Abstract
We studied the physiological response to limitation by diverse nutrients in batch and steady-state (chemostat) cultures of S. cerevisiae. We found that the global pattern of transcription in steady-state cultures in limiting phosphate or sulfate is essentially identical to that of batch cultures growing in the same medium just before the limiting nutrient is completely exhausted. The massive stress response and complete arrest of the cell cycle that occurs when nutrients are fully exhausted in batch cultures is not observed in the chemostat, indicating that the cells in the chemostat are “poor, not starving.” Similar comparisons using leucine or uracil auxotrophs limited on leucine or uracil again showed patterns of gene expression in steady-state closely resembling those of corresponding batch cultures just before they exhaust the nutrient. Although there is also a strong stress response in the auxotrophic batch cultures, cell cycle arrest, if it occurs at all, is much less uniform. Many of the differences among the patterns of gene expression between the four nutrient limitations are interpretable in light of known involvement of the genes in stress responses or in the regulation or execution of particular metabolic pathways appropriate to the limiting nutrient. We conclude that cells adjust their growth rate to nutrient availability and maintain homeostasis in the same way in batch and steady state conditions; cells in steady-state cultures are in a physiological condition normally encountered in batch cultures.
INTRODUCTION
The physiological adaptation of cells to changing environments remains one of the most mysterious and subtle behaviors of living organisms. The maintenance of internal state in the face of environmental change, called homeostasis, can be achieved by internalization of transporters (Volland et al., 1994), by direct allosteric regulation of proteins (Reichard, 2002), and by the transcriptional regulation of large sets of genes (Ogawa et al., 2000). The availability of complete genome sequences and technology such as DNA microarrays for gene expression studies offers the opportunity to study, in a comprehensive way, the coordination of the entire “system” of metabolic functions that is implicit in a cell's remarkable ability to maintain homeostasis in the face of a rapidly changing environment.
Conclusions drawn from traditional batch culture experiments are potentially confounded because the environment in batch is continually changing in ways that may not be entirely measurable or understood. The typical batch growth cycle involves multiple phases, which may each have a different gene expression profile and a different physiological response to perturbation. Most notably, at the end of batch growth a large set of genes, collectively known as the yeast stress response, is activated (Gasch and Werner-Washburne, 2002). When patterns of gene expression are followed by using DNA microarrays, many thousands of genes change their expression, thereby revealing unsuspected subtlety in secondary effects missed by more targeted assays. In some cases, these effects can be controlled for, but in others they are intrinsic to the treatment.
The chemostat (Monod, 1950; Novick and Szilard, 1950) offers a robust way to circumvent certain limitations of batch culture (Hayes et al., 2002) by growing cultures in a true steady state. In most realizations, the chemostat consists of a fermenter vessel with a nutrient feed and an effluent overflow. Fresh medium is added from the feed at a constant rate, causing effluent to leave the vessel at the same rate. In early studies that first defined precisely the phenomenon of metabolic homeostasis in microorganisms (Monod, 1950), it was observed that the culture achieves a steady state by somehow matching its growth rate exactly to the dilution rate over a very large range of dilution rates. In this situation, all intrinsic variables of the culture remain constant. The growth rate can thus be varied over more than one order of magnitude, and cultures can be maintained for months in steady-state growing at rates that are small fractions of the maximum of which the organism is capable. In most realizations, chemostat media are formulated such that the limiting nutrient is known, and thus different culture densities can be produced in otherwise identical media by changing only the concentration of the limiting nutrients.
A question that has remained open since the invention of the chemostat is the relationship between the physiology of cells growing in steady state on a defined limiting nutrient to those grown in a traditional batch culture. Recent interest in using chemostat culture to measure the physiological effects of various perturbations (Kal et al., 1999; ter Linde et al., 1999; Boer et al., 2003) has given this question new significance. Given that chemostat cultures are limited by a particular nutrient, how are they related to batch growth in limiting concentrations of the same nutrient? Are chemostat cultures in a state analogous to an early stationary phase culture, i.e., undergoing a prolonged stress response? Have they reconfigured their metabolism in a way distinct from any phase of batch growth?
We describe here the use of DNA microarrays to compare gene expression between time points of a batch culture and a steady-state chemostat limited on the same nutrient in the same medium. The genome-wide patterns of gene expression reflect, in a detailed way, the physiological state of the cell. Although levels of structural enzymes are commonly regulated at the level of translation or regulation of activity (Oppenheim and Yanofsky, 1980), the regulation of the overwhelming majority of metabolic enzymes is thought nevertheless to be reflected in transcriptional variation as well. In most cases, yeast responds to a changing environment not with a small adjustment in a key control point, but with the coherent transcriptional regulation of large sets of genes. Characterization of the phosphate and zinc regulons (Lyons et al., 2000; Ogawa et al., 2000), as well as the definition of the environmental stress response (Gasch et al., 2000), are a just a few examples of how microarrays can help elucidate the cellular response to an environmental change.
MATERIALS AND METHODS
Raw data used in this article, as well as clustered data files and searchable figures, can be downloaded from the supplementary Web site at http://microarray-pubs.stanford.edu/yeast_bc/home.shtml.
Selection of Strains
The CEN.PK background (Entian and Koetter, 1998) was selected for its growth characteristics in the chemostat (van Dijken et al., 2000). A single MATa prototrophic strain, DBY10148, kindly provided by Peter Kötter (Institut für Mikrobiologie der Johann Wolfgang Goethe-Universität, Frankfurt, Germany) was used for the phosphate and sulfate limitation experiments. Leucine and uracil limitations were implemented using auxotrophic strains that had a block in the synthesis of these key cellular building blocks. The isogenic strains DBY9492 (MATa ura3-52) and DBY9497 (MATa, leu2-3), bearing null mutations in enzymes required for uracil and leucine biosynthesis, were used for the uracil and leucine limitations, respectively.
Establishment of Media Formulation
To determine a medium in which the desired nutrient was limiting, synthetic medium completely lacking the nutrient was aliquoted into several identical flasks, and a different known concentration of the limiting nutrient was added to each. The flasks were inoculated with the same volume of starter culture and placed at 30°C for 3 d, at which point the optical density was assayed. A plot of final optical density against added nutrient shows two phases. At low concentrations, the final density is proportional to the amount of added nutrient. At high concentrations, the final density is insensitive to addition of nutrient. At concentrations below the transition, the culture is said to be limited on the nutrient. The limitation plots for all four nutrients are depicted in Supplementary Figure 1 (http://microarray-pubs.stanford.edu/yeast_bc/home.shtml).
The formulation used for each batch and chemostat limitation is described in Table 1 under media composition. For phosphate and sulfate, essentially the same medium was used in batch and chemostat. Strain DBY9497, bearing the leu2-3 mutation, was found to exhibit very different final densities in batch and chemostat when grown on the same medium. This difference was not due to a difference in cell viability (unpublished data). Therefore, the leucine concentration in the chemostat medium was adjusted to make the final densities approximately equal.
Table 1.
Media composition
| Media composition | Batch Medium | Chemostat medium |
|---|---|---|
| Phosphate | 13.3 mg/l KH2PO4 (98 uM) | 13.3 mg/l KH2PO4 (98 uM) |
| 0.1 g/l Calcium chloride | 0.1 g/l Calcium chloride | |
| 0.1 g/l Sodium chloride | 0.1 g/l Sodium chloride | |
| 0.5 g/l Magnesium sulfate | 0.5 g/l Magnesium sulfate | |
| 0.1 g/l Potassium chloride | 0.1 g/l Potassium chloride | |
| 5 g/l Ammonium sulfate | 5 g/l Ammonium sulfate | |
| 10 g/l Glucose | 10 g/l Glucose | |
| 1 ml/l 1000× Vitamins | 1 ml/l 1000× Vitamins | |
| 1 ml/l 1000× Metals | 1 ml/l 1000× Metals | |
| Sulfate | 3 mg/l Ammonium sulfate (22.7 uM) | 3 mg/l Ammonium sulfate (22.7 uM) |
| 0.1 g/l Calcium chloride | 0.1 g/l Calcium chloride | |
| 0.1 g/l Sodium chloride | 0.1 g/l Sodium chloride | |
| 0.412 g/l Magnesium chloride | 0.412 g/l Magnesium chloride | |
| 1 g/l KH2PO4 | 1 g/l KH2PO4 | |
| 4.05 g/l Ammonium chloride | 4.05 g/l Ammonium chloride | |
| 10 g/l Glucose | 10 g/l Glucose | |
| 1 mL/l 1000× Vitamins | 1 mL/l 1000× Vitamins | |
| 1 mL/l 1000× Metals | 1 mL/l 1000× Metals | |
| Leucine | 40 mg/l Leucine (305 uM) | 15 mg/l Leucine (114 uM) |
| 0.1 g/l Calcium chloride | 0.1 g/l Calcium chloride | |
| 0.1 g/l Sodium chloride | 0.1 g/l Sodium chloride | |
| 0.5 g/l Magnesium sulfate | 0.5 g/l Magnesium sulfate | |
| 1 g/l KH2PO4 | 1 g/l KH2PO4 | |
| 5 g/l Ammonium sulfate | 5 g/l Ammonium sulfate | |
| 10 g/l Glucose | 10 g/l Glucose | |
| 1 mL/l 1000× Vitamins | 1 mL/l 1000× Vitamins | |
| 1 mL/l 1000× Metals | 1 mL/l 1000× Metals | |
| Uracil | 6 mg/l Uracil (53.5 uM) | 5 mg/l Uracil (44.6 uM) |
| 0.1 g/l Calcium chloride | 0.1 g/l Calcium chloride | |
| 0.1 g/l Sodium chloride | 0.1 g/l Sodium chloride | |
| 0.5 g/l Magnesium sulfate | 0.5 g/l Magnesium sulfate | |
| 1 g/l KH2PO4 | 1 g/l KH2PO4 | |
| 5 g/l Ammonium sulfate | 5 g/l Ammonium sulfate | |
| 10 g/l Glucose | 10 g/l Glucose | |
| 1 mL/l 1000× Vitamins | 1 mL/l 1000× Vitamins | |
| 1 mL/l 1000× Metals | 1 mL/l 1000× Metals | |
| Stock solutions | ||
| Trace metals | 500 mg Boric acid | |
| Stock solution, 1000× | 40 mg Copper sulfate. 5H2O | |
| 100 mg Potassium iodide | ||
| 200 mg Ferric chloride. 6H2O | ||
| 400 mg Maganese sulfate. H2O | ||
| 200 mg Sodium molybdate. 2H2O | ||
| 1 l Deionized water, q.s. | ||
| Vitamins | 1 mg Biotin | |
| Stock solution, 1000× | 200 mg Calcium pantothenate | |
| 1 mg Folic acid | ||
| 1000 mg Inositol | ||
| 200 mg Niacin (nicotinic acid) | ||
| 100 mg P-aminobenzoic acid | ||
| 200 mg Pyridoxine HCl | ||
| 100 mg Riboflavin | ||
| 200 mg Thiamine HCl | ||
| 500 mL Water |
Preconditioning for Sulfate Limitations
Phosphorus and sulfur comprise a large fraction of the elemental composition of yeast, exceeded only by carbon, nitrogen, and the components of water. Even so, significant growth was observed in minimal medium devoid of sulfate after a rich medium overnight. This suggests that the cell has significant stores of sulfur in excess of that required for a few cell divisions. To ensure reproducible growth, and increase the likelihood of comparability between batch and chemostat, the cells were preconditioned by overnight growth in the sulfate-limited medium before inoculation. This dependence of growth behavior on preconditioning was not observed for the other limitations and may be a consequence of the relatively low abundance of sulfur in yeast.
Batch Growth Conditions
Cells were grown in either YP 2%D (phosphate) or minimal medium supplemented with the limiting nutrient in 5-ml overnight cultures. The overnight cultures were spun down, washed, and used to inoculate limiting medium to a starting density of ∼0.01 as measured by absorbance at 600 nm. Aliquots (50 ml) were then distributed into 500-ml Erlenmeyer flasks. The flasks were then grown on a platform shaker at 30°C, 300 rpm. At regular intervals, single flasks were harvested and assayed for residual medium components, cell morphology, and transcript abundance.
Chemostat Growth Conditions
An overnight YP 2%D culture was washed and used to inoculate a 300-ml working volume chemostat to a starting optical density of .05 as measured by absorbance at 600 nm. The chemostat was grown in batch mode for ∼12 h then switched to continuous mode with a dilution rate D of ∼0.18 per hour. After ∼2 d, cell optical density and count stabilized, at which point the chemostats were harvested and assayed for residual medium components, cell morphology, and transcript abundance.
Culture Sampling
Yeast cultures were harvested by vacuum filtration followed by quickfreezing of the filter in liquid nitrogen. Immediately before filtration, a few milliliters of culture were set aside for cell assays. The filtered medium was frozen in a -20°C freezer for later chemical assays.
Cell Density, Volume, and Morphology
Cell density and volume were assayed regularly using a Z2 automated cell counter (Beckman Coulter, Fullerton, CA). Culture was diluted into Isotone II buffer so that an appropriate number of cells, between 5000 and 20,000, could be counted.
Cell morphology was assayed using light microscopy. Cells were scored as large budded if the daughter bud was more than one-half the diameter of the mother, and small if the daughter was less than one-half the diameter of the mother.
Phosphate Assay
Phosphate was measured using the method of Chen et al. (1956). This assay relies upon the reaction of phosphate with molybdate to evolve a purple color that can be assayed by absorbance at 840 nm. A calibration curve found the assay to be very linear in the range from 2 to 180 μM. Within the linear range, on the calibration curve the assay was accurate to with 5%, fading into the background at ∼2 μM.
Sulfur Assay
Elemental sulfur concentration was assayed using a TJA IRIS Advantage/1000 Radial ICAP spectrometer. This technique vaporizes the medium in a hot plasma, which destroys all molecular structure and ionizes many of the elements. The elemental composition is then assayed by spectroscopy of the radiation emitted by relaxation of the electron back to the ground state. We seemed to be near the threshold of sensitivity for this machine, and readings below ∼5 μM were unreliable.
Microarray Data Acquisition
Microarrays were produced, processed, hybridized, and scanned according to DeRisi et al. (1997). Briefly, open reading frames PCR amplified from the yeast genome were robotically spotted onto polylysine-coated slides. Total RNA was prepared using phenol-chloroform extraction and enriched for mRNA using the Oligotex Midi kit (catalog no. 70042; QIAGEN, Valencia, CA). For each microarray, the two samples to be compared were labeled and hybridized to the microarray either by direct incorporation of labeled nucleotides or by incorporation of aminoallyl-dUTP followed by coupling to a succinimidylester-conjugated dye, by using protocols described in the LabelingProtocols.pdf document in the Supplementary Materials (http://microarray-pubs.stanford.edu/yeast_bc/home.shtml). After scanning and gridding with GenePix (Axon Instruments, Foster City, CA), quantitated microarray data were loaded into the Stanford Microarray Database (SMD) (Gollub et al., 2003) and normalized using the regression correlation method. As described in the SMD online documentation, the regression correlation method first selects spots that are not flagged and have a regression correlation of >0.6. It then calculates the average of the natural log of the channel 2 to channel 1 ratio for these spots and sets the normalization value to be this average raised to the power e. The raw channel 2 intensities are divided by the normalization value to yield the normalized channel 2 intensity.
Microarray Data Processing
There are a total of 13 time courses, designated L1, L1R, L2, L2R, P1, P2, P3, S1, S2, S4, U1, U2, and U3. The time courses beginning with L are derived from leucine limitation time courses, those with P from phosphate time courses, S from sulfate limitation time courses, and U from uracil limitation time courses. The “R” appended to two leucine time courses indicates that they were replicates, run with the same batch samples, but a different chemostat reference. P1, P2, S1, S2, and S4 were run with the batch samples labeled with Cy5, and the chemostat samples labeled with Cy3. P3, U1, U2, U3, L1, L1R, L2 and L2R were run with the batch samples labeled with Cy3 and the chemostat samples labeled with Cy5.
Median fold change ratios were downloaded as raw data by spot. For the time course data, the ratio of the batch culture value to the chemostat culture value was calculated. The per-spot data was then log transformed, clustered by time course, analyzed for spatial biases, reflagged, and collapsed by open reading frame.
There are 24 chemostat comparisons, corresponding to four sets of three pairs of dye swaps. Each of the four sets corresponds to a different chemostat pair, designated LS, PU, SP, and UL, with the first letter indicating the chemostat represented by red, and the second the chemostat represented by green. The data were processed as described above, with the additional step of inverting all the ratios for the dye swapped comparisons.
Megacluster Generation
The collapsed data files generated after processing were analyzed to determine which had the least noisy clusters. The least coherent time course for each limitation was then thrown out, and the remaining data were used to create the megacluster. The nine time courses used for the megacluster were L1R, L2, L2R, P1, P2, S1, S4, U2, and U3. The megacluster was constructed using agglomerative hierarchical clustering, as described in Eisen et al. (1998).
Gene Ontology Enrichment
This is a technique for assigning a biological process to a list of genes. The Gene Ontology project (Ashburner et al., 2000) has come up with a standard classification of biological processes. The Saccharomyces Genome Database (Dwight et al., 2002) maintains gene assignments to those processes. Using these resources together with GO::TermFinder, a Perl package by Gavin Sherlock, which is freely available off the Comprehensive Perl Archive Network (http://search.cpan.org/dist/GO-TermFinder/, submitted to Bioinformatics), we were able to ascertain statistically significant overlap between the list of genes in a cluster and the genes annotated to a given biological process.
Analysis of Ring Design
A ring design experiment is suitable when there are a small, prescribed number of conditions for which the investigator wishes to determine accurate pairwise values with the minimum number of arrays (Dobbin and Simon, 2002; Yang and Speed, 2002; Townsend, 2003). In a ring design, the conditions to be investigated are arranged in a circle, and all ratios going around the circle are measured. However, the experimenter is generally interested not in the ratio of particular conditions to each other, but whether a gene is more highly expressed in a subset of the conditions. For this purpose, it is useful to transform the data from ratios to normalized values.
If we represent the absolute expression level in a condition by ei, and the ratio of two subsequent levels as ri = ei + 1/ei, then we can write all the absolute expression levels in terms of the first one. This makes sense, because we know the ratio of expression levels between all the conditions; if only we knew the absolute expression of one! This problem can be resolved by introducing a normalization constraint. If we set the average absolute expression level to be 1, then we have the constraint that the sum of the absolute expression levels must add to the total number of conditions. This additional constraint allows us to solve for the normalized expression level of each condition purely in terms of the measured ratios.
The six estimates of each ratio from the dye swapped chemostat comparisons were averaged together to produce a best estimate of each pairwise ratio. These ratios were then transformed to absolute levels as described above. For example, to calculate the absolute level in the leucine chemostat, the following two estimates were averaged:
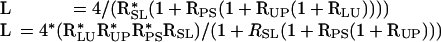 |
Rij corresponds to the ratio of the i to j chemostat. The two estimates correspond to rewriting the terms in the constraint equation, L + S + P + U = 4, by going around the ring in opposite directions.
Of interest is whether, for each gene, the normalized levels are significantly different from equally distributed among the various conditions. The variance of the normalized levels was selected as a statistic that captures the desired behavior. A null distribution of the variance was built up by randomizing the ratios within genes for the entire data set 20 times. Genes were then selected that had a higher variance in their levels than 99% of the randomized data.
RESULTS
To determine the relationship, if any, between the physiological state achieved during steady-state chemostat growth and that during batch culture, strain DBY10148 was grown using both culture techniques under both phosphate and sulfate limitation. In preliminary experiments, we found a concentration of phosphate or sulfate below which the final density of batch cultures was linearly dependent on the amount of added nutrient; a convenient limiting concentration was chosen from within this linear range. From each of the batch and chemostat cultures, samples for gene expression, cell size, culture density and cell morphology were taken and the residual limiting nutrient concentration was monitored.
Correspondence of Chemostat and Batch under Phosphate and Sulfate Limitation
To assess the degree of transcriptional correspondence between chemostat and batch cultures, samples of the batch cultures were collected at regular intervals. The chemostat reference for each time series came from a single chemostat that was harvested well after it reached a steady state, as determined by the optical density of the effluent. The batch time points and chemostat reference were then labeled and competitively hybridized to a cDNA microarray (DeRisi et al., 1997). In the comparisons below, the batch values are given as the red channel and the chemostat values are green, regardless of which dyes were actually used; further details (e.g., dye swaps) are given in Materials and Methods.
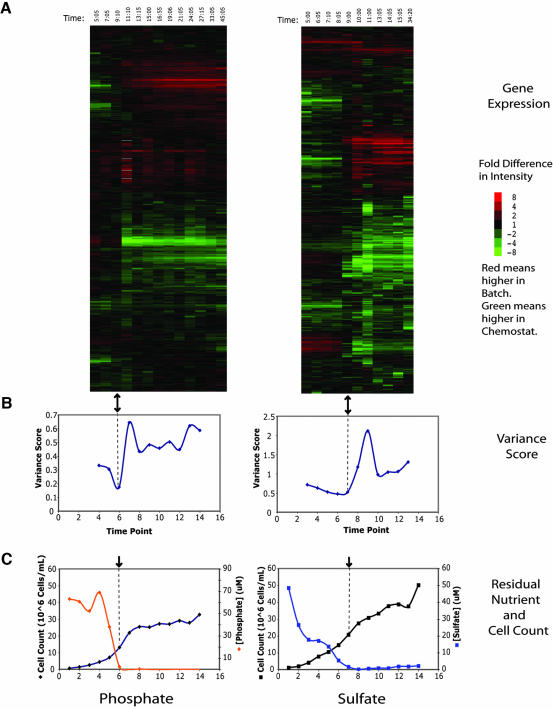
A heatmap representing gene expression data from a single phosphate and sulfate batch growth time course compared with its cognate chemostat is presented in Figure 1A. The rows have been ordered by hierarchical clustering using an uncentered Pearson correlation (Eisen et al., 1998). The clustering analysis clearly identifies two major patterns in the data: induced gene expression at the top of the heatmap and repressed gene expression at the bottom. As described below, most of these genes are part of a stereotyped environmental stress response (Gasch et al., 2000). Notably, the induction and repression values (in logarithmic units) cross zero at the same time. Because the data are not centered, this implies that there is a particular point in each time course, indicated by the black arrow, that has ratios that are close to 1.
Figure 1.
Global cluster analysis of phosphate and sulfate expression data. (A) Clustergram of genome-wide microarray data for phosphate (P1) and sulfate (S4) time courses. The array indicated by the black arrow has similar representation of transcripts in both channels. It should be noted that the data have not been centered, thus allowing the accurate representation of a constant bias in expression, but also increasing sensitivity to systematic bias. Time points were taken at intervals of approximately 2 h. Each row corresponds to a single gene and each column to a single array. The columns are arranged in order of increasing time. Red values indicate higher expression in the batch, and green values indicate higher expression in the chemostat. The intensity of the color is determined by the fold change and is indicated by the color bar to the right. (B) Variance score has a minimum near the array identified by cluster analysis, shown here with the black arrow. The variance score is a measure of the deviation of the array from equal representation of all transcripts in both channels and is calculated as the average of the square of the log ratios. (C) The limiting nutrient becomes undetectable in the filtered media near the time that the transcriptional state becomes comparable.
The identification of a single array that seemed to have equal representation of most mRNA species prompted a further analysis. The variance score of an array is the average square of the log-ratios from the array; if it is large, there are many genes with large fold changes; if it is small, the majority of genes are equally represented in both samples. The variance score for each time point thus represents the mean transcriptional difference between the batch and chemostat at that time. The plot of variance scores as a function of time (Figure 1B) shows a dip near the array, which is black in Figure 1A, indicating that the batch is most comparable with the chemostat at that point and that the black array is not an artifact of clustering. The variance never goes to zero due to both experimental noise and the exact timing of the sampling relative to the point at which the chemostat and batch are comparable.
Concentration of Limiting Nutrient
Phosphate and sulfate concentrations were measured during the course of the batch limitations as described in Materials and Methods (Figure 1C). The results indicate that the nutrients become undetectable in the culture media near the time at which we found comparable patterns of gene expression. The residual concentration in the sulfate chemostat was ∼8 μM sulfate, and in the phosphate chemostat ∼30 μM phosphate. In general the residual concentration in the batch medium is somewhat lower than the residual concentration in steady-state growth. This difference may be accounted for by internal storage forms, which would be in equilibrium in the chemostat, but not in batch culture.
Correspondence of Chemostat and Batch under Leucine and Uracil Limitation
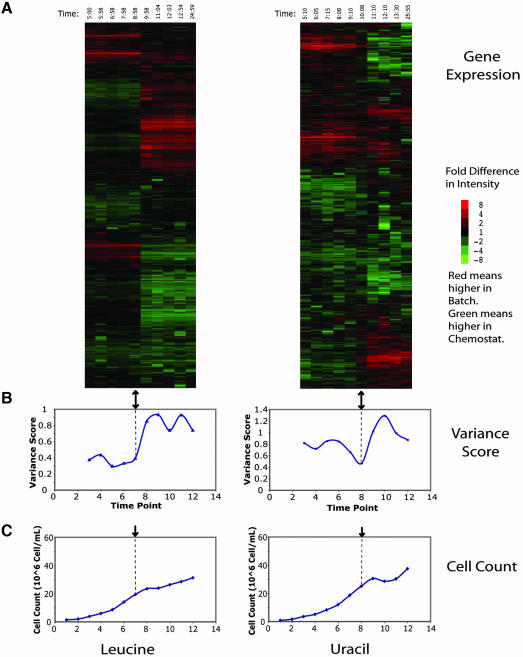
To assess the generality of the result for the “natural” nutrient limitations sulfate and phosphate, gene expression during leucine and uracil limitations was investigated using isogenic auxotrophs bearing null mutations in LEU2 and URA3, respectively (Figure 2). The primary identifiable patterns are once again the induction and repression of the massive stress-related response. Although there seems to be more noise in the data, these patterns are, as might be expected, similar to those found in the sulfate and phosphate data sets. There is again a particular array, indicated by the black arrow, in which the batch gene expression is most comparable with the chemostat. The plot of variance scores shows that there is a minimum at or near the comparable array (Figure 2B). These results illustrate a close correspondence, as measured by global pattern of transcription, between chemostat growth and the late phase of batch growth under auxotrophic limitation.
Figure 2.
Global cluster analysis of leucine and uracil expression data. Clustergram of genome-wide microarray data for leucine (L2R) and uracil (U2) time courses. Figure was produced as described in Figure 1. Again, there is an array in which the gene expression is most comparable, indicated by the black arrow, and a corresponding minimum in the variance plot.
Cell Cycle Arrest Is Uniform in Phosphate and Sulfate Limitations
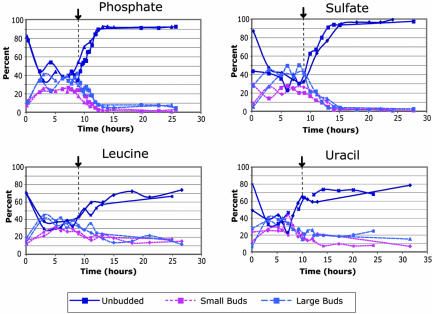
The exhaustion of phosphate and sulfate is followed by a cessation of cell division within a few cell cycles (Figure 1C). To further examine the nature of this arrest, we characterized the cell morphology of the cultures as a way of estimating the positions of cells in their cell cycles (Hartwell, 1974) (Figure 3). During the exponential phase of batch growth, the cultures are unsynchronized, with ∼40% unbudded cells. In stationary phase sulfateand phosphate-limited batch cultures, nearly 95% of the cultures consisted of unbudded cells, indicating a highly coordinated arrest of the culture at G0/G1. For comparison, the cell morphology of the phosphate chemostat was 64% unbudded, 14% small budded, and 22% large budded (n = 200), and the cell morphology in the sulfate chemostat was 66% unbudded, 16% small budded, and 17% large budded (n = 263). This is significantly different from the morphology distribution of either exponential phase or stationary phase batch cultures, as judged by a two-class T-test of budded and unbudded, indicating that the cell morphology in the chemostat is intermediate to the exponential and fully starved batch phase cultures.
Figure 3.
Cell morphology during nutrient-limited batch time courses. Cells were taken at the indicated time points and assayed for cell morphology by light microscopy, as described in Materials and Methods. Cells grown under phosphate and sulfate limitation displayed a more uniform final morphology than under leucine and uracil limitation.
Cell Cycle Arrest Is Less Uniform in Leucine and Uracil Limitations
We assayed cell morphology in these cultures to determine whether a similarly coordinated arrest was occurring. The fraction of unbudded cells in fully starved leucine and uracil limited cultures was higher than in the corresponding exponential phase, but rarely exceeded 75%. Thus, there seems to be a difference between natural starvations (i.e., for phosphate or sulfate) and “unnatural” starvations for supplements required for growth only by auxotrophic mutants. The former produce a highly organized cell cycle arrest; in the latter this arrest, if it happens at all, is much less uniform.
The Stress Response Is Not Induced in the Chemostat
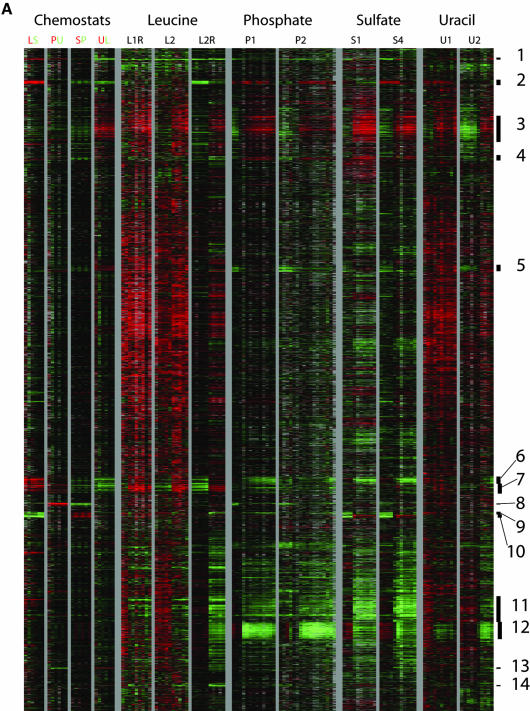
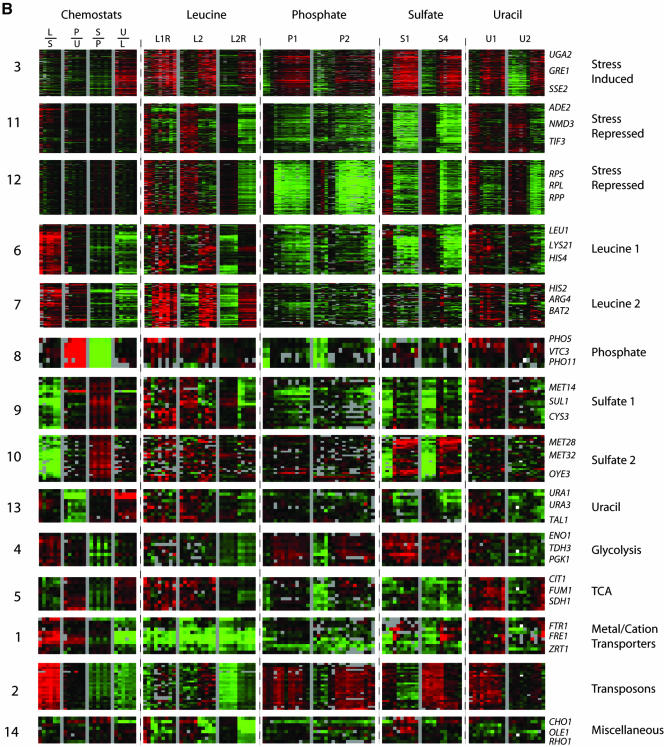
To identify the processes represented by the induced and repressed genes and assess their generality, a megacluster analysis was performed. The complete expression data for two time courses for each limitation were extracted. The values for these genes were then hierarchically clustered using an uncentered metric to reveal patterns of common and specific regulation that are shared by large sets of genes. The complete clustergram is displayed in Figure 4A. The data for each cluster are grouped by theme in Figure 4B. A gene list was extracted for each cluster and checked for statistical enrichment of Gene Ontology process terms (Ashburner et al., 2000; Harris et al., 2004) by using the GO TermFinder package (http://search.cpan.org/dist/GO-TermFinder/). The top few Gene Ontology terms for each cluster are listed in Table 2. Although there are an imposing number of clusters, we found that they fall into a small number of easily comprehensible classes.
Figure 4.
Megacluster analysis reveals clusters with distinct regulation. (A) Gene expression data were extracted for nine time courses and four pair-wise chemostat comparisons and clustered using an uncentered Pearson correlation. To the right of the clustergram are indicated 14 clusters of gene with coherent patterns of expression. (B) Summary of clusters identified in megacluster analysis. The log base 2 of the expression ratio for the 14 clusters was plotted as a function of time. The 14 clusters have been grouped by theme. Clusters 3, 11, and 12 show common regulation across the time courses. Clusters 6, 7, 8, 9, 10, and 13 show regulation specific to a particular nutrient, as indicated by name. Clusters 4, 5, 1, 2, and 14 show diverse regulation.
Table 2.
GO term enrichment in megacluster analysis
| Cluster | Size | Gene ontology terms |
|---|---|---|
| 1. “Cation transporters” | 11 | Siderochrome transport (3.557e-09, 4 of 9) |
| Transition metal ion transport (5.396e-09, 5 of 32) | ||
| Di-, tri-valent inorganic cation transport (1.996e-08, 5 of 41) | ||
| 2. “Transposons” | 24 | Biological process unknown (p = 3.821e-06, 21 of 2797) |
| Ty element transposition (p = 0.0721, 2 of 63) | ||
| DNA transposition (p = 0.1001, 2 of 75) | ||
| 3. “Stress induced” | 196 | Response to stress (p = 4.745e-10, 35 of 344) |
| Response to external stimulus (p = 1.573e-05, 23 of 247) | ||
| Response to biotic stimulus (p = 1.643e-05, 11 of 55) | ||
| 4. “Glycolysis, phosphate induced” | 10 | Glycolysis (p = 8.133e-20, 8 of 18) |
| Glucose catabolism (p = 2.007e-18, 8 of 25) | ||
| Hexose catabolism (p = 2.007e-18, 8 of 25) | ||
| 5. “TCA cycle, phosphate induced” | 10 | Tricarboxylic acid cycle (1.347e-20, 8 of 15) |
| Main pathways of carbohydrate metabolism (p = 1.527e-14, 8 of 68) | ||
| Energy derivation by oxidation of organic compounds (p = 2.520e-13, 9 of 180) | ||
| 6. “Leucine induced, sulfate repressed” | 50 | Amino acid metabolism (p = 1.478e-32, 27 of 145) |
| Amine metabolism (p = 9.000e-31, 27 of 167) | ||
| Branched chain family amino acid biosynthesis (p = 3.971e-16, 9 of 13) | ||
| 7. “Leucine induced” | 56 | Organic acid metabolism (p = 4.215e-07, 13 of 225) |
| Carboxylic acid metabolism (p = 4.215e-07, 13 of 225) | ||
| Water-soluble vitamin metabolism (p = 0.0005, 6 of 68) | ||
| 8. “Phosphate induced” | 6 | Vacuole fusion (non-autophagic) (p = 3.557e-4, 2 of 15) |
| Membrane fusion (p = 1.670e-3, 2 of 32) | ||
| Phosphorus metabolism (p = 0.04236, 2 of 163) | ||
| 9. “Sulfate induced, sulfur related” | 20 | Sulfur metabolism (p = 6.503e-21, 12 of 50) |
| Sulfur amino acid metabolism (p = 2.099e-16, 9 of 29) | ||
| Methionine metabolism (p = 5.535e-13, 7 of 20) | ||
| 10. “Sulfate induced, miscellaneous” | 24 | Sulfur amino acid metabolism (p = 0.03689, 2 of 29) |
| Sulfur metabolism (p = 0.1069, 2 of 50) | ||
| Biological process unknown (p = 0.6016, 13 of 2797) | ||
| 11. “Stress repressed, ribosome assembly” | 196 | Ribosome biogenesis (p = 7.216e-17, 31 of 157) |
| Transcription from Pol I promoter (p = 5.047e-15, 29 of 155) | ||
| rRNA processing (p = 2.028e-10, 22 of 127) | ||
| 12. “Stress repressed, ribosomal mRNA” | 126 | Protein biosynthesis (p = 3.876e-100, 114 of 759) |
| Macromolecule biosynthesis (p = 1.394e-99, 114 of 767) | ||
| Biosynthesis (p = 7.578e-80, 114 of 1120) | ||
| 13. “Uracil induced” | 10 | Pyrimidine base biosynthesis (p = 4.786e-06, 3 of 13) |
| Pyrimidine base metabolism (p = 7.604e-06, 3 of 15) | ||
| Nucleobase biosynthesis (p = 1.615e-05, 3 of 19) | ||
| 14. “Miscellaneous” | 8 | Glycerophospholipid biosynthesis (p = 4.990e-3, 2 of 30) |
| Glycerophospholipid metabolism (p = 6.422e-3, 2 of 34) | ||
| Lipid metabolism (p = 9.817e-3, 3 of 190) |
TCA, tricarboxylic acid.
By far the largest clusters arising from the analysis showed coordinated induction or repression across all time courses and were enriched for genes related to the classic stress response (Gasch et al., 2000). Cluster 3 consists of genes that are induced in all time courses, and has “response to stress” as its primary annotation. Clusters 11 and 12 are both repressed under all conditions. Cluster 12 consists almost entirely of genes encoding ribosomal proteins, whereas cluster 11 contains many genes involved in ribosomal assembly, including rRNA processing and PolI transcription. Notably, these clusters are expressed at levels comparable with the chemostat only during active batch growth, before the onset of the stress response. This is strong evidence that the stress response is not activated in cells growing at steady state in the chemostat despite the reality that they are growing at submaximal growth rates because of nutrient limitation. It also means that at the point at which the batch cells are in a physiological state comparable with that in the chemostat, the stress response has not yet been activated.
Homeostasis in Different Media Involves Distinct Sets of Genes
Aside from the large stress response clusters, the majority of the remaining clusters can be interpreted as defining genes that are changing to maintain balanced growth in the face of a particular nutrient limitation. Clusters 6, 7, 8, 9, 10, and 13 from the megacluster analysis consist of genes that respond only to particular limitations (Figure 4B). Clusters 7, 8, and 10 show induction specifically under leucine, phosphate, and sulfate limitation, respectively, and are enriched for genes related to metabolism of those molecules. Cluster 13 contains annotations to pyrimidine base biosynthesis, shows increased expression in the uracil chemostat, but it is not induced in the uracil-limited batch experiment. Clusters 6 and 9 highlight an interesting reciprocal relationship between a subset of sulfate- and leucine-regulated genes. In the sulfate batch time course, the sulfate metabolism genes of cluster 9 are induced, whereas the leucine metabolism genes of cluster 6 are repressed. The opposite is true for the leucine time course.
An independent way of determining genes specifically involved in maintaining homeostasis under diverse nutrient limitations is by direct comparison of gene expression in the several differently limited chemostats. We found a way to maximize resolution of the functional classes of these genes by using a ring design (Yang and Speed, 2002). The four steady-state chemostats were compared using cDNA microarrays as follows: sulfate limited (S) versus phosphate limited (P), P versus uracil limited (U), U versus leucine limited (L), and L versus S. These experiments were done in triplicate with dye swaps for a total of six measurements of each ratio.
To facilitate interpretation of these data, normalized levels of expression in each chemostat were derived from the ratio data according to the transformation described in Materials and Methods. A null distribution was calculated using randomized data to assess statistical significance. Interestingly, although many genes were significantly induced in specific chemostats, very few genes were significantly repressed. Four lists were produced, corresponding to genes induced specifically in the four chemstats. These lists were analyzed for enrichment of Gene Ontology terms, shown in Table 3. Data from the batch culture time courses were extracted for each of the four lists and subjected to further cluster analysis.
Table 3.
GO term enrichment of genes up-regulated in particular chemostats
| Chemostat | Size | Gene ontology terms |
|---|---|---|
| Leucine | 91 | Amino acid metabolism (p = 4.197e-12, 18 of 145) |
| Amino acid and derivative metabolism (p = 1.711e-11, 18 of 157) | ||
| Amino acid biosynthesis (p = 3.114e-11, 15 of 99) | ||
| Phosphate | 37 | Vacuole fusion (non-autophagic) (p = 1.860e-05, 4 of 15) |
| Membrane fusion (p = 4.606e-4, 4 of 32) | ||
| Pyridoxine metabolism (p = 9.326e-3, 2 of 6) | ||
| Sulfate | 36 | Sulfur metabolism (p = 3.641e-13, 10 of 50) |
| Sulfur amino acid metabolism (p = 1.021e-09, 7 of 29) | ||
| Sulfate assimilation (p = 6.286e-07, 4 of 8) | ||
| Uracil | 46 | Pyrimidine base biosynthesis (p = 3.364e-05, 4 of 13) |
| Pyrimidine base metabolism (p = 6.364e-05, 4 of 15) | ||
| Response to extracellular stimulus (p = 6.364e-05, 4 of 15) |
The results of this analysis were largely concordant with the megacluster analysis. Clustering of genes identified as up-regulated in the phosphate and sulfate chemostats by the ring design revealed one or two clusters, respectively, corresponding to clusters 8, 9, and 10 from the megacluster analysis (unpublished data; see Supplementary Material). However, clustering of genes identified as up-regulated in the leucine and uracil chemostats by the ring design analysis revealed multiple patterns, only some of which were identified in the megacluster analysis (Figures 5 and 6).
Figure 5.
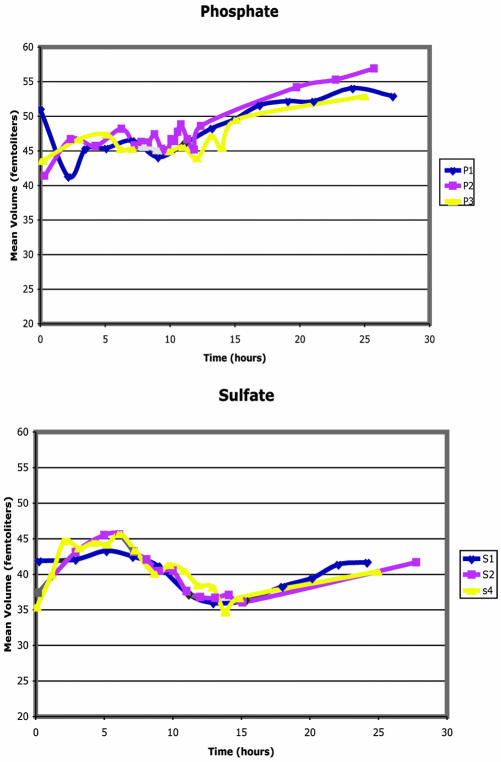
Cell size during phosphate and sulfate batch limitations. Cell suspensions taken at the indicated times were assayed for cell size using a Beckman Coulter Z2 cell counter. The two limitations consistently showed opposite changes in cell size.
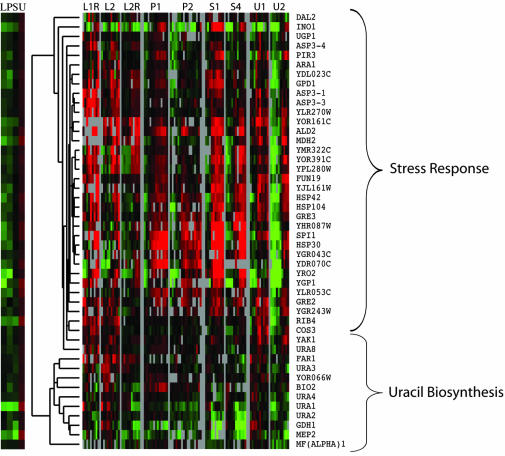
Figure 6.
Cluster analysis of genes highly expressed in uracil chemostat. Genes were called highly expressed in a particular chemostat by the ring design analysis. Data from the time-course experiments were extracted for the genes on the list and clustered to reveal two major patterns of gene expression, a consistent induction corresponding to the stress response, and no consistent regulation, corresponding to the genes involved in de novo uracil biosynthesis.
Cluster analysis showed two predominant patterns in the uracil data (Figure 5). One was induction (after exhaustion of the limiting nutrient) in all time courses and corresponded to a subset of cluster 3 (stress response) in the megacluster. The other pattern was no change in any time course, corresponding to cluster 13 in the megacluster (Figure 4); the annotations here include the genes of uracil biosynthesis (URA1, URA2, URA3, and URA4). This result reflects the expected effect of nutrient limitation on the expression of cognate biosynthetic genes; it is notable that the degree of induction in the batch is minimal.
The leucine-specific genes divided into three clusters showing induction, highly correlated induction, and a constant level of expression that was very low relative to the chemostat. Genes in the first cluster were annotated to amino acid metabolism, including leucine biosynthesis, genes in the second consisted entirely of transposons, and genes in the third had significant annotation to divalent metal cation transport. These genes were found in clusters 6 and 7, cluster 2, and cluster 1 of the megacluster, respectively. It is notable here that the leucine biosynthetic genes (ILV2, ILV3, ILV5, ILV6, LEU1, LEU4, LEU9, and BAT1) are strongly induced during both batch and chemostat limitation experiments, as expected, and in contrast to the results in the uracil pathway.
Cell Size Changes during Limitations
Opposite cell size changes were observed during phosphate and sulfate limitations (Figure 7). During the phosphate batch limitation, cell size increased dramatically from ∼45 femtoliters to nearly 55 femtoliters. Most of the increase in cell size occurred during the period after the phosphate had been exhausted. In contrast, during the sulfate limitation, cell size decreased from ∼42 femtoliters to nearly 35 femtoliters, before recovering to 40 femtoliters. This decrease in cell volume begins as early as 5 h and thus precedes the exhaustion of the limiting nutrient at ∼9 h, although the cells continue to decrease in volume until ∼12-13 h. Cell size can be thought of as a balance between growth and cell division. This suggests that under phosphate limitation, the cells can grow in size but cannot divide, perhaps due to an inability to synthesize nucleic acids, whereas under sulfate limitation the cells can divide, but do not grow in size, perhaps because the production of protein is adversely affected.
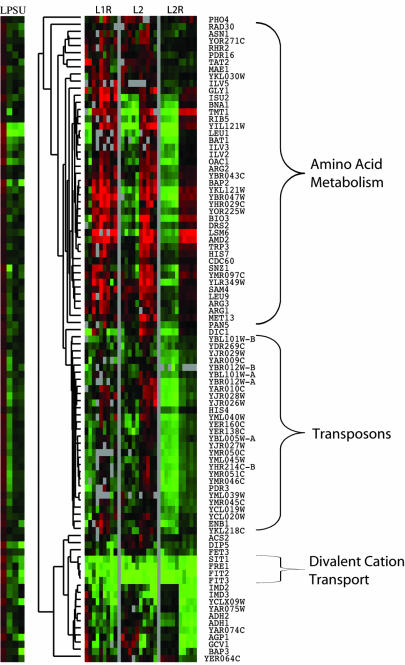
Figure 7.
Cluster analysis of genes highly expressed in leucine chemostat. Genes were called highly expresses in a particular chemostat by the ring design analysis. Data for the leucine batch time-course experiment were extracted for the genes that were up-regulated in the leucine chemostate, and clustered to reveal three major patterns of expression corresponding to amino acid biosynthesis, transposons, and divalent cation transport, as indicated.
Summary
The majority of changes in gene expression during batch growth can be accounted for as stress response and largely interpretable changes required for metabolic homeostasis. There is a particular point in batch growth at which gene expression is comparable with the chemostat. Importantly, the stress response is not activated in the chemostat, and genes that are not regulated in response to the limiting nutrient do not show coherent differences between the batch and the chemostat or between different chemostats.
DISCUSSION
At the dawn of the chemostat era, Novick and Szilard established that at low concentrations of trypotophan, Escherichia coli grew at a rate proportional to the concentration of tryptophan (Novick and Szilard, 1950). Even earlier, Monod (Monod, 1942) found that the growth rate of an exponential batch culture, mu, was related to the concentration of substrate S by the Michaelis-Menten equation,
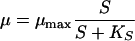 |
for bacteria grown on a variety of sugars. This equation is assumed to apply to yeast as well as bacteria for a wide variety of nutrients. It is remarkable that such a simple feedback relation of growth to nutrient availability holds for such a great diversity of limiting nutrients; it is this relation that allows the chemostat to reach a steady state at arbitrarily low dilution (i.e., growth) rates. These considerations lead to the hypothesis that cells in the chemostat have similar physiology to cells growing in batch at a suitably low concentration of the limiting nutrient. Verification of this hypothesis, for four diverse limiting nutrients, is the major result of this study. A subsidiary result is that this hypothesis seems to be true whether or not the nutrient limitation is natural (e.g., phosphate or sulfate) or imposed by an auxotrophic mutation unlikely to occur at high frequency in nature (e.g., limiting uracil for a ura3 mutant or limiting leucine in a leu2 mutant).
Relationship of Chemostat and Batch Cultures
We characterized the cell size, cell morphology, cell density, and whole genome transcript abundance of yeast in batch and chemostat cultures. As assayed by microarray analysis, the gene expression, and by extension the physiology, of cells at steady state in the chemostat is essentially the same as that of a similarly limited batch phase culture as the limiting nutrient is exhausted toward the end of batch growth. There is therefore no reason to believe that there is anything special about cells grown in the chemostat.
Our results are consistent with the notion that as cells in a batch culture exhaust the limiting nutrient, they adjust their metabolism to maintain internal homeostasis. The chemostat represents a way to maintain the cells at a particular point in that progression. In each of the four conditions (sulfate, phosphate, leucine, and uracil) the cells are “poor, not starving.” In particular, cells in the chemostat are not undergoing the environmental stress response so visible at the onset of the fully starved, or “stationary” phase of batch culture. The high level of residual nutrient in the chemostat, where measured, supports the notion that direct feedback to growth rate allows the cells to maintain homeostasis and avoid the stress response (Gasch et al., 2000).
Common Transcriptional Responses
The collection of several time-course data sets for each limitation allows further cluster analysis to discover patterns of regulation that are shared by large numbers of genes. By far the largest clusters of genes show a pattern of either coordinated induction or repression across all the time courses (clusters 3, 11, and 12). As might be expected, the genes in these clusters are primarily annotated either to the stress response (cluster 3) or protein biosynthesis (clusters 11 and 12). Interestingly, these genes did not in general show a difference in steady-state levels between different chemostats, and in many cases were expressed at similar levels in exponential phase batch as in the chemostat. These clusters likely reflect a stress response, one that is activated late in exponential batch growth and is not active in the chemostats.
For all nutrients, a cluster was identified that was significantly enriched for genes specific to the metabolism of that nutrient (clusters 6, 7, 8, 9, 10, and 13). These clusters invariably showed very high expression in the chemostat limited for the specific nutrient. With the exception of cluster 13, consisting of uracil biosynthetic genes, these clusters also showed strong induction in batch time courses limited for the specific nutrient. Together, these results suggest a common theme in yeast metabolic regulation; when a specific nutrient becomes limiting, expression of genes encoding proteins of the synthetic pathway for that nutrient are induced. Indeed, there is ample precedent for this in the literature (Denis-Duphil, 1989; Thomas and Surdin-Kerjan, 1997 #235; Persson et al., 1999; Wang et al., 1999; Kohlhaw, 2003).
The exception for the uracil pathway is interesting, in view of considerable published information indicating a regulatory role for PPR1 in the regulation of this pathway (Losson et al., 1985; Oestreicher and Scazzocchio, 1995). It is possible that this pathway happens to be fully derepressed already at the outset of the experiments. This inference will have to be tested experimentally.
Potential Novel Targets of Homeostatic Regulation
It should be noted that not all of the genes in the nutrient-specific clusters are annotated to metabolism of the relevant nutrient, but instead they may indicate novel targets of the regulatory network. For example, many of the nonamino acid biosynthesis genes in cluster 7 have nonetheless been reported as targets of the transcription factor Leu3p. In a recent review, Kohlhaw (2003) assembled a list of seven genes for which there is strong genetic and biochemical evidence of regulation by Leu3p, six of which can be found in cluster 7. The review also defined five genes identified as likely targets of Leu3p by noncoding sequence analysis (Liu and Clarke, 2002), four of which, BAT1/YHR208w, LEU9/YOR108w, OAC1/YKL120w, and MAE1/YKL029c, also are found in cluster 7. These novel targets illustrate the continuing value of transcriptional analysis under diverse growth circumstances in delineating the metabolic regulatory networks in yeast.
Adaptation in Auxotrophic Limitations
The close transcriptional correspondence on a global scale between chemostat and batch cells for a wide variety of cellular processes suggests that the cells remain near-optimally adapted to the limiting nutrient even as the concentration of the limiting nutrient changes. However, not all limitations displayed perfect adaptation. In the auxotrophic limitations, leucine- and uracil-specific clusters were induced in the chemostat that were unchanged during batch limitation.
Clustering of genes induced in the steady-state leucine-limited chemostat revealed three clusters annotated to branched chain amino acid metabolism, transposons, and divalent metal cation transporters, respectively. Of these, the branched chain amino acid metabolism and transposon clusters also were induced in the batch time course. The divalent metal cation transporters are consistently up-regulated in the chemotstat relative to the batch. Notably, no other cluster had such consistent differential regulation between the batch and the chemostat.
Analysis of the cell morphology of the starved batch cultures revealed a striking difference between the natural and auxotrophic starvations. Phosphate- and sulfate-limited cultures were found to consist of >90% unbudded cells after growth arrest, whereas the leucine and uracil cultures seldom exceeded 75%. The cell morphology of the starved auxotrophs resembled that of the cells in the chemostat, which are still undergoing exponential growth. This observation supports the possibility that natural shortages of nutrients trigger an efficient, evolved response, whereas shortages encountered only by auxotrophic mutants do not trigger an efficient response. There is indeed some evidence for mechanisms strongly linking sulfate (Patton et al., 2000) and less strongly phosphate (Carroll and O'Shea, 2002) metabolism to cell cycle control. However, this study suggests that direct links should exist for both and that our understanding of the mechanisms linking metabolism to cell cycle commitment is yet incomplete.
Implications for General Control of Amino Acid Metabolism
General amino acid control is usually taken to mean coordinated derepression of unrelated amino acid synthetic pathways in response to limitation for a specific amino acid. Our results for sulfate and leucine limitation seem inconsistent with this kind of general control. Cluster 6, enriched for branched chain amino acid biosynthesis, is induced in the leucine time course, but repressed in the sulfate time course. Cluster 9, enriched for sulfur-containing amino acid metabolism, shows the opposite relationship.
Remarkably, the repression of sulfate genes in the leucine time course and the repression of leucine genes in the sulfate time course was greater than the repression of these genes under other conditions. A stronger repression of the alternative amino acid pathway resulted from leucine or sulfate limitation than that expected from a generic stress. This suggests that under specific amino acid limitation, Saccharomyces induces expression of genes for that particular amino acid biosynthetic pathway, and represses genes for other pathways. This makes sense, because it is likely that the other amino acids are present in relative excess. This should result in feedback inhibition of the transcription of the structural enzymes. A remaining question is the physiological role of general control, because it seems to have been overridden in this case by a more specific regulation. This behavior is another instance in which global gene expression analysis suggests heretofore unsuspected complexity in the metabolic regulatory network.
Differential Regulation of Phosphate Pathway
The genes in cluster 8, induced specifically during phosphate-limited batch growth, are enriched for annotations to vacuole fusion and phosphate metabolism. This small but tightly regulated set of genes are all involved in phosphate metabolism and show much larger steady-state differences in the chemostat comparison than induction in the batch experiment. This suggests that they are already partially derepressed in the exponential phase of batch growth.
Phosphate sensing and transcription of the phosphate regulon is accomplished through differential phosphorylation of the Pho4p transcription factor. Recent results suggest that partial phosphorylation of Pho4 results in the differential induction of different subsets of the phosphate regulon (Springer et al., 2003). Our results are consistent with the hypothesis that in the batch experiment, the culture is transitioning from an intermediate to low phosphate concentration, whereas the chemostat comparisons are between cells grown in low and high phosphate conditions. In particular, PHO5 shows five- to sevenfold induction between the phosphate-limited and other chemostats, and a three- to fourfold induction between the chemostat and exponential growth, whereas PHO84 shows six- to sevenfold induction in the chemostat, but no detectable induction during the batch time course. PHO5 is sensitive to both dephosphorylation events and is thus induced by both transitions, whereas PHO84 is fully induced by the first event and shows no further induction during the batch time course. Inspection of the data for other phosphate regulated genes suggest that there may be a spectrum of sensitivity to the dephosphorylation events and that PHO5 and PHO84 represent extremes between equal response to both, and extreme sensitivity to the first.
Specific Repression of Ribosomal Proteins under Phosphate and Uracil Limitations
Clusters 11 and 12 together comprise the stress-repressed genes. However, cluster 12 showed dramatically stronger repression under phosphate and uracil limitation than under leucine and sulfate limitations. By contrast, cluster 11 is barely repressed at all in the phosphate and uracil limitations, relative to the other transcripts in the cell. Coclustering of this data set with the original stress data set revealed that no other stress elicited specific repression of cluster 12.
The interpretation of this can be sought in the membership of the clusters. Cluster 11 consists of genes involved in translation elongation and initiation, transcription from PolI promoters, and rRNA processing, all of which are supporting functions required for ribosome maturation and protein synthesis. In contrast, cluster 12 consists almost entirely of structural components of the ribosome; indeed, 123 of the 126 genes in this cluster are either ribosomal components named RPS, RPL, or RPP, or ORFs that overlap genes encoding such components.
Although much of the phosphate in the cell is inorganic, by far the majority of the organic phosphate is incorporated into RNA (Lange and Heijnen, 2001), and the majority of the RNA is in the form of ribosomal rRNA (Warner, 1999). Thus, under both low phosphate and low uracil conditions, production of ribosomal rRNA should be strongly impacted, and we suppose that there may be direct feedback to the ribosomal proteins, which must be made in stoichiometric amounts. Thus, under phosphate and uracil limitation, the cell is forced to specifically restrict ribosome production, more so than under other stresses. This strong decrease in such an abundant class of transcripts may perturb the overall normalization of the transcriptome so that less dramatic down-regulation, which may be obvious under other limitations, may become masked. It remains to be seen how this regulation intersects with the expanding number of pathways known to regulate transcription of ribosomal genes and rRNA (Zhao et al., 2003).
Comparison with Earlier Studies
There have been at least two previously published studies comparing transcriptional state between steady-state chemostats limited for different nutrients with the hope of discovering genes specific to the particular limitation (Boer et al., 2003; Wu et al., 2004). Coclustering of this data set with the other two reveals that the phosphate- and sulfate-specific clusters in this data set are also up-regulated in the phosphate- and sulfate-limited steady-state cultures from the other studies. Wu et al. (2004) also have produced profiles of fully starved cultures; comparison with the steady-state cultures confirms our observation that the stress response is activated strongly in the fully starved cultures relative to the steady state.
Implications for Functional Genomics
The steady-state environment of the chemostat may be a particularly useful system for uncovering gene function in the postgenome sequence era. Genome-wide analysis techniques provide a panoramic view of cellular processes. This inclusiveness renders them susceptible to artifacts that might be missed by a more targeted assay. An example of this is given by Hayes et al. (2002) who found that addition of a drug affects the growth rate of S. cerevisiae and that this growth rate effect seems to dominate the observed transcriptional changes. This can be avoided in the chemostat system, where in addition to the growth rate, the use of sensors and feedback allow the experimenter to specify other parameters, such as pH. Furthermore, there is evidence that gene expression profiles produced from steady-state chemostat cultures may be more reproducible than those produced from exponential phase batch cultures (Piper et al., 2002).
The results suggest that cells grown in the chemostat are in a state in which genes specific to the nutrient limitation have been induced, but in which genes encoding ribosomal proteins are still actively transcribed and the stress response has not been induced. Expression of pathways that are not related to the limitation are generally comparable between chemostat and exponential batch. This suggests that any changes observed in these pathways are likely the specific result of the perturbation and should apply equally well in chemostat and batch.
Acknowledgments
We thank Maitreya Dunham for assistance in determining the phosphate-limiting growth medium and Gavin Sherlock for assistance in constructing the companion Web site. This work was supported by a grant from the National Institutes of Health (GM-46406). A.J.S. was supported by a National Defense Science and Engineering Graduate Fellowship and the Stanford Genome Training Program (training grant NIH 5 T32 HG00044).
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E04-04-0306. Article and publication date are available at www.molbiolcell.org/cgi/doi/10.1091/mbc.E04-04-0306.
Online version of this article contains supporting material. Online version is available at www.molbiolcell.org.
References
- Ashburner, M., et al. (2000). Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 25, 25-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boer, V.M., de Winde, J.H., Pronk, J.T., and Piper, M.D. (2003). The genome-wide transcriptional responses of Saccharomyces cerevisiae grown on glucose in aerobic chemostat cultures limited for carbon, nitrogen, phosphorus, or sulfur. J. Biol. Chem. 278, 3265-3274. [DOI] [PubMed] [Google Scholar]
- Carroll, A.S., and O'Shea, E.K. (2002). Pho85 and signaling environmental conditions. Trends Biochem. Sci. 27, 87-93. [DOI] [PubMed] [Google Scholar]
- Chen, P.S., Toribara, T., and Warner, H. (1956). Microdetermination of phosphorus. Anal. Chem. 28, 1756-1758. [Google Scholar]
- Denis-Duphil, M. (1989). Pyrimidine biosynthesis in Saccharomyces cerevisiae: the ura2 cluster gene, its multifunctional enzyme product, and other structural or regulatory genes involved in de novo UMP synthesis. Biochem. Cell Biol. 67, 612-631. [DOI] [PubMed] [Google Scholar]
- DeRisi, J.L., Iyer, V.R., and Brown, P.O. (1997). Exploring the metabolic and genetic control of gene expression on a genomic scale. Science 278, 680-686. [DOI] [PubMed] [Google Scholar]
- Dobbin, K., and Simon, R. (2002). Comparison of microarray designs for class comparison and class discovery. Bioinformatics 18, 1438-1445. [DOI] [PubMed] [Google Scholar]
- Dwight, S.S., et al. (2002). Saccharomyces Genome Database (SGD) provides secondary gene annotation using the Gene Ontology (GO). Nucleic Acids Res. 30, 69-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen, M.B., Spellman, P.T., Brown, P.O., and Botstein, D. (1998). Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. USA 95, 14863-14868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Entian, K.D., and Koetter, P. (1998). Yeast mutant and plasmid collections. In: Yeast Gene Analysis, vol. 26, ed. A.J.P. Brown and M.F. Tuite, London: Academic Press, United Kingdom, 431-449. [Google Scholar]
- Gasch, A.P., Spellman, P.T., Kao, C.M., Carmel-Harel, O., Eisen, M.B., Storz, G., Botstein, D., and Brown, P.O. (2000). Genomic expression programs in the response of yeast cells to environmental changes. Mol. Biol. Cell 11, 4241-4257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasch, A.P., and Werner-Washburne, M. (2002). The genomics of yeast responses to environmental stress and starvation. Funct. Integr. Genomics 2, 181-192. [DOI] [PubMed] [Google Scholar]
- Gollub, J., et al. (2003). The Stanford Microarray Database: data access and quality assessment tools. Nucleic Acids Res. 31, 94-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris, M.A., et al. (2004). The Gene Ontology (GO) database and informatics resource. Nucleic Acids Res. Database issue 32, D258-261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartwell, L.H. (1974). Saccharomyces cerevisiae cell cycle. Bacteriol. Rev. 38, 164-198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes, A., Zhang, N., Wu, J., Butler, P.R., Hauser, N.C., Hoheisel, J.D., Lim, F.L., Sharrocks, A.D., and Oliver, S.G. (2002). Hybridization array technology coupled with chemostat culture: tools to interrogate gene expression in Saccharomyces cerevisiae. Methods 26, 281-290. [DOI] [PubMed] [Google Scholar]
- Kal, A.J., et al. (1999). Dynamics of gene expression revealed by comparison of serial analysis of gene expression transcript profiles from yeast grown on two different carbon sources. Mol. Biol. Cell 10, 1859-1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohlhaw, G.B. (2003). Leucine biosynthesis in fungi: entering metabolism through the back door. Microbiol. Mol. Biol. Rev. 67, 1-15, table of contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange, H.C., and Heijnen, J.J. (2001). Statistical reconciliation of the elemental and molecular biomass composition of Saccharomyces cerevisiae. Biotechnol. Bioeng. 75, 334-344. [DOI] [PubMed] [Google Scholar]
- Liu, X., and Clarke, N.D. (2002). Rationalization of gene regulation by a eukaryotic transcription factor: calculation of regulatory region occupancy from predicted binding affinities. J. Mol. Biol. 323, 1-8. [DOI] [PubMed] [Google Scholar]
- Losson, R., Fuchs, R.P., and Lacroute, F. (1985). Yeast promoters URA1 and URA3. Examples of positive control. J. Mol. Biol. 185, 65-81. [DOI] [PubMed] [Google Scholar]
- Lyons, T.J., Gasch, A.P., Gaither, L.A., Botstein, D., Brown, P.O., and Eide, D.J. (2000). Genome-wide characterization of the Zap1p zinc-responsive regulon in yeast. Proc. Natl. Acad. Sci. USA 97, 7957-7962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monod, J. (1942). Recherches sure la croissance des cultures bacteriennes. Paris: Herman & Cie.
- Monod, J. (1950). La Technique de culture continue. Theorie et applications. Ann. Inst. Pasteur 79, 390-410. [Google Scholar]
- Novick, A., and Szilard, L. (1950). Description of the chemostat. Science 715-716. [DOI] [PubMed]
- Oestreicher, N., and Scazzocchio, C. (1995). A single amino acid change in a pathway-specific transcription factor results in differing degrees of constitutivity, hyperinducibility and derepression of several structural genes. J. Mol. Biol. 249, 693-699. [DOI] [PubMed] [Google Scholar]
- Ogawa, N., DeRisi, J., and Brown, P.O. (2000). New components of a system for phosphate accumulation and polyphosphate metabolism in Saccharomyces cerevisiae revealed by genomic expression analysis. Mol. Biol. Cell 11, 4309-4321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oppenheim, D.S., and Yanofsky, C. (1980). Translational coupling during expression of the tryptophan operon of Escherichia coli. Genetics 95, 785-795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patton, E.E., Peyraud, C., Rouillon, A., Surdin-Kerjan, Y., Tyers, M., and Thomas, D. (2000). SCF(Met30)-mediated control of the transcriptional activator Met4 is required for the G(1)-S transition. EMBO J. 19, 1613-1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persson, B.L., Petersson, J., Fristedt, U., Weinander, R., Berhe, A., and Patti-son, J. (1999). Phosphate permeases of Saccharomyces cerevisiae: structure, function and regulation. Biochim. Biophys. Acta 1422, 255-272. [DOI] [PubMed] [Google Scholar]
- Piper, M.D., Daran-Lapujade, P., Bro, C., Regenberg, B., Knudsen, S., Nielsen, J., and Pronk, J.T. (2002). Reproducibility of oligonucleotide microarray transcriptome analyses. An interlaboratory comparison using chemostat cultures of Saccharomyces cerevisiae. J. Biol. Chem. 277, 37001-37008. [DOI] [PubMed] [Google Scholar]
- Reichard, P. (2002). Ribonucleotide reductases: the evolution of allosteric regulation. Arch. Biochem. Biophys. 397, 149-155. [DOI] [PubMed] [Google Scholar]
- Schneider, K.R., Smith, R.L., and O'Shea, E.K. (1994). Phosphate-regulated inactivation of the kinase PHO80-PHO85 by the CDK inhibitor PHO81. Science 266, 122-126. [DOI] [PubMed] [Google Scholar]
- Spellman, P.T., Sherlock, G., Zhang, M.Q., Iyer, V.R., Anders, K., Eisen, M.B., Brown, P.O., Botstein, D., and Futcher, B. (1998). Comprehensive identification of cell cycle-regulated genes of the yeast Saccharomyces cerevisiae by microarray hybridization. Mol. Biol. Cell 9, 3273-3297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Springer, M., Wykoff, D.D., Miller, N., and O'Shea, E.K. (2003). Partially phosphorylated Pho4 activates transcription of a subset of phosphate-responsive genes. PLoS Biol. 1, E28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ter Linde, J.J., Liang, H., Davis, R.W., Steensma, H.Y., van Dijken, J.P., and Pronk, J.T. (1999). Genome-wide transcriptional analysis of aerobic and anaerobic chemostat cultures of Saccharomyces cerevisiae. J. Bacteriol. 181, 7409-7413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas, D., Rothstein, R., Rosenberg, N., and Surdin-Kerjan, Y. (1988). SAM2 encodes the second methionine S-adenosyl transferase in Saccharomyces cerevisiae: physiology and regulation of both enzymes. Mol. Cell. Biol. 8, 5132-5139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas, D., and Surdin-Kerjan, Y. (1997). Metabolism of sulfur amino acids in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 61, 503-532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Townsend, J.P. (2003). Multifactorial experimental design and the transitivity of ratios with spotted DNA microarrays. BMC Genomics 4, 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dijken, J.P., et al. (2000). An interlaboratory comparison of physiological and genetic properties of four Saccharomyces cerevisiae strains. Enzyme Microbiol. Technol. 26, 706-714. [DOI] [PubMed] [Google Scholar]
- Verma, R., Annan, R.S., Huddleston, M.J., Carr, S.A., Reynard, G., and Deshaies, R.J. (1997). Phosphorylation of Sic1p by G1 Cdk required for its degradation and entry into S phase. Science 278, 455-460. [DOI] [PubMed] [Google Scholar]
- Volland, C., Urban-Grimal, D., Geraud, G., and Haguenauer-Tsapis, R. (1994). Endocytosis and degradation of the yeast uracil permease under adverse conditions. J. Biol. Chem. 269, 9833-9841. [PubMed] [Google Scholar]
- Wang, D., Zheng, F., Holmberg, S., and Kohlhaw, G.B. (1999). Yeast transcriptional regulator Leu3p. Self-masking, specificity of masking, and evidence for regulation by the intracellular level of Leu3p. J. Biol. Chem. 274, 19017-19024. [DOI] [PubMed] [Google Scholar]
- Warner, J.R. (1999). The economics of ribosome biosynthesis in yeast. Trends Biochem. Sci. 24, 437-440. [DOI] [PubMed] [Google Scholar]
- Wu, J., Zhang, N., Hayes, A., Panoutsopoulou, K., and Oliver, S.G. (2004). Global analysis of nutrient control of gene expression in Saccharomyces cerevisiae during growth and starvation. Proc. Natl. Acad. Sci. USA 101, 3148-3153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, Y.H., and Speed, T. (2002). Design issues for cDNA microarray experiments. Nat. Rev. Genet. 3, 579-588. [DOI] [PubMed] [Google Scholar]
- Zhao, Y., Sohn, J.H., and Warner, J.R. (2003). Autoregulation in the biosynthesis of ribosomes. Mol. Cell. Biol. 23, 699-707. [DOI] [PMC free article] [PubMed] [Google Scholar]