Abstract
We studied whether nucleoid exclusion contributes to the segregation and retention of Tsr chemoreceptor clusters at the cell poles. Using live time-lapse, single-cell microscopy measurements, we show that the single-cell spatial distributions of Tsr clusters have heterogeneities and asymmetries that are consistent with nucleoid exclusion and cannot be explained by the diffusion-and-capture mechanism supported by Tol-Pal complexes at the poles. Also, in cells subjected to ampicillin, which enhances relative nucleoid lengths, Tsr clusters locate relatively closer to the cell extremities, whereas in anucleated cells (deletion mutants for mukB), the Tsr clusters are closer to midcell. In addition, we find that the fraction of Tsr clusters at the poles is smaller in deletion mutants for Tol-Pal than in wild-type cells, although it is still larger than would be expected by chance. Also in deletion mutants, the distribution of Tsr clusters differs widely between cells with relatively small and large nucleoids, in a manner consistent with nucleoid exclusion from midcell. This comparison further showed that diffusion-and-capture by Tol-Pal complexes and nucleoid exclusion from the midcell have complementary effects. Subsequently, we subjected deletion mutants to suboptimal temperatures that are known to enhance cytoplasm viscosity, which hampers nucleoid exclusion effects. As the temperature was lowered, the fraction of clusters at the poles decreased linearly. Finally, a stochastic model including nucleoid exclusion at midcell and diffusion-and-capture due to Tol-Pal at the poles is shown to exhibit a cluster dynamics that is consistent with the empirical data. We conclude that nucleoid exclusion also contributes to the preference of Tsr clusters for polar localization.
Introduction
Escherichia coli chemoreceptor proteins perform multiple tasks, including assessing chemical gradients (1), thermosensing (2), and aerotaxis (3). These proteins are organized in trimer-of- dimers that form large clusters whose structure is further stabilized by the adaptor protein CheW and the histidine kinase CheA (1, 4, 5). The purpose of clustering is likely signal-processing enhancement of the receptor system (6, 7, 8, 9). The clustering process is robust, as receptors can assemble via their cytoplasmic domains even in the absence of some chemotaxis-associated proteins, such as CheW (10). Most studies agree that cluster formation occurs via an energy-free, self-assembly process known as stochastic nucleation (11, 12, 13, 14).
Chemotaxis-associated clusters preferentially locate at the cell poles (15, 16, 17), but the means by which this occurs remain unclear, given the lack of evidence for active transport mechanisms. Studies have suggested various mechanisms by which this may occur. For example, it has been suggested that the clusters first form at midcell and then attach to the cell membranes, and are dragged to the poles by cell growth after a few rounds of cell division (11, 12). It has also been suggested that the clusters diffuse freely in the cell membranes and that polar accumulation is caused by the curved shape of the poles and the ability of the clusters to match this curvature (7, 18).
Recent studies suggested that instead a diffusion-and-capture process (19) is responsible for the spatial distribution of this and several other polar proteins (20, 21, 22, 23). One study in particular (24) identified the trans-envelope Tol-Pal complex, a widely conserved component of the cell envelope of Gram-negative bacteria (25), as being responsible for capturing the clusters at the poles, since in deletion mutants for Tol-Pal this process is impaired. The existence of a diffusion-and-capture mechanism is further supported by the observation that a fairly constant fraction (∼7%) of Tsr proteins exhibit free diffusion over the entire cell surface at any given time (26).
Tsr, one of the methyl-accepting chemoreceptor proteins of the E. coli chemotaxis system (2), is a serine chemotaxis receptor protein that preferentially forms heterotrimeric membrane complexes at the poles. The mobility of Tsr labeled with fluorescent Venus proteins was recently investigated and found to be similar to that of the natural system (26). These proteins can diffuse over the entire cell surface but usually exhibit restricted diffusion, particularly at the poles, where they appear to move freely except for being restricted to the same pole for several generations (12). When the cytoskeletal protein MreB is disrupted and the cell becomes rounded, Tsr clusters at the poles tend to fragment and the fraction of mobile Tsr increases (26). This suggests that, aside from the diffusion-and-capture process made possible by Tol-Pal complexes (24), one or more additional mechanisms may contribute to the preference of the chemoreceptor clusters for a polar location.
In E. coli, the nucleoid is usually at midcell and confined within the cell cylinder. Among other components, it contains most of the DNA, RNA, and nucleoid-associated proteins of the cell. Major nucleoid-associated proteins include H-NS, HU, Fis, IHF, and StpA. The dimeric histone-like protein HU in particular is highly abundant and involved in DNA compaction, and thus can be used to assess the nucleoid’s morphology and positioning in vivo when fused with fluorescent proteins such as mCherry (27). The partitioning of replicated nucleoids in cell division involves the structural maintenance of chromosome complex MukBEF (28). The deletion of mukB causes a temperature-sensitive lethal phenotype that fails to partition the chromosome, resulting in the formation of anucleate cells.
Recent studies have reported that in addition to Tsr clusters, other types of large biomolecules in E. coli are segregated to and then retained at the poles. This is due to an energy-free volume exclusion caused by the presence of the nucleoid at midcell (29, 30) that affects plasmids (31, 32) and other large complexes (33, 34). A possible contribution of nucleoid exclusion to the distribution of chemoreceptor clusters associated with chemotaxis has not yet been considered.
Here, we explored whether nucleoid exclusion contributes to the segregation and retention of Tsr chemoreceptor clusters at the cell poles. In addition, we evaluated the contribution from other, previously proposed mechanisms, namely, Tol-Pal diffusion-and-capture and dragging by cell elongation. We used E. coli cells expressing Tsr-Venus and harboring a plasmid that expresses the nucleoid-tagging protein HupA-mCherry, and performed live single-cell studies of the spatiotemporal distribution of Tsr clusters and nucleoids. The Venus protein is a YFP variant that is derived from GFP and has a fast maturation time (35) that allows real-time imaging by fluorescence microscopy. The tagging of Tsr with Venus does not affect its spatial distribution and is not toxic to cells (35). Measurements were conducted in wild-type (WT) and isogenic mutant cells lacking the Tol-Pal complex (Δtolpal). We further studied the Tsr cluster spatial distribution in anucleoid cells (ΔmukB). Finally, we performed stochastic simulations of dynamic models and compared the long-term behaviors of the clusters, as indicated by in silico and in vivo data, to assess whether the proposed volume-exclusion mechanism could explain the empirical observations.
Materials and Methods
Chemicals
For routine cultures, M9-glucose media components, isopropyl β-D-1-thiogalactopyranoside (IPTG), 4′,6-diamidino-2-phenylindole (DAPI), formaldehyde, agarose for microscopic slide gel preparation, and antibiotics were purchased from Sigma-Aldrich (St. Louis, MO). Amino acids and vitamins were purchased from GIBCO/BRL (Grand Island, NY). SYTOX Orange and a Live/Dead BacLight Viability Kit (L7007) were purchased from Thermo Fisher Scientific/Molecular Probes (Waltham, MA).
Strains and plasmids
We used E. coli strain (SX4) expressing the chimeric gene tsr-venus under the control of the Plac promoter, which is incorporated into the chromosome (35) (a kind gift from Sunny Xie, Harvard University). We transformed this strain with a pBR322 derivative plasmid expressing HupA-mCherry (a nucleoid-tagging protein) under the control of a constitutive promoter with ampicillin resistance (36) (a kind gift from Nancy Kleckner, Harvard University). The host E. coli K-12 strain has a genotype of (lacIp4000 hsdR514 DE(araBAD)567 DE(rhaBAD)568 rph-1).
We also used E. coli strain MG1655 and its isogenic derivative strain lacking Tol-Pal components (MG1655 Δtolpal) (27) (a kind gift from Douglas Weibel, University of Wisconsin-Madison). These were transformed with a pBR322 derivative plasmid expressing the gene tsr-venus under the control of the Plac promoter. The E. coli strain MG1655 has a genotype of (F− λ− rph-1). The SX4 and MG1655 strains served as the WT for our studies.
The mutant strain AZ5372 lacking mukB (trpC9941 ΔmukB::kan), referred to as ΔmukB, was obtained from the Keio single-gene knockout collection (36) and transformed with a pBR322 derivative plasmid expressing the gene tsr-venus under the control of the Plac promoter.
All overnight liquid cultures were grown in M9-glucose medium for 14 h at 37°C with shaking (250 rpm), except for ΔmukB, which was incubated at 22°C. The M9-glucose (0.4%) medium was supplemented with amino acids and vitamins, along with kanamycin (35 μg mL−1) and ampicillin (50 μg mL−1).
We subsequently made subcultures by diluting the overnight culture into fresh M9 glucose medium containing the appropriate antibiotics. We opted for M9-glucose medium to maintain a well-defined cell culture condition and achieve low cellular autofluorescence. In addition, previous studies have shown that cells have a higher propensity to form arrays in minimal media than in richer media such as Terrific broth and Luria broth, suggesting that the arrays’ functionality is enhanced (18).
Induction of production of Tsr-Venus
Strains from overnight cultures were diluted into fresh media with the appropriate antibiotics (as described above) at an initial OD600 of 0.02 and grown until they reached an OD600 of ∼0.3, at 37°C with shaking (250 rpm). In the SX4 strain, Tsr-Venus production, controlled by Plac, was induced by IPTG at the appropriate concentrations. In the MG1655 strain and its derivative, containing the plasmid expressing Tsr-Venus, induction was performed with final concentrations of 50 μM mL−1 IPTG for 1 h at 37°C. In both cases, cells were then left in the shaker to grow until they reached an OD600 of ∼0.45–0.5 before microscopy was performed. In addition to microscopy, we also measured Tsr-Venus expression as a function of induction with a microplate fluorometer at 37°C (Fig. S3 in the Supporting Material).
Nucleoid visualization by HupA-mCherry tagging
To observe Tsr-Venus clusters and nucleoids in individual cells simultaneously over time, we used cells containing the plasmid from which HupA-mCherry is constitutively expressed (SX4-HupA-mCherry strain). These were induced with 200 μM mL−1 IPTG for 1 h at 37°C and then centrifuged, and the supernatant discarded. Then, 4 μL of cells was placed on a 1% agarose gel pad prepared in M9-glucose medium for image acquisition.
Nucleoid visualization by DAPI and SYTOX Orange staining
DAPI stains nucleoids specifically with little or no cytoplasmic labeling. Cells at an OD600 of ∼0.3 were induced with 200 μM mL−1 IPTG (SX4 strain) or 50 μM mL−1 IPTG (MG1655 and MG1655 Δtolpal strain) and left in the shaker incubator at 37°C until they reached an OD600 of ∼0.5. The cells were then fixed with 3.7% formaldehyde in phosphate-buffered saline (PBS) for 30 min and washed with PBS to remove excess formaldehyde. The pellets were suspended in PBS, and DAPI (2 μg mL−1) was added to this suspension (37). After incubation for 20 min in the dark, the cells were centrifuged and washed twice with PBS to remove excess DAPI. The cells were then resuspended in PBS and 8 μL of these samples was placed on a 1% agarose gel pad prepared in M9-glucose medium for microscopy observation.
To observe the nucleoids of ΔmukB cells, we used SYTOX Orange dye instead of DAPI, since we found that DAPI staining does not allow one to distinguish easily between cells with and without nucleoid, due to strong background autofluorescence and image blurring (38). ΔmukB cells containing the plasmid expressing Tsr-Venus were grown overnight in Luria broth medium at 22°C. Subsequently, they were subcultured and allowed to grow exponentially in M9-glucose medium at 22°C, followed by activation of Tsr-Venus (to a final concentration of 50 μM mL−1 IPTG) for 1 h at 37°C. To this culture, a SYTOX Orange solution (50μM stock concentration) was added to a final concentration of 500 nM and incubated for 10 min in the dark (38). The cells were then centrifuged twice and resuspended in fresh M9-glucose medium, and 8 μL of these samples was placed on a 1% agarose gel pad prepared in M9-glucose medium along with inducer IPTG for microscopy observation in a temperature-controlled chamber (see the “Microscopy” section below).
Measurements with ampicillin
For measurements with ampicillin, we used SX4 cells (35). Cells were grown as described in the previous section. Next, the cells were induced with IPTG (200 μM) to activate Tsr-Venus expression. Simultaneously, we introduced freshly prepared ampicillin (100 μg mL−1) in the media and left the cells in the incubator for 30 min. The nucleoid was observed by DAPI staining.
To assess the proportion of viable cells after ampicillin treatment, we performed live/dead staining according to the protocol for the Live/Dead BacLight Viability Kit. Equal volumes of SYTO 9 (reagent A) and propidium iodide (PI; reagent B) from the kit were mixed and added to the cells suspended in 0.85% (w/v) NaCl (3 μL mL−1of cells). After mixing and incubation at room temperature in the dark for 15 min, the cells were visualized by epifluorescence microscopy with the standard fluorescent long-pass filter set for simultaneous visualization of live and dead cells. The green fluorescent dye SYTO 9 permeates both intact and damaged cell membranes, whereas red PI only enters cells with significant membrane damage (39). Thus, upon ampicillin treatment, cells with damaged membranes take up the red PI dye, which saturates the green SYTO 9 dye that is taken up by all cells. The presence of red color is therefore indicative of cell death, and green indicates live cells.
Measurements at suboptimal temperatures
Tol-Pal deletion mutant cells (MG1655 Δtolpal) with a Tsr-Venus-expressing plasmid were grown as described above. Tsr-Venus expression was induced by adding 50 μM mL−1 of IPTG to the culture. The cells were then left in the incubator at the appropriate temperatures (10°C, 15°C, and 24°C) for 1 h and fixed with 3.7% formaldehyde, followed by DAPI staining and microscopy image acquisition.
Microscopy
We performed single-time-point imaging of cells with Tsr-Venus and DAPI (or) SYTOX Orange-stained nucleoids (SX4, MG1655, and MG1655 Δtolpal and ΔmukB strains), and time-lapse imaging of cells with Tsr-Venus and HupA-mCherry-tagged nucleoids (SX4-HupA-mCherry strain). With both imaging methods, phase-contrast images were acquired for cell segmentation and, in time series, for lineage construction.
In single-time-point microscopy, Tsr-Venus was induced by adding 200 μM mL−1 IPTG (SX4 strain) or 50 μM mL−1 IPTG (MG1655, MG1655 Δtolpal and ΔmukB strains) to the liquid culture. The cells were then left in the shaker incubator at 37°C for 1 h before image acquisition. For this purpose, 8 μL of cells was placed on a 1% agarose gel pad prepared in M9-glucose medium. Images were taken after the cells were placed under observation.
For time-lapse microscopy measurements, noninduced cells (SX4 strain) were placed on a microscope slide between a coverslip and M9-glucose agarose gel pad containing IPTG (200 μM mL−1). During image acquisition, the cells were constantly supplied with fresh media containing IPTG, at the same concentration, by a microperfusion peristaltic pump (Bioptechs, Butler, PA) at 1 mL min−1. Images were captured every 3 min for 1 h by confocal microscopy.
Imaging was performed using a Nikon Eclipse inverted microscope (Ti-E, Nikon, Tokyo, Japan) with a C2+ point scanning confocal system and a 100× Apo total internal reflection fluorescence objective (1.49 NA, oil). For population imaging, Tsr-Venus fluorescence was measured using a 488 nm argon laser (Melles-Griot, Rochester, NY) and HQ514/30 filter. HupA-mCherry fluorescence was measured using a 543 nm He-Ne laser (Melles-Griot) and HQ585/65 filter (Nikon). For time-lapse microscopy measurements of Tsr-Venus- and mCherry-tagged nucleoid(s) (SX4-HupA-mCherry strain), we performed highly inclined and laminated optical sheet microscopy (40) using an EMCCD camera (iXon3 897, Andor Technology, Belfast, UK) and the same lasers, along with an HQ515/30 filter and Texas Red filter (Nikon). DAPI-stained nucleoids were observed by epifluorescence microscopy using a mercury lamp with a DAPI filter (Nikon), and SYTOX Orange-stained nucleoids were observed with the EMCCD camera. SYTOX Orange was visualized using 543-nm laser excitation and a Texas Red filter (Nikon). Phase-contrast images were captured simultaneously by a CCD color camera (DS-Fi2, Nikon). Finally, for the Live/Dead BacLight viability assay, we used the CCD color camera and an LF488/LP-B-NTE filter cube (Semrock, Rochester, NY) for simultaneous visualization of SYTO 9- and PI-stained cells.
Images were acquired with the use of NIS-Elements software (Nikon). Slides were kept in a temperature-controlled chamber (FCS2, Bioptechs) at a stable temperature (37°C unless stated otherwise).
Image analysis
The image analysis procedure included cell segmentation, lineage construction, detection and characterization of fluorescent spots (Tsr clusters) and nucleoids, and characterization of the spatial distributions of clusters and nucleoids. The various steps were performed using custom-made software that integrates components from the software MAMLE (41) and CellAging (42), along with the cluster-detection method proposed in (43).
Cell segmentation was performed from phase-contrast images by MAMLE (41), followed by manual correction. Next, confocal images were aligned to the phase-contrast images as described in (44). Lineage construction, when needed, was automatically performed by CellAging (42) and then manually corrected. Detection and characterization of the size and intensity of fluorescent Tsr clusters were performed as described in (43), by defining a cluster as a connected component, with each pixel having a light intensity above a threshold. For this purpose, we assume that the background pixel intensities follow a Gaussian distribution with the same median and upper quartile as the pixels in the cell. The threshold is selected by visual inspection of the outcome. From the segmented image, the number of clusters is calculated. The area of each cluster is calculated by counting the number of pixels within.
For nucleoids, we used the same method as described above. For each cell, we determined the fluorescence levels along the major cell axis of a background-corrected cell and summed the fluorescence intensities along the minor axis. For a given cell population, we normalized the cell lengths and averaged the fluorescence intensities over all cells. This method is used for images of DAPI-stained and HupA-mCherry-tagged nucleoids. To detect the boundaries of the nucleoid in each cell, we fitted a piecewise constant probability density function with three pieces to its fluorescence intensity distribution along the major cell axis by maximum likelihood. Given the two separation points obtained from the fit, we determined the center of the nucleoid as the middle point between them.
To examine Tsr-Venus spatial distributions, we first obtained background-corrected cells by subtracting the median cell intensity from each cell pixel intensity, and summed the fluorescence intensities along the minor axis. Finally, to distinguish between midcell and poles, we defined a boundary at 0.5 (with 0 being midcell and 1 being the cell extreme) as in (24). The overall image-analysis procedure is illustrated in Fig. S1.
Stochastic modeling
We implemented stochastic models of the two mechanisms responsible for the preference for a polar localization of the Tsr clusters considered here (diffusion-and-capture by Tol-Pal complexes at the poles and nucleoid exclusion from midcell; see Fig. S2). The dynamics of the models was driven by the stochastic chemical kinetics simulator SGNS2 (45), which can implement transient compartments. We used this feature to model, among other things, the diffusion-and-capture mechanism and the volume-exclusion mechanism caused by the nucleoid. It also allowed us to obtain a spatial location for each cluster in the cell (via cell compartmentalization into blocks).
The models are two-dimensional and were used to compare the dynamics of the Tsr clusters in the presence of relatively small and large nucleoids, and in the presence and absence of Tol-Pal. A description of the models, reactions, and parameters is provided in Supporting Materials and Methods.
Results
To visualize chemoreceptors in vivo, we used 1) the SX4 strain with a chimeric tsr-venus gene inserted into the chromosome (35), 2) the MG1655 strain (WT and deletion mutant for Tol-Pal, Δtolpal), and 3) the mukB deletion mutant strain (ΔmukB). The latter two strains contain a low-copy plasmid coding for Tsr-Venus proteins (see Materials and Methods) under the control of a lac promoter. SX4 cells further contain a plasmid expressing a nucleoid-tagging protein, HupA-mCherry (Materials and Methods). We found no significant difference in the spatial distribution and kinetics of Tsr clusters between SX4 and MG1655 cells. Also, we found no significant difference in nucleoid lengths when measured by DAPI/SYTOX Orange staining and by HupA-mCherry tagging, and we saw no difference in the Tsr spatial distributions when we visualized the nucleoids with either method (Fig. S9). Further, it is worth noting that, since under normal conditions the tsr gene is highly expressed (46), small amounts of exogenous Tsr-Venus are expected to cause minimal perturbations to normal cell functioning. Finally, unless stated otherwise, the results refer to measurements obtained using the MG1655 strain.
Spatial distributions of nucleoids and Tsr protein clusters
First, in SX4 cells, we assessed whether the spatial distributions of Tsr clusters and nucleoids are consistent with a nucleoid-exclusion phenomenon affecting the clusters’ location. To do so, from images taken 1 h after induction of Tsr-Venus expression, we compared the spatial distributions of nucleoids (DAPI stained) and Tsr clusters in cells with relatively large and relatively small nucleoids.
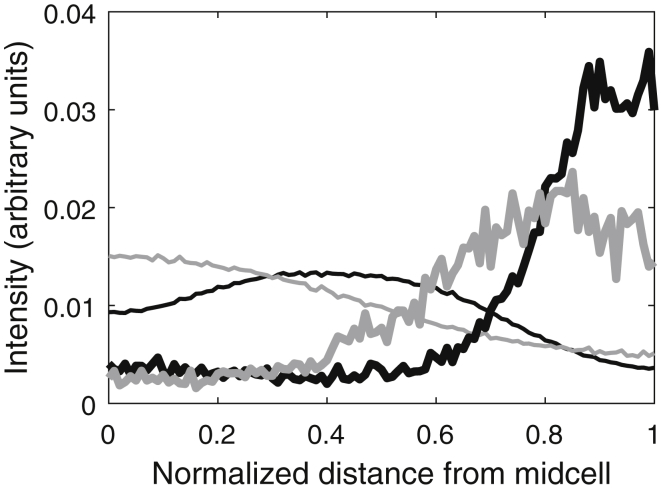
Based on the data from 1195 cells, we selected the 10% of cells with relatively larger and relatively smaller nucleoid(s) (along the major cell axis). Cells at the thresholds of nucleoid length were included in the analysis. Also, we did not exclude cells with two nucleoids, and thus we expected that at least some of the cells with larger nucleoid(s) would have two nucleoids. The results (Fig. 1, thin black line) support this expectation, as in the 10% of cells with larger nucleoid(s), the fluorescence intensity from nucleoid(s) exhibits a local minimum at midcell (as the two nucleoids are at the focal points).
Figure 1.
Averaged spatial distributions of fluorescence intensity (in arbitrary units (a.u.)) of Tsr-Venus and nucleoids along the major cell axis (collapsed axis, from midcell to cell extreme). The thin black line is the average nucleoid fluorescence intensity distribution from the 10% of cells with the largest relative nucleoid(s) (due to the existence of two nucleoids; 155 cells), and the thin gray line is the same distribution from the 10% of cells with the smallest relative nucleoid lengths (126 cells). The bold black and gray lines are the average spatial distributions along the major cell axis of Tsr-Venus fluorescence intensity in cells with relatively larger and smaller nucleoids, respectively.
As is visible in Fig. 1, the center of mass of the DAPI intensity distribution is significantly closer (in a statistical sense) to the cell center in cells with relatively small nucleoids (0.37 ± 0.01) than in those with relatively large nucleoids (0.42 ± 0.00; Kolmogorov-Smirnov (KS) test, p < 0.01). Also, the Tsr protein clusters are significantly farther away from midcell in cells with relatively larger nucleoid-occupied regions (KS test, p < 0.01), as would be expected if nucleoid exclusion affects their location. In particular, the center of mass of the Tsr fluorescent intensities is 0.72 ± 0.02 in cells with relatively larger nucleoids and 0.66 ± 0.02 in cells with relatively smaller nucleoids. The same phenomenon was observed in cells of the MG1655 strain (data not shown).
As these results could be affected by the normalization of the cell lengths (provided, e.g., that most cells with relatively larger nucleoids also had larger absolute lengths, which can be expected), we performed the same analysis on a subset of cells that exhibited similar absolute lengths (all within 2.8 ± 0.5 μm). From this subset, we again selected the 10% of cells with larger (70 cells) and smaller (70 cells) nucleoids. We then confirmed that the cell lengths of these two subsets of cells did not differ significantly (KS test, p = 0.73). Finally, we performed the same test as above and found that in the cells with larger nucleoids, the Tsr clusters were closer to the cell extremities, i.e., the center of mass of the DAPI intensity distribution was significantly closer (in a statistical sense) to the cell center in cells with relatively small nucleoids (0.34 ± 0.01) than in those with relatively large nucleoids (0.42 ± 0.01; KS test, p < 0.01). Also, the Tsr molecules were farther away from midcell in cells with relatively larger nucleoid-occupied regions (KS test, p ≈ 0.02), as would be expected if nucleoid exclusion affects the location of the clusters. The center of mass of the Tsr fluorescent intensities was 0.65 ± 0.02 in cells with relatively smaller nucleoids and 0.72 ± 0.02 in cells with relatively larger nucleoids.
The above results do not rule out the (unlikely) possibility that cells with larger nucleoids (e.g., close to dividing) also possess more Tol-Pal, indirectly causing the Tsr clusters to be located closer (on average) to the cell extremities. To investigate this possibility, from the data above, we selected cells with the same absolute nucleoid length (within a small range). Note that these cells differ in total cell length. If Tol-Pal levels are nucleoid-length dependent, these cells should all exhibit similar Tol-Pal levels. As such, if nucleoid exclusion is not relevant to the Tsr clusters’ location, these cells should not exhibit a tangible correlation between the absolute cell length and absolute distance of Tsr clusters to the nearest cell extremity. We obtained that correlation and found it to be equal to 0.73 (p < 0.01), showing that in cells with similar nucleoid lengths but different cell lengths, the clusters behave differently and in accordance with a nucleoid-exclusion phenomenon.
In addition to the tests above, we also performed microscopy time-series measurements (see Materials and Methods) in which the expression of Tsr-Venus was only activated after we started observations. From these data, we assessed the location of the Tsr clusters when they first appeared. The data in Fig. S6 show that the clusters usually first appeared (i.e., became detectable) when they were already at the poles, where they tended to remain thereafter. From this, one can conclude that most clusters remain at the poles for most, if not all, of their lifetime.
Asymmetries in the spatial distributions of nucleoids and Tsr protein clusters
Finally, in search of additional evidence of nucleoid exclusion from the midcell of Tsr clusters, we assessed whether, at the single-cell level, asymmetries in the position of the nucleoid center along the major cell axis (34, 47, 48) are correlated with asymmetries in the Tsr clusters location. From the set of SX4 cells mentioned above (1195 cells), for each cell, we obtained the fluorescence intensity from Tsr clusters at each pole (using the definition of “pole” in Materials and Methods) and the normalized location of the nucleoid center relative to the cell center along the major axis, and calculated the Pearson correlation between these variables. As would be expected if there is nucleoid exclusion of the Tsr clusters from midcell, we found a significant (p ≪ 0.01) correlation of −0.52 with a 95% confidence interval (CI) of [−0.57, −0.48].
It is possible that this result is associated with the age of the poles, since the older pole of a cell would contain more Tol-Pal complexes and, simultaneously, off-centered nucleoids would preferentially locate closer to the older pole (although no evidence for either phenomenon has been reported so far).
To assess whether the position of the nucleoid(s), when off-centered, is biased toward the newer (or older) pole (34, 49, 50), we localized the nucleoid center along the major axis at each moment during the cell’s lifetime (tracked from birth to division). Then, we obtained the fraction of times a nucleoid was located at the newer side of the cell. Next, we assessed whether the data collected from all cells could have resulted from an unbiased binomial distribution. We obtained a p-value of 0.148, consistent with the data being extracted from an unbiased binomial. We conclude that the asymmetries in nucleoid(s) positioning are not biased toward the newer (or older) pole, and thus differences in pole age are not a source of the anticorrelation in the positioning of the nucleoid and Tsr clusters in individual cells.
Similarly, we assessed whether the clusters’ choice of pole is correlated with the pole age (Supporting Materials and Methods). We found no evidence for a tangible correlation. In addition, we also found no difference in the curvatures of the cell poles that could explain the asymmetries, at the single-cell level, in cluster numbers at each pole.
Overall, we conclude that there are heterogeneities and asymmetries in the spatial distribution of Tsr clusters that are consistent with a nucleoid-exclusion phenomenon and cannot be explained solely by a diffusion-and-capture mechanism caused by Tol-Pal complexes at the poles (24).
Spatial distribution of Tsr clusters in cells subjected to ampicillin
Next, we subjected cells (SX4 strain) to ampicillin (these cells lack ampicillin resistance; see Materials and Methods), causing enhanced elongation while halting division (51, 52). We performed this test to show that when the ratio between cell and nucleoid lengths is altered by external perturbation, the spatial distribution of Tsr-Venus clusters behaves as would be expected if it is affected both by diffusion-and-capture by Tol-Pal complexes at the cell poles and by nucleoid exclusion from midcell.
Note that although SX4 cells are sensitive to ampicillin (division is halted), most appeared to remain healthy (Fig. S4). This was verified with the Live/Dead BacLight Viability Kit, which stains viable cells but not the shells of deceased cells (Materials and Methods). Also, we observed that once the drug was removed from the medium, the cells recovered and started dividing (as observed in the microscope and by spectrophotometry). As an additional precaution, during image analysis, we manually discarded (by visual inspection) cells that exhibited inclusion bodies and spheroplasts that commonly appear after ampicillin treatment, as these could affect the Tsr cluster dynamics. The remaining cells exhibited a normal rate of accumulation of Tsr clusters (as observed by inspection).
From the images of control cells and cells subjected to ampicillin, we selected cells whose length ranged from 3.5 to 4.0 μm and compared their mean relative nucleoid length (Fig. S5) and center of mass of the Tsr fluorescence intensity distribution. As shown in Table 1, we found that as the nucleoid became longer relative to the cell length, the center of mass of the distribution of Tsr-Venus moved toward the cell extremities, as would be expected from the existence of a nucleoid-exclusion phenomenon.
Table 1.
Mean Relative Nucleoid Length and Center of Mass of the Averaged Tsr Fluorescence Distribution of the Control and Ampicillin-Treated Cells
| Condition | No. of Cells | Mean Relative Nucleoid Length with 95% CI (μm) | Center of Mass of the Averaged Tsr Fluorescence Distribution with 95% CI |
|---|---|---|---|
| Control | 105 | 0.71 ± 0.01 | 0.69 ± 0.02 |
| Ampicillin | 79 | 0.75 ± 0.02 | 0.75 ± 0.03 |
Also shown are the 95% CIs and the number of cells examined in each condition.
Effects of Tol-Pal complex deletion on the spatial distribution of Tsr
We next assessed whether, in the absence of Tol-Pal, the Tsr clusters would no longer exhibit a preference for a polar localization. For this purpose, we compared the mean fraction of Tsr fluorescence intensity at the poles in control (WT) and Δtolpal cells. The results are shown in Table 2, along with the 95% CIs and the results of a test of statistical comparison between the conditions.
Table 2.
Mean Percentage of Fluorescence Intensity at the Cell Poles for WT and Δtolpal Cells
| WT | Δtolpal | Student’s t-test (WT versus Δtolpal) | |
|---|---|---|---|
| Number of cells | 176 | 138 | – |
| Mean fluorescence intensity at poles (%) | 96.2 | 88.8 | <0.01 |
| 95% CI (%) | [95.0, 97.3] | [85.3, 92.3] | – |
Shown are the number of cells observed, the percentage of total fluorescence intensity located at the poles, and the corresponding 95% CIs. Also shown is the p-value of the Student’s t-test between WT and Δtolpal.
Finally, we performed KS tests, which showed that the mean percentage of fluorescence intensity in the cells (in both WT and Δtolpal cells) was inconsistent with a normal fluorescence intensity along the major axis, which would be expected if Tsr proteins are distributed solely by means of diffusion (p-value much smaller than 0.01 for a normal distribution with a mean fluorescence at the poles equaling 50% of the total fluorescence).
As shown in Table 2, in both WT and Δtolpal cells, the mean fraction of Tsr clusters at the poles is much higher than would be expected by chance (i.e., when compared with a uniform distribution along the major cell axis). This finding and the KS tests indicate that diffusion-and-capture by Tol-Pal is not the only cause of the Tsr clusters’ preference for polar localization, even though the presence of Tol-Pal does significantly increase the fraction of clusters at the poles (the p-value from a two-tailed Student’s t-test of statistical significance is much smaller than 0.01), as expected from previous studies (24).
Effects of Tol-Pal deletion on the spatial distribution of Tsr as a function of nucleoid length
If Tsr clusters are preferentially located at the cell poles due to a diffusion-and-capture mechanism caused by Tol-Pal complexes and a nucleoid-exclusion phenomenon, we expect that if at least one of these mechanisms is present, the Tsr clusters will still preferentially locate at the poles, although not as pronouncedly as they would if both mechanisms were active.
Given that we expect the effects of nucleoid exclusion on the Tsr clusters’ spatial distribution to be gradual as a function of the nucleoid length, to study this, from the data above, we measured in each cell the relative distance of each cluster to the nearest cell extreme as well as the relative nucleoid length. Then, for WT and Δtolpal cells separately, we selected the cells with relatively smaller (≤35% of the cell length) and larger (≥65% of the cell length) nucleoids.
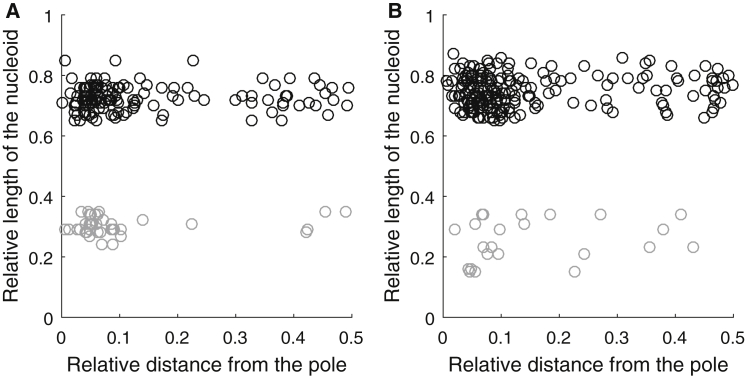
From the data, we first plotted the relative distance of each cluster to the nearest cell extreme versus the normalized nucleoid length in each condition (Fig. 2, A and B, for WT and deletion mutants, respectively). Visibly, at least for cells with relatively small nucleoids, the presence/absence of Tol-Pal complexes appears to be an influencing factor (as expected (24)), since in their absence the Tsr clusters are more uniformly scattered throughout the major cell axis (Fig. 2 B). This difference in the Tsr clusters’ behavior due to the presence/absence of Tol-Pal complexes is less clear in cells with relatively large nucleoids (Fig. 2, A and B), in agreement with our hypothesis that nucleoid exclusion contributes to the Tsr clusters’ preference for a polar localization. These results are also in line with those shown in Fig. 1.
Figure 2.
(A) Relative nucleoid length versus the relative distance of individual Tsr clusters from the closest cell extremity in WT cells: 133 Tsr clusters from 52 cells whose nucleoid length is ≥65% of the cell length are shown in black, and 39 Tsr clusters from 17 cells whose nucleoid length is ≤35% of the cell length are shown in gray. (B) Relative nucleoid length versus relative distance of individual Tsr clusters from the closest cell extremity in Δtolpal cells: 198 Tsr clusters from 88 cells whose nucleoid length is ≥65% of the cell length are shown in black, and 23 Tsr clusters from eight cells whose nucleoid length is ≤35% of the cell length are shown in gray.
To quantify the differences in the Tsr clusters’ behavior between conditions more precisely, we calculated the degree of correlation of the Tsr clusters’ relative locations along the major cell axis between all pairs of conditions (Table S2). We expected this correlation to be significant between conditions differing solely in nucleoid length (as the Tol-Pal diffusion-and-capture mechanism is present) and between cells with large nucleoids differing in the presence/absence of Tol-Pal complexes (as nucleoid exclusion is strong). Between other pairs of conditions, we expected a weaker or no correlation. The results in Table S2 confirm these expectations, showing that for the latter, the correlations become nonsignificant.
Finally, note that the degree of the correlation between WT cells with small nucleoids and Δtolpal cells with large nucleoids suggests that the two mechanisms (Tol-Pal diffusion-and-capture and nucleoid exclusion) have similar effects when they act solely. Overall, the results in Table S2 further suggest that the two mechanisms have complementary effects.
In addition, this single-cell analysis of the spatial distribution of individual clusters as a function of relative nucleoid length allows us to conclude that the effects of the phenomenon of exclusion from midcell change gradually with changing nucleoid lengths, as expected (Figs. S7 and S8).
Effects of mukB deletion on the spatial distribution of Tsr clusters
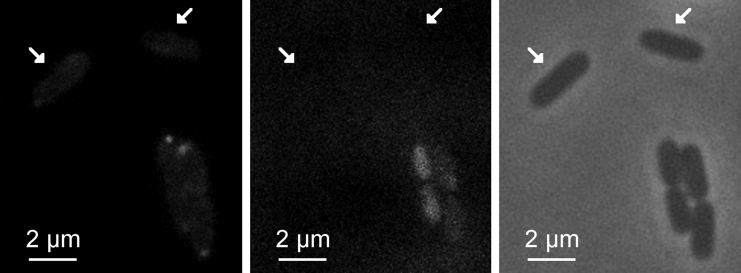
Next, to further test whether nucleoid exclusion contributes to the preferential polar localization of Tsr clusters, we studied their localization in cells where the nucleoid is absent. For this purpose, we made use of mutant cells lacking the mukB protein (ΔmukB) (Materials and Methods). A fraction of these cells lack the nucleoid (thus becoming anucleate cells) due to failures in chromosome segregation in cell division (53). We verified this spontaneous formation of anucleate cells by visually inspecting the microscope images. In ΔmukB cells, although the Tsr clusters will still preferentially locate at the poles due to the presence of Tol-Pal complexes (24), their fraction at midcell is expected to increase compared with control cells (see Fig. 3, where the two cells lacking nucleoid visibly have a larger fraction of Tsr clusters at midcell than the other cells in the image).
Figure 3.
Localization of Tsr-Venus in ΔmukB cells at 37°C. Nucleoids were visualized by SYTOX Orange dye. The arrows indicate anucleate cells. Left: green channel showing Tsr-Venus. Middle: red channel showing nucleoids. Right: phase-contrast images showing the cell borders used for cell segmentation.
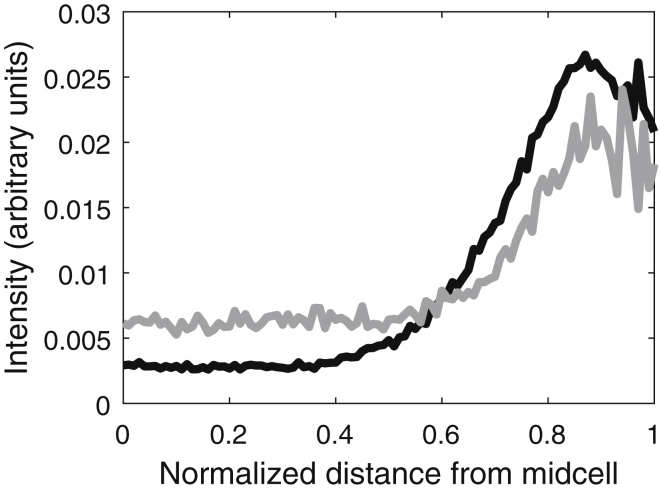
From the images, we segmented and analyzed 302 cells, 68 of which were anucleate (as determined by visual inspection of the red channel). We compared the relative positioning along the major axis of the Tsr clusters in these cells and in WT cells (1195 control cells). The results are shown in Fig. 4. Whereas in ΔmukB cells the center of mass of the fluorescence intensity distribution of Tsr is at 0.59 ± 0.03, in control cells it is at 0.70 ± 0.01, i.e., the Tsr clusters are much closer to the cell extremities in the control cells, with the two distributions differing significantly in a statistical sense (KS test, p ≪ 0.01). This supports the hypothesis that nucleoid exclusion enhances the Tsr clusters’ preference for polar localization.
Figure 4.
Spatial distributions of average fluorescence intensity (in a.u.) of Tsr-Venus proteins along the major cell axis (collapsed axis, from midcell to cell extremes). The black line is the fluorescence intensity distribution averaged over 1195 control cells, and the gray line is the same distribution averaged over 68 anucleate ΔmukB cells.
Effects of increased cytoplasm viscosity on the spatial distribution of Tsr clusters
We performed a final measurement to further strengthen the hypothesis that nucleoid exclusion from midcell enhances the Tsr clusters’ preference for polar localization. Under reduced metabolic activity or suboptimal temperatures, the cytoplasm of E. coli is known to acquire glass-like features (54) that enhance its viscosity (48). This in turn greatly reduces the effects of nucleoid exclusion on the spatial distribution of large protein complexes (48).
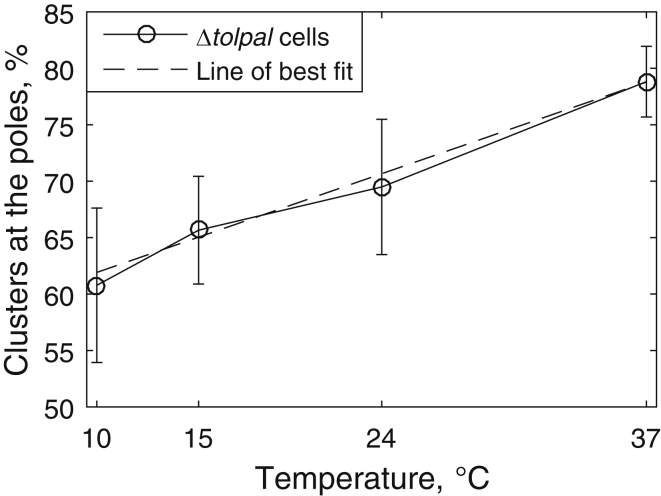
We subjected Δtolpal cells to low temperatures and assessed whether, in cells with relatively large nucleoids (i.e., occupying 65–80% of the cell length), the degree of exclusion of Tsr clusters from midcell decreases gradually as the temperature decreases. We studied Δtolpal cells alone, as the effects of lower temperatures on the functionality of Tol-Pal are unknown. The results are shown in Fig. 5.
Figure 5.
Percentage of Tsr clusters at the poles in Δtolpal cells with relatively large nucleoids (occupying 65–80% of the cell length) as a function of temperature. The error bars correspond to one standard uncertainty. A line and a polynomial of second order were fitted to the data points by weighted least-square fit. The Akaike information criterion showed that the best-fitting model is a line (dashed line).
As can be seen in Fig. 5, the fraction of Tsr clusters at the poles decreases gradually with gradually decreasing temperature, as expected from the increased cytoplasm viscosity (48). As a side note, since this decrease is best fitted by a straight line (using a weighted least-square fit (55) and the Akaike information criterion (56)), we expect that in the absence of Tol-Pal, only nucleoid exclusion is involved in the segregation of clusters to the poles.
Stochastic model of nucleoid exclusion of Tsr clusters from midcell
Next, we made use of a stochastic model (for a complete description, see Supporting Materials and Methods) to test whether a nucleoid-exclusion mechanism could reproduce the empirical data. Note that in this model, the nucleoid-exclusion phenomenon acts only on Tsr proteins that are unbound to the membrane, in accordance with (26).
The model uses empirical data regarding Tsr production and degradation rates (35, 57) and cell and nucleoid sizes (length and width) obtained from SX4 cells (Supporting Materials and Methods). Also, the clusters’ diffusion rate and unbinding rate from Tol-Pal complexes are tuned so that the in silico spatial distributions of Tsr in cells with relatively large and relatively small nucleoids fit the empirical data (Fig. 2 A).
Using this fitted model, we first tested whether the model could reproduce the observed effect of asymmetries in nucleoid positioning on the Tsr distributions along the major cell axis at the single-cell level. We found a significant (p ≪ 0.01) correlation of −0.65 with a 95% CI of [−0.68, −0.62].
Thus, the stochastic model, assuming the existence of nucleoid exclusion and asymmetries in nucleoid positioning, reproduces the empirical observations of a strong anticorrelation between Tsr clusters and nucleoid location at the single-cell level.
Next, we assessed whether imposing differences in nucleoid length, along with assuming the presence and absence of Tol-Pal, would suffice to reproduce the observed differences in the Tsr spatial distributions observed in vivo. The results are shown in Table S3.
If we compare Tables S2 and S3, we observe a strong similarity between the model and the measurements. In short, in cases where the measurements indicate strong correlations, the model is in agreement. However, in cases where the measurements show no correlation, the model suggests much weaker correlations than in the cases where the measurements indicate strong correlations. However, the detected correlations in the simulations (Table S3, line 1, columns 2 and 3) are expected (since although smaller nucleoids should be less efficient in excluding Tsr clusters from midcell, they are not expected to be entirely ineffective). As a side note, these results also agree with the empirical data from cells subjected to ampicillin (which results in cells with relatively larger nucleoids) versus control cells.
Overall, we conclude that the dynamics of Tsr clusters of a model assuming a diffusion-and-capture mechanism caused by Tol-Pal at the poles, along with a mechanism of volume exclusion from midcell caused by the presence of the nucleoid, is in agreement with the empirical data.
Discussion
Chemoreceptor proteins can assemble into large arrays, which is believed to enhance the signal-processing capabilities of the receptor system (6, 7, 8, 9), allowing proper chemotaxis (5). In agreement with this, their ability to cluster is conserved in all known prokaryotic chemotaxis systems (58).
In E. coli, these clusters preferentially locate at the cell poles. This preference for polar localization may further enhance the clustering process itself and thus the signal-processing capabilities. A recent study suggested that this preference for polar localization is due to a mechanism of diffusion-and-capture made possible by Tol-Pal complexes at the cell poles (24).
Recent observations also showed that at any given time, several Tsr clusters are not in a fixed position, but rather diffuse freely within the polar region (26) (or more rarely at midcell), and that in spherical cells (when the cytoskeletal protein MreB is disrupted) the clusters are more fragmented and the fraction of freely diffusing ones increases (26). Given these observations, and the known ability of the nucleoid to exclude macromolecules such as plasmids and large protein complexes from midcell (31, 32, 33, 34, 47), we investigated the possibility that the preference of Tsr clusters for polar localization is supported by the presence of the nucleoid at midcell. For this purpose, we performed several tests while observing at the single-cell level both clusters and nucleoids.
All measurements performed here, including perturbations of the cytoplasm and cell growth, and observations in cells lacking Tol-Pal components or a nucleoid led to the same conclusion: Tsr clusters locate at the poles due to the presence of Tol-Pal at the poles along with the presence of the nucleoid at midcell. Also, these two factors have complementary effects. In addition, two observations suggest that these are likely the only two mechanisms that ensure the Tsr clusters’ preference for polar localization. First, in cells lacking Tol-Pal components, the preference for polar localization decreases linearly with decreasing temperature, suggesting that only one mechanism (nucleoid exclusion) is being affected. Second, in cells lacking Tol-Pal components with relatively small nucleoids, the ability to retain Tsr clusters at the poles is much weakened.
Strikingly, a mechanism of volume exclusion from midcell is only expected to be efficient if the chemoreceptor proteins are able to form sufficiently large clusters (see, e.g., (34, 59, 60)). As such, we suggest that cluster formation not only enhances the signal-processing capabilities of the chemotaxis protein arrays (6, 7, 8, 9) but also is likely essential for ensuring that the clusters are located at the cell poles. Cluster formation should greatly enhance the chances that chemoreceptor proteins will reach and remain at the poles, which is expected to enhance the efficiency of the diffusion-and-capture mechanism made possible by Tol-Pal complexes at the cell extremities.
Recent studies showed that the nucleoid plays a central role in the spatial organization of plasmids (31, 32) and unwanted protein aggregates (33, 34) in the cytoplasm of E. coli, as well as in the choice of location of the cell-division septum (61). By showing how the nucleoid contributes to the spatial organization of sensory complexes, and thus to the functional response of E. coli populations to various external stresses (43, 62, 63), our study further supports the notion that the nucleoid is critical for generating heterogeneities and asymmetries in the cytoplasm of E. coli that are essential for intracellular spatial organization and, combined with cell division, for cell-to-cell diversity within lineages.
Author Contributions
R.N.-V. performed experiments. S.S. and A.S.R. generated mathematical models and simulations. T.A. and S.S. performed image and data analysis. A.S.R. conceived the study. All authors contributed to the experimental design, discussion, and writing of the paper.
Acknowledgments
This work was supported by the Tampere University of Technology President’s Graduate Programme (R.N.-V. and S.S.), the Academy of Finland (general research grant 295027 to A.S.R.), Academy of Finland Key Project Funding (305342 to A.S.R.), and the Jane and Aatos Erkko Foundation (610536 to A.S.R.).
Editor: Zemer Gitai.
Footnotes
Supporting Materials and Methods, Supporting Results, nine figures, and three tables are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(16)30951-1.
Supporting Material
References
- 1.Sourjik V., Berg H.C. Functional interactions between receptors in bacterial chemotaxis. Nature. 2004;428:437–441. doi: 10.1038/nature02406. [DOI] [PubMed] [Google Scholar]
- 2.Lee L., Mizuno T., Imae Y. Thermosensing properties of Escherichia coli tsr mutants defective in serine chemoreception. J. Bacteriol. 1988;170:4769–4774. doi: 10.1128/jb.170.10.4769-4774.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rebbapragada A., Johnson M.S., Taylor B.L. The Aer protein and the serine chemoreceptor Tsr independently sense intracellular energy levels and transduce oxygen, redox, and energy signals for Escherichia coli behavior. Proc. Natl. Acad. Sci. USA. 1997;94:10541–10546. doi: 10.1073/pnas.94.20.10541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wadhams G.H., Armitage J.P. Making sense of it all: bacterial chemotaxis. Nat. Rev. Mol. Cell Biol. 2004;5:1024–1037. doi: 10.1038/nrm1524. [DOI] [PubMed] [Google Scholar]
- 5.Parkinson J.S., Ames P., Studdert C.A. Collaborative signaling by bacterial chemoreceptors. Curr. Opin. Microbiol. 2005;8:116–121. doi: 10.1016/j.mib.2005.02.008. [DOI] [PubMed] [Google Scholar]
- 6.Kentner D., Sourjik V. Spatial organization of the bacterial chemotaxis system. Curr. Opin. Microbiol. 2006;9:619–624. doi: 10.1016/j.mib.2006.10.012. [DOI] [PubMed] [Google Scholar]
- 7.Endres R.G. Polar chemoreceptor clustering by coupled trimers of dimers. Biophys. J. 2009;96:453–463. doi: 10.1016/j.bpj.2008.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Skidmore J.M., Ellefson D.D., Maddock J.R. Polar clustering of the chemoreceptor complex in Escherichia coli occurs in the absence of complete CheA function. J. Bacteriol. 2000;182:967–973. doi: 10.1128/jb.182.4.967-973.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liberman L., Berg H.C., Sourjik V. Effect of chemoreceptor modification on assembly and activity of the receptor-kinase complex in Escherichia coli. J. Bacteriol. 2004;186:6643–6646. doi: 10.1128/JB.186.19.6643-6646.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kentner D., Thiem S., Sourjik V. Determinants of chemoreceptor cluster formation in Escherichia coli. Mol. Microbiol. 2006;61:407–417. doi: 10.1111/j.1365-2958.2006.05250.x. [DOI] [PubMed] [Google Scholar]
- 11.Greenfield D., McEvoy A.L., Liphardt J. Self-organization of the Escherichia coli chemotaxis network imaged with super-resolution light microscopy. PLoS Biol. 2009;7:e1000137. doi: 10.1371/journal.pbio.1000137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Thiem S., Kentner D., Sourjik V. Positioning of chemosensory clusters in E. coli and its relation to cell division. EMBO J. 2007;26:1615–1623. doi: 10.1038/sj.emboj.7601610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Thiem S., Sourjik V. Stochastic assembly of chemoreceptor clusters in Escherichia coli. Mol. Microbiol. 2008;68:1228–1236. doi: 10.1111/j.1365-2958.2008.06227.x. [DOI] [PubMed] [Google Scholar]
- 14.Wang H., Wingreen N.S., Mukhopadhyay R. Self-organized periodicity of protein clusters in growing bacteria. Phys. Rev. Lett. 2008;101:218101. doi: 10.1103/PhysRevLett.101.218101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maddock J.R., Shapiro L. Polar location of the chemoreceptor complex in the Escherichia coli cell. Science. 1993;259:1717–1723. doi: 10.1126/science.8456299. [DOI] [PubMed] [Google Scholar]
- 16.Sourjik V., Berg H.C. Localization of components of the chemotaxis machinery of Escherichia coli using fluorescent protein fusions. Mol. Microbiol. 2000;37:740–751. doi: 10.1046/j.1365-2958.2000.02044.x. [DOI] [PubMed] [Google Scholar]
- 17.Zhang P., Khursigara C.M., Subramaniam S. Direct visualization of Escherichia coli chemotaxis receptor arrays using cryo-electron microscopy. Proc. Natl. Acad. Sci. USA. 2007;104:3777–3781. doi: 10.1073/pnas.0610106104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang K.C., Mukhopadhyay R., Wingreen N.S. A curvature-mediated mechanism for localization of lipids to bacterial poles. PLOS Comput. Biol. 2006;2:e151. doi: 10.1371/journal.pcbi.0020151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rudner D.Z., Pan Q., Losick R.M. Evidence that subcellular localization of a bacterial membrane protein is achieved by diffusion and capture. Proc. Natl. Acad. Sci. USA. 2002;99:8701–8706. doi: 10.1073/pnas.132235899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ebersbach G., Briegel A., Jacobs-Wagner C. A self-associating protein critical for chromosome attachment, division, and polar organization in caulobacter. Cell. 2008;134:956–968. doi: 10.1016/j.cell.2008.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bowman G.R., Comolli L.R., Shapiro L. A polymeric protein anchors the chromosomal origin/ParB complex at a bacterial cell pole. Cell. 2008;134:945–955. doi: 10.1016/j.cell.2008.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ringgaard S., Schirner K., Waldor M.K. A family of ParA-like ATPases promotes cell pole maturation by facilitating polar localization of chemotaxis proteins. Genes Dev. 2011;25:1544–1555. doi: 10.1101/gad.2061811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yamaichi Y., Bruckner R., Waldor M.K. A multidomain hub anchors the chromosome segregation and chemotactic machinery to the bacterial pole. Genes Dev. 2012;26:2348–2360. doi: 10.1101/gad.199869.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Santos T.M.A., Lin T.Y., Weibel D.B. Polar localization of Escherichia coli chemoreceptors requires an intact Tol-Pal complex. Mol. Microbiol. 2014;92:985–1004. doi: 10.1111/mmi.12609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sturgis J.N. Organisation and evolution of the tol-pal gene cluster. J. Mol. Microbiol. Biotechnol. 2001;3:113–122. [PubMed] [Google Scholar]
- 26.Oh D., Yu Y., Ritchie K. Dynamics of the serine chemoreceptor in the Escherichia coli inner membrane: a high-speed single-molecule tracking study. Biophys. J. 2014;106:145–153. doi: 10.1016/j.bpj.2013.09.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fisher J.K., Bourniquel A., Kleckner N. Four-dimensional imaging of E. coli nucleoid organization and dynamics in living cells. Cell. 2013;153:882–895. doi: 10.1016/j.cell.2013.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Niki H., Jaffé A., Hiraga S. The new gene mukB codes for a 177 kd protein with coiled-coil domains involved in chromosome partitioning of E. coli. EMBO J. 1991;10:183–193. doi: 10.1002/j.1460-2075.1991.tb07935.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Winkler J., Seybert A., Bukau B. Quantitative and spatio-temporal features of protein aggregation in Escherichia coli and consequences on protein quality control and cellular ageing. EMBO J. 2010;29:910–923. doi: 10.1038/emboj.2009.412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Coquel A.S., Jacob J.P., Berry H. Localization of protein aggregation in Escherichia coli is governed by diffusion and nucleoid macromolecular crowding effect. PLOS Comput. Biol. 2013;9:e1003038. doi: 10.1371/journal.pcbi.1003038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Reyes-Lamothe R., Tran T., Tolmasky M.E. High-copy bacterial plasmids diffuse in the nucleoid-free space, replicate stochastically and are randomly partitioned at cell division. Nucleic Acids Res. 2014;42:1042–1051. doi: 10.1093/nar/gkt918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vecchiarelli A.G., Mizuuchi K., Funnell B.E. Surfing biological surfaces: exploiting the nucleoid for partition and transport in bacteria. Mol. Microbiol. 2012;86:513–523. doi: 10.1111/mmi.12017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Straight P.D., Fischbach M.A., Kolter R. A singular enzymatic megacomplex from Bacillus subtilis. Proc. Natl. Acad. Sci. USA. 2007;104:305–310. doi: 10.1073/pnas.0609073103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gupta A., Lloyd-Price J., Ribeiro A.S. In vivo kinetics of segregation and polar retention of MS2-GFP-RNA complexes in Escherichia coli. Biophys. J. 2014;106:1928–1937. doi: 10.1016/j.bpj.2014.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yu J., Xiao J., Xie X.S. Probing gene expression in live cells, one protein molecule at a time. Science. 2006;311:1600–1603. doi: 10.1126/science.1119623. [DOI] [PubMed] [Google Scholar]
- 36.Baba T., Ara T., Mori H. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2006;2 doi: 10.1038/msb4100050. 2006.0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chazotte B. Labeling nuclear DNA using DAPI. Cold Spring Harb. Protoc. 2011;2011:t5556. doi: 10.1101/pdb.prot5556. [DOI] [PubMed] [Google Scholar]
- 38.Bakshi S., Choi H., Weisshaar J.C. Nonperturbative imaging of nucleoid morphology in live bacterial cells during an antimicrobial peptide attack. Appl. Environ. Microbiol. 2014;80:4977–4986. doi: 10.1128/AEM.00989-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Boulos L., Prévost M., Desjardins R. LIVE/DEAD BacLight : application of a new rapid staining method for direct enumeration of viable and total bacteria in drinking water. J. Microbiol. Methods. 1999;37:77–86. doi: 10.1016/s0167-7012(99)00048-2. [DOI] [PubMed] [Google Scholar]
- 40.Tokunaga M., Imamoto N., Sakata-Sogawa K. Highly inclined thin illumination enables clear single-molecule imaging in cells. Nat. Methods. 2008;5:159–161. doi: 10.1038/nmeth1171. [DOI] [PubMed] [Google Scholar]
- 41.Chowdhury S., Kandhavelu M., Ribeiro A.S. Cell segmentation by multi-resolution analysis and maximum likelihood estimation (MAMLE) BMC Bioinformatics. 2013;14(Suppl 10):S8. doi: 10.1186/1471-2105-14-S10-S8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Häkkinen A., Muthukrishnan A.B., Ribeiro A.S. CellAging: a tool to study segregation and partitioning in division in cell lineages of Escherichia coli. Bioinformatics. 2013;29:1708–1709. doi: 10.1093/bioinformatics/btt194. [DOI] [PubMed] [Google Scholar]
- 43.Annila, T., R. Neeli-Venkata, and A. S. Ribeiro. 2016. Robustness to sub-optimal temperatures of the processes of Tsr cluster formation and positioning in Escherichia coli. Proc. 9th Int. Jt. Conf. Biomed. Eng. Syst. Technol. 3:137–141.
- 44.Lloyd-Price J., Startceva S., Ribeiro A.S. Dissecting the stochastic transcription initiation process in live Escherichia coli. DNA Res. 2016;23:203–214. doi: 10.1093/dnares/dsw009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lloyd-Price J., Gupta A., Ribeiro A.S. SGNS2: a compartmentalized stochastic chemical kinetics simulator for dynamic cell populations. Bioinformatics. 2012;28:3004–3005. doi: 10.1093/bioinformatics/bts556. [DOI] [PubMed] [Google Scholar]
- 46.Mowbray S.L. Bacterial chemoreceptors: recent progress in structure and function. Mol. Cells. 1999;9:115–118. [PubMed] [Google Scholar]
- 47.Neeli-Venkata R., Martikainen A., Ribeiro A.S. Robustness of the process of nucleoid exclusion of protein aggregates in Escherichia coli. J. Bacteriol. 2016;198:898–906. doi: 10.1128/JB.00848-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Oliveira S.M.D., Neeli-Venkata R., Ribeiro A.S. Increased cytoplasm viscosity hampers aggregate polar segregation in Escherichia coli. Mol. Microbiol. 2016;99:686–699. doi: 10.1111/mmi.13257. [DOI] [PubMed] [Google Scholar]
- 49.Lindner A.B., Madden R., Taddei F. Asymmetric segregation of protein aggregates is associated with cellular aging and rejuvenation. Proc. Natl. Acad. Sci. USA. 2008;105:3076–3081. doi: 10.1073/pnas.0708931105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ping L., Weiner B., Kleckner N. Tsr-GFP accumulates linearly with time at cell poles, and can be used to differentiate ‘old’ versus ‘new’ poles, in Escherichia coli. Mol. Microbiol. 2008;69:1427–1438. doi: 10.1111/j.1365-2958.2008.06372.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Spratt B.G. Distinct penicillin binding proteins involved in the division, elongation, and shape of Escherichia coli K12. Proc. Natl. Acad. Sci. USA. 1975;72:2999–3003. doi: 10.1073/pnas.72.8.2999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yao Z., Kahne D., Kishony R. Distinct single-cell morphological dynamics under beta-lactam antibiotics. Mol. Cell. 2012;48:705–712. doi: 10.1016/j.molcel.2012.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Onogi T., Yamazoe M., Hiraga S. Null mutation of the dam or seqA gene suppresses temperature-sensitive lethality but not hypersensitivity to novobiocin of muk null mutants. J. Bacteriol. 2000;182:5898–5901. doi: 10.1128/jb.182.20.5898-5901.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Parry B.R., Surovtsev I.V., Jacobs-Wagner C. The bacterial cytoplasm has glass-like properties and is fluidized by metabolic activity. Cell. 2014;156:183–194. doi: 10.1016/j.cell.2013.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chatterjee S., Hadi A.S. John Wiley & Sons; Hoboken, NJ: 2006. Regression Analysis by Example. [Google Scholar]
- 56.Akaike H. A new look at the statistical model identification. IEEE Trans. Automat. Contr. 1974;19:716–723. [Google Scholar]
- 57.Taniguchi Y., Choi P.J., Xie X.S. Quantifying E. coli proteome and transcriptome with single-molecule sensitivity in single cells. Science. 2010;329:533–538. doi: 10.1126/science.1188308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gestwicki J.E., Lamanna A.C., Adler J. Evolutionary conservation of methyl-accepting chemotaxis protein location in Bacteria and Archaea. J. Bacteriol. 2000;182:6499–6502. doi: 10.1128/jb.182.22.6499-6502.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rudner D.Z., Losick R. Protein subcellular localization in bacteria. Cold Spring Harb. Perspect. Biol. 2010;2:a000307. doi: 10.1101/cshperspect.a000307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Thompson S.R., Wadhams G.H., Armitage J.P. The positioning of cytoplasmic protein clusters in bacteria. Proc. Natl. Acad. Sci. USA. 2006;103:8209–8214. doi: 10.1073/pnas.0600919103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gupta A., Lloyd-Price J., Ribeiro A.S. Robustness of the division symmetry in Escherichia coli and functional consequences of symmetry breaking. Phys. Biol. 2014;11:066005. doi: 10.1088/1478-3975/11/6/066005. [DOI] [PubMed] [Google Scholar]
- 62.Alon U., Surette M.G., Leibler S. Robustness in bacterial chemotaxis. Nature. 1999;397:168–171. doi: 10.1038/16483. [DOI] [PubMed] [Google Scholar]
- 63.Oleksiuk O., Jakovljevic V., Sourjik V. Thermal robustness of signaling in bacterial chemotaxis. Cell. 2011;145:312–321. doi: 10.1016/j.cell.2011.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.