Abstract
The vertebrate nuclear pore complex (NPC) is a macromolecular assembly of protein subcomplexes forming a structure of eightfold radial symmetry. The NPC core consists of globular subunits sandwiched between two coaxial ring-like structures of which the ring facing the nuclear interior is capped by a fibrous structure called the nuclear basket. By postembedding immunoelectron microscopy, we have mapped the positions of several human NPC proteins relative to the NPC core and its associated basket, including Nup93, Nup96, Nup98, Nup107, Nup153, Nup205, and the coiled coil-dominated 267-kDa protein Tpr. To further assess their contributions to NPC and basket architecture, the genes encoding Nup93, Nup96, Nup107, and Nup205 were posttranscriptionally silenced by RNA interference (RNAi) in HeLa cells, complementing recent RNAi experiments on Nup153 and Tpr. We show that Nup96 and Nup107 are core elements of the NPC proper that are essential for NPC assembly and docking of Nup153 and Tpr to the NPC. Nup93 and Nup205 are other NPC core elements that are important for long-term maintenance of NPCs but initially dispensable for the anchoring of Nup153 and Tpr. Immunogold-labeling for Nup98 also results in preferential labeling of NPC core regions, whereas Nup153 is shown to bind via its amino-terminal domain to the nuclear coaxial ring linking the NPC core structures and Tpr. The position of Tpr in turn is shown to coincide with that of the nuclear basket, with different Tpr protein domains corresponding to distinct basket segments. We propose a model in which Tpr constitutes the central architectural element that forms the scaffold of the nuclear basket.
INTRODUCTION
The nuclear pore complex (NPC), formed by multiple copies of ∼30 proteins (Cronshaw et al., 2002), serves as a gateway for the exchange of material between cytoplasm and nucleus in eukaryotes. Its core structure of eightfold-radial symmetry consists of globular subunits that form a central spoke-ring complex, including a cylindrical structure surrounding the actual translocation channel (Allen et al., 2000; Vasu and Forbes, 2001; Fahrenkrog and Aebi, 2002; Suntharalingam and Wente, 2003). Coaxial ring-like structures flank the spoke-ring complex on both its cytoplasmic and nuclear side.
In addition to this NPC proper, eight fibrils of distinctive shape are attached to each of these rings. Fibers of the cytoplasmic ring look like short tufts ∼30–35 nm in length with free distal ends. In contrast, the thin fibrils emanating from the nuclear ring are ∼40–60 nm in length and laterally interconnected at their distal ends, forming another ring-like structure referred to as the terminal ring. Nuclear fibrils and terminal ring are believed to represent a structural and functional entity, called the nuclear basket (Jarnik and Aebi, 1991; Ris, 1991; Goldberg and Allen, 1992).
Several NPC proteins (nucleoporins, Nups), including Nup153 (Sukegawa and Blobel, 1993), Nup98 (Powers et al., 1995; Radu et al., 1995), Nup 50 (Guan et al., 2000), some components of the Nup160 subcomplex (Radu et al., 1994; Fontoura et al., 1999; Belgareh et al., 2001; Vasu et al., 2001), and members of the Nup93 subcomplex (Grandi et al., 1997) have been assigned to the nuclear side of the NPC in higher eukaryotes. The 267-kDa protein Tpr has been located at this side as well (Cordes et al., 1997; Zimowska et al., 1997). However, it has remained uncertain which of these proteins are architectural components of the NPC core, the associated basket fibrils, or the terminal ring.
Nup96, a stable component of the Nup160 subcomplex (Belgareh et al., 2001; Vasu et al., 2001) was reported to occur exclusively at the nuclear side of the NPC in HeLa cells, at an average distance of 100–200 nm from the nuclear envelope (NE) midplane (Fontoura et al., 1999). There it has been considered a distal part of the nuclear basket together with other members of the Nup160 subcomplex (Allen et al., 2000; Vasu and Forbes 2001). Most recently, however, Nup96 was found on both sides of the NPC periphery when using isolated rat liver NEs for immuno-electron microscopy (EM) (Enninga et al., 2003). Two other components of the Nup160 subcomplex, Nup107 and Nup133, were localized on both the cytoplasmic and nuclear side as well, but closer to the horizontal midline passing through the NPC (Radu et al., 1994; Belgareh et al., 2001).
Another Nup96 binding partner, Nup98, also was recently found on both sides of the NPC (Griffis et al., 2003), but other studies have located this Nup exclusively at the nuclear side, close to (Radu et al., 1995) or further away from the NE (Frosst et al., 2002). Nup98 has therefore been regarded as an Nup forming the basket fibers or the terminal ring (Allen et al., 2000; Vasu and Forbes, 2001; Fahrenkrog and Aebi, 2002; Harel et al., 2003). Vertebrate Nup93 again was reported to occur at two distinct sites at the nuclear NPC side, with predominant localization in the terminal ring area (Grandi et al., 1997; Fahrenkrog and Aebi, 2002). In contrast, the yeast homolog Nic96 is symmetrically arranged on both sides of the NPC close to its midplane (Rout et al., 2000).
Nup153 was located in proximity of the nuclear NPC side in somatic mammalian cells (Sukegawa and Blobel, 1993; Cordes et al., 1993; Frosst et al., 2002), but in amphibian oocytes it also has been found associated with NPC-attached filament bundles several hundred nanometers in length that go beyond the dimensions of the nuclear basket (Cordes et al., 1993; Ris, 1997). However, when isolated oocyte NEs stripped of these additional fibers were immunolabeled with antibodies against the N terminus of Nup153, it was located either at the nuclear coaxial ring (Walther et al., 2001, Fahrenkrog et al., 2002) or at the terminal basket ring 50–100 nm away from the NE midplane (Pante et al., 2000). Antibodies against central and C-terminal parts of Nup153 were reported to label both the terminal ring and cytoplasmic side of the NPC in amphibian oocytes (Pante et al., 1994; Fahrenkrog et al., 2002) but not in somatic cells (Sukegawa and Blobel, 1993).
Immuno-EM localizations of Tpr have been similarly diverse. First described as a protein located at the cytoplasmic NPC side (Byrd et al., 1994), it was later found attached to the nuclear NPC side in mammalian cells (Cordes et al., 1997; Frosst et al., 2002). In Xenopus oocytes, Tpr labeling occurred not only within the territory occupied by the nuclear basket but also at the basket-attached fibers that project further into the nucleus (Cordes et al., 1997). In insect cells, Tpr was reported to occur throughout the extrachromosomal space of the nuclear interior (Zimowska et al., 1997). An extensive Tpr filament network crossing the nuclear interior of HeLa cells has been reported as well (Fontoura et al., 2001). However, the specificity of some Tpr antibodies causing such staining patterns in mammals has been disputed (Kuznetsov et al., 2002).
Differences in localization of individual Nups might be due to cell type- or species-specific features. For example, the conspicuous masses of fibrous material attached to the nuclear basket in mature amphibian oocytes (Scheer et al., 1988; Arlucea et al., 1998) have not been identified in other cell types and may be specific for oocytes, perhaps representing a stockpile of Tpr and other proteins required during embryogenesis (Hase and Cordes, 2003). Different types of antibodies, ranging from single epitope monoclonals to polyclonals against large protein segments, and differences in specimen preparation and labeling procedures may have contributed to the diversity of results as well.
In an attempt to minimize variations due to differences in experimental procedures and to allow direct comparisons, we have reinvestigated the localization of several Nups and Tpr in parallel, performing immunogold-EM with peptide-specific antibodies by using a postembedding labeling approach. To gain additional insight into the contribution of certain Nups to NPC architecture, cells were depleted of these proteins by RNA interference (RNAi) and then studied with respect to NPC integrity and composition. The results tempt us to propose a model in which Tpr constitutes the central architectural element of the nuclear basket and other Nups are components of the NPC proper.
MATERIALS AND METHODS
Antibodies
Guinea pig (gp) antibodies against mNup62 (Cordes et al., 1991); rabbit (rb) and gp antibodies against hNup358 aa 2285–2314 (Cordes et al., 1996) and hTpr aa 2063–2084, gp α-hTpr aa 1622–1640, gp and rb α-hTpr aa 1–14, 8–21, 604–615, and 2338–2363; monoclonal antibody (mAb) 203–37 against hTpr (Cordes et al., 1997); mAb PF190 × 7A8 against rNup153 (Cordes et al., 1993); gp antibodies for hNup153 aa 22–36 and 391–404 (Ferrando-May et al., 2001), hNup107 aa 33–51, Nup205 aa 1784–1803 (accession no. D86978), hNup98 aa 206–220, and 596–618, hNup93 aa 2–203, 350–369, and 371–389, rb α-hNup50 aa 126–147, and gp and rb α-hNup96 aa 880–900 have been described previously (Hase and Cordes, 2003). The aa numbering for hNup96 relates to the full-length protein of ∼106 kDa. The corresponding gene encodes the Nup98-Nup96 precursor of Mr 195846 (accession no. AAL56659) that is autoproteolytically processed to give rise to Nup98, and Nup96 of Mr 105941. A formerly described hNup96 sequence of Mr 96196 (AF071076) represents a truncated version resulting from a sequence inversion neither present in the genomic sequence nor in other Nup98-Nup96 mRNAs. Novel gp antibodies were raised against synthetic peptides corresponding to 1) hTpr aa 636–655 and 2) aa 914–932. Peptides were coupled to keyhole limpet hemocyanin via a C-terminal 1) or N-terminal 2) cysteine residue. Affinity-purification of Igs followed standard procedures (Kuznetsov et al., 2002). Rb antibodies against hNup93 aa 2–218 (Grandi et al., 1997) had been kindly provided by Dr. Ed Hurt (University of Heidelberg, Heidelberg, Germany). mAb 414 was from Babco (Richmond, CA). Secondary antibodies coupled to horseradish peroxidase, fluorochromes, and colloidal gold were from Jackson ImmunoResearch Laboratories (West Grove, PA), DakoCytomation Denmark A/S (Glostrup, Denmark), and Sigma-Aldrich (Stockholm, Sweden).
Cell Fractionation and Immunoblotting
Preparation of total protein samples from HeLa cells grown to near confluence (∼80%), and cell fractionation was performed as described previously (Kuznetsov et al., 2002) with some modifications. Cells were first incubated with phosphate-buffered saline (PBS) plus 0.005% digitonin for 3 min, briefly washed with PBS, and then incubated with PBS plus 0.4% Triton X-100 for 3 min, and washed again. All solutions were recollected and centrifuged at 13,000 × g for 5 min followed by methanol precipitation of proteins. Residual cyto- and karyoskeletons attached to the dishes were scraped off in 95°C SDS protein sample buffer and boiled. SDS-PAGE and immunoblotting were as described previously (Hase et al., 2001).
Posttranscriptional Gene Silencing and Immunofluorescence Microscopy
Transfection of HeLa cells with small interfering RNAs (siRNAs) was as described previously (Elbashir et al., 2001; Kuznetsov et al., 2002; Hase and Cordes, 2003). Synthetic RNA oligonucleotides were from Dharmacon Research (Lafayette, CO) and DNA Technology A/S (Aarhus, Denmark). Antisense strands were complementary to nucleotide (nt) 3732–3712 of the hNup196 cDNA (AF231130), nt 530–510 of hNup107 (AJ295745), nt 978–958 of hNup93 (NM_014669) and nt 440–420 of Nup205 (D86978). Cells were analyzed 2, 3, 3.5, 4, and 5 d posttransfection. Immunofluorescence microscopy (IFM) of formaldehyde (FA)-fixed HeLa cells was as described previously (Hase and Cordes, 2003). For digitonin permeabilization of cells before or after fixation, cells were treated with 0.005 or 0.05% digitonin in ice-cold PBS for 10 or 20 min. Confocal microscopy and image analysis was performed with an LSM 510 (Carl Zeiss, Oberkochen, Germany).
Immunoelectron Microscopy and Specimen Evaluation
HeLa cells grown to near confluence in 8-cm dishes were washed with PBS and fixed in 2% FA plus 0.1% glutaraldehyde (GA) in PBS for 1 h. This low GA concentration was found to represent an acceptable compromise between a reasonable degree of NE and NPC structure preservation and maintaining intact target epitopes for most antibodies in this study. For labeling of GA-sensitive epitopes, cells were fixed with 3.7% FA in PBS for 2 h. After washes in PBS, cells were scraped off the dishes and sedimented at 9,000 × g for 20 min. The resulting pellet was cut into small pieces and dehydrated in ethanol (70, 95, 100%). Pellet pieces were then incubated in a mixture of equal parts (vol/vol) of ethanol and LR White (London Resin, Reading, United Kingdom) at 20°C for 30 min, followed by 12 h in pure resin at 4°C. After two further 30-min incubations in fresh resin, the samples were transferred into resin-filled gelatin capsules and allowed to harden in a UV polymerization unit (Agar Scientific, Stansted, United Kingdom) for 12 h. Sections of 60 to 80 nm in thickness were transferred onto nickel grids coated with Formvar film. For immunogold staining, the grids were first placed onto droplets of PBS with 5% bovine serum albumin (BSA) (PBS-BSA) for 30 min, transferred to droplets containing primary antibodies diluted in PBS-BSA, and incubated in a humid box for 2 h. After washes with PBS-BSA, the grids were placed on droplets of gold-labeled secondary antibodies diluted in PBS/0.5% BSA for 1 h, washed in PBS/0.5% BSA, and then PBS alone. Specimen were postfixed with 2% GA in PBS for 5 min, rinsed with PBS and H2O, and air-dried. Staining for contrast was in a saturated aqueous solution of uranyl acetate for 5 min, followed by lead citrate for 5 s. Immunolabelings were performed in duplicate, with four to six primary antibodies against different target proteins in parallel. Before EM analysis, all grids were encoded and only decoded again after data collection and completion of the evaluation process. Some immunolabelings were paralleled by controls without primary antibodies, and with unrelated gp and rb IgGs. Specimens were examined in a Philips CM120 EM at 80 kV. For quantitative evaluation of gold particle distribution, images were collected at a primary magnification of 53,000× with a Kodak MegaPlus charge-coupled device camera. Borders on both sides of the NPC between which gold particles were considered were set at 200-nm distance from the NE midplane. The analySIS system (Soft Imaging Software, Münster, Germany) was used for distance measurements. Mean grain diameters in different batches of commercial 10-nm gold-coupled antibodies were found to be close to 9 nm; individual grain sizes deviated from the mean by up to 25%. Magnification of the EM was calibrated using a 2160 line/mm crossed ruled grating (Agar Scientific).
RESULTS
An earlier immuno-EM study on Nup62, a component of the NPC central core, revealed striking differences in epitope accessibility and immunogold grain distribution patterns depending on the labeling procedure chosen (Grote et al., 1995). Whereas preembedding labeling resulted in a peak distribution of gold grains ∼35 nm away from the NPC midplane, postembedding immuno-EM gave labeling of the NPC center. These differences were explained by limited antibody access to certain NPC components in preembedding approaches, and better accessibility on ultrathin sections through NPCs used for postembedding labeling.
In preembedding studies on isolated NEs, detergent-permeabilized cells or thick cryostat sections, the antibodies encounter NPCs that still represent three-dimensional entities. Steric constraints are then likely to dictate the angle and preferential orientation by which some antibodies bind to certain NPC components. In fact, when using IgGs as primary antibodies, together with gold-coupled secondary antibodies for indirect immunolabelings, the mean distance between target and peak distribution of gold particles can come close to 30 nm (Kellenberger and Hayat, 1991; Mayer and Bendayan, 2001). In contrast, antibodies binding to epitopes only exposed on the surface of ultrathin sections through resin-embedded NPCs can enjoy more rotational freedom. Consequently, the mean distribution of gold grains in postembedding immuno-EM of NPCs may define the actual location of certain target proteins more accurately. This in mind, we set out to reinvestigate the localization of several Nups and Tpr at the NPC.
Postembedding Immuno-EM Locates Nup153, Components of the Nup93 and Nup160 Subcomplexes, and Nup98 at NPC Core Structures
For postembedding immuno-EM, HeLa cells were embedded in the acrylic resin LR White (Newman et al., 1983). On sections through such resin-embedded cells, only antigens exposed at the surface are labeled by gold-coupled antibodies. Penetration into the depth of the section is limited to a few nanometers at most (Bendayan et al., 1987; Stierhof and Schwarz, 1991).
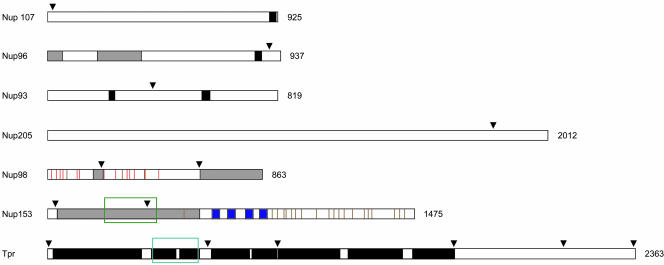
Recently, we have described peptide-antibodies specific for Nup93, 96, 98, 107, 153, and 205 (Ferrando-May et al., 2001; Hase and Cordes, 2003). Relative positions of the antibody target sites on these proteins are shown in Figure 1. Now used for postembedding immuno-EM, several of these antibodies specifically labeled central or peripheral parts of the NPC proper. Even Nup93, which in IFM can only be labeled after preextraction of nonfixed cells with strong detergents (Grandi et al., 1997; Hase and Cordes, 2003), became accessible for antibodies in sectioned NPCs.
Figure 1.
Peptide antibody target sites on Nups and Tpr. Relative positions of target sites for peptide antibodies used in this study are indicated by arrowheads. Black boxes represent sequence segments for which probability of coiled-coil formation is predicted to be >50%. Blue boxes represent the C2-C2 zinc fingers that constitute the Ran-GTP binding domain of Nup153. Brown vertical stripes in the Nup153 and Nup98 diagrams indicate individual FxFG and GFxF sequence motifs, and red verticals represent GLFG motifs and derivatives thereof present only in Nup98. Simple FG repeats are not included. The Nup98 and Nup96 diagrams represent the products of Nup196 autoproteolysis. The shaded boxes in the Nup96 diagram represent the N-terminal binding site for Nup98, and the Sec13 binding region. The shaded boxes in the Nup98 diagram stand for the binding site for the mRNA transport factor Rae1/Gle2, and the C-terminal binding region for Nup96 and Nup88. The shaded box in the Nup153 diagram represents the overlapping bindings regions for the Nup160 subcomplex (aa 39–339), Tpr (aa 228–439), Nup50 (337–611), and the RNA binding domain (250–400). The green rectangle encloses the Tpr binding domain of Nup153 and the turquoise rectangle the NPC/Nup153 binding domain of Tpr.
The antibodies against Nup93 and 205 decorated regions near the NPC center, with gold grains found on both sides of the midplane (x-axis). Grains that decorated Nup96 and 107 were located on both sides of the NPC as well. When looking at each of these gold grain pools as a whole, their patterns only slightly diverged from a mirror-symmetrical distribution (see below). In general, gold grains for Nup93 and 205 were located closer to the x-axis than those for Nup96 and 107.
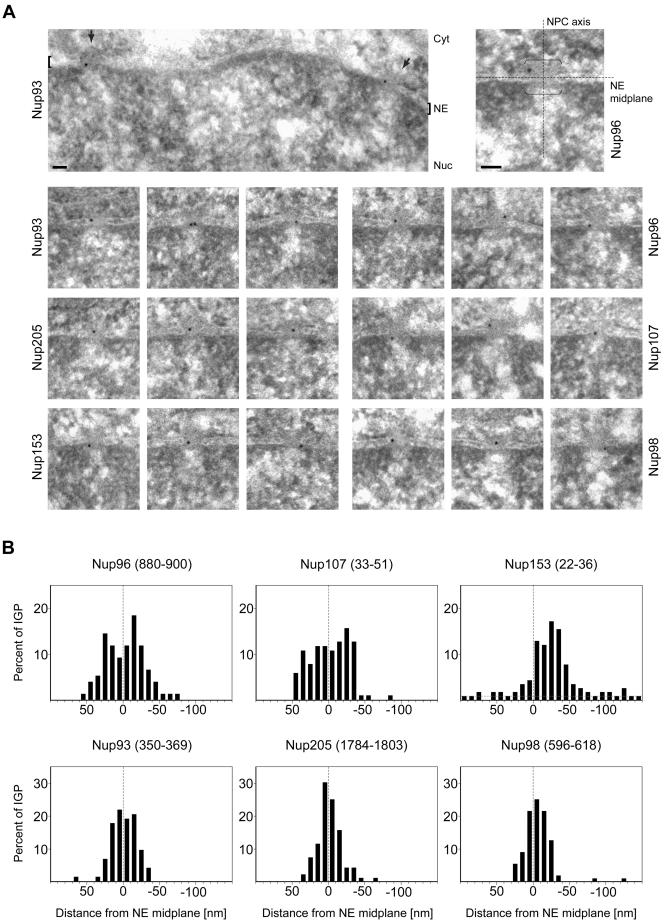
In contrast, antibodies targeting the N-terminal domain of Nup153 predominantly labeled the nuclear margin of the NPC, with far less gold grains on the cytoplasmic side. The majority of gold particles that labeled Nup98 were in turn located much closer to the x-axis and decorated both sides of the midplane. However, the calculated mean distance from the NE midplane pointed at a slight preference for Nup98 labeling at the nuclear NPC side as well (see below). Labelings with other Nup98 peptide antibodies resulted in similar gold grain distribution patterns (Supplemental Figure S1). A selection of representative micrographs is shown in Figure 2A. The histograms in Figure 2B represent gold particle distributions relative to the midplane of the NE, after normalization of values for NE width variations (see below). Calculated mean distances of gold grains from the NE midplane are listed in Table 1.
Figure 2.
Postembedding immunogold-EM of nucleoporins in HeLa cells. (A) Representative examples of LR White-embedded NEs after labeling with peptide-specific antibodies against Nup93 aa 350–369, Nup96 aa 880–900, Nup98 aa 596–618, Nup107 aa 33–51, Nup153 aa 22–36, and Nup205 aa 1784–1803, and 10-nm gold-coupled secondary antibodies. Cells were fixed with FA plus GA. Cytoplasm (Cyt) and nuclear compartment (Nuc), separated by the NE, are oriented toward top and bottom, respectively. Arrows (top left) and brackets (right) demark some NPCs in cross section. Bars, 50 nm. Same magnification for top left image and image panels below. (B) Distribution of immunogold particles (IGP) relative to the NE midplane (dashed vertical). Data collection was from randomly chosen gold-labeled NEs from encoded specimens and values were normalized for NE width variations as described in Figure 3A. Histograms include all grains detected in proximity to perpendicularly sectioned NPCs up to 100 nm from the NE midplane at the cytoplasmic and 150 nm at the nuclear side (negative values). Additional specific labeling of distinct structures deep within the nuclear interior, such as centromere labeling with α-Nup107 (Belgareh et al., 2001) and labeling of GLFG bodies with α-Nup98 (Griffis et al., 2002) was not considered. Sample sizes (n) are 75 (Nup93), 78 (Nup96), 109 (Nup98), 105 (Nup107), 116 (Nup153), and 98 (Nup205). In sporadic cases in which two gold particles <25 nm apart labeled the same NPC, their mean distance value was included, treating them as an immunogold cluster that labeled only one site. Very rare clusters of three or more grains were not considered. Use of α-Nup153 aa 22–36 on GA-fixed specimen produced some additional staining throughout the cell that was rated as background also observed far beyond the borders of the histogram. When compared at a 1:1 surface ratio, the approximate level (dotted gray horizontal) of such background in the cytoplasmic and nuclear compartments reached ∼8–11% of the peak distribution at the nuclear side of the NE.
Table 1.
Mean distances of nucleoporins from the NE midplane and the central symmetry axis of the NPC in FA/GA-fixed specimen
| Distance NE midplaneb (x-axis) | Distance NE midplane [normalized]c | Distance NE midplane [bilateral symmetry, normalized]d | Distance NPC-axis (y-axis)e | Distance NPC-axis [normalized]f | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Antibody | Sample sizea | (Mean) | (Mean) | (SE)g | (Mean ±) | (SE) | Sample sizee | (Mean) | (Mean) | (SE) |
| Nup96 (880-900) | 78/74 | 1.9 | -0.4 | 2.8 | 20.5 | 1.4 | 62 | 19.1 | 33.0 | 1.1 |
| Nup107 (33-51) | 105/102 | 1.0 | -0.8 | 2.5 | 21.3 | 1.2 | 74 | 19.3 | 32.9 | 1.0 |
| Nup98 (596-618) | 109/108 | -1.2 | -4.5* | 1.3 | 11.3* | 0.8 | 55 | 13.0 | 29.9 | 1.2 |
| Nup93 (350-369) | 75/74 | 0.5 | -2.1 | 1.8 | 13.3 | 0.9 | 53 | 15.6 | 30.2 | 1.0 |
| Nup205 (1784-1803) | 98/97 | 1.5 | -0.5 | 1.6 | 12.1 | 1.0 | 50 | 12.2 | 29.2 | 0.7 |
| Nup153 (22-36) | 116/89 | -15.6** | -17.6** | 2.2 | 51 | 23.2 | 34.3 | 1.3 | ||
Cells were fixed in 2% formaldehyde plus 0.1% glutaraldehyde.
Data collection was from randomly chosen gold-labeled NEs from encoded specimens that were decoded only after completion of the evaluation process. Boundaries between which gold grains located in proximity to perpendicularly sectioned NPCs were measured had initially been set at a distance of 100 nm from the NE midplane on its cytoplasmic and 200 nm on its nuclear side (sample size values on the left). For further calculations of mean distances only gold grains located up to 50 nm on each side of the NE were considered (sample size values on the right).
Cytoplasmic and nuclear locations are indicated by positive and negative distance values, respectively. The algebraic signs of the distance values were taken into account in these calculations. Distances are in nanometer.
Normalization of values for NE width variations are as described in Figure 3A.
Calculations of means that ignore the algebraic signs of the distance values for nuclear and cytoplasmic gold grains. Such calculations are based on the assumption that certain nucleoporins are bilateral symmetrically arranged and that immunogold grains would belong to equivalent pools on both sides of the NPC midplane.
For the calculation of the mean distance to the central eightfold rotational symmetry axis of the NPC, only such gold-labeled NPC sections were considered in which both membrane borders of the pore were discernible. Coordinates of gold grains that resided within an area up to 50 nm from the NE midplane on the cytoplasmic side, 100 nm on the nuclear side, and 60 nm from the central symmetry axis of the NPC were considered for calculations.
Normalization of values for non-diametric perpendicular section planes through NPCs as described in Figure 3B. See also considerations in Supplemental Figure S7.
Standard error.
For antibody α-Nup98 aa 206-222 (Supplemental Figure S1), the normalized mean value was -5.2 nm. The corresponding normalized mean that ignored the algebraic signs of the distance values was ±12.8 nm.
Nup153 mean values for gold grains collected only on the nuclear side of the NE up to 50 nm from the NE midplane are -22.7 nm (nonnormalized, n = 73) and -24.5 nm (normalized, n = 76).
The width of the perinuclear lumen between inner and outer NE membrane was often found to vary by several nanometers between neighboring NE segments. Because this affected the position of the NE midplane, the relative position of each gold grain was normalized for such NE width variations as described in Figure 3A. On the whole, resulting histograms (Figure 2B) did not differ much from the nonnormalized versions (Supplemental Figure S2A) but emphasized the existence of a bilateral symmetrical arrangement of some of the Nups. Gold grain positions relative to the NPC were then plotted according to their x/y coordinates onto a scheme of an idealistic NPC in cross section (Figure 4). In addition to y value normalization for NE width variations, x values were normalized for nondiametric perpendicular section planes as described in Figure 3B. Calculated mean distances of gold particles from the central symmetry-axis of the NPC are listed in Table 1.
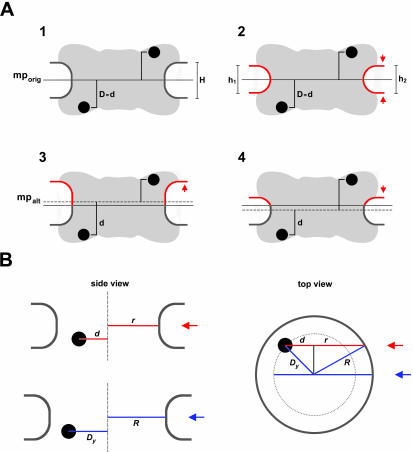
Figure 3.
Normalization of gold grain coordinates for NE width variations and nondiametric perpendicular section planes through NPCs. (A) Variations in NE width may occur naturally or as a result of the fixation and dehydration process that may cause some shrinkage of the NE's perinuclear lumen. Local swelling and increase in NE width can be seen as well, although less frequently. The dimensions of the NPC proper are less subjected to such alterations. However, because the NE midplane is used as the x-axis relative to which the distance of an individual gold grain is measured, variations in NE width can cause deviations between the relative position of a grain in the picture and when incorporated according to its measured distance d into an idealistic NPC model (A1). In principle, swelling or shrinkage (red arrows) of the NE might affect both inner and outer NE membrane positions similarly (A2). In this case, the midplane would still be the same as the original mid-axis (mporig) in the idealistic model A1. However, the inner NE membrane and its position relative to the NPC are more likely to be stabilized by interactions with the underlying lamina and heterochromatin. In this scenario, it is mainly the relative position of the outer NE membrane that will be affected by shrinkage or swelling of the perinuclear lumen (A3, A4). This alters the position of the midline (mpalt) between both membranes. To take such deviations into account, the width of the NE, including perinuclear lumen and membranes, was measured on each side of a gold-grain decorated NPC. This yielded the two values h1 and h2. The width H for the idealistic NE was set at 32 nm. The normalized distance value D for each gold grain located on the nuclear side of the midplane was then calculated by using the equation D = d + [(H – h1) + (H – h2)] 2–2; for grains on the cytoplasmic side the equation was D = d + [(H – h1) + (H – h2)] 2–2 (B) Perpendicular section planes through most gold-labeled NPCs do not pass directly through the NPC center but represent chords that are shorter than the real NPC diameter. Consequently, the measured mean distance between gold grains and apparent vertical mid-axis (y-axis) through cross-sectioned NPCs is smaller than the real radial distance of the target protein from the NPC's eightfold rotational symmetry axis. This deviation can be accounted for by measuring not only the distance between apparent mid-axis and gold grain but also the apparent diameter of this NPC in cross section. The real distance Dy between gold-labeled target and real mid-axis can then be calculated using the equation 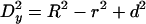 , respectively:
, respectively: 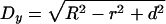 . R stands for the real NPC radius, i.e., the distance between mid axis and membrane boundary of an ideal NPC (here set at 40 nm), r for the measured distance between apparent mid-axis and membrane boundary of the cross-sectioned NPC, and d for the measured distance between apparent mid-axis and gold grain. Grains on grazing sections of the pore where r was <25 nm were not considered.
. R stands for the real NPC radius, i.e., the distance between mid axis and membrane boundary of an ideal NPC (here set at 40 nm), r for the measured distance between apparent mid-axis and membrane boundary of the cross-sectioned NPC, and d for the measured distance between apparent mid-axis and gold grain. Grains on grazing sections of the pore where r was <25 nm were not considered.
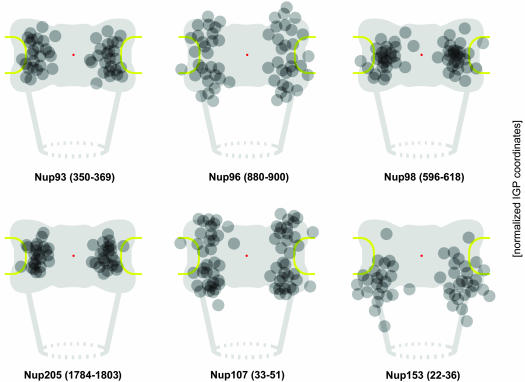
Figure 4.
Relative distributions of immunogold label for different nucleoporins with respect to an idealistic NPC in cross section. Data collection was from at least 50 randomly chosen gold-labeled NPCs in which both membrane boundaries flanking the pore channel were discernible. The coordinates for each immunogold particle (IGP) were normalized for NE width variations and nondiametric perpendicular section planes as described in Figure 3. Because of the eightfold rotational symmetry of the NPC, assignment of x values to either side of the y-axis is arbitrary. Therefore, the algebraic signs of a few x values were reversed to allocate similar numbers of grains to each side of the y-axis. To further enhance clarity of peak distributions and reduce signal spread due to extremes of antibody spreading or minor background label, sporadic grains most distant from the calculated mean position for each Nup (Table 1) were not included (cut-off ≤5%). Red dots mark the NPC center, flanked by the NPC core structures, including the coaxial rings, and the NE membrane (yellow). Attached to the nuclear ring, only two of the eight rod-like fibers of the nuclear basket, and the distal terminal ring (dashed circle) are depicted. Pore diameter in this scheme corresponds to 80 nm, NE width to 32 nm, and distance between NE midplane and terminal ring to 80 nm.
In summary, these immuno-EM data were indicative of a bilateral symmetrical arrangement of Nup96 and Nup107 on both NPC sides; a preferential location of the targeted Nup98, Nup93, and Nup205 domains within the NPC core region as well; and a peripheral position for the Nup153 target domain at the nuclear coaxial ring. These conclusions find further support by confocal IFM studies of semipermeabilized HeLa cells outlined in Supplemental Figure 3.
Congruent Locations of Tpr Protein Domains and Different Parts of the Nuclear Basket
The Tpr protein is located near the nuclear side of the NPC in mammalian cells (Cordes et al., 1997; Frosst et al., 2002), forms coiled-coil homodimers of extended rod-like shape (Hase et al., 2001), and is linked to the NPC via direct binding to Nup153 (Hase and Cordes, 2003). These findings made Tpr a likely candidate for an architectural element of the nuclear basket (Hase and Cordes, 2003). To follow up this idea, we determined where different Tpr domains are located relative to the NPC and basket, by using a panel of peptide-specific antibodies raised against various parts of Tpr. These antibodies comprise such that have been described previously (Cordes et al., 1997; Kuznetsov et al., 2002; Hase and Cordes, 2003) and novel ones raised for this study (see below).
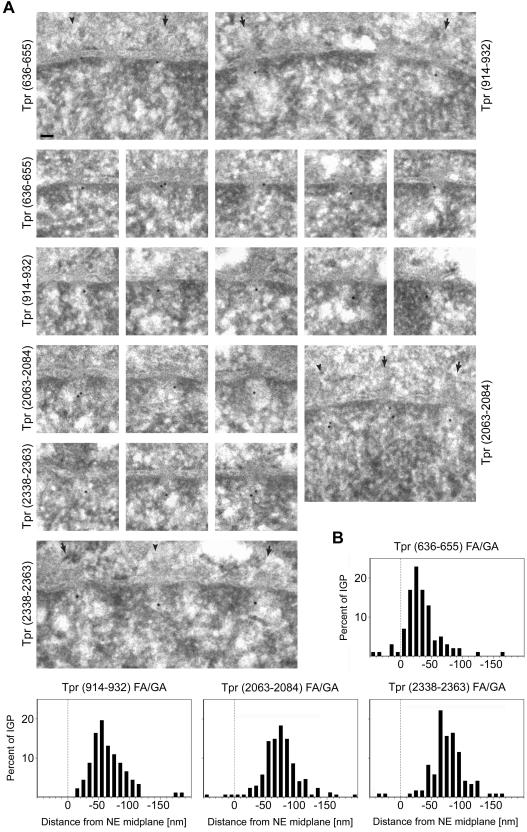
Immunolocalization of Tpr's C-terminal domain on LR White sections of GA-fixed HeLa cells was performed with antibodies against aa 2063–2084 and the actual C-terminus (aa 2338–2363). Both specifically decorated a region in proximity to the nuclear NPC side, at normalized mean distances of ∼74 nm (α-Tpr 2063–2084) and ∼76 nm (α-Tpr 2338–2363) from the NE midplane (Figures 5 and 6 and Table 2). This region coincides with the approximate position of the terminal ring (Goldberg and Allen, 1992).
Figure 5.
Postembedding immunogold localization of Tpr domains relative to the NPC in HeLa cells. (A) Representative examples of LR White-embedded NEs after labeling with peptidespecific antibodies against Tpr aa 636–655, 914–932, 2063–2084, and 2338–2363. The nuclear compartment is oriented toward the bottom. Arrows and arrowheads demark some NPCs in cross and grazing section, respectively. Bar, 50 nm, for all micrographs. Cells were fixed with FA plus GA. (B) Distribution of gold particles relative to the NE midplane. Data were from randomly chosen gold-labeled NEs from encoded specimens. Histograms include all grains detected in proximity to perpendicularly sectioned NPCs up to 50 nm from the NE midplane at the cytoplasmic and 200 nm at the nuclear side. Values were normalized for NE width variations as described in Figure 3A. Sample sizes (n) are 101 (636–655), 92 (914–932), 172 (2063–2084), and 121 (2338–2363). The histogram for α-Tpr 2063–2084 combines two highly similar data sets from separate labelings with antibodies from one animal. For α-Tpr 636–655, only one of two similar data sets are shown, obtained with different batches of Tpr 636–655 antibodies from two animals (see Figure 7).
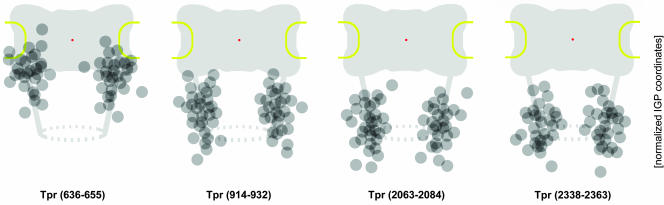
Figure 6.
Distributions of gold grains relative to NPCs after immunogold labeling of different Tpr domains. Details of schemes and presentation of gold grains collected from cells fixed with FA plus GA are as for Figure 4. The coordinates for each IGP were normalized for NE width variations and nondiametric perpendicular section planes through NPCs as described in Figure 3. To enhance clarity of peak distributions, sporadic gold particles most distant from the normalized mean position for each Tpr segment (Table 2) were not included (cut-off ≤5%).
Table 2.
Mean distances of Tpr domains from the NE midplane and the central symmetry axis of the NPC in FA/GA-fixed specimen
| Distance NE midplaneb (x-axis) | Distance NE midplane [normalized]c | Distance NPC-axis (y-axis)d | Distance NPC-axis [normalized]e | |||||
|---|---|---|---|---|---|---|---|---|
| Antibody | Sample sizea | (Mean) | (Mean) | (SE) | Sample sized | (Mean) | (Mean) | (SE) |
| Tpr (636-655)f | 101/93 | -33.0* | -35.4* | 2.3 | 62 | 23.0 | 35.0 | 1.2 |
| Tpr (914-932) | 92/90 | -58.7 | -62.4 | 2.6 | 65 | 16.1 | 32.6 | 0.8 |
| Tpr (2063-2084) | 179/165 | -70.5 | -73.9 | 1.8 | 65 | 13.7 | 30.8 | 1.0 |
| Tpr (2338-2363) | 121/117 | -73.2 | -76.1 | 2.1 | 63 | 12.6 | 27.8 | 1.0 |
Cells were fixed in 2% formaldehyde plus 0.1% glutaraldehyde.
Data collection was as described for Table 1. Boundaries between which gold grains located in proximity to perpendicularly sectioned NPCs were measured had initially been set at a distance of 100 nm from the NE midplane on its cytoplasmic side and 200 nm on its nuclear side (sample size values on the left). For further calculations of mean distances only gold grains located up to 150 nm on the nuclear side of the NE were considered (sample size values on the right).
The negative algebraic sign of an NE distance value (given in nanometer) indicates nuclear localization.
Normalization of values for NE width variations as described in Figure 3A.
For the calculation of the mean distance to the central symmetry axis of the NPC, the same parameters were applied as for the corresponding calculations in Table 1.
Normalization of values for non-diametric perpendicular section planes through NPCs as described in Figure 3B. See also considerations in Supplemental Figure S7.
Values presented are from only one of two highly similar data sets that were obtained with different batches of Tpr 636-655 antibodies from two guinea pigs. Normalized mean values for the second data set were -33.5 nm (distance to NE midplane) and 36.2 nm (distance to NPC-axis; n = 61), respectively.
Mean values for Tpr (636-655) that only include gold grains up to 50 nm on the nuclear side (n = 77) are -24.9 nm (nonnormalized) and -27.4 nm (normalized). Mean values for grains up to 100 nm on the nuclear side are -32.0 nm (nonnormalized, n = 92) and -34.6 nm (normalized, n = 93).
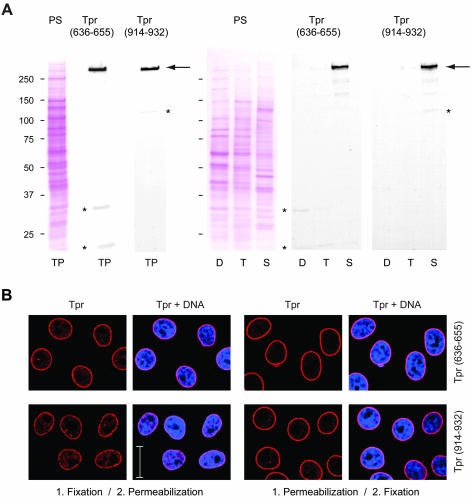
Previously, we had raised several monoclonal and peptide-specific polyclonal antibodies against epitopes that flank Tpr's NPC binding domain (NBD) between aa 400 and 600 (Hase et al., 2001; Kuznetsov et al., 2002; unpublished data). However, these antibodies were found to be of no use for postembedding immuno-EM, either because of pronounced in situ cross-reactions with Tpr-unrelated proteins (Kuznetsov et al., 2002) or because some epitopes apparently had become blocked during the fixation and embedding procedure. Therefore, we screened the Tpr segments that flank the NBD for other antigenic sites that would remain intact upon specimen preparation. This resulted in the identification of two Tpr segments against which we raised peptide-specific antibodies (Figure 7). The one site was located close to the NBD (aa 636–655) and the other more distal (aa 914–932). Tpr specificity was controlled by immunoblotting (Figure 7A). In postembedding immuno-EM, both α-Tpr 636–655 and α-Tpr 914–932 decorated regions in the proximity of the nuclear NPC side (Figure 5A). However, whereas the normalized mean distance of immunogold label with α-Tpr 914–932 was found to be ∼62 nm from the NE midplane, labeling with α-Tpr 636–655 was closer to the NPC core, at an approximate mean distance of 27–35 nm (Figures 5B and 6 and Table 2).
Figure 7.
Characterization of novel peptide-specific Tpr antibodies. (A) Immunoblotting of HeLa cell proteins by using antibodies against Tpr aa 636–655 and aa 914–932. Proteins were separated by SDS-PAGE and stained with Ponceau S (PS) after transfer to nitrocellulose filters. Lanes include total cell proteins (TP), soluble proteins first extracted with digitonin (D) and then with Triton X-100 (T), and residual cell sediment proteins (S). Lanes D, T, and S were loaded with the same percentage of each fraction. Immunodetection of Tpr by enhanced chemiluminescence reaction was on filters identical or similar to the ones stained with Ponceau S. Positions of marker proteins are given on the left margin. Note that both antibodies label one major band of >250 kDa (large arrow), in line with Tpr's mass of 267 kDa. Only weak cross-reactions of α-Tpr 914–932 with a detergent-insoluble protein of ∼135 kDa, and of α-Tpr 636–655 with detergent-soluble cytoplasmic proteins of ∼32 and ∼22 kDa (asterisks) are noted after prolonged exposure. These latter proteins were not recognized by a second batch of Tpr 636–655 antibodies raised in another guinea pig, which revealed other minor cross-reactions instead (unpublished data). (B) Confocal IFM of HeLa cells immunolabeled with α-Tpr 636–655 and 914–932. Cells were permeabilized with detergent either after or before fixation. DNA-staining with TO-PRO-3 is in blue. Bar, 20 μm.
To also determine the relative positions of the N terminus and the distal end of the rod domain, we tested a collection of antibodies against sites located between Tpr aa 1–135 and between aa 1370–1640 (Cordes et al., 1997; Kuznetsov et al., 2002; our unpublished data) but had to realize that none of the corresponding epitopes tolerated fixation with GA. However, when GA was omitted and cells were fixed with FA only, several of these antibodies were found to be useful for immuno-EM. Although the NEs in such FA-fixed cells sometimes seemed bloated, with NE membranes less well structurally preserved than in GA-fixed specimens, the direct comparison of both fixation methods, by using several GA-insensitive Tpr and Nup antibodies in parallel, revealed rather similar gold grain distributions relative to the NE midplane. Only slight differences in the calculated means were observed (Figure 8 and Table 3; our unpublished data).
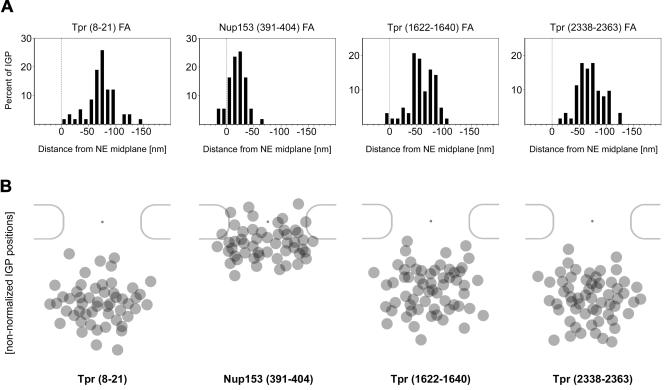
Figure 8.
Postembedding immunogold localization of Tpr domains and the Tpr-binding domain of Nup153 in FA-fixed HeLa cells. (A) Nonnormalized distributions of gold grains relative to the NE midplane, after immunogold-labeling of FA-fixed HeLa cells with antibodies against Tpr aa 8–21, 1622–1640, and 2338–2363, and against Nup153 aa 391–404. Data were collected from randomly chosen gold-labeled NEs from encoded specimens. Histograms include all gold particles detected in proximity to perpendicularly sectioned NPCs, up to 50 nm from the NE midplane at the cytoplasmic and 200 nm at the nuclear side. Because membrane boundaries and perinuclear lumen of the NE in FA-fixed specimen are often not clearly discriminable, values were not normalized for NE width variations. Sample sizes (n) are 55 (Nup153), 58 (Tpr 8–21), 63 (Tpr 1622–1640), and 62 (Tpr 2338–2363). (B) Nonnormalized distributions of gold grains relative to nuclear pores. Membrane borders of the pore are generally difficult to pinpoint in FA-fixed specimens, which usually prevents accurate measurements of pore diameters. IGP positions were therefore superimposed onto the scheme of a representative pore in cross section as described previously (Walther et al., 2002), without normalization for nondiametric perpendicular section planes and NE width variations. To enhance clarity of peak distributions, gold grains most distant from the approximate nonnormalized mean positions (Table 3) were not included (cut-off ≤5% for α-Nup153, α-Tpr 1622–1640, and α-Tpr 2338–2363; 9% for α-Tpr 8–21). Immunogold labelings were performed with several GA-insensitive Nup and Tpr antibodies in parallel, to allow double blind comparative experiments with ample encryption of specimen.
Table 3.
Mean distances of Tpr domains and the Tpr-binding domain of Nup153 from the NE midplane in FA-fixed specimen
| Distance NE midplanea (x-axis) | |||
|---|---|---|---|
| Antibody | Sample size | (Mean) | (SE) |
| Nup153 (391-404) | 55 | -22.9* | 1.8 |
| Tpr (8-21) | 58 | -72.9 | 3.4 |
| Tpr (1622-1640) | 63 | -60.9 | 2.7 |
| Tpr (2338-2363) | 62 | -70.1 | 2.9 |
Cells were fixed in 3.7% formaldehyde.
Calculations are based on gold grains collected on the nuclear side of the NE, up to 150 nm from the approximate position of the NE midplane. The negative algebraic sign of an NE distance value (in nm) indicates nuclear localization. Because membranes and perinuclear lumen of the NE in FA-fixed specimen were often not clearly discriminable, values were not normalized for NE width variations. Deviations by a few nm between alleged and actual positions of the NE midplane cannot be excluded in individual cases.
Sample size n = 49. The mean value for Nup153, including gold grains located up to 30 nm from the NE midplane on the cytoplasmic side and up to 70 nm on the nuclear side (sample size n = 55) is -19.2 nm.
Under these conditions, the peak of gold grains after labeling with α-Tpr 2338–2363 was ∼70 nm from the NE midplane (Table 3, nonnormalized data). Using a peptide antibody against Tpr aa 1622–1640, the distal end of the rod domain at aa 1630 was found closer to the midplane (∼61 nm). However, with antibodies against aa 8–21, the N terminus was localized similarly far from the midplane (∼73 nm) as the C terminus. Nonnormalized positions of these gold particles from FA-fixed specimen were then summarized by superimposing them onto schemes of nuclear pores in cross section (Figure 8B).
Recently, we have shown that Nup153 segment aa 228–439 includes the binding site for the NBD of Tpr (Hase and Cordes, 2003). To complement our immunolocalization of Tpr's NBD relative to the NPC, we labeled the corresponding Nup153 binding site for Tpr as well, by using FA-fixed cells and a peptide-specific antibody raised against Nup153 aa 391–404. This antibody specifically decorated a region corresponding to the position of the nuclear coaxial ring, at a mean distance of 19–23 nm from the NE midplane (Figure 8 and Table 3).
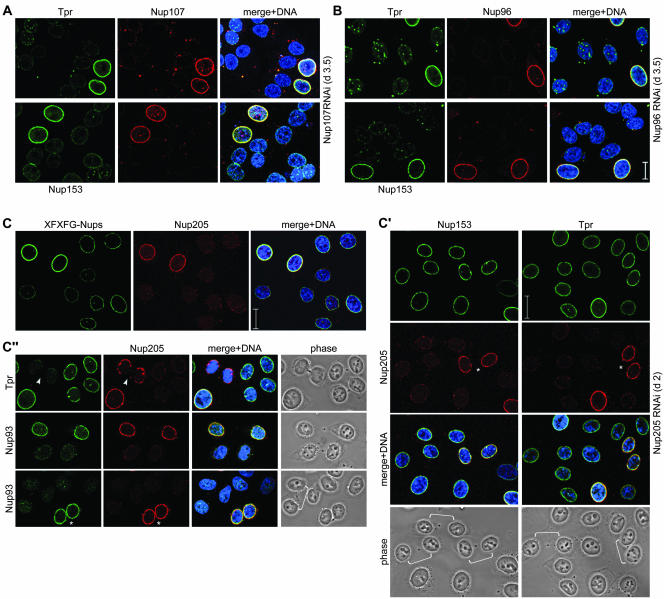
Distinct Effects on NPC Integrity and Docking of Tpr after In Vivo Depletion of Individual Nup160 and Nup93 Subcomplex Components
Recently, we have used siRNAs to posttranscriptionally silence the genes for Tpr and Nup153 in HeLa cells, revealing that Tpr is dispensable for NPC assembly and does not act as a scaffold for presumptive nucleoporins of the nuclear basket. Similarly, in vivo depletion of Nup153 had no immediate effect on NPC assembly and incorporation of various other Nups, but it caused mislocalization of both Tpr and the mobile nucleoporin Nup50, another binding partner of Nup153, to the nuclear interior (Hase and Cordes, 2003). Targeted disruption of the gene encoding Nup98 again did not prevent NPC-binding of either Nup153 or Tpr (Wu et al., 2001).
To also assess the roles of the Nup93 and 96 subcomplex components for NPC architecture and docking of Tpr and Nups to the NPC, our immuno-EM study was paralleled by RNAi experiments in which we targeted the transcripts for Nup93, 107, 205, and 96, the latter encoding the Nup98-Nup96 precursor protein of 196 kDa (Fontoura et al., 1999). The effects after in vivo depletion of Nup96 or 107 (Figure 9, A and B; Supplemental Figures S4 and S5) clearly differed from those in Tpr- or Nup153-depleted cells and pointed at essential roles in NPC assembly or maintenance, and in anchorage of Nup153 and Tpr to the NPC. Steady loss of Nup96 or 107 over a period of several days went along with seemingly simultaneous decrease in NE labeling for other Nups and Tpr. At 3–4 d after treatment with siRNAs, when transfected cells were largely depleted of the target protein, NE labeling for Tpr and Nup153 was largely diminished as well. Instead, both were occasionally found mislocalized to intranuclear and cytoplasmic aggregates.
Figure 9.
Temporally distinct loss of different NPC components after RNAi of Nup96, Nup107, and Nup205. HeLa cells were studied by confocal IFM at day 3.5 posttransfection with Nup107 (A) and Nup 96 (B) siRNAs, and at day 2 posttransfection with Nup205 siRNAs (C–C″), by using α-Nup96 aa 880–900, α-Nup107 aa 33–51, α-Nup93 aa 2–218, α-Nup205 aa 1784–1803, α-Tpr 2063–2084 (A, C′–C″), α-Nup 153 aa 22–36 (B), Nup153 mAb PF190 × 7A8 (A and C′), Tpr mAb 203–37 (B), and mAb 414 against Nup62 and other FxFG-repeat nucleoporins. Cells were permeabilized with Triton X-100 after (A and B) or before fixation (C–C″). (A and B) Cells transfected with siRNAs for Nup96 or 107 show no or only traces of the target protein at day 3.5 posttransfection; at this point, staining for Tpr and Nup153 is strongly reduced as well. Bright NE staining for these proteins is visible only in few cells that have remained untransfected. In contrast to Tpr- and Nup153-deficient nuclei that still permit different types of nucleocytoplasmic transport as well as nuclear growth during interphase (Hase and Cordes, 2003; our unpublished data), the majority of Nup96- and 107-deficient nuclei are characterized by small size indicative of nuclear growth arrest. (C) Cells transfected with siRNAs for Nup205 are already largely devoid of the target protein at day 2 posttransfection and already then often exhibit small-sized nuclei. At this point, NE staining intensity with mAb 414 in most transfected cells is also strongly diminished, whereas (C′) staining for Tpr and Nup153 is often reduced only moderately. Notably, NEs of cells in early G1 phase (some marked by brackets) can be largely depleted of Nup205 and nevertheless be clearly positive for Nup153 and Tpr, indicative that their binding to the NPC does not require stoichiometric amounts of Nup205. Two nontransfected Nup205-positive pairs of cells in early G1 (asterisks) are shown as reference. (C″) Nup205 deficiency impairs Nup93 incorporation into newly assembled NEs in early telophase (Hase and Cordes, 2003), resulting in strongly reduced Nup93 staining in postmitotic NEs (bracket). A nontransfected pair of cells in early G1 positive for both Nup205 and Nup93 (asterisk), and a Nup205-positive pair of telophase cells (arrowhead) in which late reincoporation of Tpr into the NE (Hase and Cordes, 2003) has not yet occurred are shown as reference. Bars, 20 μm; same magnification in A, B, and in C′, C″.
Cells transfected with Nup205 siRNAs were more rapidly depleted of Nup205 protein. Already 48 h after the initial transfection, many cells were essentially devoid of Nup205. At this point, NE labeling with mAb 414, an antibody against Nup62 and other FxFG-repeat Nups, was often strongly reduced as well (Figure 9C). In contrast, NE staining for Tpr and several other Nups, including Nup153 and 50, was generally much less affected (Figure 9C′). In fact, different from Nup96- and 107-depleted cells, reduction of NE staining for these Nups and Tpr trailed Nup205 reduction with obvious delay. Even at day 3.5 posttransfection, with no or only traces of Nup205 detectable, NE staining for these proteins was often still only moderately reduced (Supplemental Figure S6).
Furthermore, Tpr and Nup153 were found incorporated into the newly assembled NEs of postmitotic cells early in G1 despite these being largely devoid of Nup205. In contrast, Nup205 deficiency in such postmitotic NEs went along with a strong reduction of Nup93 staining (Figure 9C″). Although RNAi-mediated knockdown of the rather abundant Nup93 protein itself (Cronshaw et al., 2002) was less complete than for Nup205, we noted a similarly rapid reduction of mAb 414 signal from such Nup93-reduced NPCs, but again no immediate defection of Nup153 or Tpr (Supplemental Figure S6).
DISCUSSION
In this study, we have investigated the localization of several Nups and Tpr in HeLa cells by postembedding immuno-EM, complemented by cell permeabilization and RNAi experiments evaluated by confocal IFM. Here, we incorporate our data into a model of the NPC and discuss the possible contributions of these proteins to NPC and basket architecture.
The Nup160 and Nup93 Subcomplexes as Architectural Elements of NPC Core Structure
The Nup160 subcomplex consists of six proteins, Nup85, 96, 107, 133, 160, and Sec13 (Belgareh et al., 2001; Vasu et al., 2001; Harel et al., 2003), whereas the Nup93 subcomplex comprises Nup93, 188, and 205 (Miller et al., 2000). To map their positions relative to the NPC, we used peptide-specific antibodies against Nup93, 96, 107, and 205.
Our localizations of Nup96 and 107 are in line with other immuno-EM studies of cultured cells in which Nup107 and 133 were found on both the cytoplasmic and nuclear side of NPCs (Radu et al., 1994; Belgareh et al., 2001). IFM of semipermeabilized cells supports these EM data, showing Nup96 and 107 labeling at the cytoplasmic NPC side when the nuclear interior is inaccessible (Belgareh et al., 2001; this study). Further support for an NPC core position of the Nup160 subcomplex comes from our RNAi experiments, demonstrating that Nup96 and 107 are essential for NPC assembly and docking of Nup153 and Tpr. Recent studies on the roles of Nup85, 107, and 133 in NPC assembly have reported similar findings (Boehmer et al., 2003; Harel et al., 2003; Walther et al., 2003). In contrast, Nup153 and Tpr are dispensable for NPC assembly and neither acts as an NPC anchor for Nup96 or 107 (Hase and Cordes, 2003).
The distribution of the Nup96 and 107 immunogold label assigns at least parts of the Nup160 subcomplex to both of the coaxial ring structures. However, we do not exclude that the spoke complex sandwiched between these rings may harbor other domain segments of Nup96 and 107, or other members of this subcomplex.
The immuno-EM localization of Nup205 is the first reported and rates this Nup as another NPC core component. These data differ from those for Nup192p, the yeast homolog of Nup205, which has been located exclusively within the nuclear interior ∼60 nm from the NPC midplane (Kosova et al., 1999). They are, however, in line with another immuno-EM study in which Nup192p was shown symmetrically positioned on both sides of the yeast NPC (Rout et al., 2000). Moreover, the localization of Nup205 close to the NPC midplane is in harmony with the occurrence of its binding partner Nup93 at a similar position. Because Nup93 in intact NPCs is not accessible for any of the Nup93 antibodies used in this study, we believe it to be embedded deep within the NPC core, perhaps as part of the central spoke-ring complex sandwiched between the coaxial rings. This view finds support by our RNAi experiments as well as by NPC reconstitution studies (Grandi et al., 1997), demonstrating that the Nup93 subcomplex is essential for NPC stability and correct assembly of some of its parts. However, because siRNA-mediated loss of Nup93 or 205 did not prevent reincorporation of Nup153 and Tpr into NPCs in early G1, we conclude that the Nup93 subcomplex is not directly required for their mooring to the NPC. Nup153 and Tpr themselves are dispensable for NPC binding of Nup93 and 205 (Hase and Cordes, 2003).
We are aware of the variance between our Nup93 immuno-EM data and a study in which Nup93 was located exclusively at the nuclear NPC side, with peak distributions at 35–55 nm from the NE midplane (Grandi et al., 1997). However, using similar antibodies against a recombinant hNup93 polypeptide comprising aa 2–203, as well as another Nup93 peptide antibody against aa 371–389 for immuno-EM (unpublished data), we obtained staining patterns on both sides of the NPC close to the midplane that were very similar to the one obtained with the representative Nup93 peptide antibody shown in this study. None of our immunogold labelings allowed us to conclude that components of the Nup93 subcomplex may be structural components of the basket terminal ring. However, they are compatible with immuno-EM data for Nic96p, the yeast homolog of Nup93 symmetrically positioned on both sides of the NPC (Rout et al., 2000).
In future immuno-EM studies a higher resolution will be required to clarify whether the Nup93 subcomplex is located in a single plane at or near the NPC midplane or symmetrically on both sides of this axis. This might be achieved by using primary antibodies or Fab fragments coupled directly to colloidal gold, which will reduce signal spread and certain distortion effects that can arise as a consequence of secondary antibody rotation (Supplemental Figure S7).
Using an antibody against the central part of Nup98, immunogold label was mainly found within the NPC core region, on both sides of the NPC midplane. Similar gold grain distributions were found with other Nup98 peptide antibodies as well (Supplemental Figure S1), in keeping with the localization of Nup98 on both sides of the NPC by preembedding immuno-EM (Griffis et al., 2003). Future studies will need to seek an explanation that can harmonize these Nup98 immuno-EM results with reports showing that Nup98 is a protein that shuttles between nucleus and cytoplasm (Zolotukhin and Felber, 1999; Griffis et al., 2002, 2004). Immunogold label for such a mobile Nup may have been anticipated being more spread over NPC and basket, rather than mainly found in the NPC core region.
However, these and other observations unanimously speak against a direct role for Nup98 in basket formation. In HeLa cells depleted of Nup153 and Tpr, NPC binding of Nup98 is not impaired (Hase and Cordes, 2003), and in Nup98-deficient cells not only Nup93 and 96 but also Nup153 and Tpr remain bound to the NPC (Wu et al., 2001). In contrast, targeted disruption of the Nup98 gene leads to impaired NPC-binding of Nup88, 214, and 358 (Wu et al., 2001), Nups that are located only at the cytoplasmic NPC side. Although Nup98 was shown to be a binding partner of Nup88 (Griffis et al., 2003), it still remains to be understood why Nup98 deficiency selectively impairs binding of Nups to the cytoplasmic side. Clearly, however, none of these data support models in which Nup98 is located exclusively at the nuclear side of the NPC as an architectural element of the nuclear basket fibers.
Nup153 as the Proposed Link between NPC Core Structures and Nuclear Basket
By using two antibodies against the N-terminal domain (aa 22–36, 391–404), Nup153 was located at the nuclear NPC side, in an area occupied by the nuclear coaxial ring. Using the same LR White-embedded cells and mAb PF190 × 7A8 that targets an epitope between aa 439–611 of Nup153 (Hase and Cordes, 2003), we observed labeling of the same area (unpublished data).
These localizations are in harmony with a study in which Nup153 was located at the nuclear NPC side on ultrathin cryosections of BRL cells, by using antibodies against both N- and C-terminal Nup153 segments. Although the authors proposed Nup153 to be a component of basket fibers or terminal ring, the micrographs presented show gold grains mainly at the nuclear margin of the NPC proper (Sukegawa and Blobel, 1993). Furthermore, in a scanning EM study with antibodies against aa 1–149 of Xenopus Nup153 (Walther et al., 2001) and a transmission electron microscopy (TEM) study with antibodies against aa 436–655 of the same protein (Fahrenkrog et al., 2002), gold grains were directly attached to the nuclear coaxial ring on manually isolated Xenopus oocyte NEs. Interestingly, yeast Nup60p, which has been proposed to be the yeast version of Nup153 (Hase and Cordes, 2003), was located exclusively at the nuclear side of the NPC as well (Rout et al., 2000).
These localizations of Nup153 are compatible with RNAi experiments in which cellular depletion of Nup153 was shown to have no direct effect on the integration of Nup160 and Nup93 subcomplexes into the NPC but prevented NPC binding of Tpr (Hase and Cordes, 2003). Together, RNAi and immunolocalization data support a model in which Nup153 is proposed to act as the suspender of the nuclear basket at the NPC (Figure 10).
Figure 10.
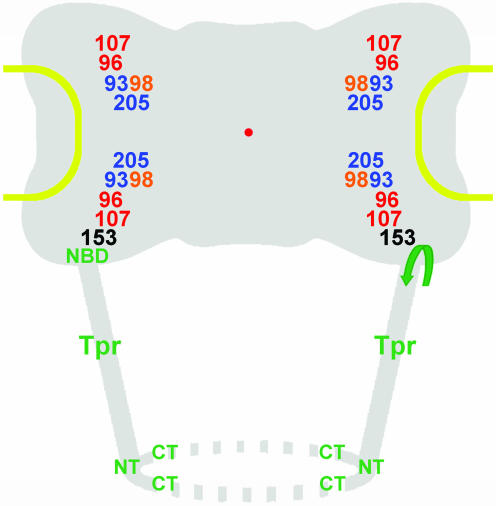
Model for nucleoporins as NPC core components and Tpr as the architectural element of the nuclear basket. Approximate mean positions of immunogold-labeled protein domains of Nups and Tpr, deduced from Figures 4 and 6, are superimposed onto the scheme of an idealistic NPC. The relative position of the Nup98 target domain, close to the Nup93 subcomplex, is shown slightly offset compared with its mean coordinates, due to spatial limitations. All Nups but 153 are regarded as bilateral symmetrically arranged on both sides of the NE midplane, without excluding that some Nups might locate in more than two planes, or that others like Nup205 might locate only in one plane closer to or equivalent to the midplane. The present degree of resolution also does not allow clarifying whether Nups located in the cytoplasmic plane are pointing into the same or opposite directions as their counterparts on the nuclear side. Furthermore, the actual distances from the eightfold rotational symmetry axis might be slightly shorter for some Nups and Tpr domains, and larger for others (Supplemental Figure S7). Relative positions of the flexible FG repeat domains of Nup98 and 153 were not a subject of this study and might locate in areas distant from the Nup98 and 153 domains targeted in this work. In this model, the basket fibers are composed of coiled-coiled Tpr homodimers or oligomers that fold back on themselves, thereby forming tetrameric or oligomeric intramolecular assemblies, with the first and second half of the rod domains arranged in an antiparallel manner. The region where Tpr bends backwards (arrow) is located close to its NBD that tethers the protein to the nuclear coaxial ring, mediated by interaction with the N-terminal domain of Nup153. Distal segments of the Tpr rod domains and the C-terminal tail domains are proposed to represent the main constituents of the terminal ring. Relative positions of Tpr N terminus (NT) and C terminus (CT) are indicated.
This notion clearly differs from our former proposal of Nup153 being a fibrous basket protein in somatic cells. That conclusion was built on a preembedding immuno-EM study in which thick mouse liver cryostat sections were exposed to mAb PF190 × 7A8, resulting in Nup153 labeling exclusively at the nuclear NPC side at a mean distance of 49 nm from the NE midplane (Cordes et al., 1993). We now interpret that gold grain distribution as affected by steric constraints that have dictated the angle by which primary and gold-coupled secondary antibodies bound to their targets. However, we cannot explain the variance between our present Nup153 immuno-EM data and a recent study in which mammalian Nup153 was located far from the NPC, at a distance of 75–84 nm from the NE midplane (Frosst et al., 2002).
Our present data do not rule out the possibility that other parts of the Nup153 protein, not targeted by the antibodies used here, may be located in different areas of the NPC. Using antibodies against the C-terminal FxFG-repeat domain for preembedding immuno-EM on manually isolated Xenopus oocyte nuclei, this Nup153 segment was located both at the terminal basket ring and the NPC proper (Fahrenkrog et al., 2002). Such FxFG-repeat sequences have little regular secondary structure and are highly flexible (Bayliss et al., 2000; Denning et al., 2003). Therefore, they are unlikely candidates for stable architectural elements but might well be capable of transiently interacting with different parts of the NPC and basket (Fahrenkrog et al., 2002). Future postembedding immuno-EM studies by using peptide-antibodies against these and other parts of Nup153 will aim at immunolocalizing these domains in somatic cells as well.
Tpr Proposed as the Central Architectural Element of the Nuclear Basket
Using peptide-antibodies against different Tpr segments, we located this protein in an area at the nuclear side of the NPC occupied by the nuclear basket. This is in line with other studies in which Tpr was found at the nuclear NPC side in human cells (Cordes et al., 1997; Frosst et al., 2002). Added to our knowledge of Tpr's ultrastructural properties and dimensions (Hase et al., 2001), the finding that Tpr binding to the NPC is mediated by Nup153 and that Tpr does not act as a scaffold onto which other Nups need to assemble (Hase and Cordes, 2003), the present immuno-EM data tempt us to outline a model in which Tpr acts as the main architectural element of the nuclear basket.
The mean positions determined for different parts of Tpr's C-terminal tail domain correlate well with the position of the basket's distal end ∼60–80 nm from the NPC midplane (Jarnik and Aebi, 1991; Goldberg and Allen, 1992; Ris, 1997). In contrast, the Tpr-binding domain of Nup153, and an epitope flanking the NBD of Tpr itself, are both located in an area ∼23–35 nm from the midplane. The distance of ∼40–50 nm between this NPC anchor site of Tpr and its C terminus correlates well with a basket fiber length of 40–60 nm (Goldberg and Allen, 1992; Ris, 1997). This distance can easily be bridged by the rod-shaped Tpr coiled-coil that can reach >100 nm when fully extended in solution in the absence of Nup153 (Hase et al., 2001; our unpublished data).
Remarkably, the mean position of Tpr's N terminus was not found at the nuclear coaxial ring but close to the basket's distal end. This tempts us to suggest that the basket fibers are formed by Tpr coiled-coils in which one part of the dimer folds back onto another part of itself, forming a shorter but thicker fiber in which both N and C termini are oriented toward the nuclear interior. Folding of the initially soluble and rectilinear Tpr dimer, with a Stokes radius of ∼160 Å and a sedimentation coefficient of ∼7.5 S (Hase et al., 2001; our unpublished data), would occur at the NBD upon binding to Nup153, and intradimer interactions would take place between the first two thirds of Tpr's rod domain. The last one-third of the rod, which tends to bifurcate (see below), would instead branch off toward the distal rod ends of adjoining Tpr polypeptides that constitute the neighboring basket fibers. Together with the C-terminal tail domains, these distal rod segments would then constitute the terminal ring structure (see below).
Apart from the immunolocalization data, also some of Tpr's structural properties speak for this model. First, the coiled-coil segment of ∼51 nm formed by the Tpr sequences upstream (aa 1–398) of the NBD (∼aa 400–600) is strictly rectilinear as are the rod segments of ∼350–400 amino acids directly downstream. In contrast to these segments that form stable homodimers, rod segments closer to the tail domain engage only in weak homodimeric interactions and often seem kinked, bent, or bifurcated. Second, the first two thirds of the rod domain tend to interact with each other, but neither of them does so with the last third (Hase and Cordes, 2001; our unpublished data).
At present, we cannot exclude the possibility that the small-sized Nups recently identified (Cronshaw et al., 2002) might include a linker protein that helps connecting the neighboring basket fibers. Future postembedding immuno-EM of these Nups will show whether any of them are basket-associated proteins. In the meantime, we favor a model in which the branched-off distal rod segments of Tpr interdigitate and can loosely catenate neighboring basket fibers without need for further linker proteins. Predicted basket flexibility and the different conformations a basket in action may adopt (Kiseleva et al., 1998) could then reflect dynamic and alternating interactions between these distal Tpr rod segments. In this model, the Tpr tail domains that do not homodimerize and are mainly composed of β-sheets, β-turns, and random coils (Hase et al., 2001) would remain flexible and have free C termini.
A basket in which the neighboring fibers are interconnected also would allow to harmonize the role of Nup153 in tethering Tpr to the NPC with studies in which the major of at least two nuclear pools of Nup153 was found to represent a highly mobile form that dynamically interacts with the NPC (Daigle et al., 2001; Griffis et al., 2004; our unpublished data). In addition to other explanations proposed (Hase and Cordes, 2003), we can imagine a scenario in which not all basket fibers need to be anchored to the NPC at the same time, once the basket has been assembled. Basket stabilization by interconnections between neighboring fibers would in fact allow high on-off rates for individual Nup153 molecules without jeopardizing NPC attachment of the basket as a whole. Rather than being cemented onto the NPC, a more dynamic nature of a Nup153-mediated conjunction between basket and NPC might even turn out to be important for basket function.
Several not mutually exclusive scenarios might explain the spread distribution of immunogold label for Tpr that cannot be explained by large antibody rotation radii alone. In one scenario, some of the Tpr polypeptides in the highly proliferating HeLa cells might simply not have been properly incorporated into the basket. For example, assuming that basket fibers are built up of Tpr dimers that need to fold back onto themselves, incorrect folding might cause exposure of free binding sites to which other Tpr molecules could bind erroneously. Accurate localizations of Tpr also may be affected if specimen fixation and embedding procedures compromise NPC and basket integrity to some extent.
In another scenario, the entire basket represents a highly flexible structure (Kiseleva et al., 1998). The gold grain distributions then reflect this flexibility and the areas within which Tpr domain segments move. Spread distribution of Tpr gold grains might therefore be directly correlated to baskets being in full action and the high NPC traffic rates in the proliferating cells used in this study. Future immuno-EM studies will need to follow up this notion, compare Tpr distributions in proliferating cells and their growth-arrested state, and investigate whether distribution patterns can be correlated to differences in cellular activity. Other comparative studies of cells of different morphogenetic origin, including both germ and somatic cells, should in turn help to clarify whether there might exist differences in certain aspects of functional NPC architecture and in the distribution of Tpr and other Nups that are cell-type specific.
In yet another scenario, a basket filament would be composed of several Tpr dimers arranged in a staggered, out-of-register manner. In that case, their N and C termini would be spread over a wider distance than those of a single Tpr homodimer in which both strands are arranged in parallel and in-register (Hase et al., 2001). However, the number of Tpr molecules that form a basket fiber still needs to be accurately determined. In Xenopus oocytes, basket fibers studied by TEM are 3–6 nm in diameter (Cordes et al., 1993; Ris, 1997). In scanning electron microscopy (SEM), the fibers generally seem slightly thicker (8–10 nm) due to metal coatings (Goldberg and Allen, 1992; Ris, 1997). Guided by fiber models for other coiled-coil proteins (Herrmann and Aebi, 1999), we estimate that a 6-nm-thick basket fiber could accommodate up to 16 rectilinear Tpr homodimers or eight dimers folded back onto themselves. A 3-nm fiber could instead hold only a tetramer or a single folded dimer. Experiments to distinguish between these possibilities are in progress.
Final Conclusions and Future Perspectives
When steric constraints such as encountered at the NPC prevent antibody binding or dictate a preferential angle of binding, accurate localizations of some target proteins can be difficult to achieve by certain preembedding immuno-TEM or -SEM techniques. To map such proteins and compare their relative positions, postembedding immuno-TEM, or studies using ultrathin cryosections, may be more suitable. However, we also are aware of the advantages of immuno-SEM and related techniques, especially when performed on isolated NEs from Xenopus oocytes (Walther et al., 2001, 2002; Fahrenkrog et al., 2002). These approaches allow visualization of immunolabeled NPC and basket substructures in three dimensions and can provide insight into certain aspects of NPC architecture not amenable by standard preembedding or postembedding TEM of ultrathin sections. These three-dimensional techniques may in fact be most appropriate for testing the basket model proposed in this study. Therefore, once a wider assortment of antibodies for Xenopus Nups and Tpr is available, such immuno-EM studies of oocyte NEs, and postembedding immuno-TEM on these and other cell types should complement each other. As a result, a further broadening of our understanding of NPC architecture can be expected.
Supplementary Material
Acknowledgments
We thank Dirk Görlich and Georg Krohne for stimulating discussions and suggestions on the manuscript. This investigation was supported by grants from the Swedish Research Council, the Swedish Heart Lung Foundation, the King Gustaf V 80th Birthday Fund, and the Marianne & Marcus Wallenberg Foundation.
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E04–03–0165. Article and publication date are available at www.molbiolcell.org/cgi/doi/10.1091/mbc.E04–03–0165.
Abbreviations used: aa, amino acid(s); FA, formaldehyde; GA, glutaraldehyde; IFM, immunofluorescence microscopy; mAb, monoclonal antibody; NE, nuclear envelope; NBD, nuclear pore complex binding domain; NPC, nuclear pore complex; Nup, nucleoporin; RNAi, RNA interference; siRNA, small interfering RNA.
Online version of this article contains supporting material. Online version is available at www.molbiolcell.org.
References
- Allen, T.D., Cronshaw, J.M., Bagley, S., Kiseleva, E., and Goldberg, M.W. (2000). The nuclear pore complex: mediator of translocation between nucleus and cytoplasm. J. Cell Sci. 113, 1651–1659. [DOI] [PubMed] [Google Scholar]
- Arlucea, J., Andrade, R., Alonso, R., and Arechaga, J. (1998). The nuclear basket of the nuclear pore complex is part of a higher-order filamentous network that is related to chromatin. J. Struct. Biol. 124, 51–58. [DOI] [PubMed] [Google Scholar]
- Bayliss, R., Littlewood, T., and Stewart, M. (2000). Structural basis for the interaction between FxFG nucleoporin repeats and importin-β in nuclear trafficking. Cell 102, 99–108. [DOI] [PubMed] [Google Scholar]
- Belgareh, N., et al. (2001). An evolutionarily conserved NPC subcomplex, which redistributes in part to kinetochores in mammalian cells. J. Cell Biol. 154, 1147–1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendayan, M., Nanci, A., and Kan, F.W.K. (1987). Effect of tissue processing on colloidal gold cytochemistry. J. Histochem. Cytochem. 35, 983–996. [DOI] [PubMed] [Google Scholar]
- Boehmer, T., Enninga, J., Dales, S., Blobel, G., and Zhong, H. (2003). Depletion of a single nucleoporin, Nup107, prevents the assembly of a subset of nucleoporins into the nuclear pore complex. Proc. Natl. Acad. Sci. USA 100, 981–985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrd, D.A., Sweet, D.J., Pante, N., Konstantinov, K.N., Guan, T., Saphire, A.C.S., Mitchell, P.J., Cooper, C.S., Aebi, U., and Gerace, L. (1994). Tpr, a large coiled coil protein whose amino terminus is involved in activation of oncogenic kinases, is localized to the cytoplasmic surface of the nuclear pore complex. J. Cell Biol. 127, 1515–1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordes, V.C., Reidenbach, S., and Franke, W.W. (1996). Cytoplasmic annulate lamellae in cultured cells: composition, distribution, and mitotic behavior. Cell Tissue Res. 284, 177–191. [DOI] [PubMed] [Google Scholar]
- Cordes, V.C., Reidenbach, S., Köhler, A., Stuurman, N., van Driel, R., and Franke, W.W. (1993). Intranuclear filaments containing a nuclear pore complex protein. J. Cell Biol. 123, 1333–1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordes, V.C., Reidenbach, S., Rackwitz, H.-R., and Franke, W.W. (1997). Identification of protein p270/Tpr as a constitutive component of the nuclear pore complex-attached intranuclear filaments. J. Cell Biol. 136, 515–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordes, V.C., Waizenegger, I., and Krohne, G. (1991). Nuclear pore complex glycoprotein p62 of Xenopus laevis and mouse: cDNA cloning and identification of its glycosylated region. Eur. J. Cell Biol. 55, 31–47. [PubMed] [Google Scholar]
- Cronshaw, J.M., Krutchinsky, A.N., Zhang, W., Chait, B.T., and Matunis, M.J. (2002). Proteomic analysis of the mammalian nuclear pore complex. J. Cell Biol. 158, 915–927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daigle, N., Beaudouin, J., Hartnell, L., Imreh, G., Hallberg, E., Lippincott-Schwartz, J., and Ellenberg, J. (2001). Nuclear pore complexes form immobile networks and have a very low turnover in live mammalian cells. J. Cell Biol. 154, 71–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denning, D.P., Patel, S.S., Uversky, V., Fink, A.L., and Rexach, M. (2003). Disorder in the nuclear pore complex: the FG repeat region of nucleoporins are natively unfolded. Proc. Natl. Acad. Sci. USA 100, 2450–2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbashir, S.M., Harborth, J., Lendeckel, W., Yalcin, A., Weber, K., and Tuschl, T. (2001). Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature 411, 494–498. [DOI] [PubMed] [Google Scholar]
- Enninga, J., Levay, A., and Fontoura, B. (2003). Sec13 shuttles between the nucleus and the cytoplasm and stably interacts with Nup96 at the nuclear pore complex. Mol. Biol. Cell 23, 7271–7284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahrenkrog, B., and Aebi, U. (2002). The vertebrate nuclear pore complex: from structure to function. Results Probl. Cell Differ. 35, 25–48. [DOI] [PubMed] [Google Scholar]
- Fahrenkrog, B., Maco, B., Fager, A.M., Koser, J., Sauder, U., Ullman, K.S., and Aebi, U. (2002). Domain-specific antibodies reveal multiple-site topology of Nup153 within the nuclear pore complex. J. Struct. Biol. 140, 254–267. [DOI] [PubMed] [Google Scholar]
- Ferrando-May, E., Cordes, V.C., Biller-Ckovric, I., Mirkovic, J., Görlich, D., and Nicotera, P. (2001). Caspases mediate nucleoporin cleavage, but not early redistribution of nuclear transport factors and modulation of nuclear permeability in apoptosis. Cell Death Differ. 8, 495–505. [DOI] [PubMed] [Google Scholar]
- Fontoura, B.M.A., Blobel, G., and Matunis, M.J. (1999). A conserved biogenesis pathway for nucleoporins: proteolytic processing of a 186-kilodalton precursor generates Nup98 and the novel nucleoporin, Nup96. J. Cell Biol. 144, 1097–1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontoura, B.M.A., Dales, S., Blobel, G., and Zhong, H. (2001). The nucleoporin Nup98 associates with the intranuclear filamentous protein network of TPR. Proc. Natl. Acad. Sci. USA 98, 3208–3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frosst, P., Guan, T., Subauste, C., Hahn, K., and Gerace, L. (2002). Tpr is localized within the nuclear basket of the pore complex and has a role in nuclear protein export. J. Cell Biol. 156, 617–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg, M.W., and Allen, T.D. (1992). High resolution scanning electron microscopy of the nuclear envelope: demonstration of a new, regular, fibrous lattice attached to the baskets of the nucleoplasmic face of the nuclear pores. J. Cell Biol. 119, 1429–1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandi, P., Dang, T., Pante, N., Shevchenko, A., Mann, M., Forbes, D., and Hurt, E. (1997). Nup93, a vertebrate homologue of yeast Nic96p, forms a complex with a novel 205-kDa protein and is required for correct nuclear pore assembly. Mol. Biol. Cell 8, 2017–2038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffis, E.R., Altan, N., Lippincott-Schwartz, J., and Powers, M.A. (2002). Nup98 is a mobile nucleoporin with transcription-dependent dynamics. Mol. Biol. Cell 13, 1282–1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffis, E.R., Xu, S., and Powers, M.A. (2003). Nup98 localizes to both nuclear and cytoplasmic sides of the nuclear pore and binds to two distinct nucleoporin subcomplexes. Mol. Biol. Cell 14, 600–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffis, E.R., Craige, B., Dimaano, C., Ullman, K.S., and Powers, M.A. (2004). Distinct functional domains within nucleoporins Nup153 and Nup98 mediate transcription dependent mobility. Mol. Biol. Cell 15, 1991–2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grote, M., Kubitscheck, U., Reichelt, R., and Peters, R. (1995). Mapping of nucleoporins to the center of the nuclear pore complex by post-embedding immunogold electron microscopy. J. Cell Sci. 108, 2963–2972. [DOI] [PubMed] [Google Scholar]
- Guan, T., Kehlenbach, R.H., Schirmer, E.C., Kehlenbach, A., Fan, F., Clurman, B.E., Arnheim, N., and Gerace, L. (2000). Nup50, a nucleoplasmically oriented nucleoporin with a role in nuclear protein export. Mol. Cell. Biol. 20, 5619–5630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harel, A., Orjalo, A.V., Vincent, T., Lachish-Zalait, A., Vasu, S., Shah, S., Zimmerman, E., Elbaum, M., and Forbes, D.J. (2003). Removal of a single pore subcomplex results in vertebrate nuclei devoid of nuclear pores. Mol. Cell 11, 853–864. [DOI] [PubMed] [Google Scholar]
- Hase, M.E., Kuznetsov, N., and Cordes, V.C. (2001). Amino acid substitutions of coiled-coil protein Tpr abrogate anchorage to the nuclear pore complex but not parallel, in-register homodimerization. Mol. Biol. Cell 12, 2433–2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hase, M.E., and Cordes, V.C. (2003). Direct interaction with Nup153 mediates binding of Tpr to the periphery of the nuclear pore complex. Mol. Biol. Cell 14, 1923–1940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrmann, H., and Aebi, U. (1999). Intermediate filament assembly: temperature sensitivity and polymorphism. Cell Mol. Life Sci. 55, 1416–1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarnik, M., and Aebi, U. (1991). Towards a more complete 3 D-structure of the nuclear pore complex. J. Struct. Biol. 107, 291–308. [DOI] [PubMed] [Google Scholar]
- Kellenberger, E., and Hayat, M.A. (1991). Some basic concepts for the choice of the methods. In: Colloidal Gold: Principles, Methods, and Applications, Vol. 3, ed. M.A. Hayat, London: Academic Press, 1–30. [Google Scholar]
- Kiseleva, E., Goldberg, M.W., Allen, T.D., and Akey, C.W. (1998). Active nuclear pore complexes in Chironomus: visualization of transporter configurations related to mRNP export. J. Cell Sci. 111, 223–236. [DOI] [PubMed] [Google Scholar]
- Kosova, B., Pante, N., Rollenhagen, C., and Hurt, E. (1999). Nup192p is a conserved nucleoporin with a preferential location at the inner side of the nuclear membrane. J. Biol. Chem. 274, 22646–22651. [DOI] [PubMed] [Google Scholar]
- Kuznetsov, N.V., Sandblad, L., Hase, M.E., Hunziker, A., Hergt, M., and Cordes, V.C. (2002). The evolutionarily conserved single-copy gene for murine Tpr encodes one prevalent isoform in somatic cells and lacks paralogs in higher eukaryotes. Chromosoma 111, 236–255. [DOI] [PubMed] [Google Scholar]
- Mayer, G., and Bendayan, M. (2001). Amplification methods for the immunolocalization of rare molecules in cells and tissues. Prog. Histochem. Cytochem. 36, 3–85. [DOI] [PubMed] [Google Scholar]
- Miller, B.R., Powers, M., Park, M., Fischer, W., and Forbes, D.J. (2000). Identification of a new vertebrate nucleoporin, Nup188, with the use of a novel organelle trap assay. Mol. Biol. Cell 11, 3381–3396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman, G.R., Jasani, B., and Williams, E.D. (1983). A simple post-embedding system for the rapid demonstration of tissue antigens under the electron microscope. Histochem. J. 15, 543–555. [DOI] [PubMed] [Google Scholar]
- Pante, N., Bastos, R., McMorrow, I., Burke, B., and Aebi, U. (1994). Interactions and three-dimensional localization of a group of nuclear pore complex proteins. J. Cell Biol. 126, 603–617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pante, N., Thomas, F., Aebi, U., Burke, B., and Bastos, R. (2000). Recombinant Nup153 incorporates in vivo into Xenopus oocyte nuclear pore complexes. J. Struct. Biol. 129, 306–312. [DOI] [PubMed] [Google Scholar]
- Powers, M.A., Macaulay, C., Masiarz, F.R., and Forbes, D.J. (1995). Reconstituted nuclei depleted of a vertebrate GLFG nuclear pore protein, p97, import but are defective in nuclear growth and replication. J. Cell Biol. 128, 721–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radu, A., Blobel, G., and Wozniak, R.W. (1994). Nup107 is a novel nuclear pore complex protein that contains a leucine zipper. J. Biol. Chem. 269, 17600–17605. [PubMed] [Google Scholar]
- Radu, A., Moore, M.S., and Blobel, G. (1995). The peptide repeat domain of nucleoporin Nup98 functions as a docking site in transport across the nuclear pore complex. Cell 81, 215–222. [DOI] [PubMed] [Google Scholar]
- Ris, H. (1991). The three-dimensional structure of the nuclear pore complex as seen by high voltage electron microscopy and high resolution low voltage scanning electron microscopy. EMSA Bull. 21, 54–56. [Google Scholar]
- Ris, H. (1997). High-resolution field-emission scanning electron microscopy of nuclear pore complex. Scanning 19, 368–375. [DOI] [PubMed] [Google Scholar]
- Rout, M.P., Aitchison, J.D., Suprapto, A., Hjertaas, K., Zhao, Y., and Chait, B.T. (2000). The yeast nuclear pore complex: composition, architecture, and transport mechanism. J. Cell Biol. 148, 635–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheer, U., Dabauvalle, M.C., Merkert, H., and Benevente, R. (1988). The nuclear envelope and the organization of the pore complexes. Cell Biol. Int. Rep. 12, 669–689. [DOI] [PubMed] [Google Scholar]
- Stierhof, Y.-D. and Schwarz, H. (1991). Yield of immunolabel compared to resin sections and thawed cryosections. In: Colloidal Gold. Principles, Methods, and Applications, Vol. 3, ed. M.A. Hayat, London: Academic Press, 87–115. [Google Scholar]
- Sukegawa, J., and Blobel, G. (1993). A nuclear pore complex protein that contains zinc finger binding motifs, binds DNA, and faces the nucleoplasm. Cell 72, 29–38. [DOI] [PubMed] [Google Scholar]
- Suntharalingam, M., and Wente, S.R. (2003). Peering through the pore: nuclear pore complex structure, assembly, and function. Dev. Cell 4, 775–789. [DOI] [PubMed] [Google Scholar]
- Vasu, S., Shah, S., Orjalo, A., Park, M., Fischer, W.H., and Forbes, D.J. (2001). Novel vertebrate nucleoporins Nup133 and Nup160 play a role in mRNA export. J. Cell Biol. 155, 339–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasu, S.K., and Forbes, D.J. (2001). Nuclear pores and nuclear assembly. Curr. Opin. Cell Biol. 13, 363–375. [DOI] [PubMed] [Google Scholar]
- Walther, T.C., Fornerod, M., Pickersgill, H., Goldberg, M., Allen, T.D., and Mattaj, I.W. (2001). The nucleoporin Nup153 is required for nuclear pore basket formation, nuclear pore complex anchoring and import of a subset of nuclear proteins. EMBO J. 20, 5703–5714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walther, T.C., Pickersgill, H.S., Cordes, V.C., Goldberg, M.W., Allen, T.D., Mattaj, I.W., and Fornerod, M. (2002). The cytoplasmic filaments of the nuclear pore complex are dispensable for selective nuclear protein import. J. Cell Biol., 158, 63–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walther, T.C., Alves, A., Pickersgill, H., Loiodice, I., Hetzer, M., Galy, V., Hülsmann, B.B., Köcher, T., Wilm, M., Allen, T., Mattaj, I.W., and Doye, V. (2003). The conserved Nup107–160 complex is critical for nuclear pore complex assembly. Cell 113, 195–206. [DOI] [PubMed] [Google Scholar]
- Wu, X., Kasper, L.H., Mantcheva, R.T., Mantchev, G.T., Springett, M.J., and van Deursen, J.M. (2001). Disruption of the FG nucleoporin NUP98 causes selective changes in nuclear pore complex stoichiometry and function. Proc. Natl. Acad. Sci. USA 98, 3191–3196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimowska, G., Aris, J.P., and Paddy, M.R. (1997). A Drosophila Tpr protein homolog is localized both in the extrachromosomal channel network and to nuclear pore complexes. J. Cell Sci. 110, 927–944. [DOI] [PubMed] [Google Scholar]
- Zolotukhin, A.S., and Felber, B.K. (1999). Nucleoporins Nup98 and Nup214 participate in nuclear export of human immunodeficiency virus type 1 Rev. J. Virol. 73, 120–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.