Abstract
Cystic fibrosis is the most widespread hereditary disease among the white population caused by different mutations of the apical membrane ATP-binding cassette transporter cystic fibrosis transmembrane conductance regulator (CFTR). Its most common mutation, ΔF508, leads to nearly complete degradation via endoplasmic reticulum-associated degradation (ERAD). Elucidation of the quality control and degradation mechanisms might give rise to new therapeutic approaches to cure this disease. In the yeast Saccharomyces cerevisiae, a variety of components of the protein quality control and degradation system have been identified. Nearly all of these components share homology with mammalian counterparts. We therefore used yeast mutants defective in the ERAD system to identify new components that are involved in human CFTR quality control and degradation. We show the role of the lectin Htm1p in the degradation process of CFTR. Complementation of the HTM1 deficiency in yeast cells by the mammalian orthologue EDEM underlines the necessity of this lectin for CFTR degradation and highlights the similarity of quality control and ERAD in yeast and mammals. Furthermore, degradation of CFTR requires the ubiquitin protein ligases Der3p/Hrd1p and Doa10p as well as the cytosolic trimeric Cdc48p-Ufd1p-Npl4p complex. These proteins also were found to be necessary for ERAD of a mutated yeast “relative” of CFTR, Pdr5*p.
INTRODUCTION
The endoplasmic reticulum (ER) is responsible for folding, modification, and delivery of secretory proteins to their site of action (Glick, 2002; Haigh and Johnson, 2002). It contains a highly active protein quality control system (QC), which scans the folding process of secretory proteins and retains those species that are unable to fold (Ellgaard and Helenius, 2003). They are eliminated by a process called ER-associated degradation (ERAD) via the ubiquitin proteasome system (Kostova and Wolf, 2003). Malfunction of these processes is the cause of many diseases (Kostova and Wolf, 2002; Rutishauser and Spiess, 2002). One of the most common hereditary diseases among the white population that is directly linked to QC and ERAD is cystic fibrosis (Kerem et al., 1989; Riordan et al., 1989). Here, a mutated ATP-binding cassette (ABC) transporter and chloride channel of the apical membrane, the cystic fibrosis transmembrane conductance regulator (CFTR) is recognized by the QC as being malfolded and degraded via the ubiquitin-proteasome system. The most frequent mutation of CFTR that causes this effect is the deletion of a phenylalanine at position 508 in the first nucleotide binding domain (ΔF508). Interestingly even wild-type CFTR is very unstable and shows a degradation rate of ∼70% (Ward and Kopito, 1994; Jensen et al., 1995). It is important to note that ΔF508 CFTR can still function as a chloride channel (Dalemans et al., 1991) but due to QC and ERAD, it fails to be transported to the apical plasma membrane. This could be overcome by lower temperature (Denning et al., 1992), high glycerol (Sato et al., 1996), or 4-phenylbutyrate (Rubenstein and Zeitlin, 2000). Because none of these treatments are of therapeutic value, other means have to be found to promote transport of mature ΔF508 CFTR to the plasma membrane. Elucidation of the components of QC and ERAD that are responsible for the elimination of ΔF508 CFTR is an important step in this direction. They may become targets for specific therapeutic manipulations to lead mutated but active CFTR to the cell surface. Previous studies have shown that the lectin calnexin interacts with CFTR in the ER lumen (Pind et al., 1994). Furthermore the bidirectional Sec61 translocon of the ER membrane was suggested to contribute to QC and ERAD (Bebök et al., 1998) as well as the cytosolic chaperones heat-shock protein Hsp70/Hsc70, Hsp90, and the cochaperone CHIP (Yang et al., 1993; Loo et al., 1998; Meacham et al., 1999; Meacham et al., 2001).
Cellular QC and ERAD are highly conserved mechanisms from yeast to human (Kostova and Wolf, 2002, 2003; Ellgaard and Helenius, 2003). The ready availability of yeast mutants defective in QC and ERAD makes this organism a preferred model for investigation of these processes. Indeed, the expression of CFTR in Saccharomyces cerevisiae has proven that the yeast components of QC and ERAD recognize this protein and degrade it via the proteasome in a ubiquitin-dependent manner (Kiser et al., 2001; Zhang et al., 2001). These studies have uncovered the yeast ubiquitin-conjugating enzymes Ubc6p and Ubc7p as well as the cytosolic Hsp70 as necessary components for the degradation of CFTR. Influence of the ubiquitin protein ligase Der3p/Hrd1p in CFTR degradation is somewhat controversial (Kiser et al., 2001; Zhang et al., 2001). Recently, the mammalian counterparts of yeast Ubc6p, Ubc7p, and Der3p/Hrd1p have been identified (Fang et al., 2001; Tiwari and Weissman, 2001; Lenk et al., 2002). Thus, S. cerevisiae proves to be an excellent model organism to further investigate the components of the QC and ERAD, which are required for the degradation of CFTR.
Mutants defective in newly discovered components of these processes have enabled testing of their involvement in CFTR degradation. These new experiments reveal that the ubiquitin protein ligases Der3p/Hrd1p and Doa10p seem to have a synergistic effect on the degradation of the CFTR protein. Furthermore, the cytosolic trimeric Cdc48-Ufd1-Npl4 complex is found to be crucially required for proteasomal elimination of the protein. In addition the QC and degradation process of CFTR is considerably disturbed in a mutant defective in the ER lumenal lectin Htm1p. Most interestingly this defect can be complemented by the expression of the mammalian EDEM protein, showing that Htm1p and EDEM are functional homologues with respect to CFTR degradation.
MATERIALS AND METHODS
Construction and Growth Conditions of Yeast Strains
Standard procedures for genetic and molecular biological techniques and for media were carried out as described in Sambrook et al. (1989), Guthrie and Fink (1991), and Ausubel et al. (1992).
All S. cerevisiae strains used are listed in Table 1. Cells were grown at 25°C. Strains YAG 153/YAG 154 (CDC48/cdc48-1) also were grown at the semipermissive temperature of 23°C in standard media. For CPY* (carboxy-peptidase yscY*) expression, cells were grown at 30°C, and starvation media for pulse-chase experiments contained only 0.1% glucose to induce CPY* expression.
Table 1.
Yeast strains used in this study
| Strain | Genotype | Reference |
|---|---|---|
| W303-1B | Matα ade2-1 ura3-1 trp1-1 leu2-3, 112 his3-11, 15 can1-100 | Chiang and Schekman, 1991 |
| W303 WTCF | W303-1B [PDR5-human WT CFTR-URA3] | This study |
| W303 ΔCF | W303-1B [PDR5-human ΔF508 CFTR-URA3] | This study |
| YWO 500 | MATa PDR1-3 ura3-1 trp1-1 leu2-3, 112 his 3-11, 15 | Mahé et al., 1996 |
| YAG 123 | YWO500 [PDR5-human WT CFTR-URA3] | This study |
| YAG 124 | YWO500 [PDR5-human ΔF508 CFTR-URA3] | This study |
| GKY 356 | PDR1-3-LEU2 leu2 ura3 [PDR5-human WT CFTR-URA3] | Kiser et al., 2001 |
| GKY 360 | PDR1-3-HIS3 his3 leu2 ura3 ubc6Δ/ubc7Δ::LEU2 [PDR5-human WT CFTR-URA3] | Kiser et al., 2001 |
| YAG 129 | YWO500 Δhtm1::kanMX [PDR5-human WT CFTR-URA3] | This study |
| YAG 171 | YWO500 Δhtm1::kanMX [PDR5-human WT CFTR-URA3/p413 TEF] | This study |
| YAG 172 | YWO500 Δhtm1::kanMX [PDR5-human WT CFTR-URA3/p413 TEF-mEDEM-HA] | This study |
| YAG 174 | YWO500 Δhtm1::kanMX [PDR5-human WT CFTR-URA3/p423 TEF-mEDEM-HA] | This study |
| YAG 198 | YWO500 Δhtm1::kanMX [HA-pdr5-26-LEU2/p423 TEF] | This study |
| YAG 199 | YWO500 Δhtm1::kanMX [HA-pdr5-26-LEU2/p423 TEF-mEDEM-HA] | This study |
| YAG 182 | W303 Δprc1::LEU2 Δhtm1::kanMX [pRS316-prc1-1-URA3/p423 TEF] | This study |
| YAG 183 | W303 Δprc1::LEU2 Δhtm1::kanMX [pRS316-prc1-1-URA3/p423 TEF-mEDEM-HA] | This study |
| YAG 60 | MATα PDR1-3-HIS3 ura3-52 trp1-1 leu2-3 leu2-112 his3Δ200 lys2-801 [PDR5-human WT CFTR-URA3] | This study |
| YAG 61 | MATα PDR1-3-HIS3 ura3-52 trp1-1 leu2-3 leu2-112 his3Δ200 lys2-801 Δhrd1::LEU2 [PDR5-human WT CFTR-URA3] | This study |
| YAG 58 | MATα PDR1-3-LEU2 ura3-52 trp1-1 leu2-3 leu2-112 his3Δ200 lys2-801 [PDR5-human WT CFTR-URA3] | This study |
| YAG 59 | MATα PDR1-3-LEU2 ura3-52 trp1-1 leu2-3 leu2-112 his3Δ200 lys2-801 Δdoa10::HIS3 [PDR5-human WT CFTR-URA3] | This study |
| YAG 97 | MATα PDR1-3-TRP1 ura3-52 trp1-1 leu2-3 leu2-112 his3Δ200 lys2-801 Δdoa10::HIS3 Δhrd1::LEU2 [PDR5-human WT CFTR-URA3] | This study |
| YAG 153 | PDR1-3-LEU2 ade2 ura3 trp1-1 leu2-3 his3-11 [PDR5-human CFTR-URA3] | This study |
| YAG 154 | PDR1-3-LEU2 ade2 ura3 trp1-1 leu2-3 lys2-801 cdc48-1 [PDR5-human CFTR-URA3] | This study |
| YAG 184 | ade2 ura3 trp1-1 leu2-3 his3-11 Δpdr5::TRP1 [HA-pdr5-26-LEU2] | This study |
| YAG 185 | ade2 ura3 trp1-1 leu2-3 lys2-801 cdc48-1 Δpdr5::TRP1 [HA-pdr5-26-LEU2] | This study |
| YAG 192 | MATa PDR1-3-LEU2 ade1-100 ura3-52 leu2-3 leu2-112 his4-519 Δpdr5::hisG [PDR5-human WT CFTR-URA3] | This study |
| YAG 193 | MATa PDR1-3-LEU2 ade1-100 ura3-52 leu2-3 leu2-112 his4-519 ufd1-1 Δpdr5::hisG [PDR5-human WT CFTR-URA3] | This study |
| YAG 188 | MATa ade1-100 ura3-52 leu2-3 leu2-112 his4-519 Δpdr5::hisG [HA-pdr5-26-LEU2] | This study |
| YAG 189 | MATa ade1-100 ura3-52 leu2-3 leu2-112 his4-519 ufd1-1 Δpdr5::hisG [HA-pdr5-26-LEU2] | This study |
| YAG 208 | YWO500 [PDR5-human WT CFTR-URA3/p413 TEF] | This study |
| YAG 209 | YWO500 [HA-pdr5-26-LEU2/p423 TEF] | This study |
| YAG 210 | W303 Δprc1::LEU2 [pRS316-prc1-1-URA3/p423 TEF] | This study |
Yeast strains were modified either by transformation with integrative or episomal plasmids, mating, and tetrad dissection or by deletion via PCR cassettes and homologous recombination (Wach et al., 1994; Longtine et al., 1998). The cdc48-1 strain DBY 2030 originally created by K. U. Fröhlich (Fröhlich et al., 1991) was crossed with strain W303-1B to obtain strains with suitable auxotrophy markers. HTM1 was deleted in YWO 500 (Mahé et al., 1996) by one step gene deletion (Wach et al., 1994; Longtine et al., 1998). The kanamycin deletion cassette with homologous sequences for the HTM1 gene was amplified by PCR with the primers 5′ HTM1 (5′ gcggtaggataatctccttgacgg 3′) and 3′ HTM1 (5′ gcgaccagcgaaatggatgagctg 3′). Gene deletion was proven by kanamycin resistance and polymerase chain reaction (PCR) with primers Δ htm1 frw (5′ ggcatctagagtgatgacg 3′) and 3′kan (5′ gaggcataaattccgtcagcc 3′) or 5′kan (5′ cgagtcggaatcgcagaccg 3′) and Δ htm1 rws (5′ tttacccctaggaatatcg 3′).
All strains used for CFTR expression were modified with the PDR1-3 mutation. The PDR1-3 plasmids with LEU and HIS marker were from Kiser et al. (2001). In addition the PDR1-3 sequence was cloned into pRS 304 (TRP) (Sikorski and Hieter, 1989) via ApaI/SacI to obtain pAGC 28. Plasmids are summarized in Table 2. All three plasmids are of integrative nature. Before transformation pGK 122 and pAGC 28 were linearized with NsiI; pGK 121 with HpaI. Insertions were verified by prototroph selection and increased resistance against cycloheximide (CHX) on YPD plates (CHX resistance assay). Expression of the test proteins was done as described below.
Table 2.
Plasmids
| Plasmid | Insert | Reference |
|---|---|---|
| pGK118 | PDR5-human WT CFTR-URA3 | Kiser et al., 2001 |
| pGK119 | PDR5-human ΔF508 CFTR-URA3 | Kiser et al., 2001 |
| pGK 121 | PDR1-3-HIS3 | Kiser et al., 2001 |
| pGK122 | PDR1-3-LEU2 | Kiser et al., 2001 |
| pAGC 28 | PDR1-3-TRP1 | This study |
| pAGC 10 | Δpdr5::hisG-URA3-hisG | Mahe et al., 1996 |
| pAGC 19 | Δpdr5::TRP1 | Bissinger and Kuchler, 1994 |
| pAGC 52 | HA-pdr5-26-LEU2 | Bissinger and Kuchler, 1994 |
| p413 TEF | Mumberg et al., 1995 | |
| p423 TEF | Mumberg et al., 1995 | |
| pAGC 54 | p413 TEF mouse EDEM-HA | This study |
| pAGC 55 | p423 TEF mouse EDEM-HA | This study |
| pCT 40 | pRS316-prc1-1-HA3-URA3 | C. Taxis (Institut für Biochemie, Stuttgart, Germany) |
CFTR Expression
CFTR was expressed from a low copy yeast expression vector with PDR5 promoter and terminator (Kiser et al., 2001). To increase the expression level of the protein a PDR1-3 gain of function mutation was introduced by homologues recombination with an integrative PDR1-3 plasmid (Kiser et al., 2001; this study). CFTR expression was proven by Western blotting and/or immunoprecipitation (IP). The antibody M3A7 (Kartner et al., 1991) was used for Western and immunoprecipitation analysis.
PDR5 Deletion and Pdr5*p Expression
Wild-type PDR5 was deleted by homologous recombination with plasmids either carrying a TRP or a hisG-URA3-hisG fragment flanked by PDR5 sequence stretches (Bissinger and Kuchler 1994; Mahé et al., 1996) as described in Rose et al. (1990). HA-PDR5* (Bissinger and Kuchler, 1994; Egner et al., 1998) was introduced on a centromeric plasmid by standard transformation (Soni et al., 1993). Deletion and insertion were analyzed by Western blotting by using antibodies either against Pdr5p (Plemper et al., 1998) for wild-type protein or against the N-terminal hemagglutinin (HA)-tag of the introduced HA-Pdr5*p (Babco, Berkeley, CA) and by CHX resistance assays.
CPY* Expression
A Δ htm1/Δ prc1 yeast strain was transformed with pCT 40 to obtain YAG 183. Expression of CPY* in pulse-chase experiments was induced with labeling media containing only 0.1% glucose. CPY antibodies were obtained from Molecular Probes (Eugene, OR).
CHX Resistance Assay
To show higher resistance of yeast cells carrying the PDR1-3 gain of function mutation, CHX was added to YPD agar at concentrations of 0, 0.1, 0.25, 0.5, and 1 μg/ml. The range was adjusted according to results of Carvajal et al. (1997). Cells were grown to an A600 of 1, washed in TE (50 mM Tris, pH 7.6, 1 mM EDTA), and 1 A600 of cells was taken up in 1 ml of TE. The cells were diluted 1:100 and 1:1000 in TE, and 3 μl of both dilutions were spotted onto YPD-CHX plates and incubated at 30°C for 2 to 3 d until colonies were clearly visible. Cells that showed stronger resistance than the respective wild-type were picked for further tests and experiments.
Deletion of PDR5 causes higher susceptibility to CHX. Δpdr5 deletion strains stop growth under CHX treatment earlier than correlated PDR5 wild-type strains. Used CHX concentrations were 0, 0.1, 0.25, and 0.5 μg/ml according to Bissinger and Kuchler (1994).
Western Blotting of CFTR and Pdr5*p
Cells were grown in selective media at 25°C to an A600 of 1.2. ER membranes were isolated from 3 A600 of cells per sample. Cells were resuspended in 100 μl of lysis buffer (50 mM Tris, pH 7.6, 1 mM EDTA) with protease inhibitor mix and broken with glass beads. Debris was removed by centrifugation at 3000 rpm, and membranes were sedimented at 13,000 rpm for 15 min at 4°C. Proteins were taken up in 50 μl of urea sample buffer (8 M urea, 5% SDS, 200 mM Tris-HCl, pH 6.8, 0.1 mM EDTA, 1.5% dithiothreitol [DTT], 0.03% bromphenol blue), incubated for 30 min at room temperature (CFTR) or 15 min at 34°C (Pdr5*p), and 20 μl of each sample were subjected to SDS-PAGE. Proteins were transferred to nitrocellulose membranes in a semidry blot at 90 mA for 90 min. Antibodies for detection were M3A7 or C-terminal CFTR antibody from R&D Systems (Minneapolis, MN) for CFTR and α N-1 (Plemper et al., 1998) or α HA (Babco) for Pdr5p and HA-Pdr5*p, respectively. Proteins were visualized by enhanced chemiluminescence (ECL) detection (Amersham Biosciences UK, Little Chalfont, Buckinghamshire, United Kingdom) according to the manufacturer's instructions.
Pulse-Chase
For pulse-chase experiments of CFTR and Pdr5*p, cells were grown as for Western blotting and thereafter starved in sulfur-free media for 1-2 h. Subsequently, they were labeled for 30 min to 1 h with 20 μCi/A600 35S-TRANS Label (ICN Biomedicals, Aurora, CA) in media lacking any nitrogen source and chased in standard CM media for 90-120 min. For CFTR and Pdr5*p, 3 A600 of cells per time point were lysed in breaking buffer (50 mM Tris, pH 7.6, 1 mM EDTA, protease inhibitors) by using glass beads by vortexing six times for 1 min each, followed by cooling down on ice for 1 min. For CFTR, 4 times concentrated cell solubilization buffer (4% SDS, 600 mM NaCl, 50 mM Tris, pH 7.5, protease inhibitors) was added to yield a concentration of 1. After addition of IP buffer (50 mM Tris, pH 7.5, 150 mM NaCl, 1% DOC, 1% Triton X-100, protease inhibitors), insoluble material was centrifuged down at 13,000 rpm. The supernatant was incubated overnight with 2 μl of M3A7 antibody at 4°C. M3A7 was precipitated with A/G Agarose (Oncogene Science, Boston, MA) and washed with RIPA (IP + 0.1% SDS). Protein was taken up in 2 times concentrated SDS sample buffer (125 mM Tris, pH 6.8, 20% glycerol, 2% SDS, 2% β-mercaptoethanol, 100 mM DTT, bromphenol blue) for 30 min at room temperature and transferred to 7% SDS-PAGE. Radioactive signals were quantified from dried gels with the PhosphorImager detection system (Molecular Dynamics, Sunnyvale, CA). The mean values of three to four experiments were taken to create the graphs.
For Pdr5*p, cell debris was removed after lysis, following the protocol from Plemper et al. (1998). Sedimented membranes were resuspended in IP buffer with 1% SDS and incubated on ice for 20 min. After addition of IP buffer without SDS containing 1.5 μl of α HA, immunoprecipitation was carried out as with CFTR.
CPY* pulse-chase was carried out as described by Hiller et al. (1996) and Plemper et al. (1999). Briefly cells were grown logarithmically in selective media and labeled for 20 min with [35S]methionine (Amersham Biosciences UK). After addition of an excess of nonradioactive methionine, samples were taken at the indicated time points, followed by cell disruption, immunoprecipitation, and SDS-PAGE. Quantification was carried out as described for CFTR.
Expression of Mouse EDEM (mEDEM)
Mouse EDEM cDNA carrying the sequence for a single HA-tag at the C-terminal end of the protein (Hosokawa et al., 2001) was cloned into the low-copy and high-copy yeast expression vectors p413TEF and p423TEF (Mumberg et al., 1995) via SpeI/XhoI. The plasmids were transformed into yeast cells by using standard methods (Soni et al., 1993). Expression of mEDEM was verified by immunoblotting of yeast membrane fractions with HA antibodies (Babco).
RESULTS
CFTR Expression Is Temperature Dependent in Yeast Cells
CFTR is a polytopic transmembrane protein that is cotranslationally inserted into the ER membrane. Translocation into the membrane as well as its retrotranslocation into the cytosol for proteasomal degradation is thought to be mediated by the Sec61 complex in both mammalian and yeast cells (Bebök et al., 1998; Kiser et al., 2001). In mammalian cells, 70% of wild-type and 100% of ΔF508 CFTR is degraded by the proteasome after polyubiquitination (Jensen et al., 1995; Ward et al., 1995). In yeast, even the wild-type form of CFTR is degraded to nearly 100% (Zhang et al., 2001). There is no different degradation level of wild-type and mutant ΔF508 CFTR detectable in yeast (Zhang et al., 2001). But, as in mammalian cells, CFTR is polyubiquitinated and degraded by the proteasome in yeast cells. Mutations in proteasomal genes or inhibition of the proteasome interferes with CFTR degradation (Kiser et al., 2001; Zhang et al., 2001).
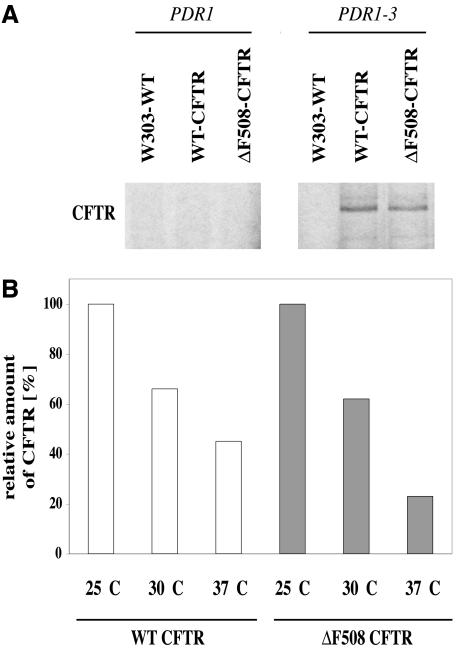
For expression of human CFTR in yeast, we used the expression vector introduced by Kiser et al. (2001). The sequence of wild-type and ΔF508 CFTR was placed onto a low-copy yeast expression vector with promoter and terminator region of the yeast ABC transporter Pdr5p (Balzi et al., 1994; Bissinger and Kuchler, 1994). As with Pdr5p levels, the introduction of a PDR1-3 gain of function mutation (Carvajal et al., 1997) increased the levels of CFTR protein to detectable amounts (Figure 1A). Because Pdr5p is reported to be induced at lower temperatures (Lashkari et al., 1997) and CFTR can partially be stabilized at lower temperatures in cell culture (Denning et al., 1992), we expressed the CFTR protein in cells growing in liquid media at 25, 30, and 37°C. As expected, the level of detectable CFTR protein increased with decreasing temperature (Figure 1B). The half-life of CFTR was dependent on the strain background and varied between 40 and 70 min. As proof for the validity of our analysis system, CFTR degradation was tested in a Δubc6/Δubc7 strain (GKY 360; Kiser et al., 2001) deficient in the two ubiquitin-conjugating enzymes Ubc6p and Ubc7p. Just as observed previously, there was a substantial decrease in the rate of CFTR degradation (unpublished data). This confirms that the expression and detection systems used function reliably.
Figure 1.
Expression of human CFTR in yeast. A total of 3 A600 per time point were labeled with 20 μCi/A600 TRANS-Label. After cell lysis, protein was immunoprecipitated and detected as described under Materials and Methods. (A) Detectable expression levels of CFTR are dependent on the PDR1-3 mutation. Wild-type CFTR (WT-CFTR) and mutated protein (ΔF508-CFTR) were detected. The strain W303-1B was taken as a control without CFTR. (B) Yeast cells grown at different temperatures show decreasing CFTR expression with increasing temperatures. The highest amount of labeled CFTR was set to 100% for WT and ΔF508 CFTR independently.
Quality Control of CFTR Requires the Lectin Htm1/EDEM
In mammalian cells, CFTR is known to be glycosylated at two positions, N894 and N900, in the ER lumen (Riordan et al., 1989). The protein acquires these oligosaccharide chains in yeast as well (Kiser et al., 2001). In mammalian and yeast cells, it has been shown that core glycosylation of carbohydrate-containing proteins plays an important role in the protein quality control process. Trimming of the three glucose residues by glucosidases I and II as well as cleavage of the mannose-9 residue by α-mannosidase I of the GlcNAc2Man9Glc3 structure are critical for the recognition of a malfolded protein and the timing of its removal (Helenius and Aebi, 2001; Kostova and Wolf, 2003). In addition lectins of the ER lumen have been found to be essential components of the folding and recognition process. Specifically, calnexin has been shown to be a partner in the folding process of glycosylated proteins (Molinari et al., 2003; Oda et al., 2003). Also, CFTR has been found to interact with calnexin in mammalian cells (Pind et al., 1994). In contrast to mammalian calnexin, no such function has been reported for Cne1p the yeast orthologue (Knop et al., 1996). Also, degradation of CFTR was not influenced by the absence of Cne1p in yeast (Zhang et al., 2001).
Recently, a newly identified yeast lectin Htm1p as well as its mammalian counterpart EDEM have been reported to trigger glycoprotein degradation (Hosokawa et al., 2001; Jakob et al., 2001). The soluble glycoprotein CPY* as well as the membrane substrates Pdr5*p and Stt3-7p but not unglycosylated Sec61-2p were shown to be dependent on Htm1p function in yeast to different extents. Overexpression of mammalian EDEM accelerated degradation of a well known mammalian model substrate, mutated α 1-anti-trypsin (Hosokawa et al., 2001).
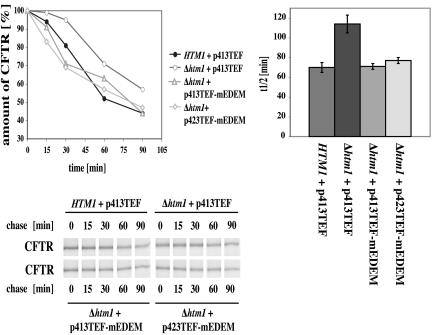
We tested the influence of a HTM1 deletion on CFTR degradation in yeast. As can be seen in Figure 2, absence of Htm1p significantly slows the degradation rate of CFTR, indicating that this lectin can indeed recognize a heterologous glycosylated protein. Confirmation of the involvement of the lectin Htm1p/EDEM in degradation of CFTR would be provided by complementation of the delayed degradation process by expression of mammalian EDEM in yeast. We therefore cloned mEDEM containing a C-terminal HA-tag (Hosokawa et al., 2001) into the low- and high-copy yeast expression vectors p413TEF and p423TEF (Mumberg et al., 1995). As can be seen, the protein could be heterologously expressed in yeast and analogous to mammalian cells, it was found in the membrane fraction (Figure 3). We tested the influence of mEDEM on CFTR degradation in the yeast Δ htm1 deletion strain YAG 174. Pulse-chase analysis revealed that mEDEM expressed from the low-copy vector p413TEF could indeed restore the Δhtm1 defect in CFTR degradation to wild-type levels. To see whether higher levels of mEDEM would further accelerate CFTR degradation in yeast, we overexpressed mEDEM from the high-copy expression vector p423TEF-mEDEM in the Δhtm1 deletion strain. Overexpression of mEDEM restored the Δhtm1 defect to the same extent as found with the p413TEF-mEDEM low copy construct (Figure 2).
Figure 2.
Influence of the yeast lectin Htm1p and its mammalian orthologue mEDEM on the degradation of CFTR. The wild-type strain HTM1 and the Δhtm1 deletion strain carried the empty yeast expression vector p413TEF as a control. mEDEM was expressed in Δhtm1 strains either from the low-copy expression plasmid p413TEF-mEDEM or the high-copy expression plasmid p423TEF-mEDEM. The Δhtm1 deletion delays the degradation rate of CFTR. Expression of mEDEM either from the low- or high-copy plasmid in the Δhtm1 strain restores the degradation rate of CFTR to wild-type levels. A total of 3 A600 cells per time point were labeled with 20 μCi/A600 TRANS-Label. Pulse-chases and quantification were done as described under Materials and Methods. Samples were separated by 7% SDS-PAGE and quantified with PhosphorImager cassettes. The protein quantity of time point t = 0 min was set to 100%. The graphs show average degradation rates of three experiments in wild-type (HTM1 + p413TEF) and deletion strains (Δ htm1) without (+ p413TEF) or with mEDEM (+ p413TEF-mEDEM/+ 423TEF-mEDEM) in a 90-min chase and average half life of CFTR protein. Error bars indicate the variations of the different experiments. As long as error bars do not overlap we can speak of statistically significant differences between the tested strains. The autoradiographs of 7% SDS gels show typical results.
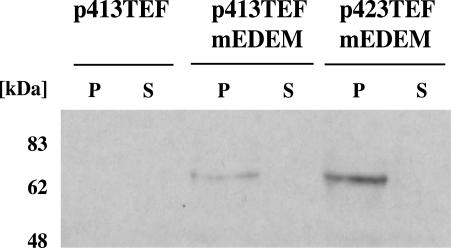
Figure 3.
Expression and membrane localization of mouse EDEM in yeast cells. mEDEM-HA was cloned into high (p423TEF) and low-copy (p413TEF) yeast expression vectors and expressed in a Δ htm1 PDR1-3 yeast strain carrying human CFTR. Cells were grown to an A600 of ∼1.2 and lysed with glass beads. Membranes were separated from supernatant and solubilized in buffer containing SDS and Triton X-100. Samples were separated by SDS-PAGE and blotted on nitrocellulose. Immunodetection was carried out with HA antibodies (Babco) and ECL (Amersham Biosciences UK). The empty vector p413 TEF was taken as a control. The protein was solely detectable in pellet fractions (P). The supernatant (S) was clear.
EDEM and Yeast ERAD Substrates
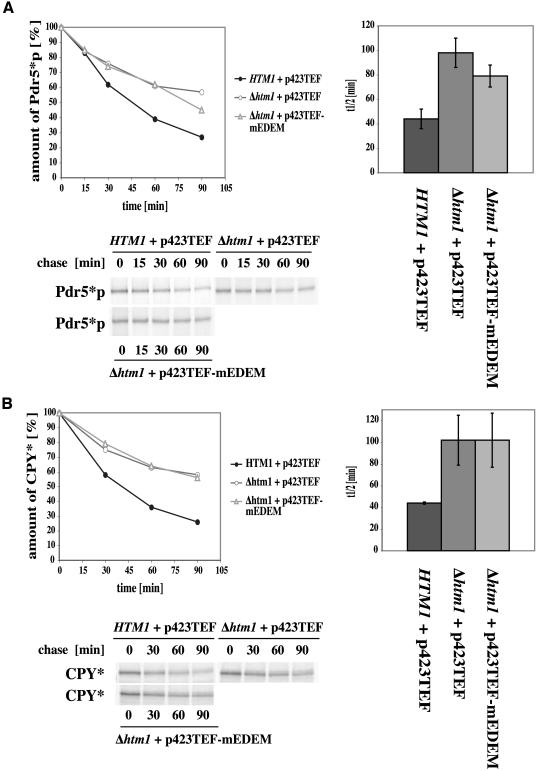
We also tested the influence of mEDEM on the yeast ERAD substrates CPY* (prc1-1) and Pdr5*p (pdr5-26) in Δhtm1 strains. Our results revealed no significant complementing capacity of mEDEM in case of the two yeast substrates (Figure 4, A and B). Degradation of the polytopic CFTR yeast orthologue Pdr5*p seems to be somewhat stimulated but is barely significant. No significant influence of mEDEM could be detected on the degradation of mutated soluble CPY*. Thus, mammalian EDEM seems to be specific for the mammalian protein CFTR.
Figure 4.
Influence of mEDEM on glycosylated yeast ERAD substrates in Δhtm1 strains. Pulse-chase analysis was done as described in text. The graphs show average kinetics of three experiments of 90-min pulse-chases and average half-life in wild-type (HTM1 + p423TEF) and Δ htm1 deletion strains with (p423TEF-mEDEM) or without (p423TEF) mouse EDEM. Error bars indicate the variations of the different experiments. The autoradiographs show typical results. (A) Pdr5*p degradation shows a barely significant increase in a strain with mEDEM expression. (B) mEDEM does not show a significant increase of CPY* degradation.
E3 Enzymes in CFTR Ubiquitination
Polyubiquitination is a prerequisite for the degradation of ERAD substrates via the proteasome (Kostova and Wolf, 2003). This has been shown to be the case for CFTR as well (Jensen et al., 1995; Ward et al., 1995). In yeast cells, the E2 enzymes Ubc6p and Ubc7p were found to be required for polyubiquitination of CFTR (Kiser et al., 2001; Zhang et al., 2001). Recent data uncovered the involvement of the mammalian orthologue of yeast Ubc6p in the polyubiquitination process (Lenk et al., 2002). The final step in ubiquitin coupling is mediated or directly carried out by ubiquitin-conjugating E3 enzymes. These enzymes are thought to have high substrate specificity. It has been shown that in mammalian cells CHIP (Meacham et al., 2001) and SCF-FBX2 (Yoshida et al., 2002) can function as E3 enzymes, which trigger degradation of CFTR to some extent. However, neither of these proteins seems to exhibit a very strong effect. This points to the fact that polyubiquitination of CFTR seems to be mediated by different E3s with overlapping specificity. Studies in yeast led to controversial results concerning the involvement of the well established E3 Der3p/Hrd1p in the degradation process (Kiser et al., 2001; Zhang et al., 2001). Recently, another ubiquitin-protein ligase, Doa10p, was found (Swanson et al., 2001).
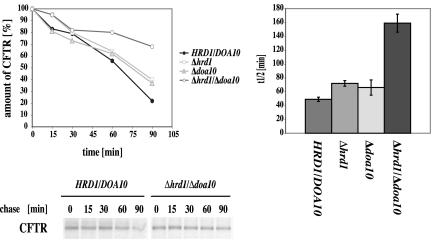
We carried out pulse-chase experiments by using mutant cells defective in the two E3 enzymes Der3p/Hrd1p and Doa10p and followed the degradation of CFTR. As can be seen in Figure 5, deletion of Der3p/Hrd1p or Doa10p leads to a slight but significant slow down of CFTR degradation in both single mutants. The double deletion of DER3/HRD1 and DOA10 results in a strong stabilization of CFTR, indicating considerable overlapping specificity of these two ubiquitinconjugating enzymes with respect to CFTR recognition.
Figure 5.
E3 ubiquitin ligases Der3p/Hrd1p and Doa10p are involved in CFTR degradation. Pulse-chase analysis was carried out as described in text. The mean values of four experiments were taken to create the kinetics and half-life graphs of CFTR protein for each, wild-type (HRD1/DOA10) and deletion strains Δ hrd1, Δ doa10 and the double deletion strain Δ hrd1/Δ doa10. The autoradiographs show typical results.
Degradation of CFTR Is Dependent on a Functional Cdc48 Complex
Very little is yet known how CFTR is delivered from the ER membrane to its final destination of degradation, the proteasome. Does the proteasome directly bind to the ER surface and tear the CFTR protein out of the membrane by virtue of its 19S cap ATPases or is the protein extracted via a chaperone-mediated mechanism? It has previously been shown that a trimeric complex in the cytosol consisting of the AAA ATPase Cdc48, Ufd1, and Npl4 is responsible for liberation of some malfolded proteins from the ER membrane (Bays et al., 2001; Ye et al., 2001; Braun et al., 2002; Jarosch et al., 2002; Rabinovich et al., 2002; Chevalier and Johnson, 2003; Taxis et al., 2003).
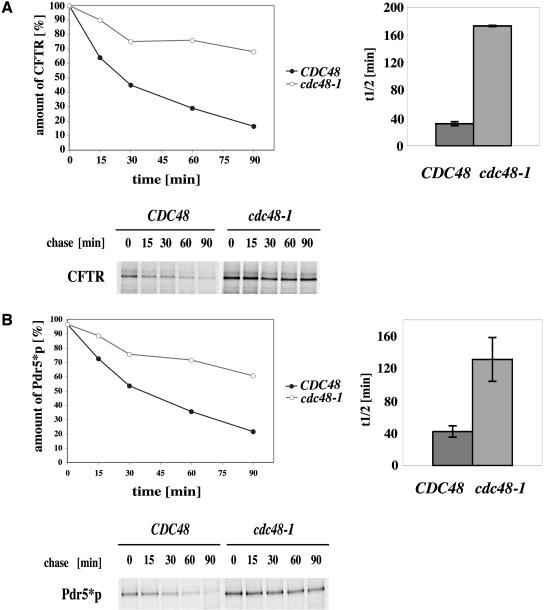
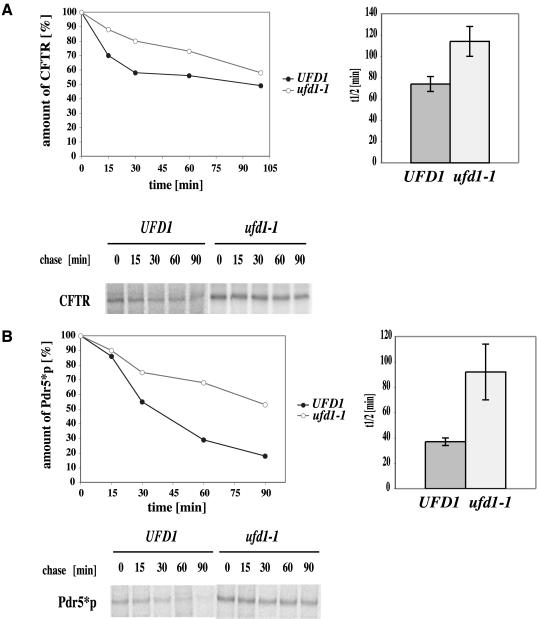
We therefore tested the involvement of this complex in the degradation of the polytopic CFTR protein as well as the mutated yeast orthologue Pdr5*p by using cdc48-1 (YAG 154) and ufd1-1 (YAG 193) mutants. Wild-type and the cold-sensitive cdc48-1 mutant strains (YAG 153; YAG 154) were grown at the permissive temperature of 25°C and the semipermissive temperature of 23°C. As can be seen, the mutant strain showed a large reduction in the rate of degradation even at the permissive temperature, which was further increased at the semipermissive temperature of 23°C (Figure 6A). The half-life of CFTR was increased from 32 min in wild-type cells to 173 min in the mutant strain. In case of Pdr5*p, the half-life increased from 42 min for wild-type and 131 min for cdc48-1 cells (Figure 6B). Because the rate of degradation is comparable between both proteins, yeast does not seem to distinguish between endogenous and heterologously expressed proteins in the use of the AAA ATPase Cdc48. This result was verified in pulse-chase experiments by using mutants defective in a second component of the trimeric Cdc48 complex, Ufd1p. As expected in a ufd1-1 mutant, CFTR as well as Pdr5*p degradation was inhibited (Figure 7, A and B). The influence of the ufd1-1 mutation on CFTR turnover was somewhat smaller than that of the cdc48-1 mutation but still significant, whereas the effect of the ufd1-1 mutation on Pdr5*p degradation was comparable with that of the strain carrying the cdc48-1 mutation. According to the proposed model which assigns the function of liberation of malfolded proteins from the ER to the trimeric Cdc48 complex (Bays et al., 2001; Ye et al., 2001; Braun et al., 2002; Jarosch et al., 2002; Rabinovich et al., 2002), this function seems to hold true also for Pdr5*p and the heterologously expressed CFTR protein.
Figure 6.
AAA ATPase Cdc48 is involved in both yeast and mammalian polytopic transmembrane protein degradation. With shift to starvation media, cells were grown at the semipermissive temperature of 23°C. A total of 3 A600 per time point were labeled with 20 μCi/A600 TRANS-Label. Pulse-chase experiments and CFTR quantification were done as described in text. (A) CFTR is strongly stabilized in a cdc48-1 mutant. The graphs show average kinetics of four experiments of degradation in the 90-min pulse-chase and average half-life of CFTR in wild-type (CDC48) and mutant strains (cdc48-1). Error bars indicate the variations of the different experiments. The autoradiograph shows a typical result. (B) same as in A but for the mutated yeast ABC transporter protein Pdr5*p, which is also strongly stabilized.
Figure 7.
An ufd1-1 mutant strain shows significant slow down of CFTR and Pdr5*p degradation. Strains were treated as described in text. (A) The graphs show average kinetics of three experiments of degradation in the 100-min pulse-chase and average half-life of CFTR in wild-type (UFD1) and mutant strains (ufd1-1). Error bars indicate the variations of the different experiments. The autoradiograph shows a typical result. (B) Same as in A but for the mutated yeast ABC transporter protein Pdr5*p.
DISCUSSION
Our studies revealed new insights into the degradation mechanism of human CFTR in yeast. By this, they strengthen the previous findings that CFTR QC and ERAD can be examined reliably in the bakers yeast S. cerevisiae. Using yeast mutants, we studied the function of newly detected components of the QC and ERAD processes on the degradation of the polytopic CFTR chloride channel.
The determining factors that decide the fate of a protein between maturation and degradation are still mostly unknown. One new factor is the yeast lectin Htm1p and its mammalian orthologue EDEM. These lectins have been shown to accelerate specifically degradation of both soluble and membrane glycoproteins in yeast and mammalian cells, respectively. As shown here, deletion of yeast Htm1p has a significant effect on degradation of human CFTR. Most interestingly, this defect can be complemented by expression of mammalian EDEM in HTM1-deleted yeast cells. Both, low- and high-copy expression of mEDEM in a Δhtm1 deletion strain restored the degradation of CFTR to HTM1 wild-type levels. This shows that this pair of orthologous lectins is indeed an important component in the recognition and degradation process of CFTR. Unlike the findings in mammalian cells (Molinari et al., 2003), overexpression of mEDEM in yeast cells did not show accelerated degradation levels compared with mEDEM expressed from a centromeric expression vector. This seems to indicate that the concentration of mEDEM expressed from the single copy vector is sufficient to bind all the available carbohydrate processed CFTR species generated in yeast.
In the case of the yeast orthologue Pdr5*p, mEDEM restored the HTM1 deficiency to a considerably lesser and barely significant extent. No effect was visible on soluble yeast CPY*. The lacking restoration ability of EDEM on the degradation of Pdr5*p and CPY* in Htm1p-deficient cells may be due to different binding properties of the glycosylated yeast proteins to EDEM. At present, the precise reason for this functional specificity of mEDEM toward mammalian CFTR remains unknown.
Proteasomal degradation of malfolded proteins requires polyubiquitination via ubiquitin conjugating enzymes (E2s) and ubiquitin protein ligases (E3s). The latter class of enzymes is thought to provide the specificity of the degradation process. Two potential E3s from different classes have been reported to partially influence CFTR degradation in mammalian cells (Meacham et al., 2001; Yoshida et al., 2002). Neither had strong effects on the degradation process. It thus seems that E3s have broader overlapping function in the degradation of malfolded proteins requiring different enzymes to work in cooperation or independently. A well-known E3 from yeast is Der3p/Hrd1p. In yeast, it showed different levels of impact on the degradation of different substrates (Bordallo et al., 1998; Plemper et al., 1998; Wilhovsky et al., 2000), including CFTR (Kiser et al., 2001; Zhang et al., 2001). Concerning CFTR expressed in yeast, the data were controversial. Therefore, we tested the effect of a DER3/HRD1 deletion on CFTR degradation. We could detect a small but significant delay of degradation of the CFTR protein. The deletion of another newly discovered yeast E3, Doa10p, led to a similar stabilization of CFTR. A double deletion of DER3/HRD1 and DOA10 strongly increased the stabilization of CFTR. This shows a cooperation of Hrd1p and Doa10p in CFTR ubiquitination. Because no complete stabilization of CFTR is found in the absence of the two enzymes tested, additional E3s involved in CFTR ubiquitination can be expected to be found. Alternatively, the contribution of additional nonproteasomal proteolytic systems cannot be excluded completely.
It had been speculated that the proteasomal ATPases might be involved in pulling CFTR from the ER and deliver it to the proteasome (Zhang et al., 2001). Such an extracting function also has been suggested for a hybrid membrane protein by Mayer et al. (1998). Proteasomes have indeed been reported to be associated with the cytoplasmic surface of the ER membrane (Rivett et al., 1992; Palmer et al., 1996). However, if there were a direct extraction of proteins from the ER membrane by the proteasome one would not expect ubiquitinated proteins in the cytosol with blocked proteasomal activity as it had been shown for CPY* (Jarosch et al., 2002) in yeast cells. Cytosolic accumulation of the protein also had been shown for MHC class I heavy chains in mammalian cells transfected with cytomegalovirus under these conditions (Wiertz et al., 1996a, b). Furthermore, the formation of aggresomes in mammalian cells containing CFTR that can occur when proteasome activity is blocked (Johnston et al., 1998; Wigley et al., 1999; Chen et al., 2000) would implicate an additional pulling force at the cytosolic surface of the ER membrane. Recently, the function of the cytosolic trimeric AAA ATPase complex Cdc48-Ufd1-Npl4 had been shown to be responsible for liberation of a variety of ERAD substrates from the ER membrane (Bays et al., 2001; Ye et al., 2001; Braun et al., 2002; Jarosch et al., 2002; Rabinovich et al., 2002; Chevalier and Johnson, 2003; Taxis et al., 2003). Therefore, we tested the involvement of Cdc48p in the degradation process of CFTR by using respective mutants. Defective Cdc48p exhibited a dramatic effect on CFTR degradation, which was comparable to the degradation block of the orthologous yeast transmembrane transporter Pdr5*p. Also, a mutation in the Ufd1p component of the complex led to decreased degradation kinetics of CFTR and Pdr5*p. In summary, the function of the trimeric Cdc48 complex in liberation of the CFTR protein from the ER is evident and supports a chaperone-mediated extraction from the ER membrane and delivery to the proteasome rather than a direct extraction by the ATPases of the 19S cap of the proteasome.
Finally, the data presented in this study demonstrate that the glycosylation of CFTR contributes to its quality control and degradation process. Ubiquitination by E3 enzymes does not exhibit the previously proposed substrate specificity but can be mediated by different enzymes probably with varying affinities. Before its degradation, CFTR is extracted from the ER membrane by involvement of the cytosolic trimeric Cdc48 complex rather than by the proteasome directly. The similarity of different QC and ERAD factors between yeast and mammalian cells is further supported by the direct complementation of yeast Htm1p by mEDEM and confirms the utility of yeast for further elucidation of bio-synthetic processing of CFTR and related membrane proteins.
Acknowledgments
We thank M. Gentzsch for strains, plasmids, antibodies, and discussion; and K. U. Fröhlich and M. Hochstrasser for strains. We also acknowledge N. Hosokawa and K. Nagata for the kind gift of cDNA of mouse EDEM. This work has been supported by the Mukoviszidose e.V., the Deutsche Forschungsgemeinschaft, and the Bundesministerium für Bildung und Forschung (Deutsch-Israelische Projektkooperation) (all Bonn, Germany).
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E04-01-0024. Article and publication date are available at www.molbiolcell.org/cgi/doi/10.1091/mbc.E04-01-0024.
References
- Ausubel, F.M., Kingston, R.E., Seidman, F.G., Struhl, K., Moore, D.D., Brent, R., and Smith, F.A. (1992). Current Protocols in Molecular Biology, New York: Greene.
- Balzi, E., Wang, M., Leterme, S., Van Dyck, L., and Goffeau, A. (1994). PDR5, a novel yeast multidrug resistance conferring transporter controlled by the transcription regulator PDR1*. J. Biol. Chem. 269, 2206-2214. [PubMed] [Google Scholar]
- Bays, N.W., Wilhovsky, S.K., Goradia, A., Hodgkiss-Harlow, K., and Hampton, R.Y. (2001). HRD4/NPL4 is required for the proteasomal processing of ubiquitinated ER proteins. Mol. Biol. Cell 12, 4114-4128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun, S., Matuschewski, A., Rape, M., Thoms, S., and Jentsch, S. (2002). Role of the ubiquitin-selective CDC48 (UFD1/NPL4) chaperone (segregase) in ERAD of OLE1 and other substrates. EMBO J. 21, 615-621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bebök, Z., Mazzochi, C., King, S.A., Hong, J.S., and Sorscher, E.J. (1998). The mechanism underlying cystic fibrosis transmembrane conductance regulator transport from the endoplasmic reticulum to the proteasome includes Sec61beta and a cytosolic, deglycosylated intermediary. J. Biol. Chem. 273, 29873-29878. [DOI] [PubMed] [Google Scholar]
- Bissinger, P.H., and Kuchler, K. (1994). Molecular cloning and expression of the Saccharomyces cerevisiae STS1 gene product. J. Biol. Chem. 269, 4180-4186. [PubMed] [Google Scholar]
- Bordallo, J., Plemper, R.K., Finger, A., and Wolf, D.H. (1998). Der3p/Hrd1p is required for endoplasmatic reticulum-associated degradation of misfolded lumenal and integral membrane proteins. Mol. Biol. Cell 9, 209-222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvajal, E., van den Hazel, H.B., Cybularz-Kolaczkowska, A., Balzi, E., and Goffeau, A. (1997). Molecular and phenotypic characterization of yeast PDR1 mutants that show hyperactive transcription of various ABC multidrug transporter genes. Mol. Gen. Genet. 256, 406-415. [DOI] [PubMed] [Google Scholar]
- Chen, E.Y., Bartlett, M.C., and Clarke, D.M. (2000). Cystic fibrosis transmembrane conductance regulator has an altered structure when its maturation is inhibited. Biochemistry 39, 3797-3803. [DOI] [PubMed] [Google Scholar]
- Chevalier, M.S., and Johnson, D.C. (2003). Human cytomegalovirus US3 chimeras containing US2 cytosolic residues acquire major histocompatibility class I and II protein degradation properties. J. Virol. 77, 4731-4738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang, H.L., and Schekman, R. (1991). Regulated import and degradation of a cytosolic protein in the yeast vacuole. Nature 350, 313-318. [DOI] [PubMed] [Google Scholar]
- Dalemans, W., et al. (1991). Altered chloride ion channel kinetics associated with the Δ F508 cystic fibrosis mutation. Nature 354, 526-528. [DOI] [PubMed] [Google Scholar]
- Denning, G.M., Anderson, M.P., Amara, J.F., Marshall, J., Smith, A.E., and Welsh, M.J. (1992). Processing of mutant cystic fibrosis transmembrane conductance regulator is temperature-sensitive. Nature 358, 761-764. [DOI] [PubMed] [Google Scholar]
- Egner, R., Rosenthal, F.E., Kralli, A., Sanglard, D., and Kuchler, K. (1998). Genetic separation of FK506 susceptibility and drug transport in the yeast Pdr5 ATP-binding cassette multidrug resistance transporter. Mol. Biol. Cell 9, 523-543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellgaard, L., and Helenius, A. (2003). Quality control in the endoplasmic reticulum. Nat. Rev. Mol. Cell. Biol. 4, 181-191. [DOI] [PubMed] [Google Scholar]
- Fang, S., Ferrone, M., Yang, C., Jensen, J.P., Tiwari, S., and Weissman, A.M. (2001). The tumor autocrine motility factor receptor, gp78, is an ubiquitin protein ligase implicated in the degradation from the endoplasmic reticulum. Proc. Natl. Acad. Sci. USA 98, 14422-14427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fröhlich, K.U., Fries, H.W., Rudiger, M., Erdmann, R., Botstein, D., and Mecke, D. (1991). Yeast cell cycle protein CDC48p shows full-length homology to the mammalian protein VCP and is a member of a protein family involved in secretion, peroxisome formation, and gene expression. J. Cell Biol. 114, 443-453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glick, B.S. (2002). The secretory pathway. In: Protein Targeting, Transport and Translocation, ed. R.E. Dalbey and G. and von Heijne, London: Academic Press, 358-376.
- Guthrie, C., and Fink, G.R. (1991) Guide to Yeast Genetics and Molecular Biology, Vol. 194, New York: Academic Press.
- Haigh, N.G. and Johnson, A.E. (2002) Protein sorting at the endoplasmic reticulum. In: Protein Targeting, Transport and Translocation, ed. R.E. Dalbey and G. von Heijne, London: Academic Press, 74-106.
- Helenius, A., and Aebi, M. (2001). Intracellular functions of N-linked glycans. Science 291, 2364-2369. [DOI] [PubMed] [Google Scholar]
- Hiller, M.M., Finger, A., Schweiger, M., and Wolf, D.H. (1996). ER degradation of a misfolded luminal protein by the cytosolic ubiquitin-proteasome pathway. Science 273, 1725-1728. [DOI] [PubMed] [Google Scholar]
- Hosokawa, N., Wada, I., Hasegawa, K., Yorihuzi, T., Tremblay, L.O., Herscovics, A., and Nagata, K. (2001). A novel ER α-mannosidase-like protein accelerates ER-associated degradation. EMBO Rep. 2, 415-422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakob, C.A., Bodmer, D., Spirig, U., Bättig, P., Marcil, A., Dignard, D., Bergeron, J.J.M., Thomas, D.Y., and Aebi, M. (2001). Htm1p, a mannosidase-like protein, is involved in glycoprotein degradation in yeast. EMBO Rep. 2, 423-430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarosch, E., Taxis, C., Volkwein, C., Bordallo, J., Finley, D., Wolf, D.H., and Sommer, T. (2002). Protein dislocation from the ER requires polyubiquitination and the AAA-ATPase Cdc48. Nat. Cell Biol. 4, 134-138. [DOI] [PubMed] [Google Scholar]
- Jensen, T.J., Loo, M.A., Pind, S., Williams, D.B., Goldberg, A.L., and Riordan, J.R. (1995). Multiple proteolytic systems, including the proteasome, contribute to CFTR processing. Cell 83, 129-135. [DOI] [PubMed] [Google Scholar]
- Johnston, J.A., Ward, C.L., and Kopito, R.R. (1998). Aggresomes: a cellular response to misfolded proteins. J. Cell Biol. 143, 1883-1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kartner, N., et al. (1991). Expression of the cystic fibrosis gene in non-epithelial invertebrate cells produces a regulated anion conductance. Cell 64, 681-691. [DOI] [PubMed] [Google Scholar]
- Kerem, B.-S., Rommens, J.M., Buchanan, J.A., Markiewicz, D., Cox, T.K., Chakravarti, A., Buchwald, M., and Tsui, L.-C. (1989). Identification of the cystic fibrosis gene, gene analysis. Science 245, 1073-1080. [DOI] [PubMed] [Google Scholar]
- Kiser, G.L., Gentzsch, M., Kloser, A.K., Balzi, E., Wolf, D.H., Goffeau, A., and Riordan, J.R. (2001). Expression and degradation of the cystic fibrosis transmembrane conductance regulator in Saccharomyces cerevisiae. Arch. Biochem. Biophys. 390, 195-205. [DOI] [PubMed] [Google Scholar]
- Knop, M., Hauser, N., and Wolf, D.H. (1996). N-Glycosylation affects endoplasmic reticulum degradation of a mutated derivative of carboxypeptidase yscY in yeast. Yeast 12, 1229-1238. [DOI] [PubMed] [Google Scholar]
- Kostova, Z. and Wolf, D.H. (2002) Protein quality control in the export pathway, the endoplasmic reticulum and its cytoplasmic proteasome connection. In: Protein Targeting, Transport and Translocation, ed. R.E. Dalbey and G. von Heijne, London: Academic Press, 180-213.
- Kostova, Z., and Wolf, D.H. (2003). For whom the bell tolls, protein quality control of the endoplasmic reticulum and the ubiquitin-proteasome connection. EMBO J. 22, 2309-2317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lashkari, D.A., DeRisi, J.L. McCusker, J.H., Namath, A.F., Gentile, C., Hwang, S.Y., Brown, P.O., and Davis, R.W. (1997). Yeast microarrays for genome wide parallel genetic and gene expression analysis. Proc. Natl. Acad. Sci. USA 94, 13057-13062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenk, U., Yu, H., Walter, J., Gelman, M.S., Hartmann, E., Kopito, R.R., and Sommer, T. (2002). A role for mammalian Ubc6 homologues in ER-associated protein degradation. J. Cell Sci. 115, 3007-3014. [DOI] [PubMed] [Google Scholar]
- Longtine, M.S., McKenzie, A., III, Demarini, D.J., Shah, N.G., Wach, A., Brachat, A., Philippsen, P., and Pringle, J.R. (1998). Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14, 953-961. [DOI] [PubMed] [Google Scholar]
- Loo, M.A., Jensen, T.J., Cui, L., Hou, Y., Chang, X.B., and Riordan, J.R. (1998). Perturbation of Hsp90 interaction with nascent CFTR prevents its maturation and accelerates its degradation by the proteasome. EMBO J. 17, 6879-6887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahé, Y., Lemoine, Y., and Kuchler, K. (1996). The ATP binding cassette transporters Pdr5 and Snq2 of Saccharomyces cerevisiae can mediate transport of steroids in vivo. J. Biol. Chem. 271, 25167-25172. [DOI] [PubMed] [Google Scholar]
- Mayer, T.U., Braun, T., and Jentsch, S. (1998). Role of the proteasome in membrane extraction of a short-lived ER transmembrane protein. EMBO J. 17, 3251-3257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meacham, G.C., Lu, Z., King, S., Sorscher, E., Tousson, A., and Cyr, D.M. (1999). The Hdj-2/Hsc70 chaperone pair facilitates early steps in CFTR biogenesis. EMBO J. 18, 1492-1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meacham, G.C., Patterson, C., Zhang, W., Younger, J.M., and Cyr, D.M. (2001). The Hsc70 co-chaperone CHIP targets immature CFTR for proteasomal degradation. Nat. Cell Biol. 3, 100-105. [DOI] [PubMed] [Google Scholar]
- Molinari, M., Calanca, V., Galli, C., Lucca, P., and Paganetti, P. (2003). Role of EDEM in the release of misfolded glycoproteins from the calnexin cycle. Science 299, 1397-1400. [DOI] [PubMed] [Google Scholar]
- Mumberg, D., Müller, R., and Funk, M. (1995). Yeast vectors for the controlled expression of heterologous proteins in different genetic backgrounds. Gene 156, 119-122. [DOI] [PubMed] [Google Scholar]
- Oda, Y., Hosokawa, N., Wada, I., and Nagata, K. (2003). EDEM as an acceptor of terminally misfolded glycoproteins released from calnexin. Science 299, 1394-1397. [DOI] [PubMed] [Google Scholar]
- Palmer, A., Rivett, A.J., Thomson, S., Hendil, K.B., Butcher, G.W., Fuertes, G., and Knecht, E. (1996). Subpopulations of proteasomes in rat liver nuclei, microsomes and cytosol. Biochem. J. 316, 401-407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pind, S., Riordan, J.R., and Williams, D.B. (1994). Participation of the endoplasmic reticulum chaperone calnexin (p88, IP90) in the biogenesis of the cystic fibrosis transmembrane conductance regulator. J. Biol. Chem. 269, 12784-12788. [PubMed] [Google Scholar]
- Plemper, R.K., Egner, R., Kuchler, K., and Wolf, D.H. (1998). Endoplasmic reticulum degradation of a mutated ATP-binding cassette transporter Pdr5 proceeds in a concerted action of Sec61 and the proteasome. J. Biol. Chem. 273, 32848-32856. [DOI] [PubMed] [Google Scholar]
- Plemper, R.K., Bordallo, J., Deak, P.M., Taxis, C., Hitt, R., and Wolf, D.H. (1999). Genetic interactions of Hrd3p and Der3p/Hrd1p with Sec61p suggest a retro-translocation complex mediating protein transport for ER degradation. J. Cell Sci. 112, 4123-4134. [DOI] [PubMed] [Google Scholar]
- Rabinovich, E., Kerem, A., Fröhlich, K.U., Diamant, N., and Bar-Nun, S. (2002). AAA-ATPase p97/Cdc48p, a cytosolic chaperone required for endoplasmic reticulum-associated protein degradation. Mol. Cell. Biol. 22, 626-634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riordan, J.R., et al. (1989) Identification of the cystic fibrosis gene: cloning and characterization of complementary DNA. Science 245, 1066-1073. [DOI] [PubMed] [Google Scholar]
- Rivett, A.J., Palmer, A., and Knecht, E. (1992). Electron microscopic localization of the multicatalytic proteinase complex in rat liver and in cultured cells. J. Histochem. Cytochem. 40, 1165-1172. [DOI] [PubMed] [Google Scholar]
- Rose, M.D., Winston, F., and Hieter, P. (1990). Methods in Yeast Genetics, a Laboratory Course Manual, Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press.
- Rubenstein, R.C., and Zeitlin, P.L. (2000). Sodium 4-phenylbutyrate down-regulates Hsc 70, implications for intracellular trafficking of DeltaF508-CFTR. Am. J. Physiol. 278, C259-C267. [DOI] [PubMed] [Google Scholar]
- Rutishauser, J., and Spiess, M. (2002). Endoplasmic reticulum storage diseases, Swiss Med. Wkly. 132, 211-222. [DOI] [PubMed] [Google Scholar]
- Sambrook, J., Fritsch, E.F., and Maniatis, T. (1989). Molecular Cloning: A Laboratory Manual, 2nd ed., Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press.
- Sato, S., Ward, C.L., Krouse, M.E., Wine, J.J., and Kopito, R.R. (1996). Glycerol reverses the misfolding phenotype of the most common cystic fibrosis mutation. J. Biol. Chem. 271, 635-638. [DOI] [PubMed] [Google Scholar]
- Sikorski, R.S., and Hieter, P. (1989). A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122, 19-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soni, R., Carmichael, J.P., and Murray, J.A.H. (1993). Parameters affecting lithium acetate-mediated transformation of Saccharomyces cerevisiae and development of a rapid and simplified procedure. Curr. Genet. 24, 455-459. [DOI] [PubMed] [Google Scholar]
- Swanson, R., Locher, M., and Hochstrasser, M. (2001). A conserved ubiquitin ligase of the nuclear envelope/endoplasmic reticulum that functions in both ER-associated and Matα 2 repressor degradation. Genes Dev. 15, 2660-2674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taxis, C., Hitt, R., Park, S.H., Deak, P.M., Kostova, Z., and Wolf, D.H. (2003). Use of modular substrates demonstrates mechanistic diversity and reveals differences in chaperone requirement of ERAD. J. Biol. Chem. 278, 35903-35913. [DOI] [PubMed] [Google Scholar]
- Tiwari, S., and Weissman, A.M. (2001). Endoplasmic reticulum (ER)-associated degradation of T cell receptor subunits. Involvement of ER-associated ubiquitin-conjugating enzymes (E2s). J. Biol. Chem. 276, 16193-16200. [DOI] [PubMed] [Google Scholar]
- Wach, A., Brachat, A., Pöhlmann, R., and Philippsen, P. (1994). New heterologous modules for classical or PCR-based gene disruption in Saccharomyces cerevisiae. Yeast 10, 1793-1808. [DOI] [PubMed] [Google Scholar]
- Ward, C.L., and Kopito, R.R. (1994). Intracellular turnover of cystic fibrosis transmembrane conductance regulator. J. Biol. Chem. 269, 25710-25718. [PubMed] [Google Scholar]
- Ward, C.L., Omura, S., and Kopito, R.R. (1995). Degradation of CFTR by the ubiquitin-proteasome pathway. Cell 83, 121-127. [DOI] [PubMed] [Google Scholar]
- Wiertz, E.J., Tortorella, D., Bogyo, M., Yu, J., Mothes, W., Jones, T.R., Rapoport, T.A., Ploegh, H.L. (1996). Sec61-mediated transfer of a membrane protein from the endoplasmic reticulum to the proteasome for destruction. Nature 384, 432-438. [DOI] [PubMed] [Google Scholar]
- Wiertz, E.J., Jones, T.R., Sun, L., Bogyo, M., Geuze, H.J., Ploegh, H.L. (1996). The human cytomegalovirus US11 gene product dislocates MHC class I heavy chains from the endoplasmic reticulum to the cytosol. Cell 84, 769-779. [DOI] [PubMed] [Google Scholar]
- Wigley, W.C., Fabunmi, R.P., Lee, M.G., Marino, C.R., Muallem, S., DeM-artino, G.N., and Thomas, P.J. (1999). Dynamic association of proteasomal machinery with the centrosome. J. Cell Biol. 145, 481-490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilhovsky, S., Gardner, R., and Hampton, R. (2000). HRD gene dependence of endoplasmic reticulum-associated degradation. Mol. Biol. Cell 11, 1697-1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, Y., Janich, S., Cohn, J.A., and Wilson, J.M. (1993). The common variant of cystic fibrosis transmembrane conductance regulator is recognized by hsp70 and degraded in a pre-Golgi nonlysosomal compartment. Proc. Natl. Acad. Sci. USA 90, 9480-9484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye, Y., Meyer, H.H., and Rapoport, T.A. (2001). The AAA ATPase Cdc48/p97 and its partners transport proteins from the ER into the cytosol. Nature 414, 652-656. [DOI] [PubMed] [Google Scholar]
- Yoshida, Y., et al. (2002). E3 ubiquitin ligase that recognizes sugar chains. Nature 418, 438-442. [DOI] [PubMed] [Google Scholar]
- Zhang, Y., Nijbroek, G., Sullivan, M.L., McCracken, A.A., Watkins, S.C., Michaelis, S., and Brodsky, J.L. (2001). Hsp70 molecular chaperone facilitates endoplasmic reticulum-associated protein degradation of cystic fibrosis transmembrane conductance regulator in yeast. Mol. Biol. Cell 12, 1303-1314. [DOI] [PMC free article] [PubMed] [Google Scholar]