Abstract
Cu+-specific P-type ATPase membrane protein transporters regulate cellular copper levels. The lack of crystal structures in Cu+-binding states has limited our understanding of how ion entry and binding are achieved. Here, we characterize the molecular basis of Cu+ entry using molecular-dynamics simulations, structural modeling, and in vitro and in vivo functional assays. Protein structural rearrangements resulting in the exposure of positive charges to bulk solvent rather than to lipid phosphates indicate a direct molecular role of the putative docking platform in Cu+ delivery. Mutational analyses and simulations in the presence and absence of Cu+ predict that the ion-entry path involves two ion-binding sites: one transient Met148-Cys382 site and one intramembranous site formed by trigonal coordination to Cys384, Asn689, and Met717. The results reconcile earlier biochemical and x-ray absorption data and provide a molecular understanding of ion entry in Cu+-transporting P-type ATPases.
Introduction
Transition metals are critical for a range of cellular processes (1, 2), yet elevated cellular concentrations of these ions are highly toxic (3, 4). Ion homeostasis is maintained by a carefully orchestrated interplay between a set of proteins that mediate transport across membranes (5, 6, 7). Breakdown of this machinery results in irregularities in the levels of heavy metals, often with severe physiological effects (8). For instance, a range of mutations in the human Cu+-transporting class IB P-type (PIB-1) ATPases ATP7A and ATP7B give rise to the severe Menkes’ and Wilson’s diseases, which manifest as copper deficiency and overload, respectively (8). The recent crystal structures of a bacterial homolog from Legionella pneumophila (LpCopA) (9, 10) and the closely related Zn2+-transporting PIB-2-ATPases (11) have provided a structural framework for elucidating transition-metal P-type (PIB) ATPase transport and related diseases. However, the Cu+- and Zn2+-ATPase crystal structures and conserved sequence motifs show significant differences in the intramembranous ion-binding sites (12, 13, 14, 15, 16), which suggests that the ion-entry mechanisms and determinants of ion specificity are likely unique to each P-type ATPase heavy-metal transporter. Here, we address the central question of how the transported Cu+ ion is delivered to the protein interior for further ATP-dependent transport.
All P-type ATPase proteins share a common structural core of intracellular A- (actuator), P- (phosphorylation), and N- (nucleotide binding) cytosolic domains and a transmembrane (TM) domain (17) (Fig. 1 A). The Post-Albers [E1]-[E1P]-[E2P]-[E2] scheme (where P denotes phosphorylated) is used to describe four structurally distinct reaction intermediate cornerstones associated with P-type ATPase-mediated transport (18, 19) (Fig. S1 in the Supporting Material). Here, E1 and E2P denote states in which the TM domains are open toward the cytoplasm and noncytoplasmic space, respectively. Conformational changes associated with autophosphorylation (from E1 to E1P) and autodephosphorylation (E2P to E2) in the A-, P-, and N-domains are coupled with changes in accessibility to intramembranous ion-binding residues in the TM portion of the protein, which results in ion transport (16). Additionally, intermediate states other than the cornerstone [E1]-[E1P]-[E2P]-[E2] occur during the transport cycle of P-type ATPases and have been extensively described elsewhere (20). In addition to TM helices M1-M6 present in all P-type ATPases, the heavy-metal PIB-ATPases also contain two N-terminal class IB-specific structural features: 1) TM helices MA and MB, and 2) an N-terminal heavy-metal-binding domain (HMBD). Furthermore, a recent study using a combined crystallography/computational approach indicated that the PIB-specific TM helices contribute to a Cu+-release pathway regulated by unique structural rearrangements (10). In contrast, the HMBD binds metal ions but does not seem to be strictly required for ion translocation, which suggests a regulatory function (9, 21, 22).
Figure 1.
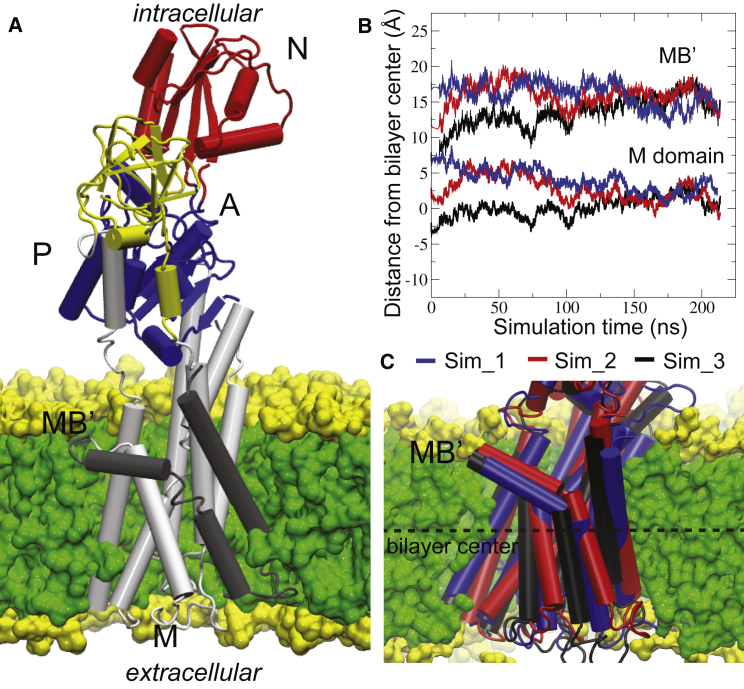
Lipid partitioning of the LpCopA membrane domain. (A) The LpCopA E2.Pi crystal structure (PDB: 3RFU) with the soluble N, P and A cytoplasmic soluble domains inserted into a DOPC lipid bilayer. The membrane M domain contains two PIB-ATPase specific and six P-type ATPase common TM helices. (B) The center-of-mass (COM) for the M-domain and MB’ helix, respectively, for the three Cu+-free E2.Pi simulations. (C) The average structures corresponding to > 150 ns displaying the docked positions in the membrane. To see this figure in color, go online.
Soluble chaperone proteins coordinate Cu+ to maintain very low intracellular concentrations and are responsible for supplying ions to Cu+-ATPases for subsequent extrusion (23). CopZ chaperones were shown to deliver Cu+ to the TM domain of Archaeoglobus fulgidus CopA (AfCopA), whereas the HMBD lacked such capability (21). Based on the LpCopA crystal structures, the C-terminal part of helix MB (referred to as MB') was assigned a putative role as a docking platform for such chaperones (9). In agreement with this notion, mutations of an electronegative patch of a CopZ chaperone abolished Cu+ transport in AfCopA (24), which suggests that positively charged amino acids on the MB' helix are the target for chaperone interactions with the surface of the protein core. In the docked position, the chaperone Cu+-binding site presumably is located close to the putative initial Cu+ receivers Met148, Glu205, and Asp337 (residue numbers refer to LpCopA throughout), as observed for Ca2+ in the related Ca2+-transporting P-type ATPase SERCA1a (9, 25). In SERCA1a, the ions are then transferred to two internal Ca2+-binding sites (26, 27); however, the exact number, location, and chemical nature of the entry pathways in CopA proteins are unknown because the available structural information is limited to E2 states (9, 10), where copper already has been deposited on the noncytosolic side. Mutagenesis and copper binding experiments have suggested two intramembranous ion-binding sites for AfCopA formed by a set of invariant residues (site I: Cys382, Cys384, and Tyr688; site II: Tyr688, Asn689, Met717, and Ser721) (16). Similarly, a two-ion stoichiometry per protein was observed in plant Cu+ ATPases (28). However, a recent study of LpCopA suggested a combination of a transient site partially exposed to the cytoplasmic membrane-water interface and a subsequent single TM high-affinity metal-binding site (14). Although the critical residues between the enzymes are similar, in agreement with the notion of a common Cu+ ATPase transport mechanism, significant alterations are found in the respective HMBD, and consequently we cannot exclude the possibility that these or subtle changes in the TM domain contribute to the observed differences in distinct metal sites. Structural information from x-ray absorption experiments indicates that the high-affinity site in LpCopA coordinates Cu+ in a trigonal sulfur-only site consisting of residues Cys382, Cys384, and Met717 (14). Therefore, residues Asn689 and Ser721 appear unlikely to constitute stable Cu+ ligands in LpCopA, although a more transient role in trafficking of the ion cannot be excluded.
Hence, although the overall structural features involved in Cu+ entry have been sketched out, the molecular details are still missing. Furthermore, because crystal structures are trapped within a static lattice and typically do not contain structural information about the surrounding lipid bilayer, alternative methods are needed to understand the dynamical features involved in Cu+ membrane transport. Molecular-dynamics (MD) simulations can describe the structural dynamics of a protein embedded in a lipid environment under physiological conditions and therefore provide a unique atomistic view of membrane protein transport. However, parameterization of force fields will introduce uncertainties, in particular when describing metal-ion interactions. In addition, enhanced sampling techniques often have to be introduced to enable simulation of biological events, such as ions entering a protein. Therefore, it is critical to verify the theoretical predictions with experimental data such as the in vitro and in vivo mutational activity results obtained in this work.
Here, we characterized the proposed MB' docking platform and Cu+ entry mechanism in LpCopA embedded in a membrane environment using MD simulation techniques and mutational activity assays. Partitioning of the LpCopA protein in the membrane resulted in a reorientation of the amphipathic MB' helix relative to the crystal structure that exposed positively charged side chains of Lys135, Lys142, and in particular Arg136 to the surrounding bulk solvent, providing antennas that could guide the docking of a negatively charged HMBD and/or copper-delivering chaperones. In vitro and in vivo activity measurements confirmed the functional importance of these positive charges. Among the three proposed entry-site residues, Met148 showed significant flexibility toward the internal Cys382 in simulations of the E2.Pi structure, and the Met148-Cys382 residue pair constituted components of a transient binding site in simulations in the presence of Cu+ ions in an E1 structural homology model. Several Met148 mutants showed impaired activities both in vitro and in vivo, establishing Met148 as the mediator of ion transfer between the entry and high-affinity binding sites. Furthermore, the E1 simulations suggest that Cys382 relays Cu+ to a single high-affinity binding site formed by residues Cys384, Asn689, and M717, which refines recent biophysical observations.
Materials and Methods
Building and simulation of Cu+-free systems
The crystal structure of the copper-transporting class IB P-type ATPase lpg1024 (LpCopA) from L. pneumophila trapped in an E2.Pi state (PDB: 3RFU) (9) was used as the starting structure for Cu+-free MD simulations. The protein was inserted into a lipid bilayer consisting of 550 1,2-dioleoyl-sn-glycero-3-phosphocholine (DOPC) lipid molecules built using the CHARMM-GUI membrane builder (29). Three positions of the protein with respect to the vertical z axis of the membrane were chosen: the position predicted by the Orientation of Proteins in Membranes (OPM) database (30) (Sim_2), 5 Å below (−z) the OPM position (Sim_1), and 5 Å above (+z) the OPM position (Sim_3) (Fig. S2). Lipids within 0.6 Å of the protein (25 lipids on each leaflet) were removed to avoid protein-lipid steric clashes. After solvation by 77,637 water molecules and addition of seven Na+ counterions to achieve electrical neutrality, each system consisted of a total of 311,957 atoms.
Three independent >200 ns atomistic MD simulations were performed. Each simulation system was relaxed using a 10,000-step conjugate-gradient energy minimization followed by gradual heating from 0 to 310 K over 120 ps. Equilibrated positions of lipids and water molecules were obtained by two consecutive 1 ns simulations at constant temperature (310 K) and volume (NVT ensemble), where the harmonic restraints of these groups were released successively. The protein was then progressively released from its initial configuration over seven 1 ns simulations, followed by a production run at constant temperature (310 K) and pressure (1 atm) (NPT ensemble). The MD simulations were run with the NAMD 2.9 software package (31). The CHARMM22 force field including CMAP correction (32) and the CHARMM36 force field (33) were used for protein and lipids, respectively, and the TIP3P model (34) was used for water molecules. We note that the new CHARMM36 protein force field (35) resulted in very similar simulated MD during a 35 ns control run (Fig. S3). In addition, the CHARMM36 force field is expected to affect protein folding (e.g., helicity) rather than the protein-lipid, protein-water interactions and side-chain dynamics investigated here. A time step of 1 fs was used to integrate the equations of motion, and a reversible multiple time step algorithm of 4 fs and 2 fs was used for electrostatic forces and short-range, nonbonded forces, respectively. The smooth particle mesh Ewald method (36, 37) was used to calculate electrostatic interactions. The short-range interactions were cut off at 12 Å. All bond lengths involving hydrogen atoms were held fixed using the SHAKE (38) and SETTLE (39) algorithms. A Langevin dynamics scheme was used for thermostating and Nosé-Hoover-Langevin pistons were used for pressure control (40, 41).
Building and simulation of the Cu+ systems
Biochemical and structural studies suggest that the overall conformational changes of P-type ATPases are well conserved (20, 42). As a consequence, individual soluble A-, N-, and P-domains of zinc- and copper-transporting PIB-ATPases superimpose almost perfectly with the corresponding calcium-pumping PIIA-ATPase states, and the same holds true for sodium/potassium and proton transporters (PIIC- and PIIIA-ATPases). Therefore, the E1 LpCopA model was built based on the high-resolution E2P structure of LpCopA (PDB: 4BBJ) (10), with a SERCA1a E1 crystal structure (PDB: 4H1W) (25) as the template, using the software Coot (43). First, the soluble A-, P-, and N-domains of 4BBJ and 4H1W were aligned using secondary-structure-matching (SSM) superpositioning (44). The resulting positions of the A- and P-domains were then used as anchoring points for the second SSM procedure, positioning TM helices M2 and M3, as well as M4 and M5, which then functioned as anchors for placement of M1 and M6 in a third SSM superpositioning. Finally, TM helices MA and MB were placed in similar positions relative to M1 as observed in 4BBJ (with 4BBJ and the E1 model aligned on M1 and M2 using SSM). The connecting linkers of the soluble domains were modeled using a similar SSM approach. In contrast, the short connecting loops between the TM helices were maintained as in the structurally determined E2 states of LpCopA.
To construct a new simulation system for modeling Cu+ interactions, we superimposed the E1 model on an average structure corresponding to the equilibrated (>150 ns) part of the Sim_2 trajectory and removed 12 lipids within 0.5 Å from the protein to avoid clashes. This was followed by a 10,000-step conjugate-gradient energy minimization and a short (10 ns) equilibration using the CHARMM22 and CHARMM36 force fields for protein and lipids, respectively (32, 33), with backbone restraints to equilibrate lipids around the protein. We now considered this system a good starting point for a new, independent simulation addressing Cu+ interactions, and therefore converted the atoms to be consistent with the ffGromos53a6 force field, which contains Cu+ parameters (45), by renaming the atom types and deleting undefined protein and lipid hydrogens. Water molecules were described by the simple point charge water model (46). To initiate this new simulation, we first performed a 50,000-step steepest-descent energy minimization, followed by a 100 ps and 100 ns equilibration in the NVT and NPT ensembles, respectively, with backbone restraints to equilibrate lipid and water interactions as well as side-chain configurations while maintaining the modeled E1 state. The Ramachandran plots of the 4BBJ crystal structure and equilibrated E1 model were more or less identical (Fig. S4). A Cu+ ion was positioned 2.5 Å away from residue E205, which has been assigned a putative role in ion entry and is located above the formed water cavity. After equilibration, the GROMACS pull code (47) with a 1000 kJ mol−1 nm−2 force constant was applied on Cu+ in the z dimension directed toward the extracellular side. In two separate simulations, pull rates of 1.25 and 2.5 Å/ns were applied for 10 and 5 ns, respectively, with the protein backbone atoms as reference. Ramachandran plots before and after the pull showed minor perturbation to the structure (Fig. S4). A 2 fs time step was used and short-range nonbonded interactions were cut off at 1.4 nm, with long-range electrostatics calculated with the particle mesh Ewald algorithm (36, 37). The temperature of the system was maintained by separate coupling of the protein and nonprotein atoms to external temperature baths at 310 K with a Nose-Hoover thermostat (48, 49), and a Parrinello-Rahman barostat (50, 51) was used to isotropically regulate pressure. All bond lengths in the protein were constrained with the LINCS algorithm (52). In the two-ion simulations, pull code protocols were applied in identical fashion as for the single Cu+ ion trajectories, except with the binding Cu+ ion positioned in either the transient entry site or the internal binding site.
Computational analyses
Molecular graphics and simulation analyses were generated with the VMD 1.9.1 software package (53). All alignments between different CopA structures were based on the TM region defined by residues 74–203 (TM helices MA–M2), 335–390 (M3–M4), and 679–730 (M5–M6). The MB' helix was defined as residues 129–144. The criteria for MB' residue interactions with water, lipid carbonyl oxygens, or lipid phosphates were a 3.5 Å distance and 40° angle, and Cu+ interactions were set to protein residues within 3.5 Å.
Protein expression and purification
Overproduction and purification of LpCopA were performed as described in Gourdon et al. (9). Briefly, the recombinant protein was overproduced in Escherichia coli followed by cell lysis in a high-pressure homogenizer. The membranes were isolated and solubilized with C12E8 at a final concentration of 18.6 mM. The LpCopA protein was subsequently purified by nickel affinity and size-exclusion chromatography.
In vitro characterization
The Baginski method employing arsenic detection under aerated conditions was used to monitor ATPase activity (54). The reaction mixture contained 4 μM LpCopA, 40 mM MOPS-KOH pH 6.8, 150 mM NaCl, 5 mM KCl, 5 mM MgCl2, 20 mM (NH4)2SO4, 20 mM l-cysteine, 5 mM NaN3, 0.25 mM Na2MoO4, 1.2 mg/mL L-α-phosphatidylcholine lipids from soybean extract, and 3.7 mM C12E8 in a total volume of 50 μL. To prevent activity from background copper and silver contaminants, 10 μM ammonium tetrathiomolybdate was added, followed by incubation for 15 min with 1 mM CuCl2 (reduced to Cu+ by the cysteine present in the sample). Reactions were started by addition of 5 mM ATP, followed by incubation at 120 rpm for 25 min at 37°C, and stopped by addition of 50 μL of freshly made stop solution containing 0.1% sodium dodecyl sulfate (SDS), 0.4 M HCl, 0.14 mM L-ascorbic acid, and 4.7 mM (NH4)6Mo7O24•4H2O. After 10 min of incubation at 19°C, 75 μL of stabilizing solution (15 mM NaAsO2, 70 mM Na3C6H5O7•2H2O, 350 mM CH3COOH) was added, followed by a final 30 min incubation at 19°C, and absorbance was measured at 860 nm. To allow comparison of wild-type (WT) and mutant LpCopA activities, the relative protein concentrations were determined by quantification and comparison of individual SDS-PAGE band intensities using ImageJ (55). A second SDS-PAGE gel was generated with roughly equal protein amounts, as calculated from the first gel (Fig. S5). The protein amounts were requantified by ImageJ, and information on the variation between these amounts was then used to scale the results of the Baginski assay accordingly. The protein concentration of the D426N mutant was determined with light absorbance at 280 nm and used as reference in the activity assay. These data were normalized to 0–100% according to the activities of the LpCopA D426N dead mutant (or no protein) and the WT proteins, respectively. Each reported measurement was based on nine replicates.
In vivo characterization
The CopA-deleted E. coli strain CN2328, its progenitor strain CN1709, and the plasmid pCN2345 containing the native E. coli CopA promoter and gene were obtained from Lisbeth Birk Møller (56). As described previously (10), the pCN2345 plasmid was mutated to allow LpCopA WT or mutant genes to replace the E. coli CopA gene while preserving the native E. coli CopA promoter. The plasmids were transformed into CN2328 cells, and an empty pET22b(+) vector was transformed into CN1709 cells and used as a control. Precultures were grown overnight and their OD600nm was adjusted to 1.0. Then, 10 μL of cell culture was used to inoculate 190 μL of LB media containing 100 μg/mL ampicillin, and the CuCl2 concentrations were adjusted to 0 and 3.5 mM. The inoculated media were grown in 96-well plates at 200 rpm and 37°C, and the OD560nm was measured at hourly intervals. The OD560nm of cells grown in 3.5 mM CuCl2 for 309 min is shown in Fig. S6. Each reported measurement was based on three replicates.
Results
Lipid partitioning of the Cu+ ATPase membrane domain
To sample the structural dynamics of membrane proteins using MD techniques, the protein must be embedded in a lipid environment. Traditionally, the initial position of the TM domain with respect to the surrounding lipid bilayer is achieved by identification of membrane-spanning helices and subsequent alignment to the lipid bilayer center-of-mass (COM). Because crystal structures typically do not provide information about the lipid bilayer boundaries, the definition of TM helices becomes somewhat speculative. The OPM database (30) utilizes transfer energy minimization between water and the membrane to predict the docking of the membrane-spanning parts into lipid bilayers. Because our study investigates the structural environment involved in ions entering the TM domain, a well-equilibrated position of the protein with respect to the membrane is critical. Therefore, to search for the optimal location of the putative MB' ion docking platform in the membrane-water interface, we monitored the partitioning process by comparing three atomistic simulations of the LpCopA: one in which the TM region was aligned with the membrane according to predictions from the OPM database, and two additional simulations in which the protein TM COM was either 5 Å above or below the aligned position (Sim_1, Sim_2, and Sim_3, respectively; see Fig. S2 and Table S1). The partitioning process was considered to have reached convergence at 150 ns, as determined by tracking the COM for the TM domain and the MB' helix for each system (Fig. 1 B). In principle, a set of independent simulations that fulfill a criterion of convergence (such as COM alignment in the z dimension) should report similar structural properties (such as internal water density). To test this, the water density profiles across the lipid bilayer were calculated for the converged parts (>150 ns) of the three trajectories, and the overall similarity of the obtained profiles validated the assumption of convergence (Fig. S7 A). Therefore, the three average structures from >150 ns can be considered equal and equilibrated (Fig. 1 C). We compared the protein insertion position achieved by our atomistic simulation protocol with that reported in the MemProtMD database (57), which relies on a coarse-grained MD simulation lipid partitioning protocol, and observed very similar results (Fig. S7 B).
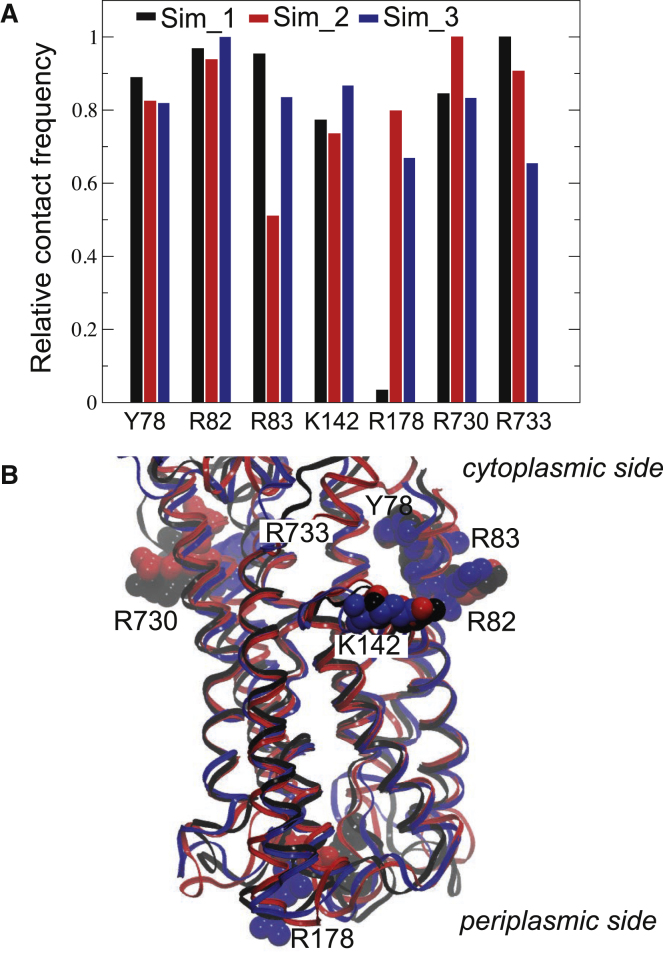
To determine the key amino acids anchoring the membrane domain to the lipid bilayer, we tracked all amino acid residues within 2 Å from a lipid phosphate during the equilibrated part in each trajectory (>150 ns). The seven most prominent lipid-interacting residues were common in-between trajectories (Fig. 2 A), confirming that the simulated lipid partitioning process was equilibrated. Interestingly, almost all residues that showed tight interactions with the surrounding lipids were situated on the cytoplasmic side (Fig. 2 B). The exception was the periplasmic residue Arg178, which was registered as highly interacting in only two of the trajectories. The anchoring residues contained one residue located on the putative MB' docking platform, Lys142. These results suggest that membrane docking of the LpCopA protein is achieved by almost exclusively cytoplasmic residues, which may reflect higher demands of a carefully defined lipid-protein-water interface on the cytoplasmic side, where ion uptake takes place, compared with the periplasmic side.
Figure 2.
The lipid partitioning of the membrane domain. (A) Relative contact frequency of interactions < 2 Å in-between amino acid side chains and lipid phosphates in the three Cu+-free E2.Pi simulations. (B) The identified lipid-interacting residues of simulations 1-3 displayed on the respective averages from > 150 ns. To see this figure in color, go online.
Exposed positive charges are important for copper transport
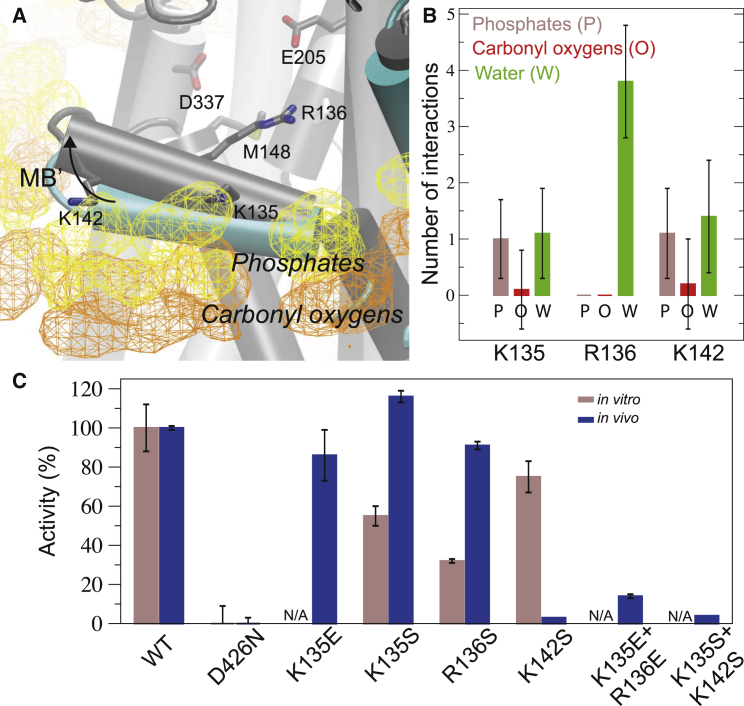
Because the MB' platform has been assigned a putative central role in Cu+ delivery at the cytoplasmic membrane interface, we monitored the structural dynamics displayed by this structural component in the lipid environment. Interestingly, the simulated structure showed a significant structural shift in MB' relative to the original crystal structure (Fig. 3 A). This structural rearrangement suggested an increased cytoplasmic exposure of all the docking platform positive charges (Lys135, Arg136, Lys142) relative to the calculated isodensity surfaces corresponding to lipid phosphates and carbonyl oxygens in the equilibrated (>150 ns) portion of the trajectory (Fig. 3 A). Because positively charged amino acid residues at the membrane interface frequently form strong interactions as membrane anchors with negatively charged lipid phosphate groups (58), exposed positive charges might be indicative of functional roles. We therefore quantified the interactions of the MB' positive charges with lipid phosphates, lipid carbonyl oxygens, and water molecules in the three trajectories. Indeed, residues Lys135, Lys142, and in particular Arg136 (which made no specific lipid interactions) showed significant exposure to bulk water molecules (Fig. 3 B). Therefore, the overall prediction is that the putative MB' docking platform is characterized by significant exposure of positive charges, in agreement with a charged-based mechanism of chaperone and/or HMBD docking to the MB' platform.
Figure 3.
The chemical environment of the putative MB' platform. (A) The transition from the LpCopA E2.Pi structure to the average protein structure representing >150 ns simulation is indicated by the arrow. Isodensity surfaces at 5% occupancy mark the average positions of lipid phosphates and carbonyl oxygens. (B) The number of interactions between Lys135, Arg136, Lys142 and, in turn, lipid phosphates, carbonyl oxygens and water molecules, from the three E2.Pi simulations. (C) Relative in vitro (first columns) and in vivo (second columns) activities of LpCopA MB' mutants as compared to wild-type (WT) protein and the inactive Asp426Asn mutant. To see this figure in color, go online.
To probe the importance of positive charges on the MB' half-helix, we characterized a set of single and double mutants using in vitro and in vivo activity assays (Fig. 3 C). Inorganic phosphate generated by ATPase turnover by the purified LpCopA protein in a lipid-detergent solution was used as a proxy for in vitro activity. The in vivo activity was assessed by the copper resistance of WT and mutant LpCopA forms under control of a native E. coli CopA promoter in a CopA-deficient E. coli strain. In both assays, the activity was plotted on a scale of 0–100% as defined by the catalytically inactive Asp426Asn mutant and WT protein activity, respectively. First, we assessed single-residue charges by individually mutating each charged residue (Lys135, Arg136, and Lys142) to a serine. The measured in vitro activities were significantly reduced, particularly with regard to Arg136Ser, which achieved only 32% of the WT activity. This corroborates the findings from the simulations in which Arg136 was fully solvated in all three trajectories, whereas Lys135 and Lys142 both interacted with surrounding lipids to some extent. However, when we probed activity in a biological context, the Lys135Ser and Arg136Ser mutants displayed unaffected growth rates under copper stress, whereas Lys142Ser cells showed almost no growth. The observed differences may arise from complex compensatory effects such as partial mutant protein production to ensure cell survival, differences in membrane composition or protein stability, or combinations of these factors. Furthermore, we note that no copper-delivering chaperone has been identified for LpCopA or in E. coli. Therefore, Cu+ ions in the in vivo system might be delivered by the HMBD, a hitherto unknown chaperone, or an alternative delivery system. The effect of quenching charges on MB' was cumulative since the Lys135Ser+Lys142Ser double mutant showed almost no activity in vivo. In addition to characterizing absent charge, we also introduced glutamic acid mutations to probe for responses to charge reversal. Compared with the Ser substitution, the Lys135Glu mutant displayed a somewhat reduced in vivo activity. An additional reversed charge in the Lys135Glu+Arg136Glu double mutant again showed a cumulative reduction of in vivo activity. Together, these activity measurements highlight the importance of positive charges on the MB' docking platform in both in vitro and in vivo settings.
Side-chain dynamics in the putative ion-entry site suggest a Cu+ transport pathway
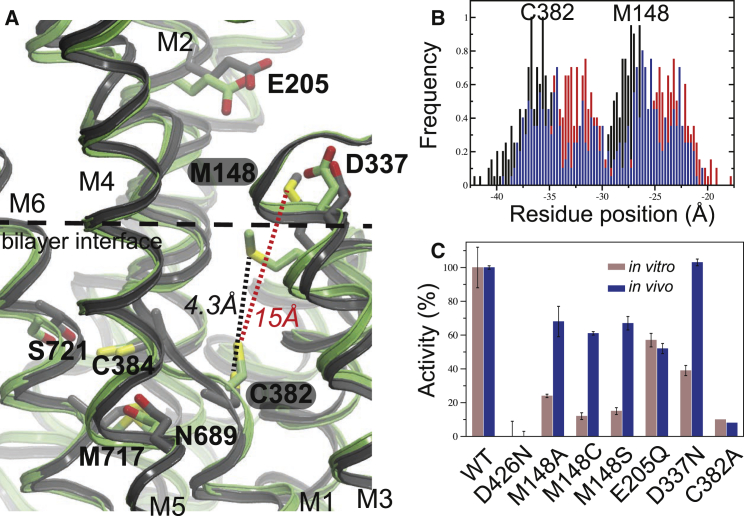
A putative entry site has been assigned to the conserved residues Met148, Glu205, and Asp337, which are located at the cytoplasmic membrane interface (Fig. 4 A). Cys382, on the other hand, belongs to the proposed internal ion-coordinating residues, and this is the residue that is most likely to first accept Cu+ from the entry site, due to the proximal location of its copper-binding sulfur atom to the cytoplasm. Therefore, to identify residues that might be involved in ion passage from the entry site into the membrane domain, we measured how the simulated distances between the three entry-site residues and Cys382 changed relative to the distances in the E2.Pi structure. The minimal distance to the Cu+ acceptor Cys382 was significantly lower for Met148 compared with residues Glu205 and Asp337 (Fig. S8). In addition, the Met148-Cys382 distance was highly dynamic and varied within an 11 Å range (Fig. S8) corresponding to interresidue distances between 15 and 4.3 Å (Fig. 4 A), the latter of which would in principle allow direct copper transfer. However, the average sulfur-to-sulfur Met148-Cys382 distance of 9 ± 2 Å is in agreement with the notion that major conformational changes are required for copper transfer from the copper entry site to the high-affinity binding residues, as anticipated from the Post-Albers cycle (Fig. 4 B; Table S1).
Figure 4.
Structural dynamics in the proposed copper entry site. (A) The dynamic side-chain reorientation between ion entry site Met148 and the internal ion acceptor Cys382 visualized by two superimposed structures that represent extreme configurations with respect to the Cys382-Met148 distance (minimal (4.3 Å) and maximal (15 Å)). (B) The simulated variations in center-of-mass of residues Met148 and Cys382 in the three Cu+-free simulations. (C) Relative in vitro (first columns) and in vivo (second columns) activities of LpCopA entry/transition site mutants as compared to wild-type (WT) protein and the inactive Asp426Asn mutant. To see this figure in color, go online.
To test the predicted importance of the Met148-C382 interplay, we again performed in vitro and in vivo mutational analyses (Fig. 4 C). Strikingly, the Met148Ala, Met148Cys, and Met148Ser mutants displayed a significantly impaired transport capability in vitro, in agreement with previous indications regarding the functional role of Met148 (14). The effect was similar to the activity reduction in the Ala mutation of Cys382, the closest high-affinity-site residue to the entry site. To test whether this was also true in a cellular environment, we assayed the mutant activities in vivo, and observed a 40% reduction in the growth rate. However, although the Asp337Asn and Glu205Gln mutants also showed a reduction of in vitro activities, it was less pronounced compared with the Met148 mutations. In addition, whereas Glu205Gln showed approximately half the activity of the protein in vivo, the Asp337Asn mutant showed no observable impairment. These observations support the idea that Met148 plays an important functional role, most likely in the early stages of metal transfer to the protein, whereas Glu205 and in particular Asp337 seem to be less important (as also underscored by their distal location to Cys382).
A putative Cu+ entry mechanism
To further characterize the molecular basis of copper transfer to the high-affinity-binding residues, we needed access to structural information about the reaction cycle states that are associated with copper uptake and hence are open to the cytoplasm. Given the lack of crystal structures, we turned to modeling based on structural homology. We constructed an LpCopA E1 model by applying E2P-to-E1 conformational changes similar to those observed in SERCA1a (PDB: 4H1W (25) and 3B9B (59), respectively) to the LpCopA E2P crystal structure (PDB: 4BBJ) (10). The strict conservation of conformational changes in the A-, P-, and N-domains observed in all available P-type ATPase crystal structures enables this modeling approach.
During equilibration of the LpCopA E1 model, water molecules entered into a cytoplasmic cavity that extended toward internal amino acid residues that have been proposed to participate in ion binding (14, 16), which is in accordance with an E1 state exposing its internal binding site to the cytoplasm (Fig. S9 A). The Cu+ ion was placed 2.5 Å from the putative entry-site residue Glu205, which showed enhanced sensitivity compared with Asp337 in our functional analyses, and a force directed toward the extracellular side was applied on the ion. In two separate simulations, pull rates of 1.25 and 2.5 Å/ns were applied for 10 and 5 ns (referred to as pull_1 and pull_2, respectively), resulting in ∼12.5 Å moves along the membrane vertical (Fig. 5). In both cases, the Cu+ ion did not engage in protein interactions until it reached Met148. The Cu+ ion maintained its interaction with Met148 while entering into the water-filled cytoplasmic opening in the modeled E1 state (red, transparent isodensity surface in Fig. S9 A), and Met148 subsequently shuttled the Cu+ ion toward Cys382, which was observed to be the primary ion acceptor, thereby confirming the prediction from the Cu+-free simulations. The average sulfur-to-sulfur distance between Met148 and Cys382 was 5 ± 1 Å in both simulations (Figs. 5 A and S9 B; Table S1), which is 4 Å shorter than that observed in the Cu+-free simulations. In addition, the minimum Met148-Cys382 interresidue distance reached ∼3 Å in several instances, which is in agreement with facilitated copper transfer to Cys382 in the E1 state (Fig. S8).
Figure 5.
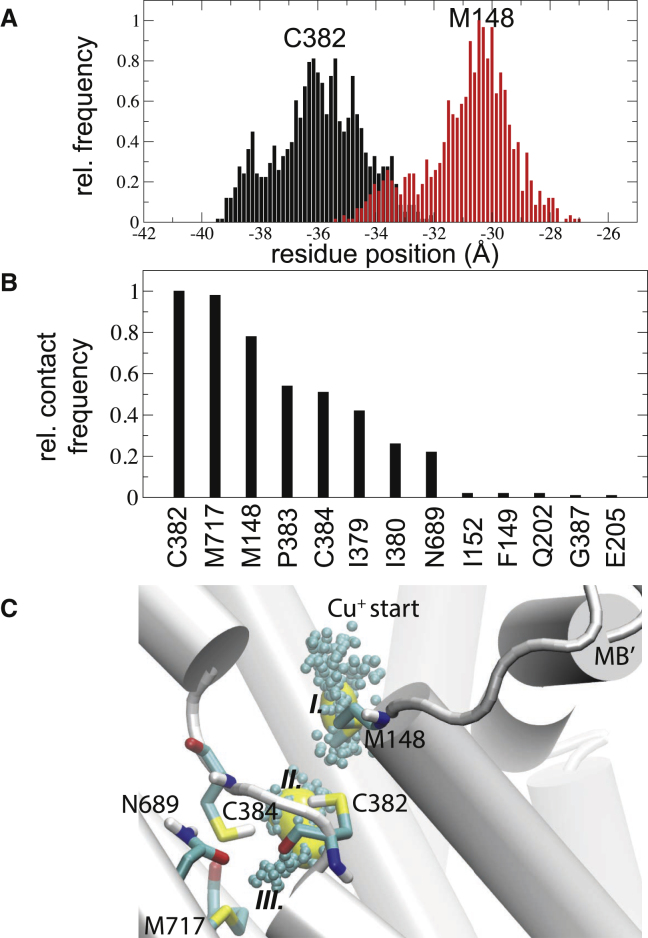
Simulated copper-entry dynamics in the pull_1 simulation. (A) Structural rearrangements of Met148 and Cys382 side chains in the presence of Cu+ depicted as center-of-mass (COM) fluctuations. (B) Contact analysis showing the relative number of instances the Cu+ was < 3.5 Å from any protein residue. (C) Copper ions (spheres) extracted from the simulation with preferred Cu+ positions shown as an isodensity surface at 40% occupancy (Sites I and II). Site III marks the end of the ion pathway. To see this figure in color, go online.
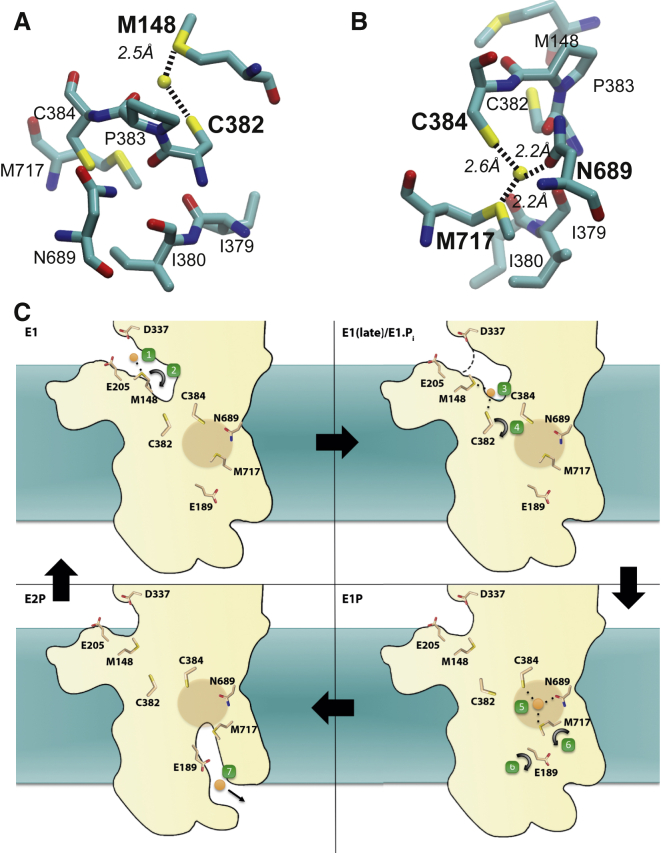
To further characterize the entry-path dynamics, we calculated the contact frequencies of residues within 3.5 Å of the Cu+ ion. Here, we focus on the results from the slower pull, since increased sampling along the putative pathway allows better discrimination among the amino acid contributions. The residues Cys382, Met717, and Met148 generated the highest scores in the contact analyses, which confirms Met148 as the primary Cu+ acceptor and Cys382 as the component that enables shuttling into the protein interior (Fig. 5 B). To further refine preferred ion locations along this putative entry pathway, we calculated an isodensity map for the Cu+ ion in the pull_1 simulation, and observed two major high-density regions: one at Met148 (referred to as site I) and the other in-between Cys382 and Cys384 (referred to as site II; Fig. 5 C). We observed that the ion came to a halt when it encountered Met717 (referred to as site III), which correlates with its assigned role as an internal ion-binding residue (16). The faster pull rate reproduced these overall results (Fig. S9). In both simulations, the Cu+ ion was transiently coordinated in-between Met148 and Cys382 (Fig. 6 A), after which it was shuttled into the internal ion-binding site consisting of residues Met717, Cys384, and Asn689, which bind Cu+ with trigonal coordination (Fig. 6 B). Of note, the adjacent, invariant, and functionally important residues Tyr688 and Ser721 did not partake in copper binding, in agreement with recent biophysical measurements (14).
Figure 6.
Proposed copper entry and binding mechanism. (A) Representative snapshot showing Cu+ shuttling between Met148 and Cys382. (B) Representative snapshot displaying Cu+ binding to the putative internal binding site comprising residues Asn689, Cys384, and Met717. (C) Schematic describing the proposed structural basis for Cu+ transport. To see this figure in color, go online.
Finally, we used a similar protocol to simulate the entry of a second Cu+ ion with either the transient (Fig. S9 A) or internal (Fig. S9 B) binding site occupied by Cu+. In both cases, the second Cu+ ion took alternative routes either into the protein (transient site occupied) or peripheral to the protein (internal site occupied). Because none of these alternative routes involved interactions with the internal acceptor Cys382 (Table S2), our results suggest the entry and binding of a single Cu+ ion.
Discussion
In this work, we characterized membrane anchoring and ion entry into LpCopA, a prototypical Cu+-transporting P-type ATPase from L. pneumophila. By combining computational and experimental techniques, we characterized exposed charges on the docking platform (helix MB') and identified the location and chemical characteristics of one transient and one internal binding site, as well as the structural dynamics involved in our proposed ion-transfer mechanism. Thus, we built on our recent characterization of the release mechanisms of Cu+ (10) and Zn2+ (11) heavy-metal-transporting P-type ATPases.
It is well established that biological membranes are not just inert matrices for membrane proteins, but rather should be viewed as allosteric modulators of protein function (60). For instance, protein-lipid interactions for the sugar transporter lactose permease (LacY) differ between phosphoethanolamine and phosphocholine lipids (61), which might explain the requirement for phosphoethanolamine lipids (62, 63). Critical lipid dependencies have also been established for P-type ATPases. The Ca2+-transporting P-type ATPase SERCA1a is unstable in the absence of lipids (64), requires an ∼30-Å-thick lipid bilayer for optimal function (65, 66), and induces local deformations in the surrounding lipid bilayer (67). Likewise, the function of copper-transporting P-type ATPases is lipid dependent, since proteins solubilized in detergent are inactive in the absence of supplemented lipids (9, 68, 69). However, the molecular details governing lipid partitioning of Cu+ ATPases have not been established.
To extract structural information from simulations of membrane protein transporters, one must have a properly positioned TM domain. In this work, we explored the membrane anchoring of the Cu+ ATPase using one of the existing crystal structures, the E2.Pi state (PDB: 3RFU), and the CHARMM36 force field, which have shown excellent agreement with experimental observables for DOPC lipids (33). By measuring the convergence of the protein position with respect to the surrounding lipids in three MD simulations with different starting positions (Fig. S2), we established an atomistic structural description of LpCopA in the membrane (Fig. 1 C). The simulated protein position closely resembled that obtained using the coarse-grained MemProtMD protocol (57). Because the OPM database (30) outputs the position of the two planes, one cannot expect a perfect agreement. Nevertheless, the predicted position of the periplasmic plane was close to our atomistic simulations, and there were larger discrepancies on the cytoplasmic side (Fig. S7 B). When we tracked protein-lipid interactions in the equilibrated part of the trajectories, we found that a majority of the amino acids with close connections to the lipids were situated on the cytoplasmic side. Therefore, it is possible that a specific anchoring in the lipid bilayer is required for optimal function of Cu+ ATPases, in particular on the cytoplasmic side where metallochaperones and/or the regulatory HMBD docks to deliver Cu+ ions (24).
Positive charges on the MB' half-helix have been proposed to interact with Cu+-carrying metallochaperones and/or the HMBD (24). The MB' is oriented parallel to the membrane mediated by a double-glycine motif and has been identified as a putative docking platform for ion delivery (9). Interestingly, the simulated E2.Pi crystal structure underwent a rearrangement in the incline of helix MB' relative to the membrane (Fig. 3 A), which may represent the optimal configuration of the CopA docking platform in the lipid interface. The starting configurations of the TM helices MA-MB and MB' represent those of the outward-facing E2P state, and we do not claim that our simulations reached an E1 state. Rather, since the SERCA1a TM helices M7–M10 (70) and the β and γ subunits in the Na+,K+-ATPase (71) show very limited rearrangements in-between the E2 and E1 states, the corresponding difference in the LpCopA MA-MB and MB' TM helices may also be characterized by similar restricted rearrangements in the E2-E1 transition. In that case, the three positively charged residues on helix MB' (Lys135, Arg136, and Lys142) were likely solvated by water to a high extent throughout the reaction cycle. This would be especially true for Arg136, which was almost exclusively exposed to water molecules (Fig. 3 B).
To verify this finding, we mutated each of the three identified residues into a serine, which resulted in significantly reduced ATPase activity (with the largest effects on Arg136) in an in vitro assay (Fig. 3 C) in which added soluble cysteine has been shown to imitate the Cu+ delivery of intracellular chaperones (68). The results from the in vivo assay only reported a significant reduction of activity in the Lys142Ser single mutant. However, the activities of the Lys135Glu+Arg136Glu and Lys135Ser+Lys142Ser double mutants were drastically lowered compared with the WT protein. Although the context of the biological assay is complex and we cannot fully explain the phenotypes associated with positions 135 and 136, it is clear that deleting positive charges has severe effects on protein activity. Together, these results corroborate previous observations in a study of AfCopA, in which deleting all positive charges on MB' prevented enzyme activation (24), and provide a putative molecular explanation where a highly flexible MB' platform results in positive charges to solvent due to a highly flexible MB' platform. The importance of the Lys135, Arg136, Lys142 triplet is also reflected in the sequence conservation patterns. Although none of the three residues belong to the invariant amino acids in the platform region, positive charges are very common in the MB' helix. In fact, in almost 90% of ∼1700 sequences of Cu+-transporting PIB-ATPases, more than three (average 3.8) arginines, lysines, or histidines are found in the short stretch from the double glycine motif to Met148 at the intracellular beginning of TM1 (Table S3) (9). These residues are typically separated by three or four other residues, which suggests a conserved charge directionality in the helix, almost certainly toward the intracellular side.
In AfCopA, it was shown that metallochaperones, rather than the HMBD, deliver Cu+ (21). Since no metallochaperone specific to the LpCopA protein has yet been identified and the HMBD structure is atypical and unknown (72), we had to limit our study to characterization of a putative ion-entry path, i.e., subsequent to ion delivery. The differences between the AfCopA and LpCopA proteins were reviewed in (14). The Met148, Glu205, Asp337 triplet has been assigned a putative role as an ion entry site, which was proposed to provide Cu+ to the internal ion-binding residues within the TM domain (9). In experiments with AfCopA, Ala mutants of the equivalent residues showed equal reductions in ATPase activity (24), but these experiments did not reveal how the ions are shuttled into the protein. Therefore, we monitored the dynamics of the putative entry-site residues, first in the E2.Pi crystal structure and then in an E1 model. In the E2.Pi state, Met148 was observed to reorient toward the closest ion acceptor in the internal binding sites, Cys382, to a higher degree than Glu205 and Asp337 (Figs. 4, A and B, and S8). This is consistent with our in vitro data showing that the mutations Glu205Gln and Asp337Asn had a significantly smaller effect on LpCopA activity than the Met148Ala, Met148Cys, and Met148Ser mutations. The in vivo results suggested a less critical role for Asp337, whereas mutating Met148 or Glu205 resulted in a significant loss of activity. This agrees with the observation that Asp337 is more distant from the proposed docking platform and suggests that this residue might serve an alternative function, such as assisting chaperone/HMBD docking, whereas the more proximal Met148, and perhaps also Glu205, might be more directly involved in ion delivery to the TM domain.
The sulfur-sulfur distance between Met148 and the likely internal receiver, Cys382, is 9.3 Å in the Cu+-free E2.Pi state crystal structure (9), and although both residues showed significant flexibility in the E2.Pi simulations, large-scale conformational changes are likely required to enable ion transfer, as observed in other P-type ATPase proteins (70). Therefore, although so far we had relied on the accuracy of the crystal structure, to characterize a putative ion-entry mechanism we had to accept the possible uncertainties of modeling an inward-facing LpCopA E1 state. In addition, although the CHARMM36 force field is excellent for modeling atomistic lipid interactions that are essential to establish the lipid partitioning of LpCopA, it does not contain Cu+ parameters. Since parameterization of transition metal ions requires a major effort, we instead shifted to the ffGromos53a6 force field (45), which proved to be a successful approach for determining the structural dynamics and qualitative free energies associated with ion release from a Cu+ ATPase (10). Still, the inherent difficulty of performing a force-field parameterization of metal ions should be acknowledged, and other copper-protein models have been described (73, 74). Although efforts have been made to develop polarizable force fields (75, 76), a complex membrane protein system is not the ideal test case, and such force fields might introduce artificial effects that would be hard or impossible to detect. Nevertheless, our previous simulations show that it is possible to obtain realistic results using nonpolarizable force-field parameters (10, 11). In addition, a Cu+-bound E1 state, high-resolution structure would add significantly to our understanding of the ion-entry process and internal coordination. However, because such structures might not be amenable to structural characterization, alternative approaches combining biophysical and biochemical techniques are needed to provide mechanistic insights and identification of key residues, and hence help decipher the molecular basis of membrane protein transport.
To introduce the ion into the protein interior, we applied a force on the Cu+ ion toward the extracellular side, with the aim of tracking the preferred pathway into the protein. The starting position of the ion was admittedly arbitrary. However, we do not claim to characterize ion delivery, but rather the entry path, and each time the ion entered the protein the exact same pathway was explored. In the simulation of the E1 state model, the average Met148-Cys382 sulfur-sulfur distance was significantly reduced and Met148 was observed to shuttle Cu+ to Cys382, which indicates that Met148 is mainly responsible for shuttling Cu+ from the entry site. Two significant binding sites were identified in addition to the first transient Met148 contact (site I in Fig. 5 C): one near the cytoplasm between Met148 and Cys382 (Fig. 6 A and site II in Fig. 5 C) and one internal, with a trigonal coordination involving residues Cys384, Asn689, and Met717 (Fig. 6 B and site III in Fig. 5 C). A recent study using extended x-ray absorption fine structure (EXAFS) spectroscopy suggested a partially exposed ion-binding site in the vicinity of Met148 (14), which agrees with our observations. In addition, the EXAFS study assigned Cys382, Cys384, and Met717 to the internal binding site (14). The simulation data showed a similar trigonal coordination, but including Asn689 instead of Cys382, which could not be excluded from the EXAFS data (and is also supported by charge transfer measurements) (14). Our simulation instead assigns a relay point function to Cys382 to transiently coordinate the Cu+ ion, followed by shuttling to the internal binding site. Furthermore, in simulations where either of the two sites was occupied by a Cu+ ion, a second ion failed to enter the proposed entry pathway. Although this suggests a single-ion stoichiometry, we have not explored the effects of different cysteine protonation states that have been shown to affect Cu+ uptake (77). Together, our results reconcile earlier experimental observations and offer a molecular view of the ion-entry process in Cu+-transporting P-type ATPases.
Conclusions
Our work suggests that Cu+-specific P-type ATPase membrane protein transporters anchor predominantly in the cytoplasmic leaflet accompanied by subtle structural differences compared with available crystal structures that result in exposed positive charges on the putative MB' docking platform. The ion-entry molecular path is proposed to involve two ion-binding sites: one transient Met148-Cys382 site and one intramembranous site formed by trigonal coordination to Cys384, Asn689, and Met717. Together, the results presented here provide a molecular understanding of ion entry in Cu+-transporting P-type ATPases.
Author Contributions
C.G. and O.S. performed the activity assays; P.G., E.L., and M.A. designed the research; M.A. performed and analyzed the computational simulations; and all authors wrote the manuscript.
Acknowledgments
We thank Profs. Poul Nissen and Stephen H. White for invaluable advice and discussions throughout this project.
This work was funded by a Marie Curie Career Integration Grant (FP7-MC-CIG-618558); grants from the Magnus Bergvalls Stiftelse (2014-00170), Stiftelsen Olle Engkvist Byggmästare (2015/768), and Åke Wibergs Stiftelse (M15-0148); and a senior postdoctoral fellowship from the Branches Cost-Sharing Fund of the Institute of Complex Adaptive Matter, supported in part by the National Science Foundation, to M.A.; and grants from the Swedish e-Science Research Center and the Swedish Research Council (2013-5901) to E.L. This work was also supported by Lundbeck Foundation, Wallenberg Academy, and Michaelsen fellowships, a Project Research Grant for Junior Researchers from the Swedish Research Council and grants from The Carlsberg, Novo Nordisk, Agnes og Poul Friis, and Brødrene Hartmanns Foundations to P.G., as well as by National Institutes of Health grant GM074637 to Stephen H. White, University of California, Irvine. Computational resources were provided by the Swedish National Infrastructure for Computing (2014/11-33) and the U.S. National Science Foundation through TeraGrid (now XSEDE) resources provided by the Texas Advanced Computing Center at the University of Texas at Austin.
Editor: Carmen Domene.
Footnotes
Ten figures and three tables are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(16)30947-X.
Supporting Material
References
- 1.Mattie M.D., Freedman J.H. Copper-inducible transcription: regulation by metal- and oxidative stress-responsive pathways. Am. J. Physiol. Cell Physiol. 2004;286:C293–C301. doi: 10.1152/ajpcell.00293.2003. [DOI] [PubMed] [Google Scholar]
- 2.Prohaska J.R. Role of copper transporters in copper homeostasis. Am. J. Clin. Nutr. 2008;88:826S–829S. doi: 10.1093/ajcn/88.3.826S. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kehrer J.P. The Haber-Weiss reaction and mechanisms of toxicity. Toxicology. 2000;149:43–50. doi: 10.1016/s0300-483x(00)00231-6. [DOI] [PubMed] [Google Scholar]
- 4.Jomova K., Valko M. Advances in metal-induced oxidative stress and human disease. Toxicology. 2011;283:65–87. doi: 10.1016/j.tox.2011.03.001. [DOI] [PubMed] [Google Scholar]
- 5.Osman D., Cavet J.S. Copper homeostasis in bacteria. Adv. Appl. Microbiol. 2008;65:217–247. doi: 10.1016/S0065-2164(08)00608-4. [DOI] [PubMed] [Google Scholar]
- 6.Kaplan J.H., Lutsenko S. Copper transport in mammalian cells: special care for a metal with special needs. J. Biol. Chem. 2009;284:25461–25465. doi: 10.1074/jbc.R109.031286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Williams L.E., Mills R.F. P(1B)-ATPases--an ancient family of transition metal pumps with diverse functions in plants. Trends Plant Sci. 2005;10:491–502. doi: 10.1016/j.tplants.2005.08.008. [DOI] [PubMed] [Google Scholar]
- 8.Gupta A., Lutsenko S. Human copper transporters: mechanism, role in human diseases and therapeutic potential. Future Med. Chem. 2009;1:1125–1142. doi: 10.4155/fmc.09.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gourdon P., Liu X.-Y., Nissen P. Crystal structure of a copper-transporting PIB-type ATPase. Nature. 2011;475:59–64. doi: 10.1038/nature10191. [DOI] [PubMed] [Google Scholar]
- 10.Andersson M., Mattle D., Gourdon P. Copper-transporting P-type ATPases use a unique ion-release pathway. Nat. Struct. Mol. Biol. 2014;21:43–48. doi: 10.1038/nsmb.2721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang K., Sitsel O., Gourdon P. Structure and mechanism of Zn2+-transporting P-type ATPases. Nature. 2014;514:518–522. doi: 10.1038/nature13618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Argüello J.M. Identification of ion-selectivity determinants in heavy-metal transport P1B-type ATPases. J. Membr. Biol. 2003;195:93–108. doi: 10.1007/s00232-003-2048-2. [DOI] [PubMed] [Google Scholar]
- 13.Raimunda D., Subramanian P., Argüello J.M. A tetrahedral coordination of zinc during transmembrane transport by P-type Zn(2+)-ATPases. Biochim. Biophys. Acta. 2012;1818:1374–1377. doi: 10.1016/j.bbamem.2012.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mattle D., Zhang L., Meloni G. A sulfur-based transport pathway in Cu+-ATPases. EMBO Rep. 2015;16:728–740. doi: 10.15252/embr.201439927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rosenzweig A.C., Argüello J.M. Toward a molecular understanding of metal transport by P(1B)-type ATPases. Curr. Top. Membr. 2012;69:113–136. doi: 10.1016/B978-0-12-394390-3.00005-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.González-Guerrero M., Eren E., Argüello J.M. Structure of the two transmembrane Cu+ transport sites of the Cu+ -ATPases. J. Biol. Chem. 2008;283:29753–29759. doi: 10.1074/jbc.M803248200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kühlbrandt W. Biology, structure and mechanism of P-type ATPases. Nat. Rev. Mol. Cell Biol. 2004;5:282–295. doi: 10.1038/nrm1354. [DOI] [PubMed] [Google Scholar]
- 18.Post R.L., Hegyvary C., Kume S. Activation by adenosine triphosphate in the phosphorylation kinetics of sodium and potassium ion transport adenosine triphosphatase. J. Biol. Chem. 1972;247:6530–6540. [PubMed] [Google Scholar]
- 19.Albers R.W. Biochemical aspects of active transport. Annu. Rev. Biochem. 1967;36:727–756. doi: 10.1146/annurev.bi.36.070167.003455. [DOI] [PubMed] [Google Scholar]
- 20.Bublitz M., Poulsen H., Nissen P. In and out of the cation pumps: P-type ATPase structure revisited. Curr. Opin. Struct. Biol. 2010;20:431–439. doi: 10.1016/j.sbi.2010.06.007. [DOI] [PubMed] [Google Scholar]
- 21.González-Guerrero M., Argüello J.M. Mechanism of Cu+-transporting ATPases: soluble Cu+ chaperones directly transfer Cu+ to transmembrane transport sites. Proc. Natl. Acad. Sci. USA. 2008;105:5992–5997. doi: 10.1073/pnas.0711446105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mitra B., Sharma R. The cysteine-rich amino-terminal domain of ZntA, a Pb(II)/Zn(II)/Cd(II)-translocating ATPase from Escherichia coli, is not essential for its function. Biochemistry. 2001;40:7694–7699. doi: 10.1021/bi010576g. [DOI] [PubMed] [Google Scholar]
- 23.O’Halloran T.V., Culotta V.C. Metallochaperones, an intracellular shuttle service for metal ions. J. Biol. Chem. 2000;275:25057–25060. doi: 10.1074/jbc.R000006200. [DOI] [PubMed] [Google Scholar]
- 24.Padilla-Benavides T., McCann C.J., Argüello J.M. The mechanism of Cu+ transport ATPases: interaction with CU+ chaperones and the role of transient metal-binding sites. J. Biol. Chem. 2013;288:69–78. doi: 10.1074/jbc.M112.420810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Winther A.M., Bublitz M., Buch-Pedersen M.J. The sarcolipin-bound calcium pump stabilizes calcium sites exposed to the cytoplasm. Nature. 2013;495:265–269. doi: 10.1038/nature11900. [DOI] [PubMed] [Google Scholar]
- 26.Toyoshima C., Nakasako M., Ogawa H. Crystal structure of the calcium pump of sarcoplasmic reticulum at 2.6 A resolution. Nature. 2000;405:647–655. doi: 10.1038/35015017. [DOI] [PubMed] [Google Scholar]
- 27.Jensen A.M., Sørensen T.L., Nissen P. Modulatory and catalytic modes of ATP binding by the calcium pump. EMBO J. 2006;25:2305–2314. doi: 10.1038/sj.emboj.7601135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Blaby-Haas C.E., Padilla-Benavides T., Merchant S.S. Evolution of a plant-specific copper chaperone family for chloroplast copper homeostasis. Proc. Natl. Acad. Sci. USA. 2014;111:E5480–E5487. doi: 10.1073/pnas.1421545111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jo S., Lim J.B., Im W. CHARMM-GUI Membrane Builder for mixed bilayers and its application to yeast membranes. Biophys. J. 2009;97:50–58. doi: 10.1016/j.bpj.2009.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lomize M.A., Lomize A.L., Mosberg H.I. OPM: orientations of proteins in membranes database. Bioinformatics. 2006;22:623–625. doi: 10.1093/bioinformatics/btk023. [DOI] [PubMed] [Google Scholar]
- 31.Phillips J.C., Braun R., Schulten K. Scalable molecular dynamics with NAMD. J. Comput. Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mackerell A.D., Jr., Feig M., Brooks C.L., 3rd Extending the treatment of backbone energetics in protein force fields: limitations of gas-phase quantum mechanics in reproducing protein conformational distributions in molecular dynamics simulations. J. Comput. Chem. 2004;25:1400–1415. doi: 10.1002/jcc.20065. [DOI] [PubMed] [Google Scholar]
- 33.Klauda J.B., Venable R.M., Pastor R.W. Update of the CHARMM all-atom additive force field for lipids: validation on six lipid types. J. Phys. Chem. B. 2010;114:7830–7843. doi: 10.1021/jp101759q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jorgensen W., Chandrasekhar J., Klein M. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983;79:926–935. [Google Scholar]
- 35.Best R.B., Zhu X., Mackerell A.D., Jr. Optimization of the additive CHARMM all-atom protein force field targeting improved sampling of the backbone φ, ψ and side-chain χ(1) and χ(2) dihedral angles. J. Chem. Theory Comput. 2012;8:3257–3273. doi: 10.1021/ct300400x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Darden T., York D., Pedersen L. Particle mesh Ewald: an N⋅log(N) method for Ewald sums in large systems. J. Chem. Phys. 1993;98:10089–10092. [Google Scholar]
- 37.Essmann U., Perera L., Pedersen L. A smooth particle mesh Ewald method. J. Chem. Phys. 1995;103:8577–8593. [Google Scholar]
- 38.Ryckaert J.-P., Ciccotti G., Berendsen H. Numerical integration of the Cartesian equations of motion of a system with constraints: molecular dynamics of n-alkanes. J. Comput. Phys. 1977;23:327–341. [Google Scholar]
- 39.Miyamoto S., Kollman P. Settle: an analytical version of the SHAKE and RATTLE algorithm for rigid water models. J. Comput. Chem. 1992;13:952–962. [Google Scholar]
- 40.Feller S., Zhang Y., Brooks B. Constant pressure molecular dynamics simulation: the Langevin piston method. J. Chem. Phys. 1995;103:4613–4621. [Google Scholar]
- 41.Martyna G., Tobias D., Klein M. Constant pressure molecular dynamics algorithms. J. Chem. Phys. 1994;101:4177–4189. [Google Scholar]
- 42.Palmgren M.G., Nissen P. P-type ATPases. Annu. Rev. Biophys. 2011;40:243–266. doi: 10.1146/annurev.biophys.093008.131331. [DOI] [PubMed] [Google Scholar]
- 43.Emsley P., Lohkamp B., Cowtan K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Krissinel E., Henrick K. Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2256–2268. doi: 10.1107/S0907444904026460. [DOI] [PubMed] [Google Scholar]
- 45.Oostenbrink C., Villa A., van Gunsteren W.F. A biomolecular force field based on the free enthalpy of hydration and solvation: the GROMOS force-field parameter sets 53A5 and 53A6. J. Comput. Chem. 2004;25:1656–1676. doi: 10.1002/jcc.20090. [DOI] [PubMed] [Google Scholar]
- 46.Berendsen H.J.C., Postma J.P.M., van Gunsteren W.F., Hermans J. Interaction models for water in relation to protein hydration. In: Pullman B., editor. Intermolecular Forces. Reidel; Dordrecht: 1981. pp. 331–342. [Google Scholar]
- 47.Hess B., Kutzner C., Lindahl E. GROMACS 4: algorithms for highly efficient, load-balanced, and scalable molecular simulation. J. Chem. Theory Comput. 2008;4:435–447. doi: 10.1021/ct700301q. [DOI] [PubMed] [Google Scholar]
- 48.Nosé S. A unified formulation of the constant temperature molecular dynamics methods. J. Chem. Phys. 1984;81:511–519. [Google Scholar]
- 49.Hoover W.G. Canonical dynamics: equilibrium phase-space distributions. Phys. Rev. A Gen. Phys. 1985;31:1695–1697. doi: 10.1103/physreva.31.1695. [DOI] [PubMed] [Google Scholar]
- 50.Nosé S., Klein M.L. Constant pressure molecular dynamics for molecular systems. Mol. Phys. 1983;50:1055–1076. [Google Scholar]
- 51.Parrinello M., Rahman A. Polymorphic transitions in single crystals: a new molecular dynamics method. J. Appl. Phys. 1981;52:7182–7190. [Google Scholar]
- 52.Hess B., Bekker H., Fraaije J. LINCS: a linear constraint solver for molecular simulations. J. Comput. Chem. 1997;18:1463–1472. doi: 10.1021/ct700200b. [DOI] [PubMed] [Google Scholar]
- 53.Humphrey W., Dalke A., Schulten K. VMD: visual molecular dynamics. J. Mol. Graph. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. 27–38. [DOI] [PubMed] [Google Scholar]
- 54.Baginski E.S., Foà P.P., Zak B. Microdetermination of inorganic phosphate, phospholipids, and total phosphate in biologic materials. Clin. Chem. 1967;13:326–332. [PubMed] [Google Scholar]
- 55.Schneider C.A., Rasband W.S., Eliceiri K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Petersen C., Møller L.B. Control of copper homeostasis in Escherichia coli by a P-type ATPase, CopA, and a MerR-like transcriptional activator, CopR. Gene. 2000;261:289–298. doi: 10.1016/s0378-1119(00)00509-6. [DOI] [PubMed] [Google Scholar]
- 57.Stansfeld P.J., Goose J.E., Sansom M.S. MemProtMD: automated insertion of membrane protein structures into explicit lipid membranes. Structure. 2015;23:1350–1361. doi: 10.1016/j.str.2015.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Andersson M., Freites J.A., White S.H. Structural dynamics of the S4 voltage-sensor helix in lipid bilayers lacking phosphate groups. J. Phys. Chem. B. 2011;115:8732–8738. doi: 10.1021/jp2001964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Olesen C., Picard M., Nissen P. The structural basis of calcium transport by the calcium pump. Nature. 2007;450:1036–1042. doi: 10.1038/nature06418. [DOI] [PubMed] [Google Scholar]
- 60.Lee A.G. How lipids affect the activities of integral membrane proteins. Biochim. Biophys. Acta. 2004;1666:62–87. doi: 10.1016/j.bbamem.2004.05.012. [DOI] [PubMed] [Google Scholar]
- 61.Andersson M., Bondar A.N., White S.H. Proton-coupled dynamics in lactose permease. Structure. 2012;20:1893–1904. doi: 10.1016/j.str.2012.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bogdanov M., Heacock P.N., Dowhan W. A polytopic membrane protein displays a reversible topology dependent on membrane lipid composition. EMBO J. 2002;21:2107–2116. doi: 10.1093/emboj/21.9.2107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bogdanov M., Heacock P., Dowhan W. Plasticity of lipid-protein interactions in the function and topogenesis of the membrane protein lactose permease from Escherichia coli. Proc. Natl. Acad. Sci. USA. 2010;107:15057–15062. doi: 10.1073/pnas.1006286107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lund S., Orlowski S., Møller J.V. Detergent structure and associated lipid as determinants in the stabilization of solubilized Ca2+-ATPase from sarcoplasmic reticulum. J. Biol. Chem. 1989;264:4907–4915. [PubMed] [Google Scholar]
- 65.Starling A.P., East J.M., Lee A.G. Effects of phosphatidylcholine fatty acyl chain length on calcium binding and other functions of the (Ca(2+)-Mg2+)-ATPase. Biochemistry. 1993;32:1593–1600. doi: 10.1021/bi00057a025. [DOI] [PubMed] [Google Scholar]
- 66.Caffrey M., Feigenson G.W. Fluorescence quenching in model membranes. 3. Relationship between calcium adenosinetriphosphatase enzyme activity and the affinity of the protein for phosphatidylcholines with different acyl chain characteristics. Biochemistry. 1981;20:1949–1961. doi: 10.1021/bi00510a034. [DOI] [PubMed] [Google Scholar]
- 67.Sonntag Y., Musgaard M., Thøgersen L. Mutual adaptation of a membrane protein and its lipid bilayer during conformational changes. Nat. Commun. 2011;2:304. doi: 10.1038/ncomms1307. [DOI] [PubMed] [Google Scholar]
- 68.Yang Y., Mandal A.K., Argüello J.M. Activation of Archaeoglobus fulgidus Cu(+)-ATPase CopA by cysteine. Biochim. Biophys. Acta. 2007;1768:495–501. doi: 10.1016/j.bbamem.2006.09.013. [DOI] [PubMed] [Google Scholar]
- 69.Chintalapati S., Al Kurdi R., Kühlbrandt W. Membrane structure of CtrA3, a copper-transporting P-type-ATPase from Aquifex aeolicus. J. Mol. Biol. 2008;378:581–595. doi: 10.1016/j.jmb.2008.01.094. [DOI] [PubMed] [Google Scholar]
- 70.Møller J.V., Olesen C., Nissen P. The sarcoplasmic Ca2+-ATPase: design of a perfect chemi-osmotic pump. Q. Rev. Biophys. 2010;43:501–566. doi: 10.1017/S003358351000017X. [DOI] [PubMed] [Google Scholar]
- 71.Nyblom M., Poulsen H., Nissen P. Crystal structure of Na+, K(+)-ATPase in the Na(+)-bound state. Science. 2013;342:123–127. doi: 10.1126/science.1243352. [DOI] [PubMed] [Google Scholar]
- 72.Sitsel O., Grønberg C., Gourdon P. Structure and function of Cu(I)- and Zn(II)-ATPases. Biochemistry. 2015;54:5673–5683. doi: 10.1021/acs.biochem.5b00512. [DOI] [PubMed] [Google Scholar]
- 73.Click T.H., Ponomarev S.Y., Kaminski G.A. Importance of electrostatic polarizability in calculating cysteine acidity constants and copper(I) binding energy of Bacillus subtilis CopZ. J. Comput. Chem. 2012;33:1142–1151. doi: 10.1002/jcc.22944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ponomarev S.Y., Click T.H., Kaminski G.A. Electrostatic polarization is crucial in reproducing Cu(I) interaction energies and hydration. J. Phys. Chem. B. 2011;115:10079–10085. doi: 10.1021/jp2051933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yu H., van Gunsteren W. Accounting for polarization in molecular simulation. Comput. Phys. Commun. 2005;172:69–85. [Google Scholar]
- 76.Lopes P.E., Roux B., Mackerell A.D., Jr. Molecular modeling and dynamics studies with explicit inclusion of electronic polarizability. Theory and applications. Theor. Chem. Acc. 2009;124:11–28. doi: 10.1007/s00214-009-0617-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Dumay Q.C., Debut A.J., Saier M.H., Jr. The copper transporter (Ctr) family of Cu+ uptake systems. J. Mol. Microbiol. Biotechnol. 2006;11:10–19. doi: 10.1159/000092815. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.