Abstract
Replication blocks and DNA damage incurred during S phase activate the S-phase and intra-S-phase checkpoint responses, respectively, regulated by the Atrp and Chk1p checkpoint kinases in metazoans. In Saccharomyces cerevisiae, these checkpoints are regulated by the Atrp homologue Mec1p and the kinase Rad53p. A conserved role of these checkpoints is to block mitotic progression until DNA replication and repair are completed. In S. cerevisiae, these checkpoints include a transcriptional response regulated by the kinase Dun1p; however, dun1Δ cells are proficient for the S-phase-checkpoint-induced anaphase block. Yeast Chk1p kinase regulates the metaphase-to-anaphase transition in the DNA-damage checkpoint pathway via securin (Pds1p) phosphorylation. However, like Dun1p, yeast Chk1p is not required for the S-phase-checkpoint-induced anaphase block. Here we report that Chk1p has a role in the intra-S-phase checkpoint activated when yeast cells replicate their DNA in the presence of low concentrations of hydroxyurea (HU). Chk1p was modified and Pds1p was transiently phosphorylated in this response. Cells lacking Dun1p were dependent on Chk1p for survival in HU, and chk1Δ dun1Δ cells were defective in the recovery from replication interference caused by transient HU exposure. These studies establish a relationship between the S-phase and DNA-damage checkpoint pathways in S. cerevisiae and suggest that at least in some genetic backgrounds, the Chk1p/securin pathway is required for the recovery from stalled or collapsed replication forks.
INTRODUCTION
Checkpoints mediate arrest of the cell cycle to ensure genomic stability by allowing time for the successful completion of DNA replication and repair of damaged DNA. Checkpoint deficiencies can result in genomic instability as well as susceptibility to cancer in higher organisms (Nyberg et al., 2002). In Saccharomyces cerevisiae, replication blocks and DNA damage activate the S-phase (or S/M) and intra-S-phase checkpoint responses, respectively. The S-phase checkpoint is elicited in response to delayed DNA replication induced by exposure of cells to hydroxyurea (HU), an inhibitor of ribonucleotide reductase (Rnrp) that causes nucleotide depletion. The intra-S-phase checkpoint detects DNA damage that occurs during S phase. The S-phase and intra-S-phase checkpoints are referred to jointly below as the S-phase-checkpoint pathways.
The S-phase-checkpoint pathways are regulated by the kinase Mec1p, which acts upstream of the kinase Rad53p to stabilize replication forks, prevent firing of late replication origins, and prevent anaphase entry (Allen et al., 1994; Weinert et al., 1994; Sun et al., 1996; Fay et al., 1997; Desany et al., 1998; Santocanale and Diffley, 1998; Lopes et al., 2001). In addition, Rad53p initiates a transcriptional response through activation of the kinase Dun1p in response to replication delay induced by HU and DNA damage (Zhou and Elledge, 1993; Allen et al., 1994; de la Torre Ruiz and Lowndes, 2000). The best-characterized targets of Dun1p are the ribonucleotide reductase (RNR) genes (Zhou and Elledge, 1993; Kiser and Weinert, 1996; Huang et al., 1998; de la Torre Ruiz and Lowndes, 2000). dun1Δ cells have increased gross chromosomal rearrangements in the absence of exogenous sources of damage, indicating that Dun1p has an additional role in maintaining genome stability (Myung et al., 2001a, 2001b). dun1Δ cells are proficient for the anaphase block induced by the S-phase-checkpoint pathways and are less sensitive than rad53Δ mutants to HU (Zhou and Elledge, 1993). The sensitivity of rad53 cells to HU is not reversible, and it has been proposed that this phenotype is a result of the role of Rad53p in preventing the irreversible collapse of replication forks (Sogo et al., 2002; Tercero et al., 2003).
Mec1p also regulates the DNA-damage checkpoint activated in late S/G2, which blocks mitotic progression through activation by phosphorylation of the kinases Chk1p and Rad53p (Weinert et al., 1994; Sanchez et al., 1996; Sun et al., 1996). In this response, Chk1p blocks the transition from metaphase to anaphase (M-A), and Rad53p has a redundant role with Chk1p in regulating anaphase entry and a role in inhibiting the mitotic exit network (MEN) via Dun1p (Sanchez et al., 1999; Hu et al., 2001; Agarwal et al., 2003). Failure to inactivate the MEN results in anaphase and mitotic exit in the presence of a DNA damage signal in both rad53 and dun1Δ cells. Thus, Dun1p also has a role in the preanaphase arrest in response to DNA damage (Pati et al., 1997; Gardner et al., 1999).
Previous studies have shown that Chk1p is not phosphorylated in response to high concentrations of HU in wild-type cells (Sanchez et al., 1999) and that neither Chk1p nor Dun1p is required for cell cycle arrest induced by HU (Zhou and Elledge, 1993; Sanchez et al., 1999). However, loss of both Chk1p and Dun1p causes increased sensitivity to DNA damage (Sanchez et al., 1999) as well as lethality during growth on HU (this study). This synthetic effect between Dun1p and Chk1p indicates that Chk1p may have a redundant role in the S-phase checkpoint pathways, or that Chk1p is somehow activated via the DNA-damage checkpoint pathway in dun1Δ cells. In addition, Pds1p (securin), a target of Chk1p, has been implicated in the response to HU; however, this role of Pds1p did not require Chk1p (Clarke et al., 1999; Clarke et al., 2001). Therefore, we set out to determine whether the sensitivity of chk1Δ dun1Δ cells to HU is due to a defect in (1) cell cycle arrest, (2) DNA repair, or (3) recovery from replication blocks. The results allowed us to establish a relationship between the S-phase and DNA-damage checkpoint pathways in S. cerevisiae.
MATERIALS AND METHODS
Yeast Strains and Plasmids
Strains used in this study are listed in Table 1. Yeast strains were generated using standard genetic techniques. BY4741 and isogenic mrc1Δ, rad9Δ, and sml1Δ strains were obtained from Open Biosystems (Huntsville, AL). pML207.1 (HA-CHK1 in pRS425, a 2 μ LEU2 plasmid) was generously provided by Stephen Elledge. Plasmid pJP1 was generated by cloning a ClaI-KpnI fragment of CHK1 containing the N-terminal HA epitope from pML207.1 into the same sites in plasmid pRS406 (a URA3 integration plasmid; Sikorski and Heiter, 1989) that had been modified to remove the BamHI site. pJP1 was linearized with BamHI and the DNA was transformed into Y300 to generate strain YJP328 expressing endogenous HA-Chk1p.
Table 1.
Yeast strains
| Strain | Genotype | Source |
|---|---|---|
| 512* | As BY4741 sml1Δ :: KanMX | Open Biosystems |
| 3468* | As BY4741 mrc1Δ::KanMX | Open Biosystems |
| 3578* | As BY4741 rad9Δ::KanMX | Open Biosystems |
| BY4741 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 | Open Biosystems |
| Y81 | MATα ade2-1 trp1-1 ura3-1 leu2-3,112 his3-11,15 can1-100 | Zhou et al. (1993) |
| Y300 | MATa ade2-1 trp1-1 ura3-1 leu2-3,112 his 3-11,15 can1-100 | Allen et al. (1994) |
| Y301 | As Y300 rad53-21 | Sanchez et al. (1999) |
| Y438 | As Y300 rad9Δ::HIS3 | Sanchez et al. (1999) |
| Y578 | As Y300 dun1Δ::HIS3 | Desany et al. (1998) |
| Y663 | As Y300 mec1-21 | Sanchez et al. (1999) |
| Y801 | As Y300 chk1Δ::HIS3 | Sanchez et al. (1999) |
| Y808 | As Y300 PDS1::3XHA-PDS1-URA3 | Sanchez et al. (1999) |
| Y857 | As Y300 chk1Δ::URA3 dun1Δ::HIS3 | Sanchez et al. (1999) |
| YJP1 | As Y300 chk1Δ::HIS3 PDS1::3XHA-PDS1-URA3 | This studya |
| YJP2 | As Y300 dun1Δ::HIS3 PDS1::3XHA-PDS1-URA3 | This studya |
| YJP3 | As Y300 sml1Δ::KanMX | This studyb |
| YJP4 | As Y300 chk1Δ::HIS3 sml1Δ::KanMX | This studyb |
| YJP5 | As Y300 dun1Δ::HIS3 sml1Δ::KanMX | This studyb |
| YJP6 | As Y300 chk1Δ::HIS3 dun1Δ::HIS3 sml1Δ::KanMX | This studyb |
| YJP7 | As Y300 chk1Δ::HIS3 dun1Δ::HIS3 PDS1::3XHA-PDS1-URA3 | This studya |
| YJP8 | As Y300 chk1Δ::HIS3 dun1Δ::HIS3 | This study |
| YJP9 | As Y81 chk1Δ::HIS3 dun1Δ::HIS3 | This study |
| YJP321 | As Y300 dun1Δ::HIS3 rad9Δ::HIS3 | This study |
| YJP328 | As Y300 CHK1::HA-CHK1-URA3 | See text |
| YJP341 | As Y300 dun1Δ::HIS3 CHK1::HA-CHK1-URA3 | This studyc |
| YKLS12 | As Y300 chk1Δ::URA3 rad52Δ::URA3 | This study |
| YKLS13 | As Y300 dun1Δ::HIS3 rad52Δ::URA3 | This study |
| YKLS14 | As Y300 chk1Δ::URA3 dun1Δ::HIS3 rad52Δ::URA3 | This study |
| YKLS16 | As Y300 cdc15-2 dun1Δ::HIS3 | This study |
| YKLS17 | As Y300 cdc15-2 chk1Δ::URA3 dun1Δ::HIS3 | This study |
| YKLS18 | As Y300 cdc15-2 rad53-21 | This study |
| YKLS26 | As Y300 rad52Δ::URA3 | This studyd |
| YKLS27 | As Y300 cdc15-2 | This studye |
| YKLS29 | As Y300 dun1Δ::HIS3 mrc1Δ::KanMX CHK1::HA-CHK1-URA3 | This studyf |
Open Biosystems clone ID number.
Generated by crossing YJP9 with Y808.
Generated by crossing 512 with YJP9.
Generated by crossing Y578 with YJP328.
Generated by crossing Y857 with KRY70 (rad52Δ::LEU2), generously provided by Thomas Petes.
Generated by crossing Y81 with K1993 (cdc15-2), generously provided by Stephen Elledge.
Generated by crossing 3468 with YJP341.
Growth Conditions
Cells were grown on ordinary or modified YPD rich medium or on SC-Leu medium (Guthrie and Fink, 1991) as indicated. To examine sensitivity to HU on plates, cells were streaked out or serial dilutions of cells were spotted on YPD containing HU ranging in concentration from 0 to 100 mM, incubated at 30°C for 3 d, and then photographed.
Reversibility Assays
Cells were grown to log phase in YPD medium at 30°C, and HU was added to the medium to a final concentration of 50 or 200 mM for the indicated lengths of time. Cells were then collected, washed with YPD medium, plated onto prewarmed YPD plates, and incubated at 30°C for 3 d. In other experiments, cells in midlog phase in YPD were arrested for 3-5 h in YPD adjusted to pH 3.9 and containing 10 μg/ml α-factor (Protein Chemistry Core Laboratory, Baylor College of Medicine, Houston, TX); cells were then released by washing and resuspending in YPD. At various times after release, 1 ml of culture was treated with 200 mM HU for 1 h. One sample of culture was then fixed in 3.7% formaldehyde and counted with a hemocytometer to determine total cells per milliliter, and a second sample was diluted, plated on YPD, and incubated at 30°C for 2 d. Percent viability was defined as (colony-forming units [CFUs] per ml)/(total cells per ml)/plating efficiency. Plating efficiency was defined as (CFUs per ml)/(total cells per ml) for the respective strain at the time of release from α-factor.
Visualization of Spindles and Nuclei
Cells synchronized with α-factor as above were released into YPD with or without 50 mM HU. A cdc15 mutation was used to block cells in telophase; thus, the temperature was raised to 37°C for 1 h before release from α-factor, and then the cells were released into medium at 37°C. Cells were fixed in 3.7% formaldehyde for 2 h, permeabilized in 70% ethanol overnight, and rehydrated in PBS. Cells were spheroplasted at 37°C for 1 h in 0.1 M potassium phosphate buffer, pH 7.5, containing 1.2 M sorbitol, 1.5 μl/ml β-mercaptoethanol, and 2.25 μg/μl zymolyase (ICN Biomedicals, Costa Mesa, CA). Spheroplasted cells were stained with rat antitubulin antibody (YOL1/34, Harlan, Indianapolis, IN), FITC-conjugated anti-rat-IgG secondary antibody (Sigma), and 0.1 mg/ml 4′,6-diamidino-2-phenylindole dihydrochloride (DAPI, Sigma, St. Louis, MO). Cells were then mounted on slides coated with 0.1% poly-l-lysine for microscopy. Images of spindles and DNA were captured using a Zeiss AxioCam with accompanying imaging software by Zeiss (Thornwood, NY).
Western Analyses
Protein extracts from cells expressing HA-CHK1 and 3XHA-PDS1 were prepared as previously described (Foiani et al., 1994), separated on 10% acrylamide/0.066% bis-acrylamide gels, and transferred to nitrocellulose membranes. HA-Chk1p and HA-Pds1p were detected by Western analysis using anti-HA antibody (16B12, Covance, Madison, WI).
RNA Isolation and Microarray Analysis
RNA was isolated as described by Spellman et al. (1998). For microarray analysis, cDNAs were synthesized from 20 μg of total RNA using an indirect amino allyl labeling method as described by Guo et al. (2004), using an oligo(dT)-primed reverse-transcriptase reaction. Microarray analysis was carried out as described by Guo et al. (2004). Microarray slides of printed S. cerevisiae cDNAs (version 6.4k) were purchased from University Health Network (Toronto, Canada).
Real-time Quantitative PCR
Quantitative PCR (QPCR) was carried out using SYBR Green (Molecular Probes, Eugene, OR) as described by Guo et al. (2004) and a DNA Engine Opticon 2 System (MJ Research, Waltham, MA). A single RNA sample per treatment was used to give three measurements per gene per experimental comparison. Yeast ACT1 provided an internal standard RNA. The following oligonucleotide primers were used: ACT1 (forward) 5′-TGTCACCAACTGGGACGATA-3′ and (reverse) 5′-GGCTTGGATGGAAACGTAGA-3′; RAD5 (forward) 5′-GCACAACGGAACCTATGGAT-3′ and (reverse) 5′-TTCGTTTGTACCAATGCCAA-3′; and RNR3 (forward) 5′-ATTGTTTCCGTTGGAACTGC-3′ and (reverse) 5′-CCGTCTCAGAATTGGATCGT-3′. These primers were tested for specificity by determining that the PCR reactions produced single bands of the predicted sizes using a DNA 1000 DNA chip from Agilent Technologies (Palo Alto, CA) on the Agilent 2100 Bioanalyzer, model G2942AA. In the negative controls, in which no template was added to the ACT1 primers, no PCR products were detected after 40 cycles. These PCR reactions were carried out using an amplification kit from Stratagene (La Jolla, CA;, cat. no. 600548).
Approximately 20 μg of total RNA was used as template for cDNA synthesis as described by Guo et al. (2004). The product of the cDNA synthesis was diluted 1:200, and a 10-μl sample was used as template for the QPCR, with the Opticon Monitor protocol set at 40 cycles. The average cycle threshold (CT) values for the RNR3 and RAD5 genes and the ACT1 reference were obtained using the Opticon Monitor software version 2.02. These values were used in the differential gene expression ratio calculations. One-way ANOVA (analysis of variance) was used for each gene to determine the degree of significance of the differences between samples from treated and untreated cultures. The p-values were corrected using the Bonferroni method.
Pulsed Field Gel Electrophoresis
Cells were synchronized with α-factor as above and released into YPD medium containing either 50 or 200 mM HU plus 10 μg/ml nocodazole. After 60 min, cells were pelleted, washed, and resuspended in YPD medium with 10 μg/ml nocodazole (Desany et al., 1998). In other experiments, synchronized cells were released into YPD medium containing either 50 or 100 mM HU without nocodazole. In these experiments, after 60 min, cells were resuspended in YPD, pH 3.9, containing 10 μg/ml α-factor. For the remainder of the experiment, 10 μg/ml α-factor was added every 90 min. In each experiment, cells were collected from equal volumes of culture (1 ml) at various times and fixed in 70% EtOH overnight at 4°C. DNA-embedded agarose plugs were prepared following a protocol slightly modified from that of Schwartz and Cantor (1984). Pulsed field gel electrophoresis (PFGE) was carried out in a Bio-Rad CHEF Mapper XA apparatus (Richmond, CA) for 18 h in auto-algorithm mode with a switch angle of 120°. Chromosomes were visualized by staining with 0.5 μg/ml EtBr for 30 min and then destaining in dH2O for 15 min. Gels were photographed using a NucleoVision UV Transilluminator and the EtBr signals were quantified using the accompanying GelExpert software by Nucleotech (San Mateo, CA) determining the intensity of each band relative to the background intensity. The intensity of each chromosomal band was divided by the intensity of the band for that chromosome at time zero. There are many factors, such as incomplete cell wall digestion, that affect the reproducibility of signal strength or effective DNA loading within and between experiments of this type; however, the conclusions drawn below are based on multiple repetitions that showed the same trends.
Flow Cytometry
Flow cytometry analysis was carried out as previously described (Sanchez et al., 1999).
RESULTS
Response of chk1Δ dun1Δ Double Mutant Cells to the Replication Inhibitor HU
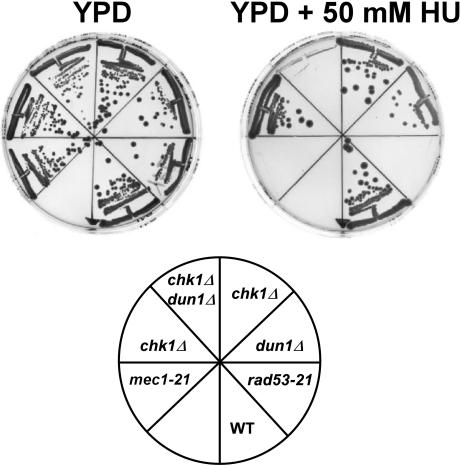
Although neither Chk1p nor Dun1p is required for the S-phase-checkpoint-activated cell-cycle arrest in response to HU (Zhou and Elledge, 1993; Sanchez et al., 1999), we found that the sensitivity of chk1Δ dun1Δ double mutants was comparable to the sensitivity of rad53-21 or mec1-21 cells (Figure 1). In contrast to the single mutants, chk1Δ dun1Δ cells failed to grow on plates containing 50 mM HU.
Figure 1.
chk1Δ and dun1Δ mutations are synthetic lethal when cells are exposed to hydroxyurea (HU). Wild-type (Y300) and mec1-21 (Y663), rad53-21 (Y301), dun1Δ (Y578), chk1Δ (Y801), and chk1Δ dun1Δ (Y857) mutants were streaked onto YPD medium with or without 50 mM HU and grown at 30°C for 3 d.
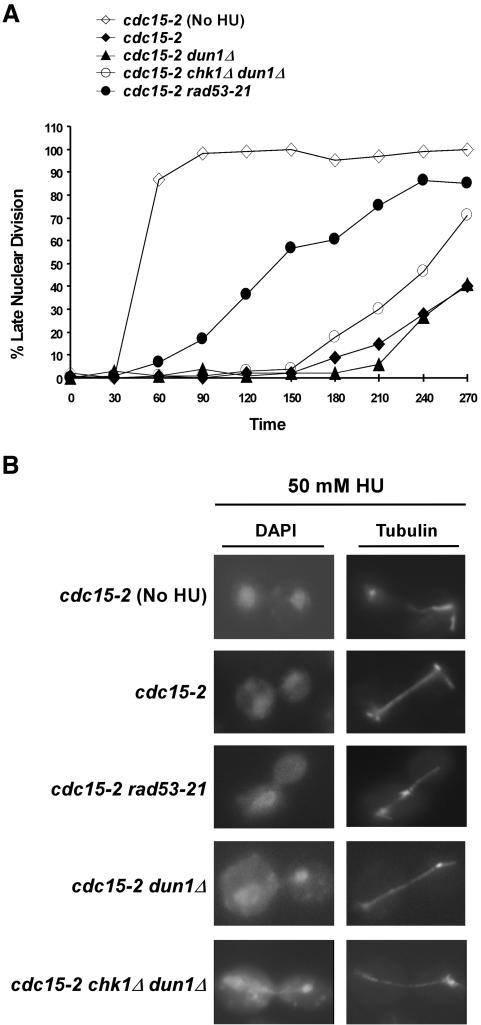
Chk1p is the primary regulator of anaphase in the DNA-damage checkpoint pathway. However, Rad53p, but not Chk1p, is required to prevent anaphase during replication blocks, which activate the S-phase checkpoint (Allen et al., 1994; Weinert et al., 1994). To determine whether chk1Δ dun1Δ cells were proficient in the preanaphase arrest when the S-phase checkpoint is activated, we examined the kinetics of spindle elongation in the double mutant cells when released into the cell cycle in the presence of 50 mM HU. Because wild-type and dun1Δ cells replicate their DNA with different kinetics in the presence of HU (unpublished data), we introduced a cdc15-2 mutation, which causes cells to arrest at a late nuclear division step (Wood and Hartwell, 1982), into the chk1Δ dun1Δ background. Failure to prevent spindle elongation in the presence of HU would thus result in the accumulation of elongated, telophase-like spindles at the restrictive temperature for cdc15-2. The cdc15-2 and dun1Δ cdc15-2 cells showed a preanaphase arrest for the first 3 h of the experiment (Figure 2A), indicating that dun1Δ cells, although defective in the up-regulation of Rnrp, were proficient in the anaphase block in response to 50 mM HU. The cdc15-2 rad53-21 cells began to elongate their spindles (indicative of a checkpoint arrest defect as shown previously; Allen et al., 1994) ∼1 h after release from the G1 block, and by 4 h 85% of the cells exhibited elongated spindles. However, the majority of chk1Δ dun1Δ cdc15-2 cells exhibited preanaphase spindles for 3 h, and only ∼ 45% of the cells exhibited elongated spindles after 4 h in 50 mM HU (Figure 2A). This finding indicates not only that the Rad53p-dependent cell cycle-arrest branch was proficient in the chk1Δ dun1Δ cdc15-2 cells, but also that these cells were unable to maintain a checkpoint arrest under prolonged replication stress. We observed an increase in aberrant mitoses evidenced by elongated spindles with nuclei displaying stretched chromosomal material or uneven distribution of DNA between the mother and daughter in the chk1Δ dun1Δ cells (22% vs. 0% in dun1Δ cells at 210 min), suggesting that these cells were entering anaphase with either incompletely replicated DNA or unresolved chromosomal structures that arose from replication in HU (Figure 2B).
Figure 2.
chk1Δ dun1Δ cells initiate a preanaphase arrest, preventing spindle elongation, in response to replication blocks. (A) Determination of chk1Δ dun1Δ proficiency in the S-phase checkpoint response. cdc15-2 (YKLS27), cdc15-2 dun1Δ (YKLS16), cdc15-2 chk1Δ dun1Δ (YKLS17), and cdc15-2 rad53-21 (YKLS18) strains were synchronized in G1 with α-factor at 22°C and then raised to 37°C (restrictive temperature for cdc15-2) for 1 h. Cells were then released into medium at 37°C with 50 mM HU unless otherwise indicated. The spindles were visualized with antitubulin antibodies, and the DNA was visualized with DAPI. The percentage of cells displaying elongated spindles indicative of late nuclear division at each time point was quantified and plotted. (B) Examples of cells in late nuclear division 240 min after release from the G1 block in the experiment of panel A.
These data suggest that the Chk1p pathway has a role in the response to lesions incurred during replication stress, perhaps collapsed replication structures, which could be different from its role in the DNA-damage checkpoint operating at the metaphase-to-anaphase transition. Therefore, we set out to test this model by analyzing the modification of Chk1p after HU treatment and the requirement for CHK1 in the recovery from replication blocks.
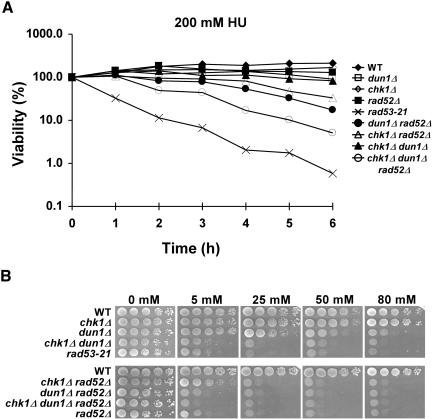
The Sensitivity of chk1Δ dun1Δ Cells to HU is Reversible
Mutant cells, such as rad53, that have defects in preventing irreversible fork collapse (Sogo et al., 2002; Tercero et al., 2003) show sensitivity to HU after even transient exposure to the drug (Allen et al., 1994). However, the HU sensitivity of cells with mutations in genes that play key roles in recombination/repair, such as RAD52, RAD54, RAD55, and RAD57, is reversible (Allen et al., 1994). To test whether the chk1Δ dun1Δ cells were defective in preventing irreversible fork damage after an S-phase block, we examined the ability of chk1Δ, dun1Δ, chk1Δ dun1Δ, rad53-21, and rad52Δ cells to reverse the effects of a replication block induced by transient exposure to 200 mM HU. Although the chk1Δ dun1Δ and rad53-21 cells showed similar sensitivity to HU on plates (Figures 1 and 3B), the sensitivity of the chk1Δ dun1Δ cells, unlike that of rad53 cells, was mostly reversible (Figure 3A). The sensitivity of chk1Δ dun1Δ and rad52Δ cells to transient HU treatment was similar (Figure 3A), and dun1Δ rad52Δ cells had already been shown to be more sensitive to chronic exposure to HU than dun1Δ or rad52Δ single mutant cells (Fasullo et al., 1999). Therefore, we set out to determine whether CHK1 and RAD52 belonged to the same epistasis group by comparing the recovery and growth of dun1Δ rad52Δ, chk1Δ dun1Δ, chk1Δ rad52Δ, and chk1Δ dun1Δ rad52Δ cells after either transient or chronic exposure to HU. Deletion of RAD52 enhanced the sensitivity of dun1Δ, chk1Δ, and chk1Δ dun1Δ cells to transient exposure to HU (Figure 3A) as well as that of dun1Δ and chk1Δ dun1Δ cells to chronic HU exposure (Figure 3B), indicating that the Chk1p and Dun1p kinases operate in different pathways than Rad52p.
Figure 3.
HU sensitivity of the chk1Δ dun1Δ cells is reversible. (A) To test for defects in the repair of lesions encountered during a prolonged S-phase block, mutant strains were tested for their ability to recover from the effects of replication interference induced by HU. Wild-type (Y300), rad53-21 (Y301), rad52Δ (YKLS26), dun1Δ (Y578), chk1Δ (Y801) mutants, and various double and triple mutants (Y857, YKLS12, YKLS13, YKLS14) were grown to log phase at 30°C and then treated with 200 mM HU for 0-6 h. Cells were then plated on YPD medium and incubated at 30°C for 2 d. (B) The same strains were analyzed for their sensitivity to chronic exposure to HU by growing them to log phase in YPD and then spotting serial dilutions onto YPD medium containing the indicated concentrations of HU.
Modification of Chk1p and Pds1p in dun1Δ Cells After Treatment with HU
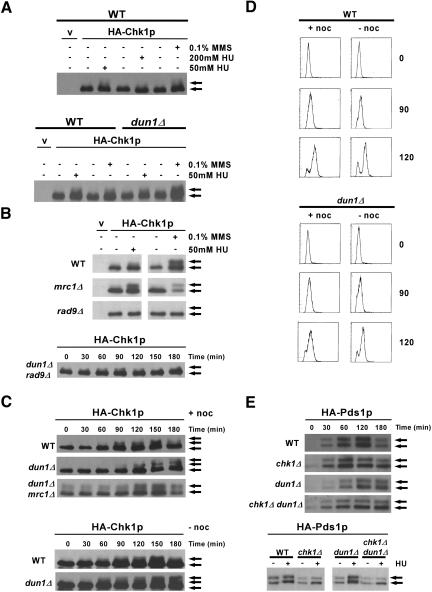
Chk1p has been shown to be phosphorylated in response to DNA damage in a Mec1p-dependent manner; however, Chk1p was not phosphorylated in wild-type cells in response to 200 mM HU (Sanchez et al., 1999). Our model predicts that Chk1p will be modified in dun1Δ cells in the absence of exogenous damage and further modified in the presence of HU. To test this, an epitope-tagged version of Chk1p was used to study Chk1p modification in different genetic backgrounds and in response to different treatments. Treatment of wild-type cells with 200 mM HU did not lead to modification of Chk1p, as shown previously (Figure 4A). We investigated whether Chk1p would be modified when cells were treated with a lower dose of HU that would slow down DNA replication but would allow cells to progress slowly through S phase. Treatment of wild-type and dun1Δ cells with 50 mM HU resulted in the appearance of slower migrating forms of Chk1p, suggesting its activation (Figure 4A). These results are reminiscent of the interaction between the kinases Chk1p and Cds1p in the fission yeast Schizosaccharomyces pombe (Lindsay et al., 1998), in which Chk1p becomes activated in response to replication blocks only when the cells lack the Cds1p kinase.
Figure 4.
Chk1p and Pds1p are modified in response to replication in the presence of 50 mM HU. After the indicated manipulations, proteins were separated by SDS-PAGE, and modification of Chk1p or Pds1p was monitored by Western analysis using anti-HA antibodies. Arrows indicate the unmodified (bottom) and modified (top) species. (A) Exponentially growing wild-type (YJP328) and dun1Δ (YJP341) cells expressing an endogenous HA-Chk1p were either not treated or treated with 50 mM HU, 200 mM HU, or 0.1% methylmethane sulfonate (MMS) for 2 h. The control lane (v) contains proteins from wild-type (Y300) cells transformed with a control vector (pRS425) and not expressing endogenous HA-Chk1p. (B, top) Exponentially growing wild-type (BY4741), mrc1Δ (3468), and rad9Δ (3578) cells containing either control vector pRS425 (v) or a high-copy plasmid encoding HA-Chk1p (pML207.1) were either not treated or treated with 50 mM HU or 0.1% MMS for 2 h. Bottom: dun1Δ rad9Δ cells (YJP321) containing plasmid pML207.1 were synchronized in G1 with α-factor, then released into medium containing 50 mM HU and nocodazole. (C) Wild-type (YJP328), dun1Δ (YJP341), and dun1Δ mrc1Δ (YKLS29) cells expressing endogenous HA-Chk1p were synchronized in G1 with α-factor and released into medium containing 50 mM HU with (top) or without (bottom) 10 μg/ml nocodazole for the indicated times. Cell cycle stage was monitored by DNA content using FACS analyses (D). (E, top) Wild-type (Y808), chk1Δ (YJP1), dun1Δ (YJP2), and chk1Δ dun1Δ (YJP7) cells expressing endogenous HA-Pds1p were synchronized in G1 with α-factor and released into medium containing 50 mM HU for the indicated times. (Bottom) Samples from untreated cultures (-, 30 min postrelease) run side by side with samples from cells released into medium with HU (+, 120 min).
The amplification of Chk1p and Rad53p signal in response to DNA damage in G2 is dependent on the Rad9p protein (Navas et al., 1996; Sanchez et al., 1996, 1999; Sun et al., 1998). The S-phase checkpoint counterpart of Rad9p is Mrc1p, which is associated with replication forks and has been shown to be required for amplification of Rad53p signal in the intra-S-phase checkpoint (Alcasabas et al., 2001; Katou et al., 2003; Osborn and Elledge, 2003). However, Rad9p has been shown to have a role in slowing DNA replication in the presence of DNA damage (Paulovich et al., 1997), and it has also been shown to localize to replication forks in cells lacking Mrc1p (Katou et al., 2003). Cells that lack Mrc1p have reduced Rad53p phosphorylation in response to HU, and in these cells the DNA-damage checkpoint is activated, as evidenced by Rad9p and Chk1p phosphorylation in response to HU (Alcasabas et al., 2001). We also observed that Chk1p was modified in mrc1Δ cells after treatment with 50 mM HU (Figure 4B). The modification of Chk1p under these conditions in both wild-type and dun1Δ cells was dependent on Rad9p (Figure 4B), suggesting that the modification of Chk1p seen in our assays was in response to DNA damage sensed during S phase.
The modification of Chk1p that we observed when cells replicated their DNA in the presence of HU could be due to 1) activation of the DNA-damage checkpoint during S phase, a novel role for Chk1p in S. cerevisiae; 2) activation of the DNA-damage checkpoint at the end of S phase or G2 that regulates the metaphase-to-anaphase transition and that requires Chk1p; or 3) activation of the Chk1p pathways when cells attempt to segregate either incompletely replicated chromosomes or chromosomes with lesions from collapsed forks at anaphase. To differentiate among these possibilities, we examined the modification of Chk1p in wild-type and dun1Δ cells released from a G1 block into media containing 50 mM HU. Activation of the spindle checkpoint with nocodazole was used to block anaphase in order to determine whether the modification of Chk1p required passage through mitosis. The modification of Chk1p began to appear 60-90 min after the release, when the cells were in S phase as determined by fluorescence-activated cell sorting (FACS) analyses, and the modification did not require passage through mitosis, because it was still observed in cells released into a nocodazole block (Figure 4, C and D). These data are consistent with our observation that the HU sensitivity of chk1Δ dun1Δ cells was not suppressed when the cells were plated on medium containing both HU and benomyl, a microtubule-destabilizing drug that also activates the spindle checkpoint (unpublished data). Although the modification of Chk1p occurred in S phase, it did not require Mrc1p (Figure 4C). These data indicate that the Chk1p pathway is activated in S phase, possibly due to collapsed forks, when replication is slowed by HU.
A target of Chk1p in the DNA-damage checkpoint is securin (Pds1p). Therefore, we examined the levels and phosphorylation status of Pds1p in synchronized cells released into the cell cycle under the same conditions in which we had observed Chk1p modification. We observed that Pds1p accumulated in wild-type cells and that Pds1p was transiently phosphorylated between 120 and 180 min after release into HU, as evidenced by slower migration of both bands corresponding to Pds1p (Figure 4E), followed by a decrease in Pds1p levels. Pds1p accumulated later in dun1Δ cells probably due to the fact that these cells replicate their DNA more slowly in HU than do wild-type cells. However, Pds1p was also phosphorylated in dun1Δ cells under these conditions, and its levels remained elevated for the remainder of the experiment compared with wild-type cells. The phosphorylation of Pds1p in this response was dependent on Chk1p (Figure 4E). These data show that the Chk1p pathway is transiently activated in response to lesions that accumulate when S phase is slowed in wild-type cells by 50 mM HU. The modification of Chk1p was concomitant with phosphorylation of Pds1p, indicating that the defect in chk1Δ dun1Δ cells could be due to lack of activation of the Chk1p/Pds1p pathway, which had been shown to function in the DNA-damage checkpoint that regulates the metaphase-to-anaphase transition. These data also suggest that the role of Chk1p in dun1Δ cells might be related to its role in the DNA-damage checkpoint.
The HU Sensitivity of chk1Δ dun1Δ Cells is Not Related to Dun1p Regulation of Ribonucleotide Reductases
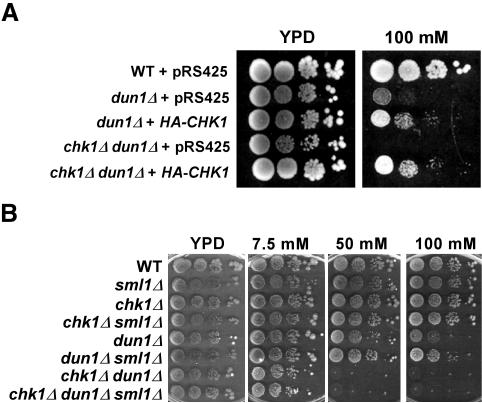
Overexpression of Chk1p in S. cerevisiae does not lead to cell-cycle arrest, unlike what has been observed in S. pombe (Walworth et al., 1993). Therefore, we examined whether overproduction of Chk1p would suppress the sensitivity of dun1Δ cells to higher concentrations of HU. dun1Δ and chk1Δ dun1Δ cells containing either the CHK1 gene on a high-copy plasmid or an empty vector were grown on SC-Leu medium and spotted on YPD with or without HU. The mutant cells expressing high levels of Chk1p grew on 100 mM HU, a concentration at which dun1Δ cells carrying an empty vector grew poorly and chk1Δ dun1Δ failed to grow (Figure 5A). We also observed that overproduction of Rad53p did not suppress the HU-sensitivity of either dun1Δ or chk1Δ dun1Δ cells (unpublished data).
Figure 5.
CHK1 overexpression suppresses the HU-sensitive phenotype of dun1Δ mutants. (A) Wild-type (Y300), dun1Δ (Y578), and chk1Δ dun1Δ (Y857) cells transformed with either a high-copy vector (pRS425) or a high-copy HA-CHK1 plasmid (pML207.1) were grown to log phase in SC-Leu at 30°C. Serial dilutions of each strain were spotted onto YPD medium or onto YPD containing the indicated concentration of HU and grown at 30°C for 3 d. (B) Serial dilutions of wild-type (Y300), sml1Δ (YJP3), chk1Δ (Y801), chk1Δ sml1Δ (YJP4), dun1Δ (Y578), dun1Δ sml1Δ (YJP5), chk1Δ dun1Δ (YJP8), and chk1Δ dun1Δ sml1Δ (YJP6) cells growing in YPD medium were spotted onto YPD containing the indicated concentrations of HU and grown at 30°C for 3 d.
Deletion of the Rnr protein inhibitor Sml1p, a target of Dun1p, has also been shown to suppress the HU sensitivity of dun1Δ cells (Zhao and Rothstein, 2002). Thus, we next investigated whether the requirement for Chk1p in dun1Δ cells was due to the low activity of Rnr proteins in dun1Δ cells (Zhou and Elledge, 1993). Deletion of SML1 allowed growth of dun1Δ cells at HU concentrations up to 80 mM (Figure 5B and unpublished data) and weak growth, at 100 mM HU, as shown previously (Figure 5B; Zhao and Rothstein, 2002). However, deletion of SML1 suppressed the sensitivity of chk1Δ dun1Δ cells only to low (up to 7.5 mM) concentrations of HU but not to the higher concentrations (up to 50 mM) on which chk1Δ and dun1Δ single mutant cells grew (Figure 5B). These data indicate that the HU sensitivity of chk1Δ dun1Δ cells was due to a role of Dun1p distinct from the regulation of ribonucleotide reductases.
Recent studies have shown a synthetic HU-sensitive phenotype between dun1Δ and mutations affecting two components of a poly(A)-nuclease complex, Pan2p and Pan3p. The synthetic sensitivity of dun1Δ pan2Δ and dun1Δ pan3Δ cells to HU was also not suppressed by deletion of SML1 (Hammet et al., 2002). Because dun1Δ pan2Δ and dun1Δ pan3Δ cells had very high levels of RAD5 mRNA, compared with wild-type cells when grown on HU, these studies suggested roles for Dun1p, along with Pan2p/Pan3p, in the negative regulation of RAD5 mRNA stability when cells were grown in HU (Hammet et al., 2002). Rad5p is involved in postreplication repair, which suggested that the role of Dun1p and Pan2p/Pan3p was to regulate the mechanism of repair of the lesions that occur during replication stress. We set out to determine whether the role of Chk1p in dun1Δ cells was to regulate postreplication repair through posttranscriptional regulation of RAD5. For this, we examined the levels of RAD5 mRNA by two approaches, microarray analyses and quantitative PCR, using RNA isolated from wild-type, dun1Δ, chk1Δ, and chk1Δ dun1Δ cells treated or not with 150 mM HU, the concentration of HU that had been used to show the mis-regulation of RAD5 mRNA in cells lacking Dun1p and Pan2p/Pan3p (Hammet et al., 2002). We observed that although the increase of RNR3 mRNA in response to HU was defective in dun1Δ cells, as shown previously (Zhou and Elledge, 1993), the levels of RAD5 mRNA were unchanged in the dun1Δ, chk1Δ, and chk1Δ dun1Δ cells (Table 2). These data indicate that increased levels of RAD5 mRNA cannot explain the synthetic phenotype in the chk1Δ dun1Δ cells.
Table 2.
RAD5 RNA levels are not significantly increased in chk1Δ dun1Δ cells
| Gene
|
||||||||
|---|---|---|---|---|---|---|---|---|
| RAD5
|
RNR3
|
ACT1
|
||||||
| Strain | Fold-changea | p-valueb | Fold-changea | p-valueb | Fold-changea | p-valueb | ||
| Microarraydata | ||||||||
| Wild type | (Y300) | −1.4 | 0.31 | 4.6 | 0.003 | −1.2 | 0.4 | |
| chk1Δ | (Y801) | 1.0 | >0.5 | 4.9 | 0.002 | 1.0 | >0.5 | |
| dun1Δ | (Y578) | −1.2 | >0.5 | 1.3 | >0.5 | −1.3 | 0.1 | |
| chk1Δ dun1Δ | (YJP8) | 1.0 | >0.5 | 1.2 | >0.5 | 1.1 | >0.5 | |
| Quantitative PCR | ||||||||
| Wild type | (Y300) | 2.6 | 0.004 | 58 | <0.0001 | 1.0 | >0.5 | |
| chk1Δ | (Y801) | 2.5 | 0.005 | 78 | <0.0001 | −1.3 | 0.5 | |
| dun1Δ | (Y578) | −1.1 | >0.5 | 2.2 | 0.1 | −1.7 | 0.01 | |
| chk1Δ dun1Δ | (YJP8) | 1.4 | >0.5 | 2.1 | 0.1 | −2.0 | 0.002 | |
The indicated strains were grown to mid-log phase in YPD and treated or not with 150 mM HU for 3 h. RNA was isolated and converted to cDNA, which was subsequently used for microarray and quantitative PCR analyses.
Fold-change indicates the change in transcript abundance of the HU-treated sample relative to the untreated sample. The fold-change for the microarray analyses was calculated as described in Guo et al. (2004). A negative number indicates that the measured value for that particular gene was higher in the untreated than in the treated sample. ANOVA and the statistical program SAS were used to calculate fold-change for the QPCR, which is equal to 2ΔCT in which CT is the cycle threshold (see Materials and Methods and Guo et al., 2004).
Fold changes with a p-value < 0.005 are considered significant.
chk1Δ dun1Δ Cells Are Defective in the Recovery from HU-induced Replication Blocks
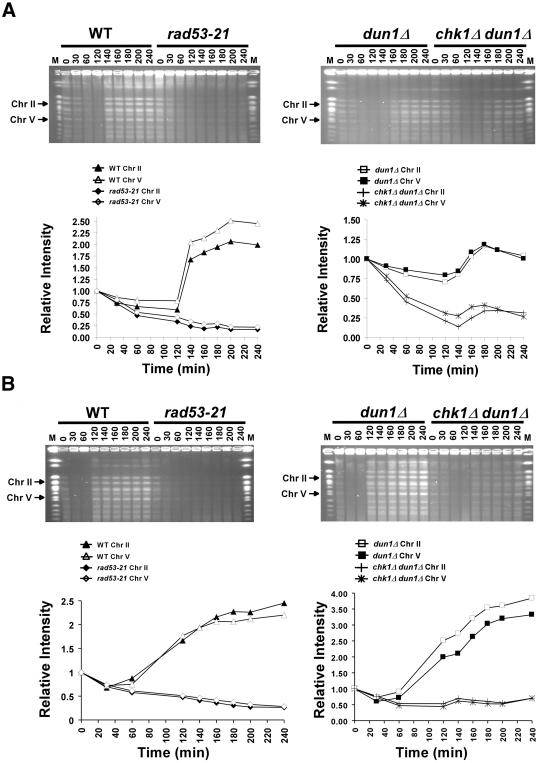
dun1Δ cells have been shown to have a prolonged S phase, which could be due to both lower levels and lower activity of Rnrp proteins (Zhao and Rothstein, 2002). In addition, dun1Δ cells have increased genomic instability, which suggests errors during replication or postreplication repair (Myung et al., 2001b). To determine whether the absence of Chk1p exacerbated the S-phase defect and/or the genomic instability of dun1Δ cells, we examined the ability of chk1Δ dun1Δ cells to complete replication of their chromosomes and repair all aberrant structures after transient HU treatment during early S phase by measuring the ability of intact chromosomes to be resolved by PFGE. In the absence of complete replication and repair, chromosomes contain structures such as replication forks and bubbles that prevent migration of chromosomal DNA into the gel.
Cells synchronized in G1 were released into the cell cycle in medium containing HU. After 60 min, the cells were washed and resuspended in medium containing nocodazole to prevent them from entering mitosis and initiating a new round of DNA replication (Desany et al., 1998). The chromosomes from cells collected during and after HU treatment were then separated by PFGE. As shown previously, wild-type cells recover from transient exposure to 200 mM HU and complete DNA replication, as evidenced by the resolution of the chromosomes by PFGE and a constant signal after 140 min, whereas rad53-21 cells fail to complete DNA replication under these conditions (Desany et al., 1998; Figure 6A). dun1Δ cells completed DNA replication with slightly slower kinetics than wild-type and chk1Δ cells, as evidenced by the decreased signal from the chromosomes resolved in the gel after release from the HU-induced block (Figure 6A and unpublished data). However, deletion of CHK1 in dun1Δ cells significantly delayed the timing of the appearance of completely replicated chromosomes after transient HU treatment. Moreover, the bands representing the chromosomes that appeared after recovery in chk1Δ dun1Δ cells had reduced signal intensity, suggesting either that fewer cells were completing DNA replication or that more cells had accumulated abnormal structures during recovery, such as collapsed forks, which prevented the chromosomes from entering the gel (Figure 6A). Similar results were obtained using a concentration of 50 mM HU, except that the chromosomes recovered somewhat earlier in the wild-type and dun1Δ strains (Figure 6B). Thus, the chk1Δ dun1Δ defect can also be observed at a dose of HU that causes DNA replication only to slow down.
Figure 6.
chk1Δ dun1Δ cells are defective in recovery from replication blocks induced by HU. Wild-type (Y300), dun1Δ (Y578), rad53-21 (Y301), and chk1Δ dun1Δ (Y857) cells were synchronized in G1 with α-factor and then released (at time = 0) into YPD containing 200 mM (A) or 50 mM (B) HU and 10 μg/ml nocodazole. Sixty minutes after release, cells were resuspended in YPD medium with nocodazole. Cells were collected at the indicated times during and after HU treatment, fixed in 70% EtOH, and used to prepare DNA-embedded agarose plugs for PFGE. Chromosomes were visualized by staining the gels with ethidium bromide, and the EtBr signals for chromosomes II and V (arrows) were quantified and graphed. M, molecular weight markers composed of S. cerevisiae chromosomes (New England Biolabs, Beverly, MA). The results shown are representative of five independent repetitions of each experiment. Despite some experiment-to-experiment variability (see Materials and Methods), the greater defect of the chk1Δ dun1Δ cells (relative to dun1Δ cells) was apparent in all trials. Note that the dun1Δ cells in panel A, unlike the wild-type cells, may not have reached a twofold increase in band intensity during the experiment because of this variability, because of residual unresolved DNA structures that prevent some chromosomes from entering the gel after transient treatment with the higher concentration of HU, or both.
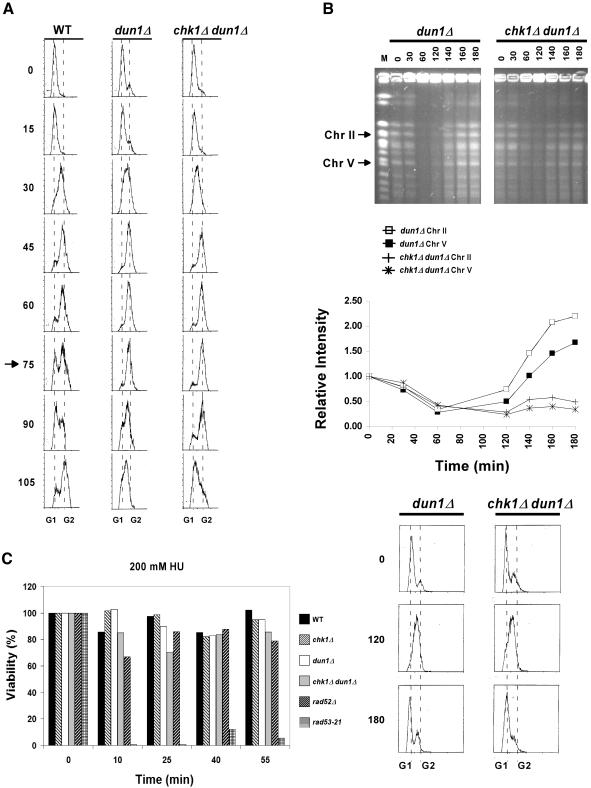
This striking difference between the dun1Δ and chk1Δ dun1Δ cells in the recovery of completely replicated/undamaged chromosomes seems inconsistent with the retention of viability observed after transient exposure to the same concentration of HU (Figure 3). There are three possible explanations for this discrepancy: 1) the length of time that it takes for the chk1Δ dun1Δ cells to recover from HU is longer compared with dun1Δ cells because the double mutant cells have a longer S phase, and the slower recovery is not observed in the reversibility assay; 2) the repair of lesions in chk1Δ dun1Δ cells requires that the cells progress through mitosis; and 3) the slight loss of viability represented a brief window of time (S phase) in which the chk1Δ dun1Δ cells were sensitive to HU, and this brief window was underrepresented in the asynchronous population used in the reversibility assay. To distinguish among these possibilities, we first addressed whether dun1Δ and/or chk1Δ dun1Δ cells had slower kinetics of progression through S phase in the absence of replication blocks by comparing the rates of cell cycle progression of wild-type, dun1Δ, and chk1Δ dun1Δ cells by FACS analyses. We observed that the dun1Δ and chk1Δ dun1Δ cells entered S phase with the same kinetics as wild-type cells but had a longer interval between the end of S phase/G2 and the next G1 (Figure 7A). These data suggested that both the dun1Δ and chk1Δ dun1Δ cells may require more time to produce completely replicated chromosomes that could be resolved by PFGE. We then performed the PFGE experiments in the absence of nocodazole to analyze the chromosomes from cells that were allowed to progress through mitosis. We carried this experiment out to 180 min after release from the transient HU block to ensure that we allowed enough time for cells to complete S phase and enter mitosis as evidenced by FACS analyses. These cells were blocked from reentering the cell cycle by adding back α-factor at 60 min. We did not observe the expected increase in the signal for chk1Δ dun1Δ chromosomes in the later time points, despite the fact that the majority of cells had reached G1 as evidenced by FACS (Figure 7B), indicating that the chromosomes in chk1Δ dun1Δ cells still contain lesions that prevent them from being resolved in the gel in the following G1. These data also suggest that the chk1Δ dun1Δ cells have a defect in the recovery from the replication interference, but that progression through mitosis is not required for the lesions to be repaired. The reversibility assays (Figure 3), however, suggest that at some point most cells are able to deal with these lesions or reverse the effects of transient HU treatment, perhaps in the subsequent cell cycle.
Figure 7.
Reversibility of the HU sensitivity of chk1Δ dun1Δ cells reaches a minimum in early S phase. (A) Similar kinetics of progression through mitosis and G2 delay in dun1Δ and chk1Δ dun1Δ cells. Wild-type (Y300), dun1Δ (Y578), and chk1Δ dun1Δ (YJP8) cells were synchronized in G1 with α-factor and released into YPD medium. Aliquots of cells were removed at the indicated times, fixed with ethanol, and stained with propidium iodide. The DNA contents were then determined by FACS and plotted to determine cell cycle distributions. Dotted lines, positions of G1 and G2 peaks; arrow, time at which the wild-type cells had reentered a new cell cycle (as judged by the renewed population of G1 cells) but the dun1Δ and chk1Δ dun1Δ cells had not. (B) Failure of chk1Δ dun1Δ cells to resolve chromosome structures during recovery from transient HU treatment in S phase. dun1Δ (Y578) and chk1Δ dun1Δ (Y857) cells were treated as in Figure 6A except that they were released into medium without nocodazole. After 1 h in HU, cells were resuspended in YPD containing α-factor for the remainder of the experiment. PFGE and FACS were then performed as in Figure 6 and panel A. (C) Wild-type (Y300), chk1Δ (Y801), dun1Δ (Y578), chk1Δ dun1Δ (YJP8), rad52Δ (YKLS26), and rad53-21 (Y301) cells were arrested in G1 by α-factor and released into YPD medium. At the indicated times after release, HU was added to each culture to a concentration of 200 mM. After 1 h in HU, cells were washed and plated on YPD medium. For each sample, the ratio of colony-forming units per ml to total cells per ml was determined and normalized to the plating efficiency for each strain to obtain the percent viability.
chk1Δ dun1Δ Cells Exhibit Increased Sensitivity to HU in Early S phase
Finally, we investigated the third possible explanation, that the HU sensitivity of the chk1Δ dun1Δ mutants was cell cycle specific, by examination of the recovery of cells from exposure to 200 mM HU for 1 h at different times after release from a G1 arrest, which would more closely resemble the treatment conditions used for the PFGE analyses. The cells were treated with HU at different times up to 50 min after release from G1. The FACS analyses had shown that by 60 min, cells from all strains had reached the end of S phase (Figure 7A). This treatment of cells with HU after 50 min would result in cells blocking replication in the next cell cycle and would be the equivalent of treatment immediately after release. Using this approach, we observed that the reversibility of chk1Δ dun1Δ cells from treatment with 200 mM of HU was reduced during S phase (25 min: Figure 7C), indicating that when treated at this time, many cells were unable either to recover from the HU-induced replication block or repair DNA damage caused by replication stress. These findings suggest that most of the irreversible damage causing lethality in the asynchronous population assay (Figure 3) was occurring in the cells exposed to HU in early S phase. These data agree with our findings using PFGE analyses that showed an inability of these cells to recover all signal from their replicated chromosomes after transient exposure to HU. However, the modest loss of viability even when the cells are treated during S phase suggests that the majority of chk1Δ dun1Δ cells are able to cope with the aberrant structures from the transient HU treatment in the following cell cycles.
DISCUSSION
Checkpoint pathways are activated after damage to DNA or when there are problems with DNA replication. Such information is transmitted via protein-protein interactions and protein modifications leading to alteration of gene expression and other events that regulate cell division or cell death. A key component in this system is the evolutionarily conserved protein kinase Chk1p (Chek1). When DNA damage occurs as a result of endogenous or environmental factors, Chk1p regulates mitotic progression in both yeast and mammals. In vertebrates, Chk1p is also required for the S-phase checkpoint, which monitors DNA replication, coordinates completion of S phase with mitosis, and allows the recovery of stalled replication forks (Guo et al., 2000; Feijoo et al., 2001; Zachos et al., 2003).
Dun1p has a role in the arrest in mitosis in response to DNA damage (Pati et al., 1997; Gardner et al., 1999; Sanchez et al., 1999; Hu et al., 2001); however, dun1 mutants are not as sensitive to HU as rad53 mutants, which are defective in blocking mitosis in response to both DNA-damage and S-phase-checkpoint signals. We have shown that Chk1p is the primary regulator of anaphase in response to DNA damage (Sanchez et al., 1999); however, Rad53p, not Chk1p, is required to prevent anaphase during replication blocks (Allen et al., 1994; Weinert et al., 1994). Although S. cerevisiae Chk1p is not required for the S-phase checkpoint, we show here that this protein is modified when cells replicate their DNA in the presence of low concentrations of HU and that Dun1p or Chk1p is required for cells to recover from replication blocks.
Relationship Between the S-phase and DNA-damage Checkpoint Pathways in S. cerevisiae
The work described here demonstrates that Chk1p has a role in the DNA-damage checkpoint that is activated after replication blocks, which is reminiscent of the known interaction between the kinases Chk1p and Cds1p in the fission yeast S. pombe (Boddy et al., 1998; Lindsay et al., 1998). In this organism, Chk1p and Cds1p have overlapping roles in the regulation of mitosis in response to different checkpoint signals via phosphorylation of the phosphatase Cdc25 (Furnari et al., 1997; Boddy et al., 1998; Lindsay et al., 1998; Zeng et al., 1998). Deletion of cds1, which encodes a kinase that mediates recovery from replication blocks, leads to activation of Chk1p in the presence of HU (Lindsay et al., 1998). A simple explanation for our results would be that Dun1p and Chk1p have overlapping roles in controlling mitosis after DNA damage or replication blocks in S. cerevisiae. However, unlike Cds1p and Chk1p in S. pombe, which are required for the response to replication blocks and DNA damage, respectively, Chk1p and Dun1p are both required for the cell-cycle arrest in response to DNA damage, but not for the block to anaphase in response to replication blocks. Even in the absence of both kinases, elongation of the mitotic spindles occurred only after extended exposure to HU (4 h), unlike the checkpoint defective rad53-21 cells, which elongated their spindles 1 h after exposure to HU. These results indicate that the chk1Δ dun1Δ cells were able to initiate a block to anaphase in response to HU, but were unable to maintain the arrest after several hours. These data also indicate that the HU sensitivity of chk1Δ dun1Δ cells was not due to a defect in the Rad53p-mediated preanaphase block.
There are several possible explanations for the synthetic lethality of chk1Δ and dun1Δ in HU. First, it is possible that a defect in the transcriptional response in dun1 mutants leads to the accumulation of DNA damage at stalled or collapsed replication forks. In this scenario, the cells would accumulate DNA damage in S phase, leading to activation of the Chk1p-dependent DNA-damage checkpoint mediated by Rad9 in late S/G2, and possibly the Rad53p/Dun1p checkpoint (Sanchez et al., 1999). If this model were correct, then Chk1p would be activated at the end of S phase or in G2. However, we showed that Chk1p is modified in mid-S phase when cells replicate their DNA in the presence of low concentrations of HU.
Second, Chk1p might have a redundant role with Dun1p in responding to either damage during S phase or to aberrant structures generated at replication forks. The modification of Chk1p required Rad9p, a protein that serves as a signal amplifier for the DNA-damage checkpoint that operates in late S/G2 (Navas et al., 1996; Sanchez et al., 1996, 1999; Sun et al., 1998). Rad9p has also been shown to have a role in the intra-S-phase checkpoint when cells replicate their DNA in the presence of alkylating agents (Paulovich et al., 1997). The modification of Chk1p did not require the S-phase checkpoint counterpart of Rad9p, Mrc1p, a protein associated with replication forks (Katou et al., 2003; Osborn and Elledge, 2003). In addition, Rad9p has been shown to localize to replication forks in cells lacking Mrc1p (Katou et al., 2003), supporting a role for Rad9p in responding to either damage during S phase or to aberrant structures generated at replication forks (Katou et al., 2003). It is possible, then, that the modification of Chk1p is through this role of Rad9p.
The modification of Chk1p was concomitant with the phosphorylation of the securin Pds1p, indicating that the defect in dun1Δ cells led to the activation of the Chk1p/Pds1p pathway, which has been shown to function in the DNA-damage checkpoint that regulates the metaphase-to-anaphase transition (Gardner et al., 1999; Sanchez et al., 1999). Although a role for Pds1p in late S phase had been proposed earlier, that role appeared to be independent of Chk1p (Clarke et al., 1999, 2001).
The Requirement for Chk1p Is Independent of the Defect in RNR Regulation in dun1Δ Cells
Dun1p has two roles in the regulation of deoxyribonucleotide pools, through the transcriptional up-regulation of the RNR genes (Zhou and Elledge, 1993; Huang et al., 1998), and by increasing the activity of Rnr proteins through targeting of their inhibitor Sml1p (Zhao and Rothstein, 2002). In addition, Dun1p has a role in chromosome stability, as dun1Δ mutants accumulate gross chromosomal rearrangements (GCR) during an unperturbed cell cycle (Fasullo et al., 1999; Myung et al., 2001b). dun1Δ cells showed a 208-fold induction of chromosomal aberrations, similar to that observed in mec1 mutants and greater than that observed for either chk1 or rad53 mutants (Myung et al., 2001b), suggesting that Dun1p contributes to chromosome stability in both Rad53p-dependent and Rad53p-independent mechanisms. The HU sensitivity (Zhao and Rothstein, 2002), but not the GCR defects of dun1Δ cells, was suppressed by deletion of SML1 (Myung et al., 2001b). We have shown here that deletion of SML1 caused suppression of the sensitivity of chk1Δ dun1Δ cells to low concentrations of HU, but did not restore growth at the higher concentrations at which chk1Δ and dun1Δ single mutant cells grew. These data indicated that the GCR defects of dun1Δ cells and the HU sensitivity of chk1Δ dun1Δ cells are due to a role of Dun1p distinct from the regulation of ribonucleotide reductases.
Because Dun1p also has a role in the regulation of transcription, the possibility remains that the defect underlying the synthetic phenotype is in the transcriptional response to HU. Two groups have shown that new protein synthesis is not required for cells to recover from HU (Desany et al., 1998; Tercero et al., 2003), although new protein synthesis was required to increase replication rates when cells were recovering from blocks (Tercero et al., 2003). These findings suggest that the possible role for transcription in the recovery of chk1Δ dun1Δ cells could be to increase the levels of factors required to complete DNA replication or carry out DNA repair.
It is also possible that Chk1p could have a role in the recombination-mediated repair of the lesions caused by HU in the dun1Δ mutants. Because chk1Δ cells are not sensitive to HU, this would be a role of Chk1p in S phase that is redundant with the role of Dun1p and may be different from its role in the DNA-damage checkpoint at M phase, and would only be uncovered during prolonged replicational stress. Should Chk1p mediate the repair of lesions incurred during replication interference, it would be through a pathway in addition to, or different from, that regulated by Rad52p, because we observed that a rad52 mutation enhanced the HU sensitivity of chk1Δ dun1Δ cells.
Role of Chk1p Could Be Due to the Requirement for Cohesion in the Repair of Lesions from DNA Replication in HU
chk1Δ dun1Δ cells were delayed in completing replication and/or in the repair of structures resulting from blocked replication forks when recovering from transient exposure to HU, as evidenced by the inability of their chromosomes to be resolved by PFGE, which suggested that the chromosomes contained abnormal structures, such as collapsed forks, that prevented them from entering the gel. Consistent with this possibility, we observed that the HU sensitivity of chk1Δ dun1Δ cells was minimally reversible during S phase.
Securin is an inhibitor of the caspase-like enzyme separase, which cleaves cohesins to allow chromosome separation at anaphase (Ciosk et al., 1998). Cohesins assemble on DNA during replication and have been shown to be required for proper chromosome segregation and postreplication repair in both yeast and vertebrate cells (Sjogren and Nasmyth, 2001; Sonoda et al., 2001). We showed here that the securin Pds1p, a previously known target of Chk1p in the DNA-damage checkpoint (Sanchez et al., 1999), was transiently phosphorylated in wild-type cells after prolonged replication in low concentrations of HU and that the phosphorylation of Pds1p in HU-treated cells was dependent on Chk1p. In cells lacking Dun1p, anaphase was delayed, probably because DNA replication was lengthened because of defects in the response to HU, which correlated with prolonged phosphorylation and stabilization of Pds1p. Our data indicate that at least in some genetic backgrounds, the Chk1p/Pds1p pathway is required for the recovery from stalled replication, and this could be related to the role of securin in blocking separase activation.
Chk1p is essential for survival of dun1Δ cells on low concentrations of HU and loss of Chk1p in dun1Δ cells resulted in the appearance of aberrant chromosomes after HU treatment. These observations, together with the fact that the chk1Δ dun1Δ cells displayed an initial preanaphase arrest in HU, support our hypothesis that the Chk1p pathway has a specific role in allowing or regulating the repair of lesions incurred during replication stress, perhaps collapsed replication structures. The observation that chk1Δ dun1Δ cells retained viability after transient exposure to HU indicated that Chk1p is not essential for preventing irreversible fork collapse. The role for yeast Chk1p in dun1Δ cells could represent a role conserved in the metazoan Chk1p orthologue. Studies addressing the roles of metazoan Chk1p in the S-phase checkpoint in vivo have been difficult, due to the fact that Chk1p is essential in metazoans. Our findings provide a new assay in a model system amenable to genetic manipulation, which can be utilized to study the roles of Chk1p kinases in both the S-phase-checkpoint pathways and the recovery from stalled or collapsed replication forks.
Acknowledgments
We thank members of the Sanchez lab for reading this manuscript and for helpful discussions. We thank Steve Elledge for plasmids and strains, Anil Menon for helpful discussions and use of the CHEF Mapper XA apparatus, Alvaro Puga for use of the thermocycler for Q-PCR, and Jitesh Kawedia and Beatriz Russell for help with the quantification of PFGE signals. We thank Mario Medvedovic and Maureen Sartor for expert analyses of the microarray and Q-PCR data and George Babcock and Sandy Schwemberger (Shriner's Hospital for Children) for expert flow cytometric analyses. The authors also thank the editor for valuable comments. This work was supported in part by grants from the Ohio Cancer Research Associates, American Cancer Society, Ohio Chapter, The Department of Defense DAMD 17-01-1-020, and from the Pew Scholars Program in the Biomedical Sciences to Y.S., and by Grants U01 ES11038 and P30 ES06096 from the National Institute of Environmental Health Sciences, National Institutes of Health.
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E03-11-0792. Article and publication date are available at www.molbiolcell.org/cgi/doi/10.1091/mbc.E03-11-0792.
References
- Agarwal, R., Tang, Z., Yu, H., and Cohen-Fix, O. (2003). Two distinct pathways for inhibiting Pds1 ubiquitination in response to DNA damage. J. Biol. Chem. 278, 45027-45033. [DOI] [PubMed] [Google Scholar]
- Alcasabas, A.A. et al. (2001). Mrc1 transduces signals of DNA replication stress to activate Rad53. Nat. Cell Biol. 3, 958-965. [DOI] [PubMed] [Google Scholar]
- Allen, J.B., Zhou, Z., Siede, W., Friedberg, E.C., and Elledge, S.J. (1994). The SAD1/RAD53 protein kinase controls multiple checkpoints and DNA damage-induced transcription in yeast. Genes Dev. 8, 2401-2415. [DOI] [PubMed] [Google Scholar]
- Boddy, M.N., Furnari, B., Mondesert, O., and Russell, P. (1998). Replication checkpoint enforced by kinases Cds1 and Chk1. Science 280, 909-912. [DOI] [PubMed] [Google Scholar]
- Ciosk, R., Zachariae, W., Michaelis, C., Shevchenko, A., Mann, M., and Nasmyth, K. (1998). An ESP1/PDS1 complex regulates loss of sister chromatid cohesion at the metaphase to anaphase transition in yeast. Cell 93, 1067-1076. [DOI] [PubMed] [Google Scholar]
- Clarke, D.J., Segal, M., Jensen, S., and Reed, S.I. (2001). Mec1p regulates Pds1p levels in S phase: complex coordination of DNA replication and mitosis. Nat. Cell Biol. 3, 619-627. [DOI] [PubMed] [Google Scholar]
- Clarke, D.J., Segal, M., Mondesert, G., and Reed, S.I. (1999). The Pds1 anaphase inhibitor and Mec1 kinase define distinct checkpoints coupling S phase with mitosis in budding yeast. Curr. Biol. 9, 365-368. [DOI] [PubMed] [Google Scholar]
- de la Torre Ruiz, M.-A., and Lowndes, N.F. (2000). DUN1 defines one branch downstream of RAD53 for transcription and DNA damage repair in Saccharomyces cerevisiae. FEBS Lett. 485, 205-206. [DOI] [PubMed] [Google Scholar]
- Desany, B.A., Alcasabas, A.A., Bachant, J.B., and Elledge, S.J. (1998). Recovery from DNA replicational stress is the essential function of the S-phase checkpoint pathway. Genes Dev. 12, 2956-2970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fasullo, M., Koudelik, J., AhChing, P., Giallanza, P., and Cera, C. (1999). Radiosensitive and mitotic recombination phenotypes of the Saccharomyces cerevisiae dun1 mutant defective in DNA damage-inducible gene expression. Genetics 152, 909-919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fay, D.S., Sun, Z., and Stern, D.F. (1997). Mutations in SPK1/RAD53 that specifically abolish checkpoint but not growth-related functions. Curr. Genet. 31, 97-105. [DOI] [PubMed] [Google Scholar]
- Feijoo, C., Hall-Jackson, C., Wu, R., Jenkins, D., Leitch, J., Gilbert, D.M., and Smythe, C. (2001). Activation of mammalian Chk1 during DNA replication arrest: a role for Chk1 in the intra-S phase checkpoint monitoring replication origin firing. J. Cell Biol. 154, 913-923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foiani, M., Marini, F., Gamba, D., Lucchini, G., and Plevani, P. (1994). The B subunit of the DNA polymerase α-primase complex in Saccharomyces cerevisiae executes an essential function at the initial stage of DNA replication. Mol. Cell. Biol. 14, 923-933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furnari, B., Rhind, N., and Russell, P. (1997). Cdc25 mitotic inducer targeted by Chk1 DNA damage checkpoint kinase. Science 277, 1495-1497. [DOI] [PubMed] [Google Scholar]
- Gardner, R., Putnam, C.W., and Weinert, T. (1999). RAD53, DUN1 and PDS1 define two parallel G2/M checkpoint pathways in budding yeast. EMBO J. 18, 3173-3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo, J., Sartor, M., Karyala, S., Medvedovic, M., Kann, S., Puga, A., Ryan, P., and Tomlinson, C.R. (2004). Expression of genes in the TGF-β signaling pathway is significantly deregulated in smooth muscle cells from aorta of aryl hydrocarbon receptor knockout mice. Toxicol. Appl. Pharmacol. 194, 79-89. [DOI] [PubMed] [Google Scholar]
- Guo, Z., Kumagai, A., Wang, S.X., and Dunphy, W.G. (2000). Requirement for Atr in phosphorylation of Chk1 and cell cycle regulation in response to DNA replication blocks and UV-damaged DNA in Xenopus egg extracts. Genes Dev. 14, 2745-2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guthrie, C., and Fink, G.R. (eds.) (1991). Guide to yeast genetics and molecular biology. In: Methods in Enzymology, vol. 194, ed. C. Guthrie and G.R. Fink, San Diego: Academic Press, 3-21.2005794 [Google Scholar]
- Hammet, A., Pike, B.L., and Heierhorst, J. (2002). Posttranscriptional regulation of the RAD5 DNA repair gene by the Dun1 kinase and the Pan2-Pan3 poly(A)-nuclease complex contributes to survival of replication blocks. J. Biol. Chem. 277, 22469-22474. [DOI] [PubMed] [Google Scholar]
- Hu, F., Wang, Y., Liu, D., Li, Y., Qin, J., and Elledge, S.J. (2001). Regulation of the Bub2/Bfa1 GAP complex by Cdc5 and cell cycle checkpoints. Cell 107, 655-665. [DOI] [PubMed] [Google Scholar]
- Huang, M., Zhou, Z., and Elledge, S.J. (1998). The DNA replication and damage checkpoint pathways induce transcription by inhibition of the Crt1 repressor. Cell 94, 595-605. [DOI] [PubMed] [Google Scholar]
- Katou, Y., Kanoh, Y., Bando, M., Noguchi, H., Tanaka, H., Ashikari, T., Sugimoto, K., and Shirahige, K. (2003). S-phase checkpoint proteins Tof1 and Mrc1 form a stable replication-pausing complex. Nature 424, 1078-1083. [DOI] [PubMed] [Google Scholar]
- Kiser, G.L., and Weinert, T.A. (1996). Distinct roles of yeast MEC and RAD checkpoint genes in transcriptional induction after DNA damage and implications for function. Mol. Biol. Cell 7, 703-718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsay, H.D., Griffiths, D.J., Edwards, R.J., Christensen, P.U., Murray, J.M., Osman, F., Walworth, N., and Carr, A.M. (1998). S-phase-specific activation of Cds1 kinase defines a subpathway of the checkpoint response in Schizosaccharomyces pombe. Genes Dev. 12, 382-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopes, M., Cotta-Ramusino, C., Pellicioli, A., Liberi, G., Plevani, P., Muzi-Falconi, M., Newlon, C.S., and Foiani, M. (2001). The DNA replication checkpoint response stabilizes stalled replication forks. Nature 412, 557-561. [DOI] [PubMed] [Google Scholar]
- Myung, K., Chen, C., and Kolodner, R.D. (2001a). Multiple pathways cooperate in the suppression of genome instability in Saccharomyces cerevisiae. Nature 411, 1073-1076. [DOI] [PubMed] [Google Scholar]
- Myung, K., Datta, A., and Kolodner, R.D. (2001b). Suppression of spontaneous chromosomal rearrangements by S phase checkpoint functions in Saccharomyces cerevisiae. Cell 104, 397-408. [DOI] [PubMed] [Google Scholar]
- Navas, T.A., Sanchez, Y., and Elledge, S.J. (1996). RAD9 and DNA polymerase ε form parallel sensory branches for transducing the DNA damage checkpoint signal in Saccharomyces cerevisiae. Genes Dev. 10, 2632-2643. [DOI] [PubMed] [Google Scholar]
- Nyberg, K.A., Michelson, R.J., Putnam, C.W., and Weinert, T.A. (2002). Toward maintaining the genome: DNA damage and replication checkpoints. Annu. Rev. Genet. 36, 617-656. [DOI] [PubMed] [Google Scholar]
- Osborn, A.J., and Elledge, S.J. (2003). Mrc1 is a replication fork component whose phosphorylation in response to DNA replication stress activates Rad53. Genes Dev. 17, 1755-1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pati, D., Keller, C., Groudine, M., and Plon, S.E. (1997). Reconstitution of a MEC1-independent checkpoint in yeast by expression of a novel human fork head cDNA. Mol. Cell. Biol. 17, 3037-3046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulovich, A.G., Margulies, R.U., Garvik, B.M., and Hartwell, L.H. (1997). RAD9, RAD17, and RAD24 are required for S phase regulation in Saccharomyces cerevisiae in response to DNA damage. Genetics 145, 45-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez, Y., Bachant, J., Wang, H., Hu, F., Liu, D., Tetzlaff, M., and Elledge, S.J. (1999). Control of the DNA damage checkpoint by Chk1 and Rad53 protein kinases through distinct mechanisms. Science 286, 1166-1171. [DOI] [PubMed] [Google Scholar]
- Sanchez, Y., Desany, B.A., Jones, W.J., Liu, Q., Wang, B., and Elledge, S.J. (1996). Regulation of RAD53 by the ATM-like kinases MEC1 and TEL1 in yeast cell cycle checkpoint pathways. Science 271, 357-360. [DOI] [PubMed] [Google Scholar]
- Santocanale, C., and Diffley, J.F.X. (1998). A Mec1- and Rad53-dependent checkpoint controls late-firing origins of DNA replication. Nature 395, 615-618. [DOI] [PubMed] [Google Scholar]
- Schwartz, D.C., and Cantor, C.R. (1984). Separation of yeast chromosome-sized DNAs by pulsed field gradient gel electrophoresis. Cell 37, 67-75. [DOI] [PubMed] [Google Scholar]
- Sikorski, R.S., and Heiter, P. (1989). A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122, 19-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sjogren, C., and Nasmyth, K. (2001). Sister chromatid cohesion is required for postreplicative double-strand break repair in Saccharomyces cerevisiae. Curr. Biol. 11, 991-995. [DOI] [PubMed] [Google Scholar]
- Sogo, J.M., Lopes, M., and Foiani, M. (2002). Fork reversal and ssDNA accumulation at stalled replication forks owing to checkpoint defects. Science 297, 599-602. [DOI] [PubMed] [Google Scholar]
- Sonoda, E. et al. (2001). Scc1/Rad21/Mcd1 is required for sister chromatid cohesion and kinetochore function in vertebrate cells. Dev. Cell 1, 759-770. [DOI] [PubMed] [Google Scholar]
- Spellman, P.T., Sherlock, G., Zhang, M.Q., Iyer, V.R., Anders, K., Eisen, M.B., Brown, P.O., Botstein, D., and Futcher, B. (1998). Comprehensive identification of cell cycle-regulated genes of the yeast Saccharomyces cerevisiae by microarray hybridization. Mol. Biol. Cell 9, 3273-3297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, Z., Fay, D.S., Marini, F., Foiani, M., and Stern, D.F. (1996). Spk1/Rad53 is regulated by Mec1-dependent protein phosphorylation in DNA replication and damage checkpoint pathways. Genes Dev. 10, 395-406. [DOI] [PubMed] [Google Scholar]
- Sun, Z., Hsiao, J., Fay, D.S., and Stern, D.F. (1998). Rad53 FHA domain associated with phosphorylated Rad9 in the DNA damage checkpoint. Science 281, 272-274. [DOI] [PubMed] [Google Scholar]
- Tercero, J.A., Longhese, M.P., and Diffley, J.F.X. (2003). A central role for DNA replication forks in checkpoint activation and response. Mol. Cell 11, 1323-1336. [DOI] [PubMed] [Google Scholar]
- Walworth, N., Davey, S., and Beach, D. (1993). Fission yeast chk1 protein kinase links the rad checkpoint pathway to cdc2. Nature 363, 368-371. [DOI] [PubMed] [Google Scholar]
- Weinert, T.A., Kiser, G.L., and Hartwell, L.H. (1994). Mitotic checkpoint genes in budding yeast and the dependence of mitosis on DNA replication and repair. Genes Dev. 8, 652-665. [DOI] [PubMed] [Google Scholar]
- Wood, J.S., and Hartwell, L.H. (1982). A dependent pathway of gene functions leading to chromosome segregation in Saccharomyces cerevisiae. J. Cell Biol. 94, 718-726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zachos, G., Rainey, M.D., and Gillespie, D.A. (2003). Chk1-deficient tumour cells are viable but exhibit multiple checkpoint and survival defects. EMBO J. 22, 713-723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng, Y., Forbes, K.C., Wu, Z., Moreno, S., Piwnica-Worms, H., and Enoch, T. (1998). Replication checkpoint requires phosphorylation of the phosphatase Cdc25 by Cds1 or Chk1. Nature 395, 507-510. [DOI] [PubMed] [Google Scholar]
- Zhao, X., and Rothstein, R. (2002). The Dun1 checkpoint kinase phosphorylates and regulates the ribonucleotide reductase inhibitor Sml1. Proc. Natl. Acad. Sci. USA 99, 3746-3751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, Z., and Elledge, S.J. (1993). DUN1 encodes a protein kinase that controls the DNA damage response in yeast. Cell 75, 1119-1127. [DOI] [PubMed] [Google Scholar]