Abstract
The Agatston score, computed from ECG-gated computed tomography (CT), is a well established metric of coronary artery disease. It has been recently shown that the Agatston score computed from chest CT (non ECG-gated) studies is highly correlated with the Agatston score computed from cardiac CT scans. In this work we present an automated method to compute the Agatston score from chest CT images. Coronary arteries calcifications (CACs) are defined as voxels contained within the coronary arteries with a value greater or equal to 130 Hounsfield Units (HU). CACs are automatically detected in chest CT studies by locating the heart, generating a region of interest around it, thresholding the image in such region and applying a set of rules to discriminate CACs from calcifications in the main vessels or from metallic implants. We evaluate the methodology in a large cohort of 1500 patients for whom manual reference standard is available. Our results show that the Pearson correlation coefficient between manual and automated Agatston score is ρ = 0.86 (p < 0.0001)
Index Terms: Agatston score, object detection, computed aided detection, segmentation, heuristics
1. INTRODUCTION
Every year, 62 million CT scans are performed in the United States alone. One third of them (31%) are chest CT studies performed to inspect the lungs. Patients suffering from lung disease are also at risk of cardiovascular disease [1]. With the latest generations of CT scanners, the artifacts due to heart-movement are minimal, and the evaluation of coronary artery disease (CAD) in such patients is possible. The extent of CAD disease in ECG-gated CT studies is measured with a well established metric, the Agatston score [2]. This score is computed by finding the coronary artery calcifications (CACs) in the image, for each of them computing a score by multiplying their volume by a factor related to the maximum intensity of the CAC, and finally adding the individual scores. Recent studies have shown excellent correlation between the Agatston score computed in cardiac ECG-gated CT and in no ECG-gated chest CT [3].
There is a large interest in automating calcium scoring. Such automation is challenging due to several issues. First, the heart needs to be accurately located. Anatomy-based approaches [4] and atlas-based location strategies [5] have been previously used towards that end. Second, CAC are defined as voxels within the coronary arteries with a value greater than 130 Hounsfield Units (HU). There are many bright structures in the images, with values greater than 130 HUs, such as bones, calcifications in the greater vessels, calcifications in the heart valves, or even metallic implants. Only a small subset of bright structures should be included in the computation of the Agatston score. Several approaches exist to eliminate non-CAC bright structures: in [6] calcifications on the mitral and aortic valves are explicitly located and removed, [7] proposes a classification scheme using location, texture and size as features and [8, 9] combine atlas-based coronary artery segmentation with discriminative learning schemas for ECG-gated CT. These later methods exhibit excellent person correlation coefficients with the reference standard Agatston score. However, they require a large dataset of positive and negative calcifications in order to generate their discriminative models.
This study presents an automated CAC estimator for non ECG-gated chest CT that produces high correlation with manual reference standard in a large cohort of patients. The main difference with previous work is that our method uses the Agatston reference standard itself to optimize the parameters that control the acceptance or rejection of calcifications, instead of a dataset of positive and negative calcifications generated explicitly to train a CAC classifier. This difference is important, since the Agatston score can be obtained from routine clinical practice, while the labeled dataset of CAC requires expert input.
The presented algorithm could be used as an automated method that suggests CAC scores to the radiologists or to process large datasets to perform statistical analysis of the burden of disease and co-morbidities in the population covered by a healthcare organization.
2. MATERIALS AND METHODS
2.1. Evaluation Database
CT scans of non-hispanic White and African American individuals with a minimum history of at least 10 pack-years of smoking were acquired using multi-detector CT scanners with at least 16 detector channels. The data is a subset from the COPDGene study [10]. Volumetric data was reconstructed with sub-millimeter slice thickness. CACs were found using a CT threshold of 130 HUs, involving at least 3 contiguous voxels for identification, resulting in a minimum lesion area of 1.02 mm2. Only lesions in the coronary arteries were taken into account. For each lesion, a score was calculated using the area density method described in [2]: the lesion area is multiplied by a density factor derived from the maximal HU within the area. The density factor was assigned in the following manner: 1 for lesions whose maximal density was [130, 199] HU, 2 for lesions [200, 299] HU, 3 for lesions [300, 399] HU, and 4 for lesions > 400 HU. A total coronary calcium score was determined by adding individual lesion scores.
2.2. Automated Agatston Score Computation
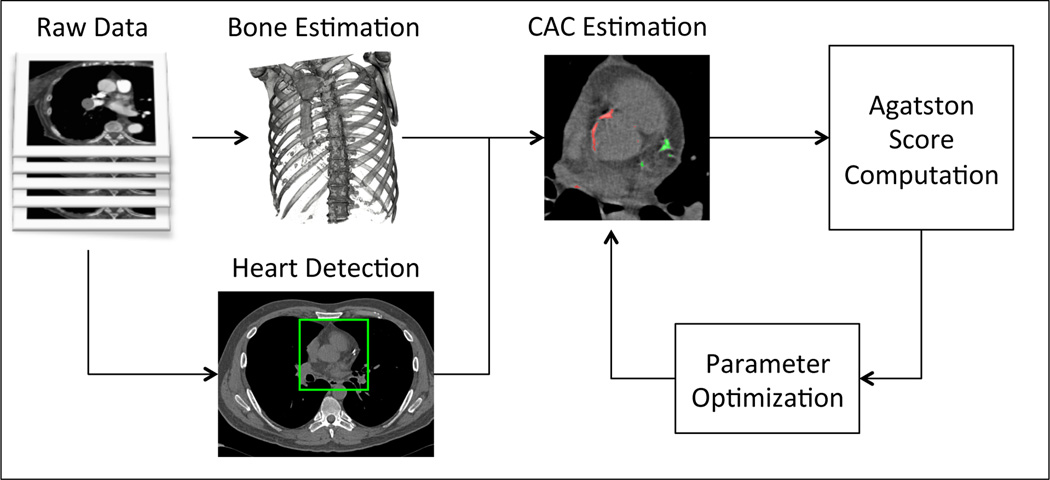
The automated computation of the CAC score is performed with the workflow shown in Figure 1. The input of the workflow is a chest CT scan and the output is a real value representing the Agatston score. The workflow is as follows: first, bone structures are detected and removed to prevent them from being misidentified as CACs. Second, the beginning of the coronary arteries is found using a 2.5D machine-learning based object detector similar to the one presented in [11], but using as 2D detector the work of [12]. A region of interest is defined around the detected object, encompassing the heart, from the pulmonary artery to the appex. Calcified voxels, defined as voxels within the region of interest with intensity greater than > 130 HUs, are identified. Such voxels are grouped in connected components. Each connected component is analyzed by a set of rules to determine if it should be included in the Agatston score computation. Finally the score is computed. In the following subsections we present the method in detail.
Fig. 1.
Method overview.
2.2.1. Bone segmentation
Bone structures are large structures, in close proximity to each other, with high HUs. To detect them, we first downsample the image to a voxel size of 2.5mm × 2.5mm × 2.5mm. We threshold the downsampled image at a value of 200 HU and perform a closing operation. Connected component analysis is performed. Bone structures are identified as the largest connected component. The bone mask is up-scaled to the size of the original image and closed to eliminate up-sampling artifacts. Since CACs are far from bone structures, they are not be included in the bone mask. An example of the bone mask can be found in Fig. 3.
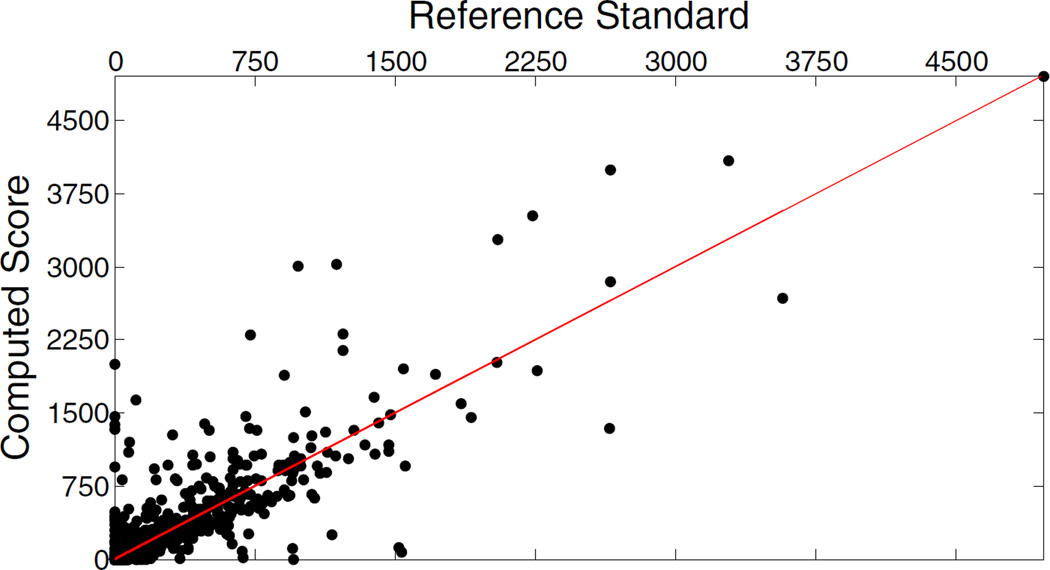
Fig. 3.
Correlation plot between the reference standard and the automatically computed score.
2.2.2. Heart Detection
We define heart detection as a standard classification problem where the input is the 3D CT image and the output is a 3D region of interest encompassing the heart. To this end, we adopted the 2.5D methodology presented in [12], adapting it to the context of chest CT scans. We trained the 2D classifier to detect the area of the great vessels of the heart, centered at the axial plane of the beginning of the coronary arteries. Such structures at that plane have very particular image properties: the pulmonary artery and the aorta have elliptical shapes and the coronary arteries form a clear winding path. Further changes to the methodology of [12] are the use of Haar wavelets as features instead of histograms of oriented gradients, the use of AdaBoost as a classifier instead of Support Vector Machines and the use of DBSCAN as a clustering algorithm instead of MeanShift. Such design choices were motivated by implementation and speed of execution constraints. Once the great vessels at the beginning of the coronary arteries structure are located, a volume surrounding them is defined. The volume width is equal to 1.5 times the width of the detection, the height is 1.5 times the height of the detection and the depth is 2 times the mean of the width and height of the detection. Such region of interest, encompassing the heart, part of the lungs and some bone structures is analyzed to find CACs.
As for any machine learning-based method, our heart detector requires positive and negative training samples. We separated 40 cases from the evaluation dataset to generate such training samples. In each case, we annotated the structure of interest, resulting in 40 positive training images. The positive training set was augmented by applying random affine transforms to the positive samples, resulting in a total of 2000 positive training images. Negative training images were defined as axial slices of the training CT cases where the structure of interest is not present, resulting in 16, 131 2D images.
2.2.3. Coronary Artery Calcification Detection
The region of interest obtained by the heart detector is analyzed to find CACs. Calcification candidates are defined as connected components whose voxels’ HUs are greater than 130 and their size is greater than three. Calcifications candidates can appear in several places of the heart, such as the heart valves, the main vessels and the coronary arteries. Furthermore, other structures such as stents or pacemakers have high HU units and can be easily confused with calcifications. We use a set of heuristics to differentiate CAC from the rest of calcifications or artifacts. The heuristics are based on four observations:
-
O1
CACs are surrounded primarily by muscle. Calcifications on the main vessels or on the cardiac valves are surrounded by blood or by blood and cardiac muscle. Blood has a standard HU range of [0, 130], while muscle is in the range [−500, 0].
-
O2
CACs have lower HU than metallic implants.
-
O3
CACs do not neighbor the lungs directly, but have some cardiac muscle surrounding them.
-
O4
CACs are small in comparison to calcifications found on the great vessels of the heart.
We analyze each calcification candidate to evaluate if it as a CAC. First, for each candidate, we analyze the voxels that are not part of the candidate and are less than 3 mm from the candidate’s border. The mean intensity value is computed for those voxels. If the mean value is greater than a threshold thBorder, the component is excluded from further analysis. Observation 2 is honored by computing the mean and maximum HU in the component. If the mean value is greater than thMean or the maximum value is greater than thMax, the component is excluded. The third observation is put into practice by generating a mask of the lungs and dilating it by 3mm. Such mask is generated by thresholding the image at a value of −500 HU. If the calcification candidate has more than thLung percent of its voxels in such mask, it is excluded from further analysis. The fourth observation implemented by defining a maximum allowed size thSize and eliminating candidates above such size.
The Agatston score is computed for all remaining calcifications by finding the maximum value in the calcification, associating a factor to it following the same equation as defined in Section 2.1, and multiplying such factor by the volume of the calcification.
Parameter optimization
The exclusion criteria depends on five different parameters:
The optimal parameter set is found by maximizing the correlation coefficient between the computed Agatston score and the reference standard on the first 452 cases of the evaluation dataset for whom the heart detector produces a valid detection using a grid search algorithm. The same 452 samples are used to perform a linear calibration between the automated score and the reference standard.
3. RESULTS
The proposed workflow is evaluated in the databased described in Section 2.1. The parameters obtained are: Θ = [10, 2100, 475, 0, 15, 2500]. The computation time per case was of 126 ± 10 seconds.
3.1. Heart Detection
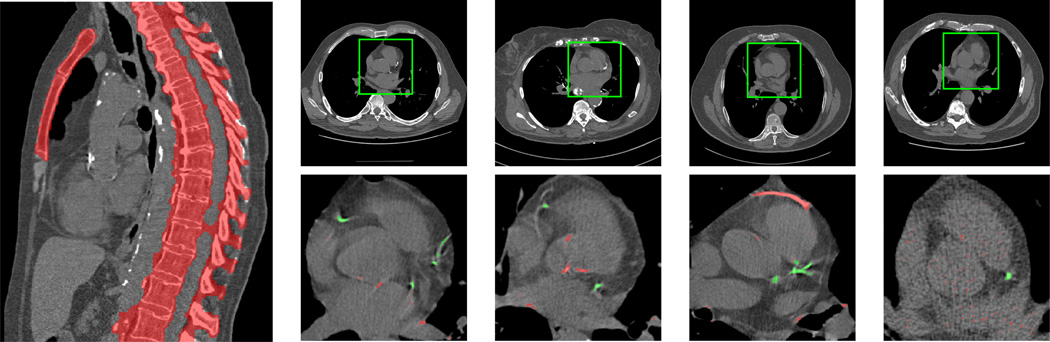
The software detected the heart in 1452 of the 1500 in which the code was run (96.8%). From the 48 wrong detections, three were in the mandible of the patient, one in the liver, and the object detector did not produce an output in the other 44 cases. Examples of correct detections are shown in Fig. 2 (top row).
Fig. 2.
Left: bone segmentation. The main structures are detected. Right-top: Correct heart detections in four different cases of the evaluation database. Right-bottom: Examples of accepted (green) and rejected (red) CAC candidates in four cases of the dataset. Rejected calcifications are in the ascending aorta, the aortic valve, the pulmonary artery, metallic implants, and in the great vessels due to noise.
3.2. Agatston Calcium Score Computation
Fig. 2 (bottom row) shows examples of the evaluation of calcification candidates by the rule-based system in four cases of the dataset. Rejected candidates are shown in red and accepted candidates are shown in green.
Fig. 3 shows the correlation between the automatically computed Agatston score and the reference standard. The pearson correlation coefficient, computed using 1000 cases, is 0.86 (p < 0.0001).
Following the same evaluation methodology performed in [13], we stratify the patients in five groups according to their Agatston score. Group I is defined as patients with Agatston score in the range [0, 10), Group II in range [10, 100), Group III in range [100, 400), Group IV in range [400, 1000) and Group V if the Agatston score is > 1000 units. Table. 1 shows the confusion matrix between the different groups, 67.7% of patients are classified correctly, 28% of patients are misclassified in a group one level above or below their correct group and 4.3% of the patients are misclassified with two or more distance groups. The risk group is over-estimated in 23.4% of the cases and under-estimated in 8.9%.
Table 1.
Confusion matrix for the different levels of severity of the calcifications.
| Automated | ||||||
|---|---|---|---|---|---|---|
| Manual | I | II | III | IV | V | |
| I | 286 | 11 | 11 | 4 | 4 | |
| II | 116 | 105 | 12 | 4 | 2 | |
| III | 9 | 65 | 164 | 19 | 2 | |
| IV | 2 | 2 | 30 | 91 | 19 | |
| V | 0 | 1 | 1 | 8 | 32 | |
The main reason for underestimation is a false negative CAC, and is specially visible in cases classified as Group I automatically while they belong to Group II (116 cases). The reasons for large over-estimation of risk group are the following: presence of stents (n=8), detection of calcifications in the aorta (n=5), inclusion of the sternum (n=7) or the vertebrae (n=2), other bright artifacts (n=4) and wrong heart detection (n=1).
4. DISCUSION
We have presented an automated method to estimate the Agatston calcium score in chest CT images that yields high pearson correlation coefficient (ρ = 0.86) against manual reference standard in a large cohort of 1000 patients. The evaluation of the method is performed in the largest dataset of Agatston score computation reported in the literature.
The correlation coefficient reported in this study is lower than the coefficients reported in [8] ρ = 0.96 and [9] ρ = 0.94. However, please note that we have evaluated the proposed algorithm in a much larger dataset (1000 cases vs. 101 and 164), and that we have not excluded patients with stents or pacemakers. To compare our results against the previous work, we selected randomly 150 cases from the 1000 test cases and computed the correlation coefficient on them. We repeated the process 1000 times. The mean correlation coefficient is ρ̂ = 0.85, and its variance is σρ = 0.063, suggesting that the result is highly variable with the selection of the 150 training samples. Furthermore, for a complete fair comparison with [9], an evaluation of the proposed method excluding patients with stents or other metallic implants is required. However, such information is not available at the time of this writing.
Several limitations exist on this study. First, one can argue that a truly 3D approach towards finding the heart in the CT images will outperform the 2.5D approach used in this paper. While such affirmation is very likely true, the 2.5D approach has shown excellent performance, is extremely fast, and leaves little margin for improvement. Second, the set of heuristics designed to eliminate calcification candidates, as well as the optimization method of the thresholds, can be considered naïve. However, we have shown slightly worse performance to state-of-the-art statistical classification methods with a much simpler and easy to reproduce method
The largest reasons for over-estimation of the Agatston score by our method is the presence of stents, calcifications in the aorta or the inclusion of the bone structures as part of the calcifications. Patients with stents can be excluded from the database, since they are known to be at risk of a cardiac event. The calcifications on the aorta could be removed by using an aorta detection method, such as the work of [14]. The inclusion of bone structures as CACs suggests the need of a better bone segmentation method.
The main advantage of the proposed method with respect to the work of [8, 9] is that the proposed method does not require labeled calcification maps, instead, we learn inclusion and exclusion rules directly from the Agatston score. This work is an example of a framework whose parameters can be inferred directly from data obtained clinically, without the need of extra labeling.
Acknowledgments
This work has been supported by grants from the National Institutes of Health R01HL116931 and R01HL116473
REFERENCES
- 1.Matsuoka Shin, Yamashiro Tsuneo, Diaz Alejandro, Estpar Ral San Jos, Ross James C, Silverman Edwin K, Kobayashi Yasuyuki, Dransfield Mark T, Bartholmai Brian J, Hatabu Hiroto, Washko George R. The Relationship between Small Pulmonary Vascular Alteration and Aortic Atherosclerosis in Chronic Obstructive Pulmonary Disease: Quantitative CT Analysis. Acad Radiol. 2011 Jan.18(1):40–46. doi: 10.1016/j.acra.2010.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Agatston Arthur S, Janowitz Warren R, Hildner Frank J, Zusmer Noel R, Viamonte Manuel, Detrano Robert. Quantification of coronary artery calcium using ultrafast computed tomography. Journal of the American College of Cardiology. 1990 Mar.15(4):827–832. doi: 10.1016/0735-1097(90)90282-t. [DOI] [PubMed] [Google Scholar]
- 3.Budoff Matthew J, Nasir Khurram, Kinney Gregory L, Hokanson John E, Barr R Graham, Steiner Robert, Nath Hrudaya, Lopez-Garcia Carmen, Black-Shinn Jennifer, Casaburi Richard. Coronary artery and thoracic calcium on noncontrast thoracic CT scans: Comparison of ungated and gated examinations in patients from the COPD Gene cohort. Journal of Cardiovascular Computed Tomography. 2011 Mar.5(2):113–118. doi: 10.1016/j.jcct.2010.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Reeves Anthony P, Biancardi Alberto M, Yankelevitz David F, Cham Matthew D, Henschke Claudia I. Heart region segmentation from low-dose CT scans: an anatomy based approach. In: Haynor David R, Ourselin Sbastien., editors. SPIE. Vol. 8314. 2012. Feb. p. 83142A. [Google Scholar]
- 5.Isgum I, Staring M, Rutten A, Prokop M, Viergever MA, van Ginneken B. Multi-Atlas-Based Segmentation With Local Decision Fusion-Application to Cardiac and Aortic Segmentation in CT Scans. IEEE Transactions on Medical Imaging. 2009 Jul;28(7):1000–1010. doi: 10.1109/TMI.2008.2011480. [DOI] [PubMed] [Google Scholar]
- 6.Xie Yiting, Cham Matthew D, Henschke Claudia, Yankelevitz David, Reeves Anthony P. Automated coronary artery calcification detection on low-dose chest CT images. SPIE. 2014 Mar.9035:90350F–90350F–9. [Google Scholar]
- 7.Isgum I, Prokop M, Niemeijer M, Viergever MA, van Ginneken B. Automatic Coronary Calcium Scoring in Low-Dose Chest Computed Tomography. IEEE Transactions on Medical Imaging. 2012 Dec.31(12):2322–2334. doi: 10.1109/TMI.2012.2216889. [DOI] [PubMed] [Google Scholar]
- 8.Shahzad Rahil, van Walsum Theo, Schaap Michiel, Rossi Alexia, Klein Stefan, Weustink Annick C, de Feyter Pim J, van Vliet Lucas J, Niessen Wiro J. Vessel Specific Coronary Artery Calcium Scoring. Academic Radiology. 2013 Jan.20(1):1–9. doi: 10.1016/j.acra.2012.07.018. [DOI] [PubMed] [Google Scholar]
- 9.Wolterink Jelmer M, Leiner Tim, Takx Richard AP, Viergever Max A, Igum Ivana. An automatic machine learning system for coronary calcium scoring in clinical non-contrast enhanced, ECG-triggered cardiac CT. SPIE. 2014;9035:90350E–90350E–8. [Google Scholar]
- 10.Regan Elizabeth A, Hokanson John E, Murphy James R, Make Barry, Lynch David A, Beaty Terri H, Curran-Everett Douglas, Silverman Edwin K, Crapo James D. Genetic Epidemiology of COPD (COPDGene) Study Design. COPD: Journal of Chronic Obstructive Pulmonary Disease. 2010 Feb.7(1):32–43. doi: 10.3109/15412550903499522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rodriguez-Lopez Sara, Jimenez-Carretero Daniel, Estepar Raul San Jose, Moreno Eduardo Fraile, Kumamaru Kanako K, Rybicki Frank J, Ledesma-Carabayo Maria Jesus, Gonzalez German. Automatic Ventricle Detection in Computed Tomography Pulmonary Angiography. Biomedical Imaging (ISBI), 2015 IEEE 12th International Symposium on; New York. 2015. Apr. pp. 1143–1146. [Google Scholar]
- 12.Viola P, Jones M. Computer Vision and Pattern Recognition, 2001. CVPR 2001. Proceedings of the 2001 IEEE Computer Society Conference on. IEEE; 2001. Rapid object detection using a boosted cascade of simple features; pp. I–511–I–518. [Google Scholar]
- 13.Erbel Raimund, Mhlenkamp Stefan, Kerkhoff Gert, Budde Thomas, Schmermund Axel. Noninvasive screening for coronary artery disease: calcium scoring. Heart. 2007 Dec.93(12):1620–1629. doi: 10.1136/hrt.2005.071258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kurugol Sila, Estepar Raul San Jose, Ross James, Washko George R. Engineering in Medicine and Biology Society (EMBC), 2012 Annual International Conference of the IEEE. IEEE; 2012. Aorta segmentation with a 3d level set approach and quantification of aortic calcifications in non-contrast chest ct; pp. 2343–2346. [DOI] [PMC free article] [PubMed] [Google Scholar]