Abstract
Histone methylation is one of the most widely studied post-transcriptional modifications. It is thought to be an important epigenetic event that is closely associated with cell fate determination and differentiation. To explore the spatiotemporal expression of histone H3 lysine 4 trimethylation (H3K4me3) and histone H3 lysine 27 trimethylation (H3K27me3) epigenetic marks and methylation or demethylation transferases in tooth organ development, we measured the expression of SET7, EZH2, KDM5B and JMJD3 via immunohistochemistry and quantitative polymerase chain reaction (qPCR) analysis in the first molar of BALB/c mice embryos at E13.5, E15.5, E17.5, P0 and P3, respectively. We also measured the expression of H3K4me3 and H3K27me3 with immunofluorescence staining. During murine tooth germ development, methylation or demethylation transferases were expressed in a spatial–temporal manner. The bivalent modification characterized by H3K4me3 and H3K27me3 can be found during the tooth germ development, as shown by immunofluorescence. The expression of SET7, EZH2 as methylation transferases and KDM5B and JMJD3 as demethylation transferases indicated accordingly with the expression of H3K4me3 and H3K27me3 respectively to some extent. The bivalent histone may play a critical role in tooth organ development via the regulation of cell differentiation.
Keywords: histone modification, methylation, post-transcriptional modification, tooth development
Introduction
Posttranslational modifications of histone proteins are thought to be important epigenetic events that are intimately associated with transcription regulation in cell fate determination and differentiation.1,2 Histones are subject to various modifications, including methylation, acetylation, phosphorylation, ubiquitination and ribosylation.3 Among them, histone methylation is one of the most widely studied posttranscriptional modifications. Prominent histone modifications include H3K4 methylation, which has been implicated in transcriptional activation and deposited by Trithorax group proteins, and H3K27 methylation, which has been implicated in transcriptional repression and deposited by Polycomb group proteins.4
Immunofluorescence studies have revealed that global patterns of histone modifications and chromatin architecture change during the early stages of development.5,6,7,8,9 Genome-wide chromatin immunoprecipitation analyses have also suggested that specific combinations of histone marks at promoters and enhancers correlate with the developmental potential and fate of cells.10,11 In undifferentiated embryonic stem cells (ESCs), pluripotency maintenance genes (e.g., Nanog, Oct4, and Sox2) are marked with high levels of H3K4 methylation at their transcriptional start sites.12,13,14 However, many developmental regulatory gene loci are marked with both H3K4 and H3K27 methylation, the so-called ‘bivalent marks'.13,15,16 The combination of the seemingly ‘conflicting' marks suggests that these genes are kept silenced by H3K27 methylation in ESCs while remaining ‘poised' for expression events that are presumably dependent upon H3K4 methylation. This poised state was proposed to be central both for the maintenance of the ground state and for the developmental potential of ESCs. Sequential chromatin immunoprecipitation has shown that H3K4me3 and H3K27me3 can co-occupy some promoters in ESCs.13,16
Interestingly, these ‘bivalent' chromatin domains often mark lineage-regulatory genes. Bivalent domains have garnered wide attention because they might contribute to the precise unfolding of gene expression programs during pluripotency and differentiation. In particular, it has been proposed that bivalent domains might repress lineage control genes (H3K27me3) during pluripotency while keeping them poised for activation upon differentiation (H3K4me3). The H3K27me3-mediated repression of developmental control genes might protect cells from the aberrant expression of lineage regulators and thus help maintain pluripotency.17 During differentiation into specific cell types, a continued association with H3K27me3 might maintain the repression of the majority of developmental control genes, though only a specific subset of regulators is activated in a given lineage. Conversely, it has been proposed that H3K4me3 might poise developmental regulators for activation upon differentiation. In this scenario, H3K4me3 might make the induction of developmental genes more efficient or more synchronous.18 H3K4me3 might also protect genes from permanent silencing, such as by repelling transcriptional repressors or blocking DNA methylation.19 Thus, it is possible that bivalent domains convey temporal and spatial precision to the expression of lineage control genes during pluripotency and differentiation.
Tooth development, similar to the organogenesis of other ectodermal appendages, is regulated by sequential and reciprocal interactions between the epithelial and mesenchymal tissues. The spatial temporal signals between these compartments are essential. Growth factor such as WNTs, FGFs and TGF-beta, and SHH families are well known for their regulating function in exerting this signalling network in organogenesis. These signalling pathways most likely are overseen by mechanisms on multiple layers both genetically and epigenetically. Tooth enamel is formed by epithelial-derived cells called ameloblasts, and the pulp dentin complex is formed by the dental mesenchyme. These tissues differentiate with reciprocal signalling interactions to form a mature tooth. In this study, we have characterized histone modification transferase and histone modification in the mouse developing first molar and further investigated the role of bivalent histone modifications on enamel organ and pulp papilla differentiation.
Materials and methods
Ethics statement
All animal samples were collected under approved guidelines set by State Key Laboratory of Oral Diseases. All human tissues were collected from legally aborted foetuses at West China Women and Children's Hospital under approved guidelines set by Sichuan University. The study was approved by Ethical Committees of West China School of Stomatology, Sichuan University and State Key Laboratory of Oral Diseases.
Animals and sample collection
Adult BALB/c mouse were purchased from Dashuo Company (Chengdu, China). The mice were maintained in a temperature-controlled room (22 °C) under artificial illumination (lights on from 800 to 1 800 h) with access to food and water ad libitum. The embryos were obtained from time-mated pregnant mice. Embryos were collected from pregnant BALB/c mice at various time points (E13.5, E15.5, E17.5, P0 and P3), with the day of observation of a vaginal plug considered to be embryonic day (E) 0.5.
Histology
Mouse heads (E13.5, E15.5 and E17.5) or mandibles (P0 and P3) were dissected in phosphate-buffered saline (PBS). For P0 and P3 mice, mandibles were first decalcified in 10% ethylenediaminetetraacetic acid (EDTA)/PBS solution. Heads and decalcified mandibles were fixed in 4% paraformaldehyde solution at 4 °C overnight, dehydrated in crescent concentrations of alcohol (50%, 70%, 80%, 90%, 95% and 100%) and treated with xylene. They were embedded in paraffin and cut into 5-µm sections or embedded in JUNG tissue freezing medium (Leica, Solms, Germany) and cut into 10-mm transversal frozen sections. We used standard hematoxylin and eosin to examine tissue morphology.
Immunohistochemistry
Paraffin blocks containing processed mouse tissue were sectioned coronally (5 mm in thickness) for immunohistochemical analysis. The slides were heated in a 60 °C oven for 30 min and subsequently deparaffinized through a series of decreasing concentrations of ethanol. The immunohistochemical staining was performed by using the SP-9001 kit (Zhongshanjinqiao, Beijing, China). Sections were subjected to epitope recovering in citrate buffer at 99 °C for 5 min three times. Once room temperature was reached, slides were washed in triethanolamine buffered saline (TBS) and nonspecific immunoglobulin binding was blocked by 5% (V/V) bovine serum albumin (BSA) for 30 min at room temperature. Sections were incubated overnight at 4 °C with the following primary antibodies: SETD7 (1∶200; Abcam, Cambridge, UK), KDM5A (1∶200; Santa Cruz Biotechnology, Santa Cruz, CA, USA), KDM5B (1∶200; Abcam, Cambridge, UK), EZH2 (1∶500; Abcam, Cambridge, UK), JMJD3 (1∶200; Santa Cruz Biotechnology, Santa Cruz, CA, USA), and UTX (1∶200; Santa Cruz Biotechnology, Santa Cruz, CA, USA). Primary antibodies were detected with a DAB Staining Kit (Zhongshanjinqiao, Beijing, China). Normal IgG was used as a negative control. The immunohistochemistry experiments were repeated at least twice, with the primary antibody Ki67 serving as a positive control. All sections were placed on one slide and processed together under the same conditions.
Immunofluorescence
Paraffin blocks containing processed mouse tissue were sectioned coronally (5 mm in thickness) for immunohistochemical analysis. The slides were heated in a 60 °C oven for 30 min and subsequently deparaffinized through a series of decreasing concentrations of ethanol. Sections were subjected to epitope recovering in citrate buffer at 99 °C for 5 min three times. Once room temperature was reached, slides were blocked in TBS 5% BSA for 30 min. Sections were incubated overnight at 4 °C with the following primary antibodies H3K4me3 (1∶4 000; Cell Signaling Technology, Boston, MA, USA) and H3K27me3 (1∶1 600; Cell Signaling Technology, Boston, MA, USA). Secondary antibody, anti-rabbit IgG FITC conjugated (1∶400; Santa Cruz Biotechnology, Santa Cruz, CA, USA), was incubated for 1 h at 37 °C and nuclear counterstaining was performed using DAPI.
RNA isolation
The whole first molar tooth germ (E17.5, 19.5, P0 and P3) containing the enamel organ and dental papilla was extracted under the stereo microscope (Olympus sz61; Olympus, Tokyo, Japan). Tooth germs were immersed by RNAlater at 4 °C overnight then preserved at −80 °C. RNA was extracted from the captured tissues with an RNeasy micro Kit (Qiagen, Duesseldorf, Germany). RNA concentration was determined using a NanoDrop ND-1000 spectrophotometer (Thermo Fisher Scientific Inc., Wilmington, DE, USA). The reactions were incubated at 16 °C for 30 min, at 42 °C for 30 min, at 85 °C for 5 min, chilled on ice for 5 min, and the cDNA was then stored at −20 °C.
Quantitative real-time polymerase chain reaction
Polymerase chain reaction (PCR) was performed in an ABI 7900 system (Applied Biosystems, Foster City, CA, USA) using a TaKaRa RNA PCR kit (TaKaRa Shuzo, Tokyo, Japan) according to the manufacturer's protocol. The protocol included an initial incubation of the reaction mixture for 1 min at 95 °C. The amplification program consisted of 40 cycles with a 95 °C denaturation for 20 s and a 55–60 °C annealing and extension for 45 s. Oligonucleotide primer sequences and annealing temperatures are displayed in Table 1. The intensities of the specific bands were analysed and normalized against glyceraldehyde-3-phosphate dehydrogenase intensity as a control. This experiment was repeated three times. The test was employed to determine significant changes at the 99% confidence level (P<0.01).
Table 1. Primers sequences for qRT-PCR.
| Gene | Primer | Annealing temperature/°C | Fragment lengt/bp | Accession number |
|---|---|---|---|---|
| SETD1A | F: 5′-AAAGGAGACATTGCCCACACA-3′ | 60 | 178 | NM_178029.3 |
| R: 5′-GGAAACAGTTTTCCGGCGTT-3′ | ||||
| SETD7 | F: 5′-TCCAAGAGTTCACACCTGCG-3′ | 60 | 167 | NM_080793.5 |
| R: 5′-TTCCCCAAACTCGGCATTCA-3′ | ||||
| KDM5A | F: 5′-GAGGGCGCAGAACTTTGAC-3′ | 59 | 161 | NM_145997.2 |
| R: 5′-CTGCTAAATCAGTCCGCCAC-3′ | ||||
| KDM5B | F: 5′-GGAAGAGACTGCGCTGTAAGT-3′ | 60 | 71 | NM_152895.2 |
| R: 5′-GTGGGGGACTGAGTAGGGAG-3′ | ||||
| EZH2 | F: 5′-TTCCATGCAACACCCAACACAT-3′ | 61 | 156 | NM_001146689.1 |
| R: 5′-TGGGCGTTTAGGTGGTGTCT-3′ | ||||
| JMJD3 | F: 5′-CAATCCCCGCAGAGCTTACC-3′ | 60 | 101 | NM_009483.1 |
| R: 5′-TTCTACTGGAGGTGGTGCATT-3′ | ||||
| UTX | F: 5′-TCTGCTGTAGCCCATAGGAC-3′ | 60 | 96 | NM_001017426.1 |
| R: 5′-CAGCCAGATGGAGTTGGAAGT-3′ |
qRT-PCR, real time quantitative polymerase chain reaction. F, forward; R, reverse.
Results
Spatial–temporal pattern of histone methylation transferase during tooth germ development
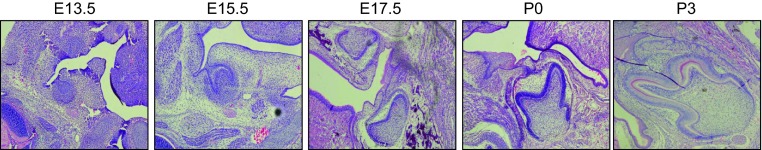
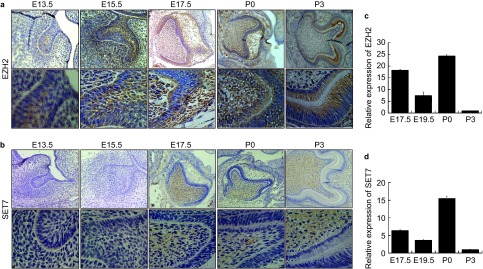
Different stages of tooth development were characterized by hematoxylin and eosin staining (Figure 1). We compared the expression pattern of EZH2 as H3K4 methylation transferase and SET7 as H3K27 methylation transferase in the developing tooth organs between E13.5 and P3 by whole-mount paraffin section immunohistochemistry. EZH2 was expressed in the enamel epithelium, stellate reticulum and dental papilla in E13.5 and E15.5-day-old mouse tooth germs, as bud stage and cap stage, respectively (Figure 2a). However, the tooth germ in the bud and cap stage was seemingly negative for expression of SET7 (Figure 2b). EZH2 was expressed in the odontoblast layer, dental papilla and enamel organ, consisting of inner enamel epithelium, outer enamel epithelium, stellate and stratum intermedium at E17.5 and P0-day-old mouse foetal cadaver tooth germs, as bell stage and early enamel formation stage, respectively (Figure 2a). Moreover, EZH2 was dramatically expressed in the odontoblast layer and inner enamel epithelium at P3-day-old mouse foetal cadaver tooth germs, as in the enamel formation stage (Figure 2a). Meanwhile, SET7 showed significant positive expression in dental papilla at the bell stage, early enamel formation stage and enamel formation stage (Figure 2b). The results showed that the histone methylation transferases express more dominantly in developing tooth germ than other adjacent tissues. qPCR examination verified the expression of both SET7 and EZH2 in tooth organ (Figure 2c and 2d). EZH2 and SET7 were expressed in the whole dental germ in bell stage, early enamel formation stage and enamel formation stage.
Figure 1.
H&E staining in different stages between cap stage and enamel formation stage of mouse tooth germ development. H&E, hematoxylin and eosin.
Figure 2.
The spatialtemporal expression of histone methylation transferase EZH2 and SET7 during tooth germ development. Protein signals of EZH2 and SET7 were detected by immunohistochemistry in mouse tooth germ from E13.5 to P3. (a) EZH2 was expressed in the enamel epithelium, stellate reticulum and dental papilla in E13.5 and E15.5 respectively and was expressed in both the dental papilla and enamel organ at E17.5 and P0. At P3, EZH2 was expressed in the odontoblast layer and inner enamel epithelium. (b) qPCR results showed that gene level of EZH2 was expressed in the whole dental germ in bell stage, early enamel formation stage and enamel formation stage. (c) SET7 showed significant positive expression in dental papilla E17.5, P0 and P3. (d) qPCR results showed that gene level of SET7 was expressed in the whole dental germ in bell stage, early enamel formation stage and enamel formation stage. qPCR, quantitative polymerase chain reaction.
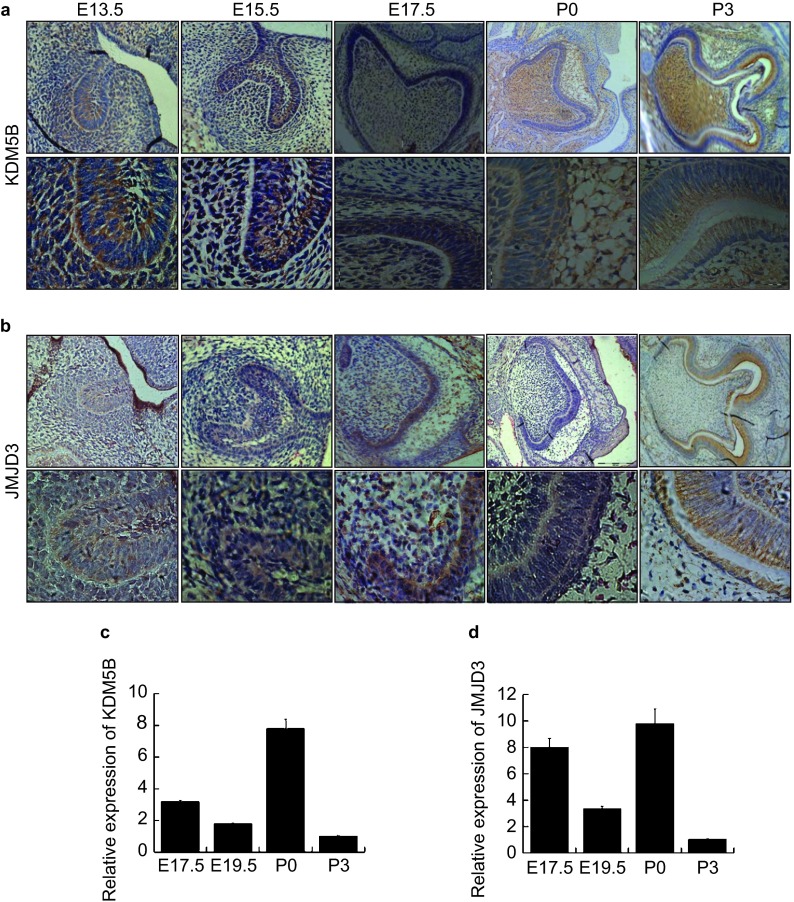
Figure 3.
The spatial–temporal expression of histone demethylation transferase KDM5B and JMJD3 during tooth germ development. Protein signals of KDM5B and JMJD3 were detected by immunohistochemistry in mouse tooth germ from E13.5 to P3. (a) KDM5B was expressed in tooth germ including the enamel organ and dental papilla between E13.5 to P3. (b) qPCR results showed that gene level of KDM5B was expressed in the whole dental germ in bell stage, early enamel formation stage and enamel formation stage. (c) JMJD3 was appeared in the enamel organ between E13.5 to P0 and in the odontoblast layer in P3. (d) qPCR results showed that gene level of JMJD3 was expressed in the whole dental. qPCR, quantitative polymerase chain reaction.
Spatial–temporal pattern of histone demethylation transferase during tooth germ development
We then compared the expression patterns of KDM5B as H3K4 demethylation transferase and JMJD3 (KDM6B) as H3K27 methylation transferase in the developing tooth organs between E13.5 and P3 by whole-mount paraffin section immunohistochemistry. KDM5B was expressed in tooth germ, including the enamel organ and dental papilla throughout the bud stage, cap stage, bell stage, early enamel formation stage and enamel formation stage (Figure 3a). The expression of JMJD3 that appeared in the enamel organ consisted of inner enamel epithelium, outer enamel epithelium, stellate and stratum intermedium, particularly in the inner enamel epithelium and the odontoblast layer in enamel formation stage (Figure 3b). The gene expression of KDM5B and JMJD3 was shown in the whole dental germ in the bell stage, early enamel formation stage and enamel formation stage by quantitative real-time PCR analysis (Figure 3c and 3d).
Histone methylation marks in developing tooth is bivalent
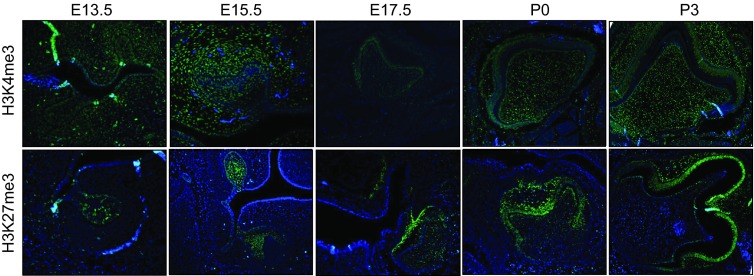
We compared histone modification status, included H3K4me3 and H3K27me3 as major histone methylation marks in the developing tooth organs between E13.5 and P3 by whole-mount paraffin section immunofluorescence (Figure 4). H3K4me3 was expressed in the enamel organ and dental papilla between the bud stage and enamel formation stage (Figure 4a). In bud stage and cap stage, H3K27me3 was expressed only in the enamel organ. H3K27me3 was expressed in the odontoblast layer and obvious in enamel organ consisted with inner enamel epithelium, outer enamel epithelium, stellate intermedium and stratum intermedium in bell stage, early enamel formation stage and enamel formation stage (Figure 4b).
Figure 4.
The spatial–temporal expression of H3K4me3 and H3K27me3 during tooth germ development. Protein signals of H3K4me3 and H3K27me3 were detected by immunofluorescence in human dental papilla cells. (a) H3K4me3 was expressed in the enamel organ and dental papilla between E13.5 to P3. (b) H3K27me3 was expressed in the enamel organ between E13.5 to P3 and in the odontoblast layer in P0 and P3.
Discussion
Epigenetic modification has been proposed to be a very complex field. Histone methylation transferase and histone demethylation transferase cooperate with one another to regulate histone modifications, suggesting that their functional interaction is actually complex. ESCs possess an open and highly dynamic chromatin landscape, which underlies their plasticity and ultimately maintains ESC pluripotency. Remarkably, there is increasing evidence that the remodelling of chromatin structure and the alteration of epigenetic marks, including histone methylation, can cause committed cells to convert from one fate to another, and such converted cells are functional when transplanted in vivo.20
Previous work documented the unexpected colocalisation of H3K4me3 and H3K27me3 in mouse ES cells in a highly conserved region consisting of approximately 2.5% of the genome, and it was suggested that this bivalent state was essential for genes in pluripotent cells to maintain their pluripotency.3,21 In the pluripotent state, many developmental loci are marked with both activating H3K4me3 and repressing H3K27me3 and are thus termed “bivalent”.3,15 This bivalent histone currently is being investigated intensively. Previous studies have shown that bivalent histone promoters are present in progenitor and adult stem cell populations, including neural progenitors, hematopoietic stem cells and mesenchymal stem cells and that these ultimately resolve to either active or inactive upon differentiation.22 In the present study, we uncovered the histone modification status in developing tooth germ. Our study provided the first spatiotemporal expression of active (H3K4me3) and repressed (H3K27me3) epigenetic marks during tooth development.
It is of interest to determine whether methylation transferase or histone demethylation transferase alters whether histone modifications occur in certain contexts to enforce changes. The dynamics of enhancer chromatin signatures mark the transition from pluripotency to cell specification during organ development. Gene expression is controlled by the concerted interactions between transcription factors and chromatin regulators. Many studies have shown that extensive epigenetic remodelling indeed occurs early in development and later during terminal cell differentiation. Teeth are organs that develop by inductive interactions between dental epithelium and subjacent mesenchyme. The generation of distinctive cell types that form different cells and organs requires the precise, temporal and spatial control of gene expression, especially in early tooth development. In particular, odontoblasts are post-mitotic polarized cells that differentiate in mouse molars at E18 bell stage, and they are involved in production of predentin–dentin matrix. In addition, ameloblasts differentiate at late bell stage (E19–20) and synthesize and secrete enamel matrix.
Although there is vast information available regarding the genes involved in tooth initiation and morphogenesis, odontoblast and ameloblast differentiation remain as complex and unclear processes.23 Epigenetic mechanisms contribute to the establishment and maintenance of cell type-specific gene expression patterns. We focused on the functions of histone lysine methylation in the context of epigenetic gene regulation during developmental transitions. In a previous study, it was shown that functions of histone lysine methylation marks in gene activation and repression are important for normal development. There has been no previous report of histone protein expression in either murine or human tooth germs. Establishment and maintenance of epigenetic profiles are essential steps of development during which stem cells, despite identical genetic information, will acquire different and selective gene expression patterns, specific for their fate.24 This highly complex programming process involves mechanisms that are not yet completely understood, although it has been established over the past few years that chromatin modifier enzymes play essential roles in the establishment of transcriptional programs accompanying cell differentiation in different stages of tooth development.
This study will focus on describing a potential model of choice to understand how epigenetic changes can drive specific cell differentiation to better understand tooth developmental processes. We sought to characterize the epigenetic changes that occur during tooth organ differentiation from tooth germ by performing protein mapping of two histone modifications, H3K4me3 and H3K27me3, at five key tooth developmental time-points. First, the spatio-temporal assay showed that bivalent histone consisting of H3K4me3 and H3K27me3 was expressed specificity in murine tooth germ from bud stage to enamel formation stage. It showed that the histone states of H3K4me3 and H3K27me3 express more strikingly in developing tooth germ than other tissues around it in maxillofacial development. It can be concluded that the bivalent histone takes part in the regulation of tooth development, although the mechanism is not completed understood. However, the results of the immunohistochemistry assays in this study showed that H3K4me3 is expressed in the enamel organ and dental papilla between the bud stage and enamel formation stage and that that H3K27me3 was expressed only in the enamel organ in the bud stage and cap stage, with little expressed in the odontoblast layer in bell stage, early enamel formation stage and enamel formation stage. This finding may explain the mechanism of how bivalent histone determines the fate of cells through H3K4me3 being expressed in both poorly differentiated cells (such as stem cell or cells in the early embryo) and in highly differentiated cells, whereas H3K27me3 is distinctively expressed in highly differentiated cells such as the inner enamel epithelium cell, outer enamel epithelium cell, stellate reticulum and odontoblast in enamel formation stage.
Over the past few years, knockout mice strains for several HMTases have been established and characterized. The functional implications of histone lysine methylation for cell-type identity and regulation of developmental transitions have been reported. The functional analysis of different HMTases has revealed that histone lysine methylation has important roles in facilitating normal development; however, many open questions remain. A more detailed functional analysis in different tissues is required to better understand their functional implications in developmental processes.
Furthermore, in light of the recent findings that histone methylase transferase SET7-trimethylated H3K4 is often associated with transcriptionally active genes, the results of the immunohistochemistry assays in this study found that SET7 was expressed in odontoblast layers and dental papilla tissues from the early bell stage of human tooth development. The mesenchymal stem cells in dental papilla have more potential ability to differentiation than do the cells in enamel organ. These results indicate that the expression of H3K4me3, which was expressed both in the enamel organ and dental papilla during H3K27me3, was absent in the dental papilla. Interestingly, EZH2 served as histone demethylase transferase that was primarily expressed in almost all tooth germ and was dramatically expressed in the odontoblast layer and enamel organ. Based on a mapping of its expression, it exhibits similarity to H3K27me3.
It is likely that KDM5B strengthens H3K4me3 demethylase's ability to serve as a transcriptional coactivator and its ability to promote differentiation, presumably due to redundancy with genes such as the JARID1 family members. Although H3K27 methylation is usually associated with stable Polycomb group-mediated transcriptional repression, the H3K27 methylation levels at specific genes have been shown to rapidly decrease during differentiation of mouse stem cells into neuronal lineages.25 Recently, given the opposing roles of H3K4 and H3K27 in gene regulation, it is tempting to speculate that UTX and JMJD3 proteins might constitute functional H3K27 demethylases. Mouse JMJD3 is upregulated during embryonic stem-cell differentiation when H3K27 marks seem to be dynamically regulated. Interestingly, based on our work, KDM5B and JMJD3 were similarly expressed in all stages, as observed in the enamel organ and dental papilla throughout mouse tooth development. Our studies also verified that these histone methylase transferase and histone demethylase transferase mRNA could be amplified from human tooth germ from as early as the bell stages. However, crosstalk between these factors and chromatin modifier enzymes occurs at several molecular levels. There is interplay and dependency between many different epigenetic mechanisms. The future challenge in epigenetic research will be to understand this complex network of regulatory mechanisms.
In summary, our study is the first to identify the spatio-temporal expression of bivalent histone modifications in tooth development. In addition, four histone methylation transferases or histone demethylation transferases were verified to be involved in histone modification in tooth germ, which lays a foundation and an initial framework for future analysis of the mechanism of epigenetics in tooth development.
Conclusions
These experiments provide the initial spatiotemporal expression of active (H3K4me3) and repressed (H3K27me3) epigenetic marks and their methylation or demethylation transferases in mouse tooth organ development. The bivalent histone may play a critical role in tooth development.
Acknowledgments
This study was supported by National Science Foundation of China (Grant No. 81371136) to Xue-Dong Zhou, National Science Foundation of China (Grant No. 81200760 and 81470711) to Li-Wei Zheng.
References
- Niwa H. How is pluripotency determined and maintained? Development 2007; 134(4): 635–646. [DOI] [PubMed] [Google Scholar]
- Jaenisch R, Young R. Stem cells, the molecular circuitry of pluripotency and nuclear reprogramming. Cell 2008; 132(4): 567–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannister AJ, Kouzarides T. Regulation of chromatin by histone modifications. Cell Res 2011; 21(3): 381–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouzarides T. Chromatin modifications and their function. Cell 2007; 128(4): 693–705. [DOI] [PubMed] [Google Scholar]
- Meshorer E, Yellajoshula D, George E et al. Hyperdynamic plasticity of chromatin proteins in pluripotent embryonic stem cells. Dev Cell 2006; 10(1): 105–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed K, Dehghani H, Rugg-Gunn P et al. Global chromatin architecture reflects pluripotency and lineage commitment in the early mouse embryo. PLoS One 2010; 5(5): e10531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuzyuk T, Fakhouri TH, Kiefer J et al. The polycomb complex protein mes-2/E(z) promotes the transition from developmental plasticity to differentiation in C. elegans embryos. Dev Cell 2009; 16(5): 699–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wongtawan T, Taylor JE, Lawson KA et al. Histone H4K20me3 and HP1α are late heterochromatin markers in development, but present in undifferentiated embryonic stem cells. J Cell Sci 2011; 124(Pt 11): 1878–1890. [DOI] [PubMed] [Google Scholar]
- Burton A, Torres-Padilla ME. Epigenetic reprogramming and development: a unique heterochromatin organization in the preimplantation mouse embryo. Brief Funct Genomics 2010; 9(5/6): 444–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rada-Iglesias A, Bajpai R, Swigut T et al. A unique chromatin signature uncovers early developmental enhancers in humans. Nature 2011; 470(7333): 279–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creyghton MP, Cheng AW, Welstead GG et al. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc Natl Acad Sci U S A 2010; 107(50): 21931–21936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen TS, Ku M, Jaffe DB et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature 2007; 448(7153): 553–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan G, Tian S, Nie J et al. Whole-genome analysis of histone H3 lysine 4 and lysine 27 methylation in human embryonic stem cells. Cell Stem Cell 2007; 1(3): 299–312. [DOI] [PubMed] [Google Scholar]
- Zhao XD, Han X, Chew JL et al. Whole-genome mapping of histone H3 Lys4 and 27 trimethylations reveals distinct genomic compartments in human embryonic stem cells. Cell Stem Cell 2007; 1(3): 286–298. [DOI] [PubMed] [Google Scholar]
- Azuara V, Perry P, Sauer S et al. Chromatin signatures of pluripotent cell lines. Nat Cell Biol 2006; 8(5): 532–538. [DOI] [PubMed] [Google Scholar]
- Bernstein BE, Mikkelsen TS, Xie X et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell 2006; 125(2): 315–326. [DOI] [PubMed] [Google Scholar]
- Vastenhouw NL, Schier AF. Bivalent histone modifications in early embryogenesis. Curr Opin Cell Biol 2012; 24(3): 374–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boettiger AN, Levine M. Synchronous and stochastic patterns of gene activation in the Drosophila embryo. Science 2009; 325(5939): 471–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fouse SD, Shen Y, Pellegrini M et al. Promoter CpG methylation contributes to ES cell gene regulation in parallel with Oct4/Nanog, PcG complex, and histone H3 K4/K27 trimethylation. Cell Stem Cell 2008; 2(2): 160–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkins RD, Hon GC, Yang C et al. Dynamic chromatin states in human ES cells reveal potential regulatory sequences and genes involved in pluripotency. Cell Res 2011; 21(10): 1393–1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vastenhouw NL, Schier AF. Bivalent histone modifications in early embryogenesis. Curr Opin Cell Biol 2012; 24(3): 374–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulka H, St John JC, Fulka J et al. Chromatin in early mammalian embryos: achieving the pluripotent state. Differentiation 2008; 76(1): 3–14. [DOI] [PubMed] [Google Scholar]
- Thesleff I, Sharpe P. Signalling networks regulating dental development. Mech Dev 1997; 67(2): 111–123. [DOI] [PubMed] [Google Scholar]
- Vincent A, van Seuningen I. Epigenetics, stem cells and epithelial cell fate. Differentiation 2009; 78(2/3): 99–107. [DOI] [PubMed] [Google Scholar]
- Wang J, Lunyak VV, Jordan IK. Chromatin signature discovery via histone modification profile alignments. Nucleic Acids Res 2012; 40(21): 10642–10656. [DOI] [PMC free article] [PubMed] [Google Scholar]