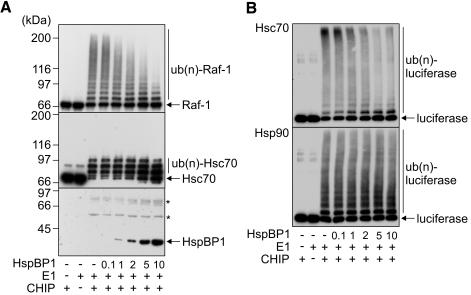
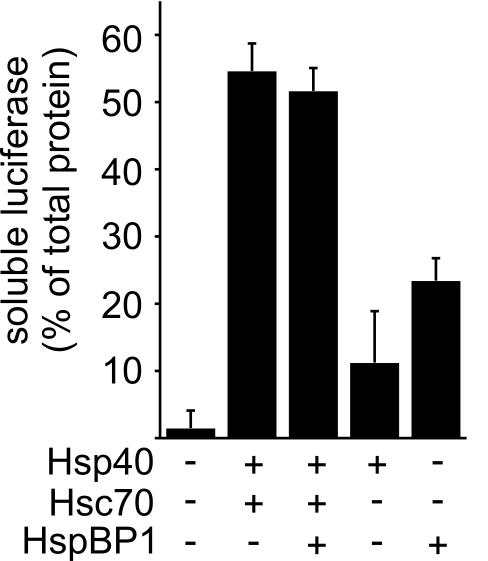
To elucidate the functional consequences of the observed interaction, effects of HspBP1 on CHIP-mediated ubiquitylation were investigated using a previously established in vitro assay (Demand et al., 2001; Alberti et al., 2002). In the presence of the ubiquitin-activating enzyme E1 and the ubiquitin-conjugating enzyme UbcH5b, CHIP mediated the ubiquitylation of the protein kinase Raf-1 when bound to Hsc70 (Figure 2A). Addition of HspBP1 strongly inhibited the ubiquitylation of the protein kinase. Similarly, the CHIP-mediated ubiquitylation of firefly luciferase, heat denatured in the presence of Hsc70, was attenuated by HspBP1 (Figure 2B). The amount of high molecular mass polyubiquitylated forms of luciferase was strongly reduced in the presence of HspBP1 and at the same time the level of unmodified luciferase increased. Remarkably, HspBP1 itself is not recognized as a substrate protein by the CHIP ubiquitin ligase (Figure 2A). A competition of HspBP1 and a substrate for limited resources during ubiquitylation can thus be excluded as the molecular basis for inhibition. An alternative explanation may invoke an only inefficient recognition of substrate proteins by Hsc70 in the presence of HspBP1. However, HspBP1 did not interfere with the Hsc70-mediated stabilization of luciferase during heat treatment (Figure 3), arguing against such a hypothesis. Furthermore, HspBP1 also inhibited the CHIP-induced ubiquitylation of Hsc70 (Figure 2A), which is itself a cellular target of the CHIP ubiquitin ligase (Jiang et al., 2001). Because HspBP1 and CHIP bind to Hsc70 in a cooperative manner (see above), HspBP1 apparently attenuates the ubiquitin ligase activity of CHIP when the two cochaperones are simultaneously associated with Hsc70. In contrast to the inhibitory activity that HspBP1 displayed in conjunction with Hsc70, HspBP1 did not significantly affect the CHIP-mediated ubiquitylation of luciferase bound to Hsp90 (Figure 2B). Complex formation between HspBP1, Hsc70 and CHIP is apparently necessary for HspBP1 to exert its CHIP-inhibiting activity.
Figure 2.
HspBP1 inhibits the ubiquitin ligase activity of CHIP when complexed with Hsc70. (A) In vitro ubiquitylation reactions were performed in the presence of purified CHIP, the ubiquitin-activating enzyme E1, and increasing concentrations of HspBP1, ranging from 0.1- to 10-fold the concentration of CHIP, as indicated. All reactions received UbcH5b and Hsc70. Raf-1, Hsc70, ubiquitylated forms of both proteins [ub(n)-Raf-1, ub(n)-Hsc70], and HspBP1 were detected using specific antibodies. Notably, Hsc70 that carried more than three ubiquitin moieties was not recovered by SDS-PAGE analysis. The stars denote polypeptides that cross-react with the anti-HspBP1 antibody. Numbers represent the molar ratio between HspBP1 and CHIP. (B) Luciferase was heat-denatured in the presence of Hsc70 (top) and Hsp90 (bottom), followed by addition of the CHIP ubiquitin conjugation system and increasing amounts of HspBP1. Luciferase was detected with a specific antibody.
Figure 3.
HspBP1 does not interfere with the stabilization of heat-denatured luciferase mediated by Hsc70. Luciferase was heat denatured in the presence of Hsp40, Hsc70 and HspBP1 as indicated and the amount of soluble luciferase was determined after removal of protein aggregates by centrifugation. Values were averaged from three independent experiments. Bars represent SEs.
HspBP1 Interferes with the Chaperone-assisted Degradation of CFTR
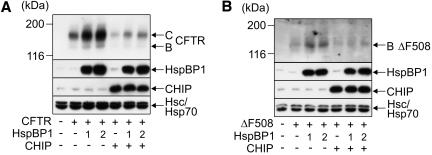
Immunofluorescence analysis revealed that a fraction of HspBP1 is localized at the ER membrane in mammalian cells (unpublished data). We therefore investigated whether HspBP1 participates in the ER-associated folding and maturation of the CFTR ion channel, which was previously identified as a substrate of CHIP (Meacham et al., 2001). In fact, elevating the cellular concentration of HspBP1 significantly increased the steady-state levels of coexpressed CFTR in HEK293 cells (Figure 4A). The increase was observed for the immature ER-localized B form of CFTR as well as for the maturely glycosylated C form that is located in the plasma membrane. HspBP1 apparently stimulates the maturation of CFTR. In contrast, overexpression of CHIP resulted in a strong decrease of CFTR levels (Figure 4A), which reflects the degradation-inducing activity of the ubiquitin ligase (Meacham et al., 2001). Notably, coexpression of HspBP1 counteracted the destructive activity of CHIP during CFTR biogenesis. This is apparently in agreement with a function of HspBP1 as an inhibitor of the CHIP ubiquitin ligase. Similar findings were obtained for CFTRΔF508, a form of the ion channel that is largely unable to proceed to the C form and is expressed in ∼70% of patients suffering from cystic fibrosis (Kopito, 1999). Overexpression of HspBP1 led to a significant increase of the cellular levels of CFTRΔF508 (Figure 4B). To verify that HspBP1 affects the degradation of CFTR, pulse-chase experiments were performed. HspBP1 slowed down CFTR turnover and attenuated the degradation-inducing activity of CHIP (Figure 5). Furthermore, we confirmed that HspBP1 inhibits the CHIP-mediated ubiquitylation of CFTR. For this purpose, CFTR was cotranslationally translocated into ER microsomes, followed by addition of the CHIP ubiquitin conjugation machinery and increasing amounts of HspBP1. HspBP1 significantly interfered with the CHIP-induced ubiquitylation of CFTR (Figure 6). Apparently, HspBP1 stabilizes CFTR by inhibiting chaperone-assisted degradation pathways. This results in a more efficient sorting of the ion channel to the plasma membrane as revealed by the increased detection of surface-biotinylated CFTR after overexpression of HspBP1 in HEK293 cells (Figure 7). Finally, we investigated how a decrease of the endogenous HspBP1 levels affected CFTR maturation. For this purpose, HEK293 cells were transfected with a pSUPER construct for expression of a HspBP1 shRNA, which is subsequently cleaved into siRNA in transfected cells. Transient transfection of this plasmid led to a considerable reduction of HspBP1 levels (Figure 8). At the same time, CFTR levels were significantly reduced, consistent with an essential role of HspBP1 in CFTR maturation. When HspBP1 was overexpressed, shRNA coexpression no longer affected cellular CFTR levels. This demonstrates that the HspBP1 shRNA does not unspecifically decrease CFTR expression. Together, the data identify HspBP1 as a cochaperone of Hsc70 and inhibitor of the CHIP ubiquitin ligase, which plays an essential role in the maturation of CFTR.
Figure 4.
HspBP1 increases the steady-state levels of coexpressed CFTR and CFTRΔF508 in HEK293 cells. (A) HEK293 cells were transiently transfected with CFTR-, HspBP1-, and CHIP-encoding plasmids in the indicated combinations. Amounts of pcDNA3.1-hspBP1 were one- and twofold of the amount of pcDNA3.1-CHIP when indicated. The ER-localized, core-glycosylated B-form (B) and the fully glycosylated, plasma membrane-localized C-form of CFTR (C) were detected by immunoblotting. Similarly, HspBP1 and CHIP levels were detected with specific antibodies. Immunodetection of Hsc70 and Hsp70 served as loading control. Each lane represents 60 μg of cellular extract. (B) Analysis of the cellular levels of CFTRΔF508 (ΔF508) in the presence of HspBP1 and CHIP as described under A.
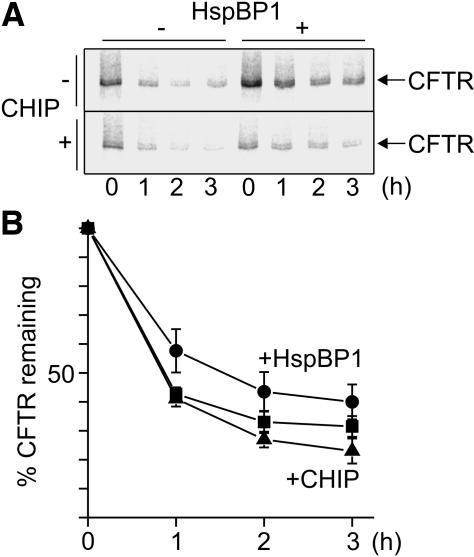
Figure 5.
HspBP1 slows down the rate of CFTR degradation. (A) Kinetics of CFTR degradation was determined in pulse-chase experiments by using HEK293 cells transiently transfected with CFTR-, CHIP-, and HspBP1-expressing plasmids as indicated. At the given time points cell extracts were prepared and radiolabeled CFTR was immunoprecipitated and deglycosylated. (B) Data obtained in pulse chase experiments for control cells (squares) or upon coexpression of HspBP1 (circles) and CHIP (triangles) were quantified. Values for CFTR turnover in the presence of CHIP and HspBP1 were omitted because degradation kinetics was indistinguishable from the control reaction in the absence of cochaperones. Presented data were averaged from four independent experiments. Error bars represent SEs.
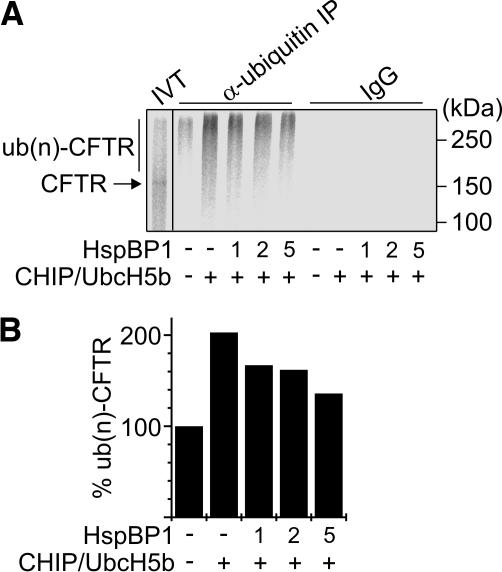
Figure 6.
HspBP1 inhibits the CHIP-induced ubiquitylation of CFTR in vitro. (A) CFTR was in vitro translated in the presence of microsomes (IVT), followed by the addition of CHIP, UbcH5b, and HspBP1 at a one-, two-, and fivefold molar excess over CHIP as indicated. Samples were incubated for 1 h followed by immunprecipitation with an anti-ubiquitin antibody that specifically binds ubiquitin conjugates (α-ubiquitin IP). Control reactions received unrelated immunoglobulins (IgG). `IVT' represents 20% of the sample used for immunoprecipitation. (B) Densitometric quantification of ubiquitylated CFTR [ub(n)-CFTR] immunoprecipitated under A. Values were averaged from two independent experiments.
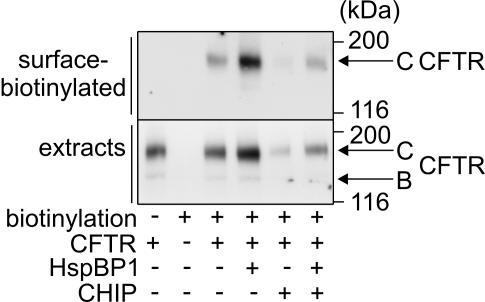
Figure 7.
HspBP1 stimulates the sorting of CFTR to the plasma membrane. Glycosylated CFTR exposed on the surface of HEK293 cells was biotinylated and subsequently isolated by avidin affinity chromatography. Surface-biotinylated proteins and total cell extracts were analyzed by SDS-PAGE and immunoblotting with an anti-CFTR antibody. Cells were processed 24 h after transfection with CFTR-, HspBP1-, and CHIP-expressing plasmids in the indicated combinations. To monitor CFTR expression, 40 μg of cell extracts was analyzed by immunoblotting.
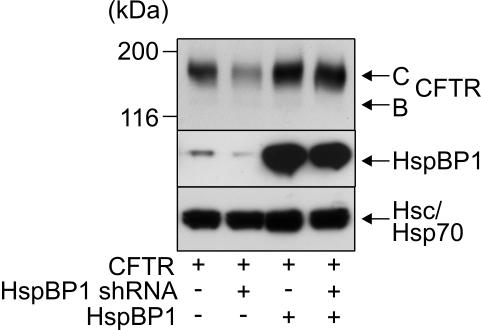
Figure 8.
HspBP1 is essential for CFTR maturation. HEK293 cells were transiently transfected with CFTR-, HspBP1 shRNA-, and HspBP1-expressing plasmids as indicated. Expression levels were analyzed 72 h posttransfection after separation of 40 μg of cell extracts on an SDS-PAG and immunoblotting by using specific antibodies. Hsc/Hsp70 served as loading control. Cells that did not express HspBP1 shRNA received the pSUPER plasmid without an insert.
DISCUSSION
The CHIP ubiquitin ligase is tightly embedded in the cellular cochaperone network that modulates the activity of Hsc70. Previously, CHIP was found to cooperate with the Hsc70 cochaperone BAG-1 during the proteasomal sorting of the glucocorticoid hormone receptor (Demand et al., 2001) (Figure 9). The characterization of HspBP1, which competes with BAG-1 in binding to the Hsc70/CHIP complex, now points to an unprecedented regulatory mechanism that confines the destructive activity of CHIP. HspBP1 inhibits CHIP-mediated ubiquitylation within an assembled Hsc70 chaperone complex (Figure 9). This inhibition similarly affects a client protein bound to Hsc70 as well as Hsc70 itself, which is a physiological substrate of the CHIP ubiquitin ligase (Jiang et al., 2001). Association of HspBP1 with the chaperone complex seems to shield Hsc70 and the bound client against CHIP-mediated ubiquitylation or may induce conformational changes of the chaperone complex that remove ubiquitin attachment sites from the reach of the CHIP ubiquitin ligase. In this way, HspBP1 directly modulates Hsc70-mediated protein quality control, as is evident from the role of the cochaperone in CFTR maturation. A main cause for misprocessing of CFTR is the inefficient folding of the first of two cytoplasmically exposed nucleotide binding domains (NBD1) of the membrane protein (Sato et al., 1996; Qu et al., 1997). The disease-causing ΔF508 mutation localizes to NBD1 and further decreases the folding propensity of this domain. During the cotranslational insertion of CFTR into the ER membrane, Hsc70 and its cochaperone Hdj-2 bind to NBD1 and facilitate intramolecular interactions between the domain and another cytoplasmic region of CFTR, the regulatory R-domain (Strickland et al., 1997; Meacham et al., 1999). When CHIP exerts its proteasomal targeting activity at this stage, immature CFTR is diverted onto the degradation pathway (Meacham et al., 2001). In this regard, HspBP1 fulfills a key regulatory function during CFTR biogenesis. By inhibiting the CHIP-mediated ubiquitylation of CFTR, HspBP1 strongly increased the cellular concentration of wild-type and mutant CFTR and stimulated the sorting of the ion channel to the plasma membrane. It was previously noted that other cochaperones of Hsc70 such as BAG-1 and Hip do not affect CFTR maturation (Meacham et al., 2001). Thus, HspBP1 seems to play a unique role within the cellular surveillance system that monitors the folding state of CFTR and apparently represents an essential CFTR assembly factor of limited availability.
Figure 9.
Model of the regulation of CHIP activity by the Hsc70 cochaperones BAG-1 and HspBP1. CHIP binds to the carboxy terminus of Hsc70 (C) and recruits the ubiquitin-conjugating enzyme UbcH5 to the chaperone complex. The two proteins cooperate to mediate the ubiquitylation of a chaperone client bound to the peptide-binding domain of Hsc70 (P). Simultaneous association of BAG-1 with the ATPase domain of Hsc70 (A) stimulates the CHIP-induced degradation of a subset of chaperone clients. On the other hand, HspBP1 inhibits the ubiquitin ligase activity of CHIP within the assembled chaperone complex.
Notably, CHIP participates in the regulation of the ATPase cycle of Hsc70 (Ballinger et al., 1999). The HspBP1-mediated inhibition of the ubiquitin ligase activity may therefore enable CHIP to modulate the Hsc70 chaperone cycle without exerting its destructive activity. In fact, degradation-independent functions of CHIP recently emerged (Cardozo et al., 2003; Jiang et al., 2003; Kampinga et al., 2003; Dai et al., 2003). CHIP was shown to regulate the chaperone-assisted folding and sorting of the androgen receptor and of endothelial nitric-oxide synthase without inducing degradation (Cardozo et al., 2003; Jiang et al., 2003). Moreover, CHIP fulfills an essential role in the chaperone-mediated regulation of the heat shock transcription factor, independent of its degradation-inducing activity (Dai et al., 2003). It remains to be seen, however, whether HspBP1 cooperates with CHIP in the regulation of the heat shock response. Although HspBP1 interfered with the degradation of CFTR, we noted that elevating the cellular levels of HspBP1 did not influence the CHIP-mediated turnover of the glucocorticoid hormone receptor (unpublished observation). Such a client specificity may arise in part from the fact that HspBP1 inhibits the ubiquitin ligase activity of CHIP in a complex with Hsc70, but it leaves Hsp90-associated ubiquitylation unaffected. In addition, direct interactions between HspBP1 and a subset of chaperone substrates may contribute to substrate selection. This is suggested by the finding that HspBP1 on its own partially stabilized luciferase during heat treatment and may therefore possess the ability to recognize certain chaperone substrates.
Modulating chaperone-assisted folding and degradation pathways has recently emerged as a promising therapeutic strategy. For example, antitumor agents, which inhibit Hsp90, induce the proteasomal degradation of signaling proteins, such as ErbB2, that play a key role in oncogenic transformation (Xu et al., 2002; deCandia et al., 2003; Kamal et al., 2003). CHIP participates on such pharmacologically induced degradation pathways (Xu et al., 2002). Consequently, the identification of HspBP1 as a cellular inhibitor of CHIP may open new roads to modulate pathologically relevant processes that involve chaperone action.
Acknowledgments
We thank Vince Guerriero for providing pET28a-hspBP1, Cam Patterson for the anti-CHIP antibody, Jan Grupp for constructing the plasmid pCMVTag2-hspBP1, and Doug Cyr and Margarida Amaral for providing CFTR expression plasmids. Albert Haas is acknowledged for critical reading of the manuscript and Karen Himmelberg for expert technical assistance. The described work was supported by the Deutsche Forschungsgemeinschaft (Ho 1518/5--2, SFB 635 TP 8).
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E04-04-0293. Article and publication date are available at www.molbiolcell.org/cgi/doi/10.1091/mbc.E04-04-0293.
References
- Alberti, S., Demand, J., Esser, C., Emmerich, N., Schild, H., and Höhfeld, J. (2002). Ubiquitylation of BAG-1 suggests a novel regulatory mechanism during the sorting of chaperone substrates to the proteasome. J. Biol. Chem. 277, 45920-45927. [DOI] [PubMed] [Google Scholar]
- Ballinger, C.A., Connell, P., Wu, Y., Hu, Z., Thompson, L.J., Yin, L.Y., and Patterson, C. (1999). Identification of CHIP, a novel tetratricopeptide repeat-containing protein that interacts with heat shock proteins and negatively regulates chaperone functions. Mol. Cell. Biol. 19, 4535-4545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardozo, C.P., Michaud, C., Ost, M.C., Fliss, A.E., Yang, E., Patterson, C., Hall, S.J., and Caplan, A.J. (2003). C-terminal Hsp-interacting protein slows androgen receptor synthesis and reduces its rate of degradation. Arch. Biochem. Biophys. 410, 134-140. [DOI] [PubMed] [Google Scholar]
- Connell, P., Ballinger, C.A., Jiang, J., Wu, Y., Thompson, L.J., Höhfeld, J., and Patterson, C. (2001). The co-chaperone CHIP regulates protein triage decisions mediated by heat-shock proteins. Nat. Cell Biol. 3, 93-96. [DOI] [PubMed] [Google Scholar]
- Dai, Q., et al. (2003). CHIP activates HSF1 and confers protection against apoptosis and cellular stress. EMBO J. 22, 5446-5458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- deCandia, P., Solit, D.B., Giri, D., Brogi, E., Siegel, P.M., Olshen, A.B., Muller, W.J., Rosen, N., and Benezra, R. (2003). Angiogenesis impairment in Iddeficient mice cooperates with an Hsp90 inhibitor to completely suppress HER2/neu-dependent breast tumors. Proc. Natl. Acad. Sci. USA 100, 12337-12342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demand, J., Alberti, S., Patterson, C., and Höhfeld, J. (2001). Cooperation of a ubiquitin domain protein and an E3 ubiquitin ligase during chaperone/proteasome coupling. Curr. Biol. 11, 1569-1577. [DOI] [PubMed] [Google Scholar]
- Farinha, C.M., Nogueira, P., Mendes, F., Penque, D., and Amaral, M.D. (2002). The human DnaJ homologue (Hdj)-1/heat-shock protein (Hsp) 40 co-chaperone is required for the in vivo stabilization of the cystic fibrosis transmembrane conductance regulator by Hsp70. Biochem. J. 366, 797-806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frydman, J. (2001). Folding of newly translated proteins in vivo: the role of molecular chaperones. Annu. Rev. Biochem. 70, 603-647. [DOI] [PubMed] [Google Scholar]
- Hartl, F.U., and Hayer-Hartl, M. (2002). Molecular chaperones in the cytosol: from nascent chain to folded protein. Science 295, 1852-1858. [DOI] [PubMed] [Google Scholar]
- Höhfeld, J., and Jentsch, S. (1997). GrpE-like regulation of the hsc70 chaperone by the anti-apoptotic protein BAG-1. EMBO J. 16, 6209-6216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Höhfeld, J., Cyr, D.M., and Patterson, C. (2001). From the cradle to the grave: molecular chaperones that may choose between folding and degradation. EMBO Rep. 2, 885-890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen, T.J., Loo, M.A., Pind, S., Williams, D.B., Goldberg, A.L., and Riordan, J.R. (1995). Multiple proteolytic systems, including the proteasome, contribute to CFTR processing. Cell 83, 129-135. [DOI] [PubMed] [Google Scholar]
- Jiang, J., Ballinger, C.A., Wu, Y., Dai, Q., Cyr, D.M., Höhfeld, J., and Patterson, C. (2001). CHIP is a U-box-dependent E3 ubiquitin ligase: identification of Hsc70 as a target for ubiquitylation. J. Biol. Chem. 276, 42938-42944. [DOI] [PubMed] [Google Scholar]
- Jiang, J., Cyr, D., Babbitt, R.W., Sessa, W.C., and Patterson, C. (2003). Chaperone-dependent regulation of endothelial nitric-oxide synthase intracellular trafficking by the co-chaperone/ubiquitin ligase CHIP. J. Biol. Chem. 278, 49332-49341. [DOI] [PubMed] [Google Scholar]
- Kabani, M., McLellan, C., Raynes, D.A., Guerriero, V., and Brodsky, J.L. (2002). HspBP1, a homologue of the yeast Fes1 and Sls1 proteins, is an Hsc70 nucleotide exchange factor. FEBS Lett. 531, 339-342. [DOI] [PubMed] [Google Scholar]
- Kamal, A., Thao, L., Sensintaffar, J., Zhang, L., Boehm, M.F., Fritz, L.C., and Burrows, F.J. (2003). A high-affinity conformation of Hsp90 confers tumour selectivity on Hsp90 inhibitors. Nature 425, 407-410. [DOI] [PubMed] [Google Scholar]
- Kampinga, H.H., Kanon, B., Salomons, F.A., Kabakov, A.E., and Patterson, C. (2003). Overexpression of the cochaperone CHIP enhances Hsp70-dependent folding activity in mammalian cells. Mol. Cell. Biol. 23, 4948-4958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopito, R.R. (1999). Biosynthesis and degradation of CFTR. Physiol. Rev. 79, S167-S173. [DOI] [PubMed] [Google Scholar]
- Kostova, Z., and Wolf, D.H. (2003). For whom the bell tolls: protein quality control of the endoplasmic reticulum and the ubiquitin-proteasome connection. EMBO J. 22, 2309-2317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisanti, M.P., LeBivic, A., Sargiacomo, M., and Rodriguez-Boulan, E. (1989). Steady-state distribution and biogenesis of endogenous Madin-Darby canine kidney glycoproteins: evidence for intracellular sorting and polarized cell surface delivery. J. Cell Biol. 109, 2117-2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loo, M.A., Jensen, T.J., Cui, L., Hou, Y., Chang, X.B., and Riordan, J.R. (1998). Perturbation of Hsp90 interaction with nascent CFTR prevents its maturation and accelerates its degradation by the proteasome. EMBO J. 17, 6879-6887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lüders, J., Demand, J., and Höhfeld, J. (2000). The ubiquitin-related BAG-1 provides a link between the molecular chaperones Hsc70/Hsp70 and the proteasome. J. Biol. Chem. 275, 4613-4617. [DOI] [PubMed] [Google Scholar]
- Meacham, G.C., Lu, Z., King, S., Sorscher, E., Tousson, A., and Cyr, D.M. (1999). The Hdj-2/Hsc70 chaperone pair facilitates early steps in CFTR biogenesis. EMBO J. 18, 1492-1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meacham, G.C., Patterson, C., Zhang, W., Younger, J.M., and Cyr, D.M. (2001). The Hsc70 co-chaperone CHIP targets immature CFTR for proteasomal degradation. Nat. Cell Biol. 3, 100-105. [DOI] [PubMed] [Google Scholar]
- Murata, S., Minami, Y., Minami, M., Chiba, T., and Tanaka, K. (2001). CHIP is a chaperone-dependent E3 ligase that ubiquitylates unfolded protein. EMBO Rep. 2, 1133-1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu, B.H., Strickland, E.H., and Thomas, P.J. (1997). Localization and suppression of a kinetic defect in cystic fibrosis transmembrane conductance regulator folding. J. Biol. Chem. 272, 15739-15744. [DOI] [PubMed] [Google Scholar]
- Raynes, D.A., and Guerriero, V., Jr. (1998). Inhibition of Hsp70 ATPase activity and protein renaturation by a novel Hsp70-binding protein. J. Biol. Chem. 273, 32883-32888. [DOI] [PubMed] [Google Scholar]
- Riordan, J.R. (1999). Cystic fibrosis as a disease of misprocessing of the cystic fibrosis transmembrane conductance regulator glycoprotein. Am. J. Hum. Genet. 64, 1499-1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato, S., Ward, C.L., Krouse, M.E., Wine, J.J., and Kopito, R.R. (1996). Glycerol reverses the misfolding phenotype of the most common cystic fibrosis mutation. J. Biol. Chem. 271, 635-638. [DOI] [PubMed] [Google Scholar]
- Sondermann, H., Scheufler, C., Schneider, C., Höhfeld, J., Hartl, F.U., and Moarefi, I. (2001). Structure of a Bag/Hsc70 complex: convergent functional evolution of Hsp70 nucleotide exchange factors. Science 291, 1553-1557. [DOI] [PubMed] [Google Scholar]
- Strickland, E., Qu, B.H., Millen, L., and Thomas, P.J. (1997). The molecular chaperone Hsc70 assists the in vitro folding of the N-terminal nucleotide-binding domain of the cystic fibrosis transmembrane conductance regulator. J. Biol. Chem. 272, 25421-25424. [DOI] [PubMed] [Google Scholar]
- Walter, P., and Blobel, G. (1983). Preparation of microsomal membranes for cotranslational protein translocation. Methods Enzymol. 96, 84-93. [DOI] [PubMed] [Google Scholar]
- Ward, C.L., Omura, S., and Kopito, R.R. (1995). Degradation of CFTR by the ubiquitin-proteasome pathway. Cell 83, 121-127. [DOI] [PubMed] [Google Scholar]
- Xu, W., Marcu, M., Yuan, X., Mimnaugh, E., Patterson, C., and Neckers, L. (2002). Chaperone-dependent E3 ubiquitin ligase CHIP mediates a degradative pathway for c-ErbB2/Neu. Proc. Natl. Acad. Sci. USA 99, 12847-12852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, Y., Janich, S., Cohn, J.A., and Wilson, J.M. (1993). The common variant of cystic fibrosis transmembrane conductance regulator is recognized by hsp70 and degraded in a pre-Golgi nonlysosomal compartment. Proc. Natl. Acad. Sci. USA 90, 9480-9484. [DOI] [PMC free article] [PubMed] [Google Scholar]