Abstract
Epidemiological studies showed that obesity and its related non-alcoholic fatty liver disease (NAFLD) promote hepatocellular carcinoma (HCC) development. We aimed to uncover the genetic alterations of NAFLD-HCC using whole-exome sequencing. We compared HCC development in genetically obese mice and dietary obese mice with wild-type lean mice fed a normal chow after treatment with diethylnitrosamine. HCC tumor and adjacent normal samples from obese and lean mice were then subjected to whole-exome sequencing. Functional and mechanistic importance of the identified mutations in Carboxyl ester lipase (Cel) gene and Harvey rat sarcoma virus oncogene 1 (Hras) was further elucidated. We demonstrated significantly higher incidences of HCC in both genetic and dietary obese mice with NAFLD development as compared with lean mice without NAFLD. The mutational signatures of NAFLD-HCC and lean HCC were distinct, with <3% overlapped. Eight metabolic or oncogenic pathways were found to be significantly enriched by mutated genes in NAFLD-HCC, but only two of these pathways were dysregulated by mutations in lean HCC. In particular, Cel was mutated significantly more frequently in NAFLD-HCC than in lean HCC. The multiple-site mutations in Cel are loss-of-function mutations, with effects similar to Cel knock-down. Mutant Cel caused accumulation of cholesteryl ester in liver cells, which led to induction of endoplasmic reticulum stress and consequently activated the IRE1α/c-Jun N-terminal kinase (JNK)/c-Jun/activating protein-1 (AP-1) signaling cascade to promote liver cell growth. In addition, single-site mutations in Hras at codon 61 were found in NAFLD-HCC but none in lean HCC. The gain-of-function mutations in Hras (Q61R and Q61K) significantly promoted liver cell growth through activating the mitogen-activated protein kinase (MAPK) and phosphatidylinositol-4,5-bisphosphate 3-kinase (PI3K)/3-phosphoinositide-dependent protein kinase-1 (PDK1)/Akt pathways. In conclusion, we have identified mutation signature and pathways in NAFLD-associated HCC. Mutations in Cel and Hras have important roles in NAFLD-associated hepatocellular carcinogenesis.
Introduction
Hepatocellular carcinoma (HCC) is now increasingly diagnosed among obese individuals in Western countries and Asia.1, 2, 3, 4 Hepatic manifestations of obesity and the metabolic syndrome are collectively termed non-alcoholic fatty liver disease (NAFLD). The clinicopathological spectrum of NAFLD widely ranges from hepatic steatosis to more aggressive form of non-alcoholic steatohepatitis. Recently, non-alcoholic steatohepatitis has been considered to be an important causative factor in HCC.5 Large population-based cohort studies from United States, Europe and Asia demonstrate a 1.89-fold increased risk of HCC in obese cohorts compared with healthy controls.6 Moreover, type 2 diabetes has also been identified as an independent risk factor for HCC.7 Although the percentage of patients diagnosed annually with NAFLD-associated HCC (NAFLD-HCC) is still relatively low, this actual number is high due to a large global population of individuals with obesity and diabetes. Obesity-induced inflammation, insulin resistance and oxidative stress may be involved in the carcinogenesis of NAFLD-HCC. However, the underlying mechanism especially the genetic mechanism of NAFLD-associated liver carcinogenesis is still unknown.
Cel, the carboxyl ester lipase, functions in lipid metabolism, including uptake of cholesteryl ester in the liver and hydrolysis of cholesteryl esters.8 Human CEL mutation has been reported to cause a syndrome involving diabetes.9 But it has never been investigated in any kind of cancers. The Harvey rat sarcoma virus oncogene 1 (Hras) belongs to the ras superfamily. Hras is a GTPase that functions in converting GTP to GDP to transduct signal to regulate cell growth. Mutations in Ras protein coding genes including Hras have been reported in cancer.10 However, the involvement of Cel and Hras has not yet been demonstrated in NAFLD-associated liver carcinogenesis.
In the present study, we demonstrated that dietary and genetic obesity are direct promoter of NAFLD-HCC development and then examined its mechanism of action. We combined the whole-exome sequencing and cross-species oncogenomics to identify important gene mutations related to NAFLD-HCC. We demonstrated the mutation signature and eight important metabolic and cancer-related pathways in NAFLD-associated HCC. We identified inactivating mutations in Cel and activating mutations in Hras that promoted the development of NAFLD-HCC. Furthermore, we elucidated the molecular mechanisms of these mutations in NAFLD-HCC.
Results
NAFLD is associated with increased susceptibility of hepatocarcinogenesis in genetic and dietary obese mouse models
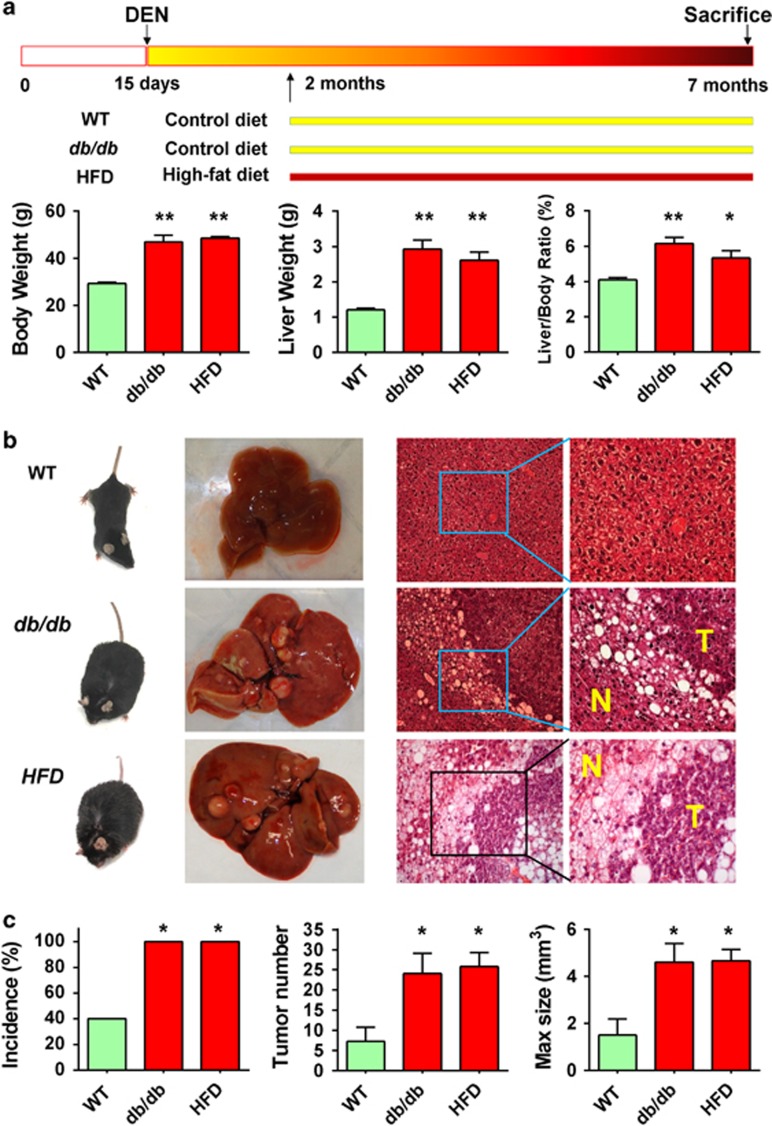
To compare HCC incidence between obese and lean mice, we treated genetically obese (db/db) mice and wild-type lean mice with the carcinogen diethylnitrosamine (DEN) by intraperitoneal injection at age of 13–15 days; dietary obese mice were established using high-fat diet (HFD) at age of 2 months after administration of DEN to wild-type lean mice (Figure 1a). At 7 months, both db/db and HFD obese mice had significantly higher body weight (47.0±9.7 and 48.6±2.3 g vs 29.3±1.7 g; P<0.001), liver weight (2.9±0.9 and 2.6±0.7 g vs 1.2±0.2 g; P<0.001) and liver-to-body weight ratio (6.2±1.2 and 5.3±1.3% vs 4.1±0.4% P<0.05) as compared with wild-type lean mice. Typical hepatic steatosis was observed in livers of both obese mice but not in wild-type lean mice (Figure 1b). HCC incidence was significantly higher in db/db mice (100%) and HFD-fed mice (100%) as compared with wild-type lean mice (40%) (P<0.05). In addition, db/db and HFD mice also developed tumor nodes significantly more in number (24.1±18.2 and 25.8±11.0 vs 7.3±11.1, P<0.05) and larger in size (4.6±2.8 and 4.7±1.6 mm3 vs 1.5±2.1 mm3; P<0.05) than wild-type mice (Figure 1c). Similar findings in HCC incidence, tumor number and size were also observed at 8 months in both obese animal models (Supplementary Figure 1). These results demonstrated that NAFLD increased the susceptibility of HCC formation.
Figure 1.
Obesity increases susceptibility of hepatocarcinogenesis in mice. (a) Schematic illustration of the treatment of wild-type (WT) control lean mice (n=10), genetically obese (db/db) mice (n=13) and dietary obese HFD mice (n=10). Mice were killed at 7 months of age. Body weight, liver weight and liver-to-body weight ratio were examined. (b) Representative gross morphology of liver tumors and microscopic features of HCC in H&E-stained liver sections were shown. (c) HCC incidence, number of HCCs per mouse and maximal size of the tumors were examined. All comparisons are performed by unpaired t tests except incidence (Fisher's exact test). *P<0.05, **P<0.001. T, tumor; N, non-tumor. Means±s.e.m. are shown in the bar charts, while means±s.d. are indicated in the corresponding ‘Result'.
Whole-exome sequencing revealed genomic alteration landscape in NAFLD-associated HCC
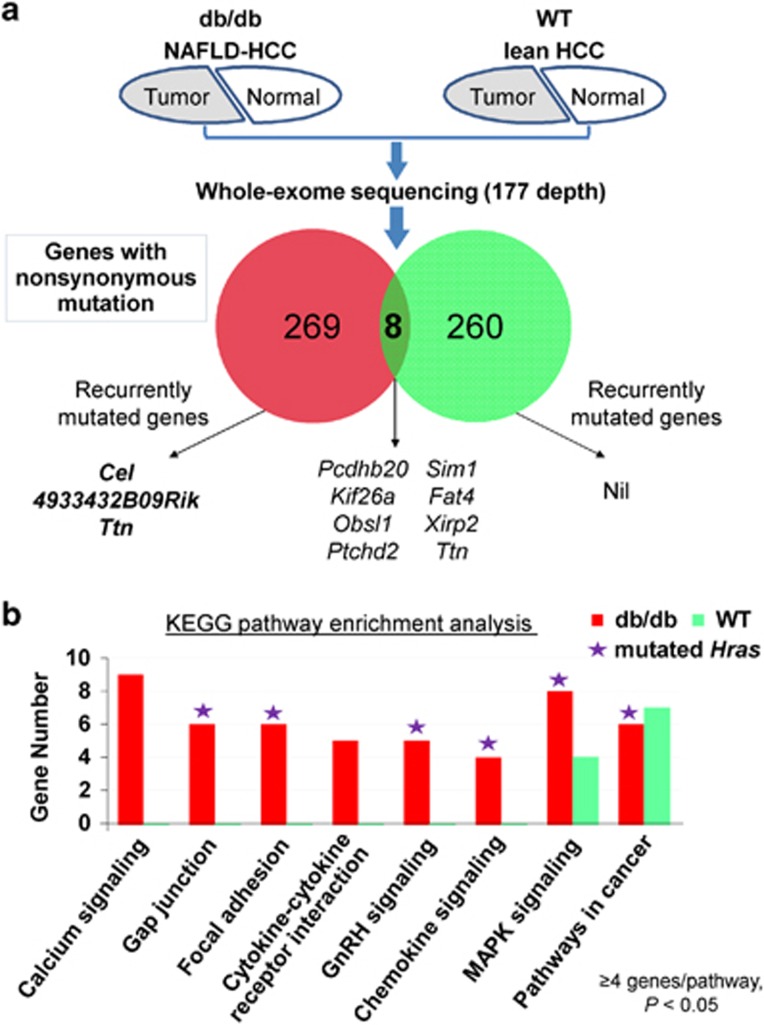
To elucidate the genetic basis of NAFLD-associated HCC, tumor and adjacent non-tumor livers from two db/db mice and two wild-type lean mice were subjected to whole-exome capture sequencing. The average sequencing depth was 177.0±31.6 ×, covering 97.7±0.1% of the targeted exome. Somatic and novel single-nucleotide variants (SNVs) and small insertions and deletions (indels) were identified by comparing tumor with adjacent non-tumor samples and filtering against dbSNP128 database. A total of 282 novel non-synonymous SNVs and 1 exonic indel covering 277 genes were identified in tumors from db/db mice, while 271 novel non-synonymous SNVs covering 268 genes were found in tumors from control lean mice (no indel) (Figure 2a). Eight mutated genes overlapped between the obese and lean HCCs. Among the mutation genes in obese mice, three genes including Cel, 4933432B09Rik and Ttn (titin) were found to be recurrently mutated in the two obese mice. Cel and 4933432B09Rik were mutated in obese mice only, while mutation in Ttn was also observed in one lean mouse (Figure 2a). Therefore, two genes (Cel and 4933432B09Rik) were identified to be recurrently and specifically mutated in NAFLD-associated HCC.
Figure 2.
Whole-exome sequencing analysis identified genomic alterations of NAFLD-HCC. (a) HCC-associated non-synonymous mutations were identified by exome sequencing in db/db mice and WT lean mice. Genes mutated in more than one case were defined as recurrently mutated genes. (b) KEGG pathway enrichment analysis identified eight important pathways of interest to be significantly dysregulated in db/db obese mice, while only two in lean mice.
Important pathways are specifically dysregulated by NAFLD-associated mutations during hepatocarcinogenesis
To understand the dysregulated molecular processes in NAFLD-associated HCC, enrichment analysis of Kyoto Encyclopedia of genes and genomes (KEGG) pathway was performed using mutated genes identified in tumors from obese mice and wild-type mice, respectively. As shown in Figure 2b and Supplementary Table 1, mutated genes in NAFLD-associated HCC were found to be enriched in eight cancer pathways (with ⩾4 genes involved in each pathway and P<0.05) including calcium signaling, gap junction, focal adhesion, cytokine–cytokine receptor interaction, gonadotropin-releasing hormone (GnRH) signaling, chemokine signaling, mitogen-activated protein kinase (MAPK) signaling and pathways in cancer, while only two of these pathways, namely MAPK signaling and pathways in cancer, were affected by genetic alterations in lean mice (⩾4 genes, P<0.05). This finding suggests that development of NAFLD-associated HCC involves genetic alterations in inflammation-related pathways (calcium signaling,11 chemokine signaling,12 cytokine–cytokine receptor interaction13 and gap junction14) besides cancer-related pathways (MAPK signaling, GnRH signaling, focal adhesion and pathways in cancer). Interestingly, mutated Hras was involved in six of the eight affected pathways in obese mice (Figure 2b), demonstrating the functional importance of the mutated Hras in the dysregulated signaling network by genetic alterations during NAFLD-HCC development.
Validation refined recurrently mutated gene Cel in NAFLD-associated HCC
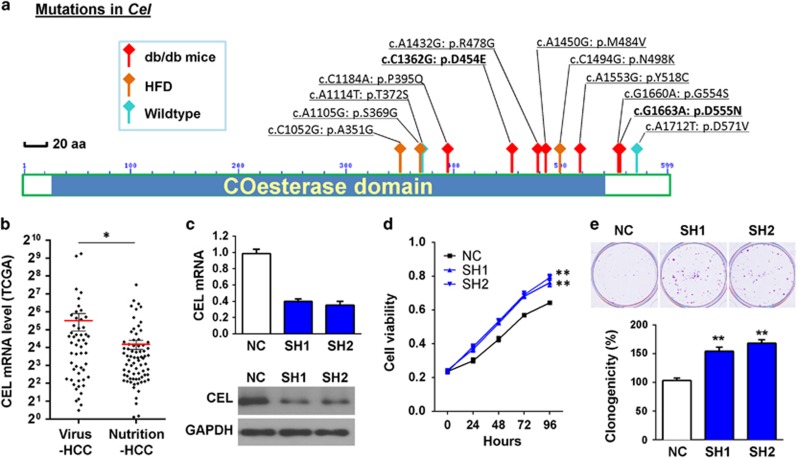
Carboxyl ester lipase, encoded by the gene Cel, functions in lipid metabolism.15 Therefore, mutations in Cel, which recurrently and specifically occurred in obese mice as revealed by exome sequencing, may take part in NAFLD-associated HCC development. We then further performed mutation examination on Cel by PCR and Sanger sequencing in tumor and adjacent non-tumor livers from 29 obese mice (16 db/db and 13 HFD) and 16 control lean mice. Somatic non-synonymous Cel mutations were found significantly more frequently in NAFLD-HCCs from obese mice (10/29, 34.5%) than in control lean HCCs (1/16, 6.3%) (P<0.05; Figure 3a). Moreover, mutations in the human ortholog CEL were only found in nutrition-associated HCC (3.75%, 3/80), but not identified in any virus infection-associated HCC (0/108) in TCGA database (Supplementary Table 2).
Figure 3.
Mutations in Cel and growth-promoting effect of loss of CEL. (a) Schematic illustration of the somatic non-synonymous mutations in Cel identified in liver tumors from obese mice (16 genetic, 13 dietary) and control lean mice (n=16). (b) The mRNA expression of CEL in nutrition-associated HCC (n=78) was significantly lower than in virus infection-associated HCC (n=56) that was available in TCGA database (independent t-test). (c) Stable knock-down of CEL in normal hepatocyte cell line MIHA by two shCEL vectors (SH1 and SH2) was confirmed by real-time PCR and western blot. (d) CEL downregulation in MIHA cells increased cell growth as shown by MTT assay. (e) CEL downregulation in MIHA cells promoted colony formation ability of MIHA cells. Data are mean±s.e.m. in (b) and mean±s.d. elsewhere. *P<0.05, **P<0.01. NC, negative control shRNA; SH1&SH2, shRNAs targeting CEL.
Downregulation of CEL promotes cell proliferation by enhancing the cholesteryl ester level
We further examined the expression level of CEL gene between nutrition-associated HCC and virus infection-associated HCC that was available in TCGA database. We found that the mRNA expression of CEL in nutrition-associated HCC (n=78) was significantly lower than in virus infection-associated HCC (n=56) (Figure 3b). To understand the role of CEL in liver cells, we knocked down expression of CEL in the immortalized normal human liver cell line MIHA using shRNAs against CEL (Figure 3c). Knock-down of CEL by two shCEL vectors (SH1 and SH2) significantly promoted cell growth as revealed by cell viability and colony formation (Figures 3d and e) assays, inferring downregulation of CEL promotes liver cell growth.
The CEL protein cholesteryl esterase functions in hydrolyzing cholesteryl esters.7 We thus examined whether knock-down of CEL would alter the homeostasis of cholesterol and cholesteryl ester. As shown in Figure 4a1, downregulation of CEL significantly increased the amount of cholesteryl ester in MIHA cells (Figure 4a1). Cholesteryl palmitate is a form of cholesteryl ester having liquid-crystalline states, thus suitable to be used in cell culture.16, 17 Treatment with this liquid cholesteryl ester significantly stimulated MIHA cell growth (Figure 4a2). On the other hand, the accumulated intracellular cholesteryl ester caused by CEL knock-down could be significantly abolished by treatment with Avasimibe, an inhibitor of the major enzyme in esterification of cholesterol, ACAT (acyl-coenzyme A:cholesterol acyltransferase) (Supplementary Figure 2A).18 This reduction in cholesteryl ester by Avasimibe subsequently reduced cell growth in CEL knock-down MIHA cells (Supplementary Figure 2B). These results collectively suggest that downregulation of CEL promotes cell proliferation by increasing intracellular accumulation of cholesteryl ester.
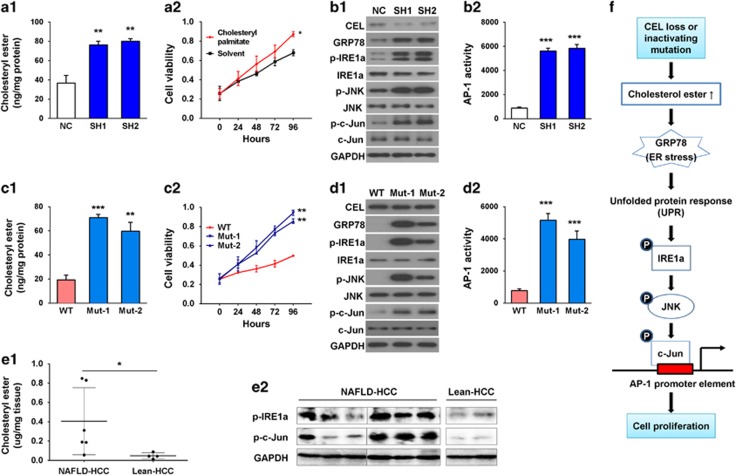
Figure 4.
Loss of CEL caused intracellular accumulation of cholesteryl ester to induce ER stress and activation of IRE1α/JNK/c-Jun/AP-1 pathway. (a1) CEL knock-down increased the level of cholesteryl ester in MIHA cells. (a2) Treatment with 25 nm cholesteryl palmitate, a form of cholesteryl ester having liquid-crystalline states, promoted cell growth as shown by MTT assay. (b1) CEL knock-down in MIHA cells activated the signaling cascade in ER stress as shown by western blot analysis of key proteins. (b2) CEL knock-down increased the transcriptional activity of AP-1 in MIHA as determined by luciferase reporter assay. (c1) MIHA cells expressing CEL mutants (Mut-1, Mut-2) caused significantly higher levels of cholesteryl ester as compared with cells overexpressing wild-type CEL. (c2) CEL mutants promoted MIHA cell growth as shown by MTT assay. (d1) CEL mutants activated the signaling cascade of ER stress in MIHA. (d2) CEL mutants increased the transcriptional activity of AP-1 in MIHA. (e) Cholesteryl ester levels (e1) and protein levels of p-IRE1a and p-c-Jun (e2) in NAFLD-HCC and lean HCC from mouse models. Proteins from two lean HCC were not adequate for both tests. (f) Mechanistic scheme of CEL knock-down or inactivating mutation in promoting liver cancer growth. Data are means±s.d. *P<0.05; **P<0.01; ***P<0.001. NC, negative control shRNA; SH1&SH2, shRNAs targeting CEL.
Knock-down of CEL causes endoplasmic reticulum stress and consequent activation of IRE1α/c-Jun N-terminal kinase/c-Jun/activating protein-1 pathway
As the synthesis of cholesteryl esters occurs in the endoplasmic reticulum (ER),19 we examined the effect of CEL knock-down on ER stress and the corresponding downstream signaling cascade. As shown in Figure 4b1, knock-down of CEL in MIHA cells remarkably enhanced the protein expression of the ER stress marker GRP78. Moreover, the level of phosphorylated inositol-requiring enzyme-1α (p-IRE1α), a key regulator of the mammalian unfolded protein response (UPR), was also markedly increased, inferring CEL depletion causes ER stress and UPR by activating IRE1α. It is known that activated IRE1α triggers c-Jun N-terminal kinase (JNK).20 Here, knock-down of CEL led to significantly enhanced levels of p-JNK and p-c-Jun (Figure 4b1). Activation of c-Jun by CEL knock-down was further confirmed by the activation of its direct downstream effector c-Jun-driven activating protein-1 (AP-1) as evidenced by luciferase reporter assay (Figure 4b2). Furthermore, treatment with Avasimibe, which reduced cholesteryl ester in CEL knock-down MIHA cells, effectively reversed the effect of CEL knock-down on ER stress and IRE1α/JNK/c-Jun/AP-1 signaling cascade (Supplementary Figure 2C), which further supporting that CEL functions via modulating the level of cholesteryl ester. Taken together, these results suggest that CEL depletion interferes with cholesterol metabolism, which promotes cell growth through enhancing ER stress and consequently activating IRE1α /JNK/c-Jun/AP-1 signaling pathway.
Loss-of-function mutations in CEL promote cell proliferation and activate IRE1α/JNK/AP-1 pathway through ER stress
To understand the effect of mutations in CEL, biological function of mutations in CEL (CELD454N and CELD555N) was evaluated in MIHA cells. As shown in Figure 4c1, ectopic expression of the mutants CELD454N and CELD555N significantly increased abnormal accumulation of cholesteryl ester as compared with wild-type CEL transfection in MIHA cells. In keeping with the induction of cholesteryl ester, CELD445N and CELD555N significantly promoted cell growth compared with wild-type CEL (Figure 4c2), suggesting that wild-type CEL possesses tumor suppressive effect through regulating cholesteryl ester homeostasis, which can be disrupted by its mutations.
We further evaluated the effect of mutant CEL on ER stress and consequent cascade. Ectopic expression of two CEL mutants remarkably increased protein levels of GRP78, p-IRE1α, p-JNK and p-c-Jun (Figure 4d1) and enhanced AP-1 activity (Figure 4d2). Furthermore, we detected an elevated level of cholesteryl ester in NAFLD-HCC compared with lean HCC tissues from mouse models (P<0.05; Figure 4e1). Increased protein levels of p-IRE1α and p-c-Jun were also detected in NAFLD-HCC compared with lean HCC from mouse models (Figure 4e2). These results indicated that CEL inactivation may promote liver cell growth by inducing ER stress and UPR-related IRE1α/JNK/AP-1 signaling cascade (Figure 4f) and ultimately contribute to tumorigenesis of NAFLD-HCC.
Mutations in Hras at codon 61 were identified in NAFLD-associated HCC
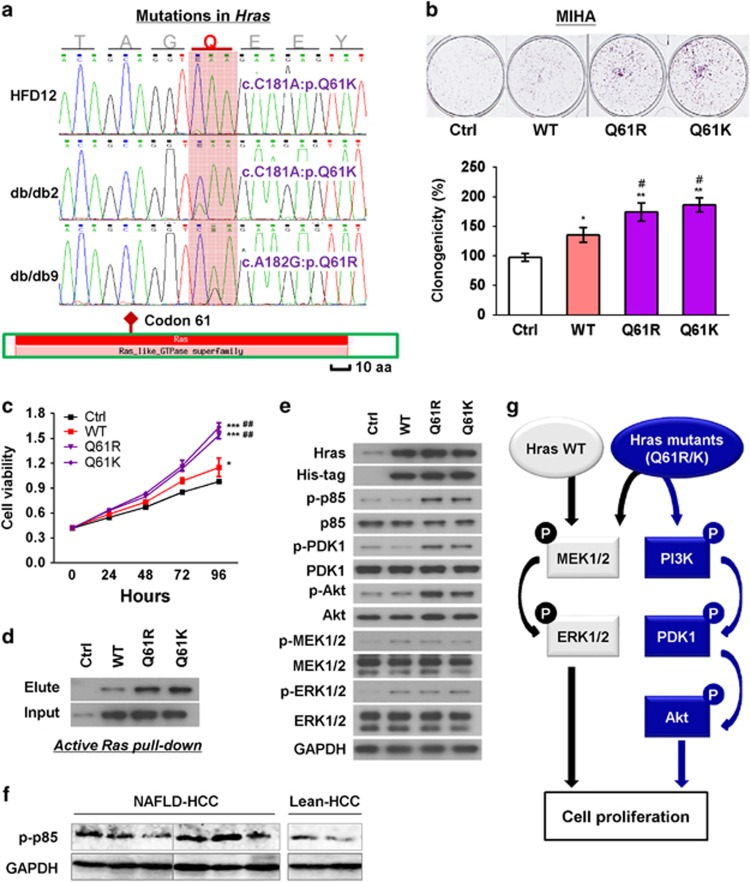
We further performed mutation examination on Hras, mutation of which was involved in six out of eight pathways dysregulated in the exome-sequenced NAFLD-HCC from obese mice (Figure 2b and Supplementary Table 1). Mutations in Hras were further found in HCCs from two db/db mice and one HFD-fed mouse (3/29, 10.3%) but none of the wild-type lean mice (0/16, 0%) by PCR and Sanger sequencing. Interestingly, mutations found in NAFLD-HCCs were all present at codon 61 (Q61R and Q61K; Figure 5a). Of note, mutations in human HRAS were observed in human cancers, with codon 61 being a mutation hot spot (Supplementary Figure 3). These findings suggest that mutations in Hras at codon 61 may have a vital role in NAFLD-associated hepatocarcinogenesis.
Figure 5.
Hras mutants promoted cell proliferation. (a) Somatic non-synonymous mutations in Hras (all located at codon 61) were found in NAFLD-HCCs of one dietary and two genetically obese mice but in none of the control lean mice. (b) Hras mutants (Q61R and Q61K) promoted cell proliferation compared with wild-type (WT) Hras and control vector (Ctrl) transfection in MIHA cells by colony formation assay. (c) Hras mutants (Q61R and Q61K) promoted cell viability compared with WT Hras and control vector transfection in MIHA cells by MTT assay. (d) Mutations of Q61R or Q61K enhanced the Ras activity of Hras. (e) Hras mutants (Q61R and Q61K) increased the protein expression of key regulators of Ras/MAPK and PI3K/AKT signaling cascades. (f) Protein level of p-p85 was examined by western blot in NAFLD-HCC and lean HCC from mouse models. The same bands for GAPDH were shown as in Figure 4e2 for the same protein samples. (g) Mechanistic scheme of signaling cascade mediated by wild-type and mutant Hras. Data were expressed as mean±s.d. *P<0.05, **P<0.01 and ***P<0.001 as compared with control vector transfection; #P<0.05 and ##P<0.01 as compared with wild-type Hras.
Hras mutants promoted cell proliferation via MAPK signaling cascade
Changing this glutamine/Q to Arginine/R or Lysine/K would potentially alter the basal activity of Hras, as this glutamine is involved in regulating the activity of GTPase. To determine the effect of these mutations, we ectopically expressed the wild-type and mutant forms of Hras (Q61R and Q61K) in MIHA cells. Expression of Hras mutants (Q61R and Q61K) significantly promoted colony formation and cell growth compared with wild-type Hras transfection as well as empty vector transfection (Figures 5b and c) MIHA cells. We further performed Ras activity assay to determine the effect of the mutations on Hras activity. Our results showed that the activities of Hras mutants Q61R and Q61K were significantly higher than wild-type Hras (Figure 5d). These results demonstrate that Hras mutations at codon 61 (Q61R and Q61K) identified in NAFLD-HCC are activating oncogenic mutations.
To determine whether the Ras/MAPK and phosphatidylinositol-4,5-bisphosphate 3-kinase (PI3K)/AKT signaling pathways, known to be downstream of RAS proteins,1 are regulated by the Hras activating mutations, we evaluated the activity of the key effectors of these pathways. As shown in Figure 5e, wild-type Hras only activated Ras/MAPK signaling by increasing p-MEK1/2 and p-ERK1/2; while mutants Q61R and Q61K activated both Ras/MAPK and PI3K/3-phosphoinositide-dependent protein kinase-1 (PDK1)/AKT (including the factors of p-p85, p-PDK1 and p-AKT) signaling cascades in MIHA cells. Of note, a relatively higher level of p-p85 was detected in NAFLD-HCC compared with lean HCC from mouse models (Figure 5f). The results therefore provide the mechanistic details for the oncogenic mutations of Hras in the pathogenesis of NAFLD-HCC (Figure 5g).
Discussion
In this study, HCC incidence, multiplicity and tumor size were significantly higher in obese mice with fatty liver as compared with lean mice, demonstrating that obesity promotes NAFLD-HCC development. This is supported by previous findings that obesity is a major HCC risk factor in human21 and in animal models.22 The DEN-induced HCC model was commonly used.22, 23, 24 Although we used the chemical carcinogen DEN to accelerate HCC development in mice, the comparison between tumor and adjacent normal tissues, as well as comparison between obese mice and control lean mice, could eliminate the direct effect of DEN. The remaining genetic alterations should be associated with NAFLD. Thus, this model is suitable for investigating the genetic alterations of NAFLD-associated HCC in this study.
We analyzed the somatic non-synonymous mutations that occurred during HCC development by comparing exome sequences of liver tumors and adjacent normal samples from genetically obese mice (db/db) or wild-type lean mice. We identified 277 mutated genes in NAFLD-HCCs from db/db mice and 268 mutated genes in lean HCCs from wild-type mice. Although similar numbers of mutated genes were identified in liver tumors from obese mice and control lean mice by exome sequencing (277 vs 268), mutated genes in NAFLD-HCC and lean HCC affected different pathways. Mutated genes in NAFLD-HCC were enriched in eight important cancer pathways while mutations in lean HCC only involved in two of these pathways as revealed by KEGG pathway enrichment analysis (Figure 2b). In NAFLD-HCC, mutations laid on genes involved in Calcium signaling pathway, which regulates a broad range of cellular events including those important in tumorigenesis;25 Gap junction, which participates in the regulation of cell growth and differentiation;26 focal adhesion, which has important roles in cell motility, proliferation, differentiation and survival;27 GnRH signaling, which functions in regulating cancer proliferation, metastasis and angiogenesis;28 chemokine signaling, which is involved in tumor survival, metastasis and neovascularization;29 MAPK signaling, which participates in cell proliferation, differentiation and migration30 and two well-known cancer-related pathways, cytokine–cytokine receptor interaction and pathways in cancer. On the other hand, only MAPK signaling and pathways in cancer were significantly affected by mutations during HCC development in lean mice. Together, our finding reveals the genomic basis of NAFLD-HCC that is different from lean HCC.
Three genes, including Cel, 4933432B09Rik and Ttn, were found to be recurrently mutated in the obese NAFLD-HCCs. Ttn mutation was also found in one control lean HCC, suggesting that Ttn mutation may be important for HCC development but may not be specifically associated with NAFLD-HCC. 4933432B09Rik encodes an uncharacterized protein. Cel is of particular interest as it has been reported to serve as a compensatory protein to other lipolytic enzymes to complete digestion and absorption of lipid nutrients.15 Malfunction of Cel may have a role in NAFLD-associated HCC development. The significantly higher mutation frequency in Cel was further validated in NAFLD-HCCs from 29 obese mice than lean HCCs from 16 control lean mice (P<0.05). Importantly, mutation in human CEL was detected in 3.75% (3/80) of nutrition-associated HCC, but not in any virus infection-associated HCC (0/108) in TCGA database (Supplementary Table 2). Collectively, these results suggested a critical role of Cel mutations in the molecular pathogenesis of NAFLD-associated HCC. Since multiple Cel mutation sites were found in NAFLD-HCCs, these mutations would lead to loss-of-function that might involve in driving the development of HCC. Indeed our results, as illustrated in Figures 3 and 4, showed that knock-down of CEL promoted cell growth by inducing cholesteryl ester accumulation and ER stress and subsequently upregulating UPR-related IRE1α/JNK/c-Jun/AP-1 pathway. Importantly, similar to knock-down of CEL, mutant CEL (D454E and D555N) also remarkably induced liver cell growth through inducing cholesteryl ester accumulation, ER stress and activating IRE1α/JNK/c-Jun/AP-1 signaling cascade (Figure 4). The importance of JNK/c-Jun/AP-1 pathway in promoting cell proliferation in the development of HCC has been well elucidated.31 Collectively, our findings suggest that dominant-negative mutations or depletion of CEL have an important oncogenic role in the pathogenesis of NAFLD-HCC by inducing the abnormal accumulation of cholesteryl esters, which results in ER stress and consequently activates IREα/JNK/c-Jun/AP-1 signaling cascade.
Our cellular studies of CEL knock-down and CEL mutations revealed that cholesteryl ester would be a key player to promote liver cell growth. Reduction of cholesteryl ester by Avasimibe, an inhibitor of the cholesterol esterification enzyme ACAT, suppressed cell proliferation in CEL knock-down cells (Supplementary Figure 2). Cholesteryl ester accumulation was reported to promote growth and invasion in PTEN-null prostate cancer cells, supporting its important role in cancer progression, but the molecular mechanism remained elusive.32 In this study, we revealed that accumulation of cholesteryl ester caused by loss or inactivation of CEL activated the UPR-related IRE1α/JNK/c-Jun signaling cascade, subsequently activating the transcriptional activity of AP-1 that regulates downstream genes involved in promoting cell proliferation.33 It has been reported that ER stress significantly contributes to HCC development.34 UPR belongs to a part of ER stress. We found that abnormal accumulation of cholesteryl esters led to activation of ER stress. Therefore, our study uncovered the mechanisms of cholesteryl ester accumulation by loss or inactivation of CEL in driving the development of NAFLD-HCC.
Hras was found to be mutated at codon 61 in only NAFLD mice by Exome sequencing. Pathway analysis showed that the mutated Hras participated in most of the pathways (6 out of 8) dysregulated by mutations in NAFLD-associated HCC. We further examined mutations in Hras in an enlarged cohort of samples by PCR and Sanger sequencing, and found it to be mutated in 10.3% of NAFLD-associated HCC (all at codon 61, Q61R or Q61K) and in 0% of lean HCC. This result demonstrated the important involvement of Hras mutation in the development of NAFLD-associated HCC. Therefore, the function of mutated Hras (Q61R and Q61K) was further investigated in in vitro experiments. As expected as the oncogenic mutation of Q61L,10 Q61R and Q61K mutants were found to contribute to a significant effect on promoting cell growth by activating two oncogenic signaling cascades. These two mutants elicited PI3K/PDK1/Akt signaling pathway in addition to the MAPK pathway, while wild-type Hras activated MAPK signaling pathway only with a moderate effect of promoting cell growth (Figure 5). Ras and ras-related proteins are often dysregulated in cancers by oncogenic mutations of Ras isoforms or its effectors in about one-third of all human cancers (http://www.sanger.ac.uk/genetics/CGP/cosmic). Furthermore, mutations in HRAS have been identified in human cancers,35, 36 with codon 61 as a mutation hot spot (Supplementary Figure S3). Alteration of the RAS-MAPK pathway has frequently been reported in human cancers as a result of activating mutations mainly in the RAS genes and results in the transcription of genes involved in controlling cell proliferation.37 PI3K/PDK1/AKT, the second best-characterized RAS effector pathway, has important roles as mediators of RAS-mediated cell survival and proliferation.38 The PI3K activation converts PIP2 (phosphatidylinositol (4,5)-bisphosphate) into PIP3 (phosphatidylinositol (3,4,5)-trisphosphate), then PIP3 propagates intracellular signaling by directly binding to proteins with PDK1 and AKT. The interaction between MAPK and PI3K/Akt pathways is important in RAS-mediated transformation of human cells.39 Activation of these pathways contributed to the enhanced oncogenic effect of Hras mutants in liver cells. Since these mutations were only found in NAFLD-associated HCC but not in lean HCC, mutations in Hras at codon 61 should be one of the driving events to promote liver tumorgenesis under obese condition (Figure 5f).
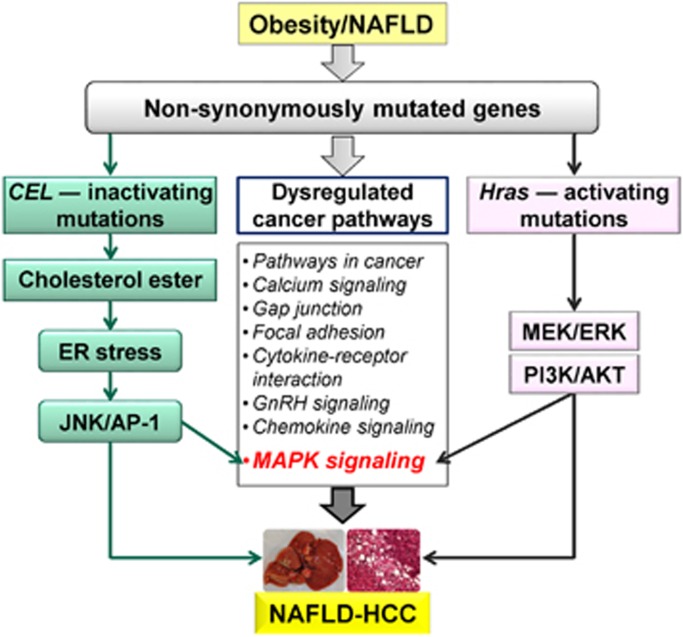
In conclusion, this study defined the important role of obesity in promoting NAFLD-HCC initiation and development in animal models. We found the specific mutational signatures and pathways of NAFLD-associated HCC by whole-exome sequencing (Figure 6). Specifically, we revealed that loss-of-function mutations in Cel and gain-of-function mutations in Hras are associated with NAFLD-HCC. Both Cel inactivating mutations and Hras activating mutations promote liver cell growth. The mechanisms by which inactivating mutations in Cel promote liver cell growth are mediated by the accumulation of cholesteryl ester, leading to induction of ER stress and consequent activation of the IRE1α/JNK/c-Jun/AP-1 signaling cascade; while activating mutations in Hras (Q61R and Q61K) promote liver cell proliferation by activating Ras/MAPK and PI3K/PDK1/Akt pathways. Interestingly, the signaling cascades activated by the mutated Cel and Hras are both associated with the MAPK signaling pathway, which is significantly dysregulated by mutated genes in NAFLD-HCC (Figure 2b and 6). Our findings illustrate a comprehensive genomic landscape and highlight the specific molecular events and signaling pathways in the pathogenesis of NAFLD-HCC.
Figure 6.
Schematic summary of this study. Genes harboring non-synonymous somatic mutations were identified in NAFLD-HCC mice, affecting eight cancer signaling pathways. Among all mutated genes, Cel and Hras are of particular interest. Mutations in these two genes both have oncogenic effects with different mechanisms.
Materials and methods
Animals and treatment
Genetically obese male mice (C57BL/6 db/db) deficient in leptin receptor activity were kept on normal diet (n=9–13/group). Dietary obese mice were established by keeping wild-type C57BL/6 male mice on HFD (D12492, Research Diets Inc., New Brunswick, NJ, USA), in which 60% of calories were derived from fat, since 2 months of age (n=9–10/group). Wild-type mice kept on normal diet were control lean mice (n=10/group). All mice received a single intraperitoneal injection of DEN (5 mg/kg body weight; Sigma Chemical Co, St. Louis, MO, USA) at age of 13–15 days, and were killed at 7 or 8 months. Body weights were recorded. Livers were rapidly excised and weighed. The presence and dimensions of surface nodules were evaluated. HCCs were confirmed histologically from either grossly or histologically evident nodules. Liver tumors and adjacent liver tissues were isolated, snap frozen in liquid nitrogen and stored in −80 °C for further experiments. Tumors and adjacent non-tumor livers from 16 db/db mice, 13 HFD-fed mice and 16 wild-type lean mice (some recruited from additional cohorts due to the low incidence of HCC in wild-type lean group) with large enough tumors were subjected to DNA extraction. All animal studies were conducted in accordance with guidelines by the Animal Experimentation Ethics Committee of the Chinese University of Hong Kong.
Whole-exome sequencing
DNA samples of paired tumor and adjacent normal tissues from genetically obese mice and control lean mice were subjected to whole-exome sequencing. Exome capture was performed using the SureSelect Target Enrichment System (Agilent, Santa Clara, CA, USA). Captured libraries were sequenced by HiSeq2000 (Illumina, San Diego, CA, USA). An average of 177x sequencing depth of target regions was obtained for each sample (Supplementary Table 3).
Reads alignment
The whole-exome sequencing reads were mapped to the UCSC mm9 reference genome (http://genome.ucsc.edu/) using the Burrows-Wheeler Aligner.40 The likely PCR duplications with the same match interval on the genomic sequence duplications were removed by Samtools.41 Local realignment around indels was done by GATK to improve alignment quality and help reducing the false discovery rate in the following mutation detection analysis.42
SNVs identification
Candidate SNVs with mutated allele frequency more than 15% in cancer tissue and no more than 2% in paired normal tissue (Fisher's exact test, P<0.05) were further filtered by the following thresholds: high-quality (Q>10) coverage ⩾10 × for each sample; reads supporting the mutated allele not a result of sequencing error (Binomial test, f=0.1, P>0.01); sequencing quality scores for mutated alleles not lower than normal alleles (Wilcoxon rank-sum test, P>0.01); mutated alleles not come from repeatedly aligned reads (Fisher's exact test, P>0.01). Mutant alleles should not be enriched in 10 bps of 5' or 3' ends of reads (Fisher's exact test, P>0.01). The resulting somatic SNVs were filtered against the Single Nucleotide Polymorphism database (mouse dbSNP128) to identify novel SNVs Annotation was performed by ANNVAR.43
Indels identification
Candidate somatic indels were first predicted by GATK SomaticIndelDetector with default parameters.42 Then predicted indels were filtered if (1) total coverage at the site <30 × ; (2) average mapping qualities of consensus indel-supporting reads <30; (3) average number of mismatches per consensus indel-supporting read ⩾2; (4) average quality of bases from consensus indel-supporting reads <20; (5) median of indel offsets from the ends/starts of the reads within 10 bp or (6) percentage of forward- or reverse-aligned indel-supporting reads <20%. Resulting somatic indels were filtered against dbSNP128. Annotation was performed by ANNVAR.43
Sanger sequencing validation
Whole coding regions of two genes of interest (Cel and Hras) were examined by PCR and Sanger sequencing in tumor and adjacent normal tissues from 16 genetically obese mice, 13 dietary obese mice and 16 control lean mice. Primer sequences were listed in Supplementary Table 4.
Plasmid construction
Mammalian expression vector pCMV-CEL was purchased (Origene, Rockville, MD, USA). CEL mutants D454E and D555N were generated by site-directed mutagenesis with primers in Supplementary Table 4. Wild-type and mutant (Q61R and Q61K) Hras were amplified from cDNA prepared from samples with wild-type or mutant Hras and then cloned into mammalian expression vector pcDNA3.1-His.
Gene expression knock-down
Commercially available vectors expressing shRNAs against CEL (TR313998) and control vector (TR30012) were purchased (Origene). Among the four shRNA plasmids against CEL provided in the kit, TR313998B (SH1) and TR313998C (SH2) achieved satisfactory knock-down effect.
Cell culture
The human immortalized liver cell line MIHA (a gift from Prof. Xinyuan Guan, Hong Kong University) was maintained as monolayer culture in high glucose Dulbecco's modified Eagle's medium with 10% of fetal bovine serum and 1% of penicillin/streptomycin at 37 °C in a humidified atmosphere of 5% CO2. Transfection was performed using Lipofectamine 2000 (Life Technologies, Carlsbad, CA, USA) according to the manufacturer's protocol.
Western blot analysis
Total protein was separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and then transferred onto PVDF membranes (GE Healthcare, Piscataway, NJ, USA). The membrane was incubated with primary antibodies overnight, and then with secondary antibody at room temperature for 1 h. Proteins of interest were visualized using ECL Plus Western blotting Detection Reagents (GE Healthcare). The antibodies used were listed in Supplementary Table 5.
Cell viability assay
Cell viability of stably transfected cells was examined using the Vybrant MTT Cell viability Assay Kit (Life Technologies) according to the manufacturer's instructions. All experiments were conducted three times in triplicates.
Colony formation assay
Cells (2 × 104/well in 6-well plates) were transfected with the corresponding plasmids, and treated with G418 (0.5 mg/ml) 48 h post transfection. After 7–10 days, cells were fixed with 70% ethanol and stained with 0.5% crystal violet solution. Colonies with more than 50 cells were counted. All experiments were conducted three times in triplicates.
Dual-luciferase reporter activity assay
AP-1-Luc luciferase reporter plasmid containing 7 × AP1 binding sites (0.1 μg/well) and pRL-CMV vector (2.5 ng/well) was co-transfected with pRL-CMV vector (2.5 ng/well) to cells (1 × 105 cell/well) in 24-well plates. Luciferase activity was measured at 48 h post transfection by the Dual Luciferase Assay System (Promega, Madison, WI, USA). The experiments were conducted three times in triplicates.
Enrichment analysis of KEGG pathways
Somatically mutated genes identified in tumors from genetic obese mice or control lean mice were subjected to KEGG pathway enrichment analysis using the Gene Set Analysis Toolkit V2 (http://bioinfo.vanderbilt.edu/webgestalt/). Hypergeometric test and BH multiple test adjustment were used. All genes from human were used as a reference. Pathways with at least four genes and adjusted P<0.05 were identified as significantly enriched.
Mutations in CEL and HRAS and expression of CEL in human HCC
Mutations in CEL were evaluated in raw sequencing data from 80 human HCC associated with nutritional factors, such as NAFLD and alcohol consumption, in comparison with those from 108 HCC associated with viruses infection (hepatitis B virus and hepatitis C virus). To discover low frequency mutations that might be ignored in the TCGA level 3 data, we have analyzed the raw TCGA level 1 data using MuTect with the following parameters: (1) minor allele frequency ⩾0.05, (2) altered allele in tumor ⩾3 and altered allele in normal <2, (3) LODT cutoff=5.2 and (4) cross individual contamination 0.005. HRAS mutation data and CEL mRNA expression data were downloaded from TCGA database at cBioPortal Cancer Genomics (http://www.cbioportal.org/public-portal/index.do).
Cholesterol and cholesteryl ester measurement
Cholesterol/Cholesteryl Ester Quantification Kit (ab65359) was purchased (Abcam, Cambridge, MA, USA) for the detection of the levels of hepatic cholesterol and cholesteryl ester. Experiments were conducted three times in duplicates according to the manufacturer's instructions.
Cholesteryl palmitate treatment
Cholesteryl palmitate (Sigma) was dissolved in solvent composed of chloroform:methanol (1:1; v/v) and 0.1% of Triton X-100. A concentration of 25 nm of Cholesteryl palmitate was used to treat cells.
Hras activity assay
Ras activity was assessed using the Active Ras Detection Kit (Cell Signaling Technologies, Boston, MA, USA), which pulls down active Ras only, according to the manufacturer's instructions. Then western blot was performed to measure the levels of pulled-down (active forms) wild-type and mutant Hras.
Statistical analysis
The chi-square test was used for the comparison of tumor incidence and prevalence of gene mutation in different groups. Numeric data were presented as mean±s.d., and compared by t-test or Mann–Whitney test. All statistical tests were performed using Graphpad Prism 5.0 (Graphpad Software Inc., San Diego, CA, USA) and Statistical Package for Social Sciences version 16.0 (SPSS, Chicago, IL, USA). A P-value of <0.05 was considered as statistically significant.
Acknowledgments
The project was supported by the RGC-GRF Hong Kong (14106415); RGC-CRF Hong Kong (CUHK3/CRF/12R; HKU3/CRF11R); National Basic Research Program of China (973 Program, 2013CB531401); RGC-Theme-based Research Scheme Hong Kong (T12-403-11); Shenzhen Virtual University Park Support Scheme to CUHK Shenzhen Research Institute. The results shown here are in part based upon data generated by the TCGA Research Network: http://cancergenome.nih.gov/.
Author contributions
JS, ESHC and XL set up animal models and validated sequencing results; HT and DL performed functional experiments; QL, AC-SY and TFC analyzed data; QL, JS and HT wrote the paper; JJYS commented on the study; JY and VWSW supervised the research.
Glossary
- ACAT
Acyl-CoA:cholesterol acyltransferase
- CEL
Carboxyl ester lipase
- DEN
diethylnitrosamine
- ER
endoplasmic reticulum
- GnRH
Gonadotropin-releasing hormone
- HCC
hepatocellular carcinoma
- HFD
high-fat diet
- Hras
Harvey rat sarcoma virus oncogene 1
- JNK
c-Jun N-terminal kinase
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- MAPK
mitogen-activated protein kinase
- NAFLD
non-alcoholic fatty liver disease
- PDK1
3-phosphoinositide-dependent protein kinase-1
- PI3K
phosphatidylinositol-4,5-bisphosphate 3-kinase
- PIP2
phosphatidylinositol (4,5)-bisphosphate
- PIP3
phosphatidylinositol (3,4,5)-trisphosphate
- SNV
single-nucleotide variant
- indel
small insertion and deletion
- Ttn
titin
- UPR
unfolded protein response.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on the Oncogene website (http://www.nature.com/onc)
Supplementary Material
References
- Turati F, Talamini R, Pelucchi C, Polesel J, Franceschi S, Crispo et al. Metabolic syndrome and hepatocellular carcinoma risk. Br J Cancer 2013; 108: 222–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borena W, Strohmaier S, Lukanova A, Bjorge T, Lindkvist B, Hallmans G et al. Metabolic risk factors and primary liver cancer in a prospective study of 578,700 adults. Int J Cancer 2012; 131: 193–200. [DOI] [PubMed] [Google Scholar]
- Schlesinger S, Aleksandrova K, Pischon T, Fedirko V, Jenab M, Trepo E et al. Abdominal obesity, weight gain during adulthood and risk of liver and biliary tract cancer in a European cohort. Int J Cancer 2013; 132: 645–657. [DOI] [PubMed] [Google Scholar]
- Wong VW. Nonalcoholic fatty liver disease in Asia: a story of growth. J Gastroenterol Hepatol 2013; 28: 18–23. [DOI] [PubMed] [Google Scholar]
- White DL, Kanwal F, El-Serag HB. Association between nonalcoholic fatty liver disease and risk for hepatocellular cancer, based on systematic review. Clin Gastroenterol Hepatol 2012; 10: 1342–1359 e1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsson SC, Wolk A. Overweight, obesity and risk of liver cancer: a meta-analysis of cohort studies. Br J Cancer 2007; 97: 1005–1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen HF, Chen P, Li CY. Risk of malignant neoplasms of liver and biliary tract in diabetic patients with different age and sex stratifications. Hepatology 2010; 52: 155–163. [DOI] [PubMed] [Google Scholar]
- Camarota LM, Chapman JM, Hui DY, Howles PN. Carboxyl ester lipase cofractionates with scavenger receptor BI in hepatocyte lipid rafts and enhances selective uptake and hydrolysis of cholesteryl esters from HDL3. J Biol Chem 2004; 279: 27599–27606. [DOI] [PubMed] [Google Scholar]
- Raeder H, Johansson S, Holm PI, Haldorsen IS, Mas E, Sbarra V et al. Mutations in the CEL VNTR cause a syndrome of diabetes and pancreatic exocrine dysfunction. Nat Genet 2006; 38: 54–62. [DOI] [PubMed] [Google Scholar]
- Prior IA, Lewis PD, Mattos C. A comprehensive survey of Ras mutations in cancer. Cancer Res 2012; 72: 2457–2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang WC. Store-operated calcium channels and pro-inflammatory signals. Acta Pharmacol Sin 2006; 27: 813–820. [DOI] [PubMed] [Google Scholar]
- Keane MP, Strieter RM. Chemokine signaling in inflammation. Crit Care Med 2000; 28: N13–N26. [DOI] [PubMed] [Google Scholar]
- Renauld JC. Class II cytokine receptors and their ligands: key antiviral and inflammatory modulators. Nat Rev Immunol 2003; 3: 667–676. [DOI] [PubMed] [Google Scholar]
- Chanson M, Derouette JP, Roth I, Foglia B, Scerri I, Dudez T et al. Gap junctional communication in tissue inflammation and repair. Biochim Biophys Acta 2005; 1711: 197–207. [DOI] [PubMed] [Google Scholar]
- Hui DY, Howles PN. Carboxyl ester lipase: structure-function relationship and physiological role in lipoprotein metabolism and atherosclerosis. J Lipid Res 2002; 43: 2017–2030. [DOI] [PubMed] [Google Scholar]
- Vogel MJ, Barrall I, E. M, Mignosa CP. Polymorphism in cholesteryl esters: cholesteryl palmitate. IBM J Res Dev 1971; 15: 52. [Google Scholar]
- Cianciola NL, Greene DJ, Morton RE, Carlin CR. Adenovirus RIDalpha uncovers a novel pathway requiring ORP1L for lipid droplet formation independent of NPC1. Mol Biol Cell 2013; 24: 3309–3325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang TY, Li BL, Chang CC, Urano Y. Acyl-coenzyme A:cholesterol acyltransferases. Am J Physiol Endocrinol Metab 2009; 297: E1–E9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du X, Yang H. Sterol-binding proteins and endosomal cholesterol transport. Front Biol 2011; 6: 190–196. [Google Scholar]
- Otoda T, Takamura T, Misu H, Ota T, Murata S, Hayashi H et al. Proteasome dysfunction mediates obesity-induced endoplasmic reticulum stress and insulin resistance in the liver. Diabetes 2013; 62: 811–824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Serag HB, Rudolph L. Hepatocellular carcinoma: epidemiology and molecular carcinogenesis. Gastroenterology 2007; 132: 2557–2576. [DOI] [PubMed] [Google Scholar]
- Park EJ, Lee JH, Yu GY, He G, Ali SR, Holzer RG et al. Dietary and genetic obesity promote liver inflammation and tumorigenesis by enhancing IL-6 and TNF expression. Cell 2010; 140: 197–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Shen B, Chu ESH, Teoh N, Cheung KF, Wu CW et al. Inhibitory role of peroxisome proliferator-activated receptor gamma in hepatocarcinogenesis in mice and in vitro. Hepatology 2010; 51: 2008–2019. [DOI] [PubMed] [Google Scholar]
- Wang J, Zhao JM, Chu ESH, Mok MTS, Go MYY, Man K et al. Inhibitory role of Smad7 in hepatocarcinogenesis in mice and in vitro. J Pathol 2013; 230: 441–452. [DOI] [PubMed] [Google Scholar]
- Swami M. Signalling: the calcium connection. Nat Rev Cancer 2010; 10: 738. [DOI] [PubMed] [Google Scholar]
- Leithe E, Sirnes S, Omori Y, Rivedal E. Downregulation of gap junctions in cancer cells. Crit Rev Oncog 2006; 12: 225–256. [DOI] [PubMed] [Google Scholar]
- Petit V, Thiery JP. Focal adhesions: structure and dynamics. Biol Cell 2000; 92: 477–494. [DOI] [PubMed] [Google Scholar]
- Limonta P, Montagnani Marelli M, Mai S, Motta M, Martini L, Moretti RM. GnRH receptors in cancer: from cell biology to novel targeted therapeutic strategies. Endocr Rev 2012; 33: 784–811. [DOI] [PubMed] [Google Scholar]
- Mohit E, Rafati S. Chemokine-based immunotherapy: delivery systems and combination therapies. Immunotherapy 2012; 4: 807–840. [DOI] [PubMed] [Google Scholar]
- Osaki LH, Gama P. MAPKs and signal transduction in the control of gastrointestinal epithelial cell proliferation and differentiation. Int J Mol Sci 2013; 14: 10143–10161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hui LJ, Zatloukal K, Scheuch H, Stepniak E, Wagner EF. Proliferation of human HCC cells and chemically induced mouse liver cancers requires JNK1-dependent p21 downregulation. J Clin Invest 2008; 118: 3943–3953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue SH, Li JJ, Lee SY, Lee HJ, Shao T, Song B et al. Cholesteryl ester accumulation induced by PTEN loss and PI3K/AKT activation underlies human prostate cancer aggressiveness. Cell Metab 2014; 19: 393–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaulian E, Karin M. AP-1 in cell proliferation and survival. Oncogene 2001; 20: 2390–2400. [DOI] [PubMed] [Google Scholar]
- Nakagawa H, Umemura A, Taniguchi K, Font-Burgada J, Dhar D, Ogata H et al. ER stress cooperates with hypernutrition to trigger TNF-dependent spontaneous HCC development. Cancer Cell 2014; 26: 331–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo G, Sun X, Chen C, Wu S, Huang P, Li Z et al. Whole-genome and whole-exome sequencing of bladder cancer identifies frequent alterations in genes involved in sister chromatid cohesion and segregation. Nat Genet 2013; 45: 1459–1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agrawal N, Frederick MJ, Pickering CR, Bettegowda C, Chang K, Li RJ et al. Exome sequencing of head and neck squamous cell carcinoma reveals inactivating mutations in NOTCH1. Science 2011; 333: 1154–1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shields JM, Pruitt K, McFall A, Shaub A, Der CJ. Understanding Ras: 'it ain't over 'til it's over'. Trends Cell Biol 2000; 10: 147–154. [DOI] [PubMed] [Google Scholar]
- Castellano E, Downward J. Role of RAS in the regulation of PI 3-Kinase. Curr Top Microbiol Immunol 2010; 346: 143–169. [DOI] [PubMed] [Google Scholar]
- Castellano E, Downward J. RAS interaction with PI3K: more than just another effector pathway. Genes Cancer 2011; 2: 261–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009; 25: 1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009; 25: 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 2010; 20: 1297–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res 2010; 38: e164. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.