Abstract
Mesenchymal stem cells (MSCs) are a promising tool in regenerative medicine due to their capacity to differentiate into multiple lineages. In addition to MSCs isolated from bone marrow (BMSCs), adult MSCs are isolated from craniofacial tissues including dental pulp tissues (DPs) using various stem cell surface markers. However, there has been a lack of consensus on a set of surface makers that are reproducibly effective at isolating putative multipotent dental mesenchymal stem cells (DMSCs). In this study, we used different combinations of surface markers (CD51/CD140α, CD271, and STRO-1/CD146) to isolate homogeneous populations of DMSCs from heterogeneous dental pulp cells (DPCs) obtained from DP and compared their capacity to undergo multilineage differentiation. Fluorescence-activated cell sorting revealed that 27.3% of DPCs were CD51+/CD140α+, 10.6% were CD271+, and 0.3% were STRO-1+/CD146+. Under odontogenic conditions, all three subsets of isolated DMSCs exhibited differentiation capacity into odontogenic lineages. Among these isolated subsets of DMSCs, CD271+ DMSCs demonstrated the greatest odontogenic potential. While all three combinations of surface markers in this study successfully isolated DMSCs from DPCs, the single CD271 marker presents the most effective stem cell surface marker for identification of DMSCs with high odontogenic potential. Isolated CD271+ DMSCs could potentially be utilized for future clinical applications in dentistry and regenerative medicine.
Keywords: dental mesenchymal stem cells, odontogenic differentiation, cell surface markers, dental pulp, fluorescence-activated cell sorting
Introduction
Embryonic stem cells (ESCs) have the ability to renew themselves and to differentiate into various specialized tissues, making them an attractive treatment modality in regenerative medicine.1 However, due to ethical concerns of isolating and using ESCs, great interest has focused on adult mesenchymal stem cells (MSCs), which are also characterized by their self-renewal and multipotent capacity.1,2,3 MSCs are believed to be responsible for growth, wound healing, and replacement of cells that are lost through daily wear and tear as well as pathological conditions, suggesting that they are important for tissue repair and maintenance.1
While MSCs derived from bone marrow (BMSCs) are the most widely recognized and studied, alternate sources for MSCs have been investigated due to the complications associated with harvesting BMSCs, such as pain, morbidity, and low cell number. Evidence has suggested that MSCs may be present in virtually any vascularized tissue in the body.4,5 In recent years, MSCs derived from craniofacial tissues including dental mesenchymal stem cells (DMSCs) have been identified as a putative alternative.6 Similar to BMSCs, these DMSCs are multipotent progenitor cells with: (1) the capability to self-renew and differentiate into a variety of cell types, (2) ease of isolation, and (3) lack of immunogenicity.7 When compared to BMSCs, DMSCs may be more advantageous for regenerating craniofacial defects because they are readily available without risk as DMSCs can be easily isolated from discarded tissues such as third molars.8 Approximately, 70% of dental patients have third molars that require extractions consequently providing an abundant source of DMSCs that can be cryopreserved for potential future use.9 These DMSCs can be obtained from dental pulp tissues (DPs), periodontal ligament, and apical papilla.10 Among these different types of DMSCs, DMSCs from DP are the most widely studied as similarities in biochemical pathways for odonto/osteogenic differentiation between BMSCs and DMSCs from DP have already been established.11
MSCs are a heterogeneous population of cells with varying magnitudes of differentiation potential among single clones of MSCs.12 It was documented that in vitro single cell cloning of human MSCs demonstrated that approximately 30% of the clonal cells were multipotent and thus “true” MSCs.12 Currently, numerous cell surface markers including STRO-1, CD29, CD44, CD73, CD90, CD105, CD106, and CD146 have been utilized to isolate homogenous and multipotent MSC populations.10,13 However, a consensus on surface markers to isolate DMSCs with high differentiation potential is lacking. As the future of successful craniofacial defect repair is dependent on the ability to isolate specific subsets of DMSCs with potent differentiation capacity into appropriate cell types, the identification of markers that isolate multipotent DMSCs effectively is critical. In this study, we assessed the effectiveness of different cell surface markers (CD51/CD140α, CD271, STRO-1/CD146) in isolating DMSCs from DP and examined odontogenic and chondrogenic potential of these isolated subsets of DMSCs.
Materials and methods
Cell isolation and culture
Primary dental pulp cells (DPCs) were isolated from DP of extracted adult third molars (IRB#13-000241-CR-00001) as previously described.14 DPCs were cultured in alpha modified Eagle's medium (α-MEM; Invitrogen, Carlsbad, CA, USA) containing 20% fetal bovine serum (FBS), non-essential amino acids, 100 units per mL penicillin, and 100 units per mL streptomycin, in a humidified 5% CO2 incubator at 37 °C (all reagents were from Invitrogen, Carlsbad, CA, USA). Media was changed every 2–3 days, and cells were passaged at 80%–90% confluency. DPCs at passages 4–8 were utilized in this study.
Fluorescence-activated cell sorting
Expression of stem cell surface markers in DPCs was determined by fluorescence-activated cell sorting (FACS) analysis. The cells were detached using trypsin in 0.25% elhylene diamine tetraacetic acid (EDTA). After neutralization, single-cell suspensions were washed with phosphate-buffered saline supplemented with 2% FBS and 0.01% NaN3 (FACS buffer). Quantities of 1 × 105 cells were incubated with the conjugated antibody for 20 min on ice in the dark. After washing, fluorescence intensity was measured on FACS Aria II cell sorter (BD Biosciences, San Jose, CA, USA). The following antibodies were used: Phycoerythrin (PE)-CD271 (Miltenyi Biotec, Auburn, CA, USA), Fluorescein Isothiocyanate (FITC)-CD90 (Biolegend, San Diego, CA, USA), allophycocyanin (APC)-CD106 (Biolegend, San Diego, CA, USA) or dual color combinations APC-STRO-1/PE-CD146 (Both from: Biolegend, San Diego, CA, USA), and PE-CD51 (Biolegend, San Diego, CA, USA)/APC-CD140α (BD Biosciences, San Jose, CA, USA). PE-IgG was used as a negative control.
Induction of odontogenic differentiation
Sorted DMSCs were plated at 1 × 105 cells per mL into 12-well plates. To induce differentiation into odontogenic lineages, sorted DMSCs were grown in odontogenic induction medium (OIM). OIM contains 90% minimum essential medium (α-MEM; Invitrogen, Carlsbad, CA, USA), 10% FBS (Invitrogen, Carlsbad, CA, USA), 50 μg·mL−1 ascorbic acid, 5 mmol·L−1 β-glycerolphosphate, and 100 nmol·L−1 dexamethasone (all from Sigma-Aldrich, St Louis, MO, USA). OIM was changed every 2–3 days. For alkaline phosphatase (ALP) staining, after odontogenic induction for 7 days, cells were fixed with 4% paraformaldehyde and incubated with a solution of 0.25% naphthol AS-BI phosphate and 0.75% Fast Blue BB (Sigma-Aldrich, St Louis, MO, USA) dissolved in 0.1 mol·L−1 tris(hydroxymethyl)aminomethane (Tris) buffer (pH 9.3). ALP activity assay was preformed using an ALP kit (Sigma-Aldrich, St Louis, MO, USA) according to the manufacturer's protocol and normalized based on protein concentrations. To detect mineralization potential, cells were induced for 2 weeks using OIM, fixed with 4% paraformaldehyde, and stained with 2% Alizarin red (Sigma-Aldrich, St Louis, MO, USA). For quantification, Alizarin red stain (ARS) was destained with 10% cetylprydiniumcholoride in 10 mmol·L−1 sodium phosphate for 30 min at room temperature. The optical absorbance was measured at 562 nm using a microplate reader with a standard calcium curve in the same solution. The final calcium level in each group was normalized with the total protein concentrations prepared from a duplicate plate.
Induction of chondrogenic differentiation
Sorted DMSCs were plated at 1 × 105 cells per mL into 12-well plates. To induce differentiation into chondrogenic lineages, sorted DMSCs were grown in chondrogenic induction medium containing α-MEM (Invitrogen, Carlsbad, CA, USA) supplemented with 10% FBS (Invitrogen, Carlsbad, CA, USA), 100 mmol·L−1 sodium pyruvate, 40 µg·mL−1 proline, 100 nmol·L−1 dexamethasone, 200 μmol·L−1 ascorbic acid (all from Sigma-Aldrich, St Louis, MO, USA), and 10 ng·mL−1 TGF-β (R&D systems, Minneapolis, MN, USA). Culture medium was changed every 2–3 days. After 4 weeks of differentiation in vitro, Alcian Blue Staining (Sigma-Aldrich, St Louis, MO, USA) was performed as previously described.15 For quantification, stained Alcian blue was eluted with 6 mol·L−1 guanidine HCl for 8 hours at room temperature. The optical absorbance was measured at 620 nm using a microplate reader.
Quantitative real-time PCR
Total RNA was isolated from DMSCs using Trizol reagents (Invitrogen, Carlsbad, CA, USA). Two-µg aliquots of RNAs were synthesized using random hexamers and reverse transcriptase according to the manufacturer's protocol (Invitrogen, Carlsbad, Hercules, CA, USA). Real-time polymerase chain reaction (RT-PCR) performed using the QuantiTect SYBR Green PCR kit (Qiagen, Valencia, CA, USA) and the Icycler IQ Multi-color Real-time PCR Detection System (Bio-Rad, Hercules, CA, USA). The primers for RUNX2 (NM_001015051.3) were: forward, 5′-TGGTTACTGTCATGGCGGGTA-3′; reverse, 5′-TCTCAGATCGTTGAACCTTGCTA-3′. The primers for DLX5 (NM_005221.5) were: forward, 5′-GCTCTCAACCCCTACCAGTAT-3′; reverse, 5′-CTTTGGTTTGCCATTCACCATTC-3′. The primers for BGLAP (NM_199173.4) were: forward, 5′-AGCAAAGGTGCAGCCTTTGT-3′; reverse 5′-GCGCCTGGTCTCTTCACT-3′. The primers for DMP1 (NM_001079911.2) were: forward, 5′-GACAGCAAGGGTGACTCTCA-3′; reverse: 5′-AGATGACAGGTTGGCCTCTT-3′. The primers for DSPP (NM_014208.3) were: forward, 5′-TCAACAGCAAGAGAAATGGG-3′; reverse: 5′-TCGTCTTCATCCTCATCTGC-3′. The primers for SOX9 (NM_000346.3) were: forward, 5′-CACACAGCTCACTCGACCTT-3′; reverse, 5′-CAAAGGGAATTCTGGTTGGT-3′. Primers were designed with GenScript Real-time PCR (TaqMan) Primer Design program. The cycling conditions were as follows: 3 min of initial denaturation at 95 °C, 50 cycles of denaturation at 95 °C for 8 s, annealing at 60 °C for 60 s, followed by a final melting curve program. The mean cycle threshold value (Ct) obtained from triplicate samples was used to calculate gene expression levels. PCR products were normalized to the level of glyceraldehyde 3-phosphate dehydrogenase (GAPDH) for each reaction. Relative gene expression levels are presented as the mean ± standard deviation from three independent experiments.
Results
Isolation of DMSCs with surface markers CD51/CD140α, CD271, and STRO-1/CD146 from DPCs
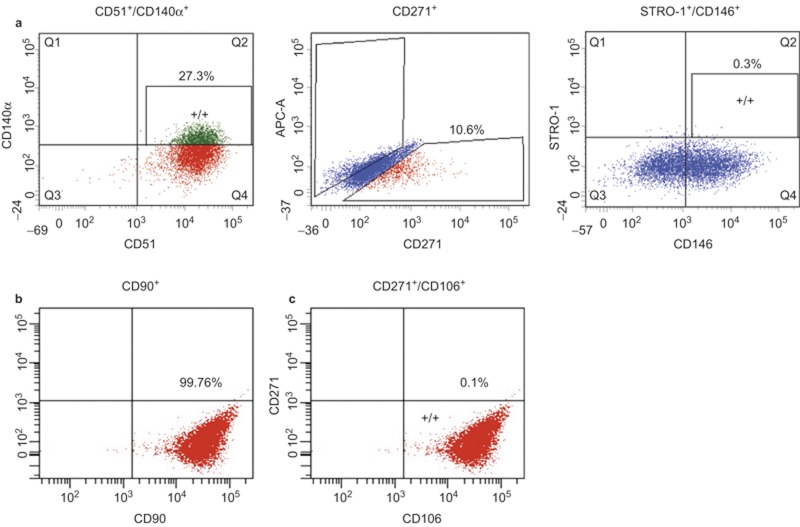
Many available MSC surface markers fail to represent MSCs in vivo as markers are not homogenously expressed across cultures.16 Recent studies showed that specific combinations of surface markers including CD51/CD140α and CD271/CD90/CD106 isolated highly enriched clonogenic cells from human bone marrow, respectively.17,18 In addition, STRO-1/CD146 combination has successfully characterized DMSCs from dental tissues such as periodontal ligament and apical papilla.19,20 However, whether these verified combination of surface markers are capable of isolating multipotent and self-renewing progenitor cells from human DP remains to be investigated. Using two surface combination of CD51/CD140α or STRO-1/CD146, FACS revealed that 27.3% of DPCs were CD51+/CD140α+ and 0.3% of DPCs were STRO-1+/CD146+ (Figure 1a). Interestingly, we found that the majority (99%) of DPCs expressed CD90 (Figure 1b), indicating that CD90 might not be useful. The combination of CD271/CD106 yielded extremely few positive cells (Figure 1c). Therefore, we decided to use single CD271 marker to isolate MSCs from DPCs and found that 10.6% of DPCs were CD271+.
Figure 1.
The expression profiles of stem cell surface markers in human primary cells from DPs determined by FACS. (a) CD51/CD140α, CD271, and STRO-1/CD146; (b) CD90; (c) CD271/CD106. Cells were isolated from dental pulp of adult third molars and stained with antibodies.
Isolated DMSCs exhibited differential odontogenic potential depending on surface markers
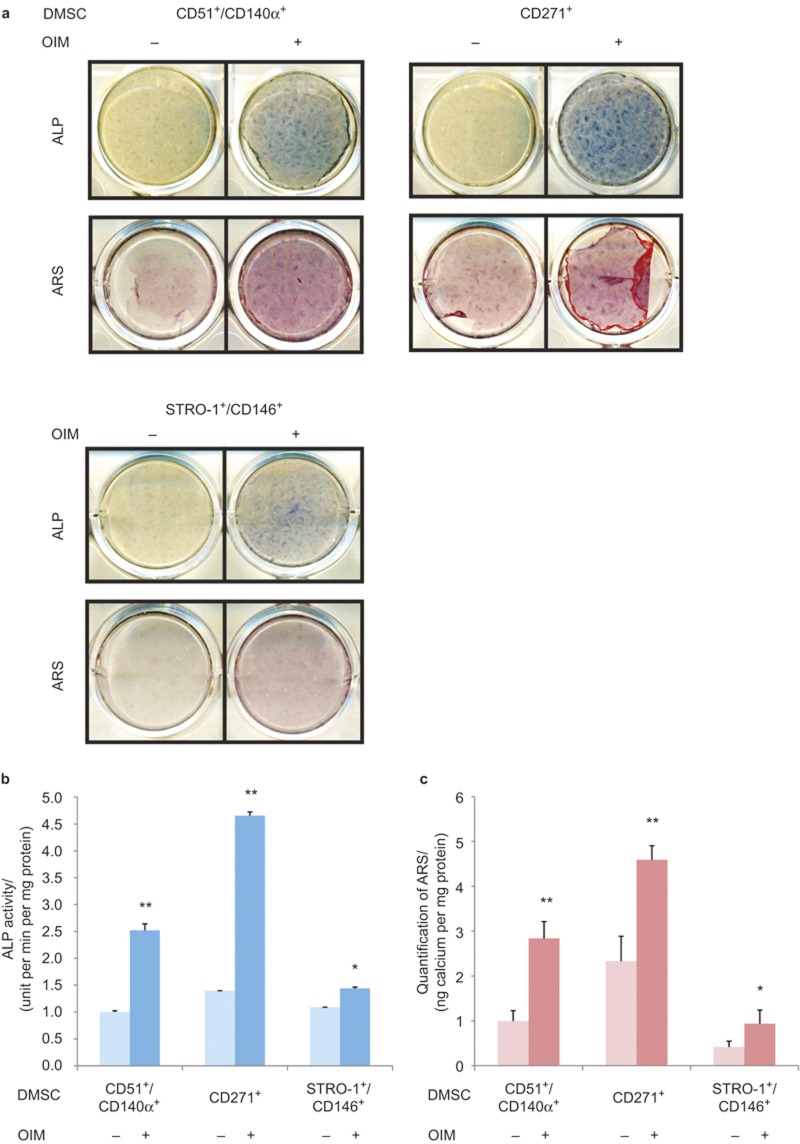
To evaluate and compare differentiation capacity, isolated CD51+/CD140α+, CD271+, and STRO-1+/CD146+, DMSCs were induced to undergo odontogenic differentiation. All three types of isolated DMSCs exhibited the capacity to differentiate into odontogenic lineage as demonstrated by ALP staining on the seventh day (Figure 2a). Quantification of ALP activity revealed the most significant increase of ∼5 folds in induced CD271+ DMSCs compared to non-induced CD51+/CD140α+. Induced CD51+/CD140α+ DMSCs exhibited 2.5-fold increase in ALP activity while induced STRO-1+/CD146+ DMSCs showed 1.25-fold increase (Figure 2b). All three isolated DMSC groups had formation of mineralized nodules after prolonged treatment with odontogenic induction media for 14 days as demonstrated by ARS (Figure 2a). The quantification of ARS also showed significant mineralization potential in CD271+ DMSCs (4.5-fold) followed by CD51+/CD140α+ DMSCs (2.75 fold) and STRO-1+/CD146+ DMSCs (1-fold; Figure 2c).
Figure 2.
Odontogenic differentiation of isolated CD51+/CD140α+, CD271+, and STRO-1/CD146+ DMSCs. (a) ALP staining and Alizarin red staining after these cells were induced to differentiate into odontogenic lineages for 7 days and 14 days, respectively. (b) Quantification of ALP activity. (c) Quantification of ARS. Values were normalized to non-induced CD51+/CD140α+ DMSCs. *P < 0.05, **P < 0.001. ALP, alkaline phosphatase; ARS, Alizarin red stain; DMSC, dental mesenchymal stem cell; OIM, odontogenic induction medium.
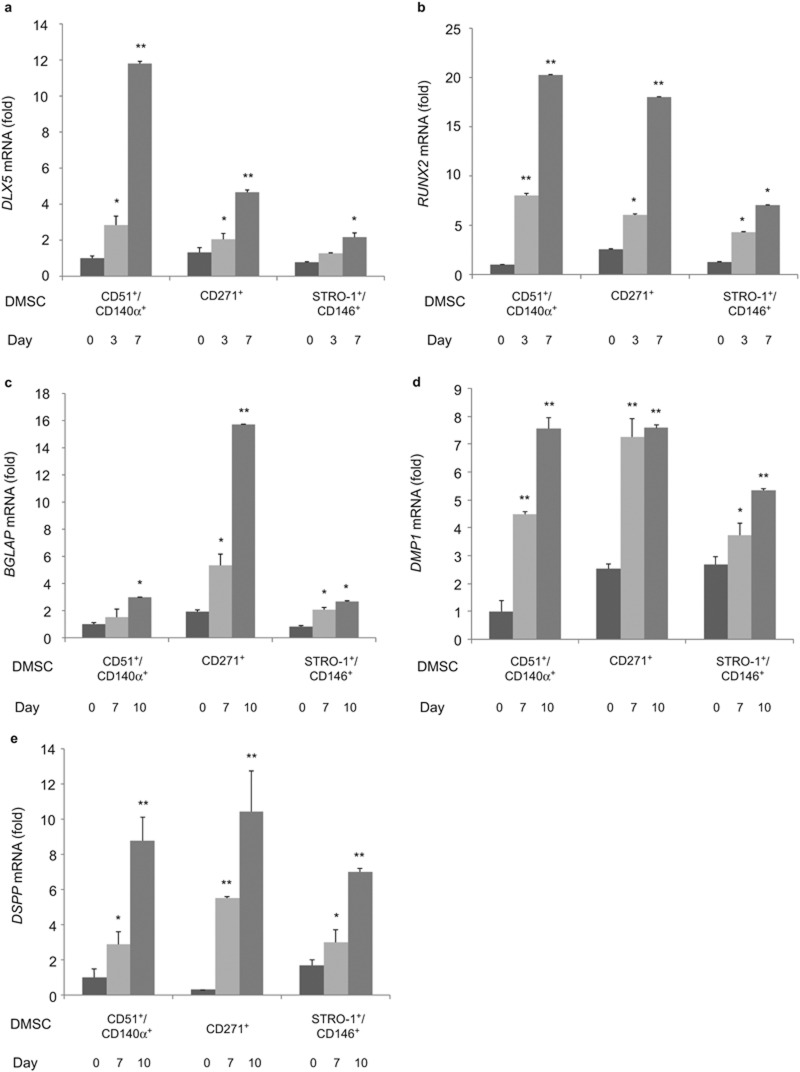
Using these isolated DMSCs, we further confirmed their odontogenic potential by examining mRNA expression of several odontogenic marker genes including DLX5, RUNX2, BGLAP, DMP1, and DSPP at different time points: 0, 3, 7, and 10 days after odontogenic induction. Consistent with ALP and ARS results, odontogenic marker gene expression was upregulated for all three subsets of DMSCs. In particular, marked increase of DLX5 expression was found in CD51+/CD140α+ DMSCs at day 7 of induction compared to CD271+ and STRO-1+/CD146+ DMSCs (Figure 3a). The expression of RUNX2 in both CD51+/CD140α+ and CD271+ DMSCs was more significantly upregulated in a time-dependent manner at day 3 and day 7 of induction (Figure 3b) compared to STRO-1+/CD146+ DMSCs. Significant BGLAP expression induction was found in CD271+ DMSCs at day 10 of odontogenic induction (Figure 3c). Lastly, in all three isolated DMSC populations, DMP1 and DSPP expression were significantly upregulated at day 10 of induction (Figure 3d and 3e).
Figure 3.
mRNA expression of odontogenic-genes after 0, 3, 7, and 10 days of odontogenic induction in CD51+/CD140α+, CD271+, and STRO-1+/CD146+ DMSCs. (a) DLX5, (b) RUNX2, (c) BGLAP, (d) DMP1, and (e) DSPP. Values were normalized to CD51+/CD140α+ DMSCs at day 0. *P < 0.05, **P < 0.001. DMSC, dental mesenchymal stem cell.
Collectively, these data suggested that homogenous populations of DMSCs isolated using FACS with different surface marker combinations are capable of differentiating into odontogenic lineages.
Isolated DMSCs exhibited differential chondrogenic potential depending on surface markers
We further evaluated and compared chondrogenic differentiation capacity of isolated DMSCs under chondrogenic conditions. All three isolated DMSC groups showed the presence of glycosaminogylcans as demonstrated by Alcian blue staining after prolonged treatment with chondrogenic induction media for 28 days (Figure 4a). The quantification of Alcian blue staining exhibited the greatest chondrogenic differentiation in CD271+ DMSCs, followed by STRO-1+/CD146+ DMSCs and CD51+/CD140α+ (Figure 4b). mRNA expression of Sox9, the master gene of chondrogenic differentiation, was significantly upregulated confirming their chondrogenic potential in all three populations of isolated DMSCs (Figure 4c)
Figure 4.
Chondrogenic differentiation of CD51+/CD140α+, CD271+, and STRO-1+/CD146+ DMSCs. (a) Alcian blue staining after cells were induced to differentiate into chondrogenic lineages for 28 days. (b) Quantification of Alcian blue. (c) mRNA expression of Sox9 after 0 and 28 days of chondrogenic induction. Values were normalized to CD51+/CD140α+ DMSCs at day 0. *P < 0.05, **P < 0.001. CMI, chondrogenic induction medium; DMSC, dental mesenchymal stem cell.
Discussion
While numerous stem cell surface markers have been routinely used to identify putative MSCs, specific markers for successful isolation of DMSCs that lead to potent differentiation into multiple lineages are lacking.10,13 In this study, we systematically used three different sets of surface markers (CD51/CD140α, CD271, and STRO-1/CD146) to isolate putative stem cell populations from primary DP cultures and compared their differentiation capacity into odontogenic and chondrogenic lineages. CD51/CD140α and CD271 have been previously used to isolate neural crest cell progenitors while STRO-1/CD146 combination has been used to isolate MSCs localized to blood vessels.17,19,21,22 In addition, the utilization of CD51/CD140α combination for DMSC isolation is not known. Our results demonstrated that although all three combinations of surface markers were able to isolate DMSCs from DP with varying magnitudes of differentiation capabilities, CD271 was the best single marker for isolating the DMSC progenitor population with greatest differentiation potential. Adipogenic potential was evaluated but found to be minimal for all three subsets of DMSCs (data not shown).
About 10.6% of primary culture DPCs were positive for CD271 (Figure 1). CD271, also known as the p75 neurotrophin receptor (p75NTR or low-affinity nerve growth factor), has been described as one of the most reliable surface markers for isolation of putative BMSCs and identified as a bona fide neural crest stem cell marker.23,24,25,26 On the contrary to our studies, CD271+ cells from CD44+/CD90+ MSCs of human deciduous DPs could not differentiate into osteoblasts and adipocytes.21 Currently, the reasons for these conflicting results are unknown. It could be due to the discrepancy in tissue sources for MSC isolation. However, CD271 was previously used in combination with THY-1/VCAM-1 in BMSCs.18 Therefore, we first examined THY-1 (CD90) in characterizing DPCs and our preliminary FACS data showed 90% of DPCs expressed THY-1 (CD90), suggesting its lack of specificity for isolating DMSCs. We further characterized DPCs with a combination of CD271 and VCAM-1 (CD106) and found that 0.1% of DPCs expressed both CD271 and VCAM-1 markers. For these reasons, the isolation of DPSCs in this study was performed with a single marker, CD271. Our study showed that CD271+ DMSCs exhibited the most significant odontogenic differentiation potential with high ALP activity, mineralization capacity, and upregulated expression of DLX5, RUNX2, BGLAP, DMP1, and DSPP (Figures 2 and 3). Similarly, during chondrogenic lineage differentiation, CD271+ DMSCs exhibited the greatest chondrogenic potential demonstrated by increased Alcian blue staining and SOX9 expression levels. Therefore, CD271 can be identified as a single specific surface marker for isolating putative DMSCs that are highly multipotent.
The use of CD51/CD140α to isolate DMSCs from DP has not been previously studied. CD51 and CD140α are highly expressed in human Nestin+ sphere forming MSCs, which are capable of hematopoetic progenitor cell expansion.17 Nestin is mainly expressed in early stages of the central nervous system and muscle development.27,28 CD51 labels the integrin V alpha subunit, which can form heterodimers with at least five distinct beta subunits.29,30 CD140α, platelet-derived growth factor receptor (PDGFR) α is a tyrosine kinase receptor and found to be expressed on mesenchymal-derived cells. 31 As such, the use of CD51/CD140α identified a large subset of perivascular Nestin+ cells highly enriched for MSCs in both human and mouse bone marrow in vivo.17 Our results demonstrated that CD51 and CD140α markers are also useful for isolating DMSCs that exhibit multilineage potential. (Figures 2, 3, 4) On average, 27.3% of DPCs were positive for both CD51 and CD140α, providing the largest proportion of DPCs among the three surface marker combinations in this study (Figure 1).
STRO-1 and CD146 are considered to be early MSC surface markers that have been identified in BMSCs and are derived from perivascular cells.32 STRO-1 was initially associated with the identification of osteogenic precursors isolated from bone marrow and later described as a promising marker for MSCs.33 However, it was suggested that the use of STRO-1 as a single marker for MSC isolation is limited; as it is not universally expressed in all types of MSCs and it can gradually decline with increasing culture passages.5,34,35 In this study, CD146 was used with STRO-1 to address these STRO-1-related concerns. The lowest proportion of DPCs, 0.3%, was STRO-1+/CD146+ (Figure 1), making it challenging to expand the STRO-1+/CD146+ DMSCs for further use. The proportion of STRO-1+/CD146+ cells was significantly smaller in these DPCs compared to periodontal ligament cells (2.6%) and apical papilla cells (13.4%).19,20 While ALP activity and mineralization capacity were increased in induced STRO-1+/CD146+ DMSCs compared to non-induced STRO-1+/CD146+ DMSCs, the enhancement was not as significant as CD271+ or CD51/CD140α DMSCs. Less significant odontogenic and chondrogenic potential was found in STRO-1+/CD146+ DMSCs compared to the other two combinations (Figures 2 and 3). Although STRO-1 is a widely utilized MSC marker, it is not the best surface marker for isolating DMSCs with high differentiation capabilities from DPCs even when combined with CD146.22,33,35
It is noteworthy that DP is originally derived from neural crest cells that have both ectodermal and mesenchymal components and can provide a pure MSC population.13,36,37,38 As some recent studies hypothesized that MSCs are neural crest derived, isolating DMSCs with surface markers of a neural crest origin such as CD51/CD140α and CD271 may be desirable.39,40 In line with this notion, our study indicated that CD271+ and CD51+/CD140α+ DMSCs exhibited more significant multipotent populations from DP compared to STRO-1+/CD146+ DMSCs. Therefore, isolation of DMSCs using surface markers of neural crest origin other than CD271 and CD51/CD140α may be important to further evaluate their potency in differentiation potential.
In conclusion, our findings demonstrated the successful isolation of homogenous populations of DMSCs from heterogenous DPCs obtained from DP with the use of specific neural surface markers and showed their ability to differentiate into odontogenic and chondrogenic lineages. Our results suggest that: (1) there are heterogeneous MSC populations in DPCs, and (2) lineage-specific differentiation potential is variable among distinct-isolated DMSC populations. Specifically, the CD51/CD140α marker isolated the largest population of DMSCs (27.3%) and showed relatively strong odontogenicand chondrogenic potential, while STRO-1+/CD146+ DMSCs were more limited in number (0.3%) and differentiation potential. The most effective marker was CD271, which isolated a relatively large population of DMSCs (10.6%) and had the strongest odontogenic and chondrogenic potential. Further studies are needed to validate whether these isolated cells may differentiate into functionally different lineages in vivo.
Acknowledgments
This study was supported by National Institute of Dental and Craniofacial Research grant T90DE022734.
References
- Wei X, Yang X, Han ZP et al. Mesenchymal stem cells: a new trend for cell therapy. Acta Pharmacol Sin 2013; 34(6): 747–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caplan AI. Adult mesenchymal stem cells for tissue engineering versus regenerative medicine. J Cell Physiol 2007; 213(2): 341–347. [DOI] [PubMed] [Google Scholar]
- Pittenger MF, Mackay AM, Beck SC et al. Multilineage potential of adult human mesenchymal stem cells. Science 1999; 284(5411): 143–147. [DOI] [PubMed] [Google Scholar]
- Caplan AI, Correa D. The MSC: an injury drugstore. Cell Stem Cell 2011; 9(1): 11–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv FJ, Tuan RS, Cheung KM et al. Concise review: the surface markers and identity of human mesenchymal stem cells. Stem Cells 2014; 32(6): 1408–1419. [DOI] [PubMed] [Google Scholar]
- Estrela C, Alencar AH, Kitten GT et al. Mesenchymal stem cells in the dental tissues: perspectives for tissue regeneration. Braz Dent J 2011; 22(2): 91–98. [DOI] [PubMed] [Google Scholar]
- Gronthos S, Brahim J, Li W et al. Stem cell properties of human dental pulp stem cells. J Dent Res 2002; 81(8): 531–535. [DOI] [PubMed] [Google Scholar]
- Jo YY, Lee HJ, Kook SY et al. Isolation and characterization of postnatal stem cells from human dental tissues. Tissue Eng 2007; 13(4): 767–773. [DOI] [PubMed] [Google Scholar]
- Zhang W, Walboomers XF, Shi S et al. Multilineage differentiation potential of stem cells derived from human dental pulp after cryopreservation. Tissue Eng 2006; 12(10): 2813–2823. [DOI] [PubMed] [Google Scholar]
- Huang GT, Gronthos S, Shi S. Mesenchymal stem cells derived from dental tissues vs. those from other sources: their biology and role in regenerative medicine. J Dent Res 2009; 88(9): 792–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi S, Robey PG, Gronthos S. Comparison of human dental pulp and bone marrow stromal stem cells by cDNA microarray analysis. Bone 2001; 29(6): 532–539. [DOI] [PubMed] [Google Scholar]
- Abdallah BM, Kassem M. Human mesenchymal stem cells: from basic biology to clinical applications. Gene Ther 2008; 15(2): 109–116. [DOI] [PubMed] [Google Scholar]
- Mao JJ, Robey PG, Prockop DJ. Stem cells in the face: tooth regeneration and beyond. Cell Stem Cell 2012; 11(3): 291–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan Z, Yamaza T, Lee JS et al. BCOR regulates mesenchymal stem cell function by epigenetic mechanisms. Nat Cell Biol 2009; 11(8): 1002–1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arpornmaeklong P, Brown SE, Wang Z et al. Phenotypic characterization, osteoblastic differentiation, and bone regeneration capacity of human embryonic stem cell-derived mesenchymal stem cells. Stem Cells Dev 2009; 18(7): 955–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianco P, Cao X, Frenette PS et al. The meaning, the sense and the significance: translating the science of mesenchymal stem cells into medicine. Nat Med 2013; 19(1): 35–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinho S, Lacombe J, Hanoun M et al. PDGFRα and CD51 mark human nestin+ sphere-forming mesenchymal stem cells capable of hematopoietic progenitor cell expansion. J Exp Med 2013; 210(7): 1351–1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mabuchi Y, Morikawa S, Harada S et al. LNGFR(+)THY-1(+)VCAM-1(hi+) cells reveal functionally distinct subpopulations in mesenchymal stem cells. Stem Cell Reports 2013; 1(2): 152–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Wang W, Kapila Y et al. Multiple differentiation capacity of STRO-1+/CD146+ PDL mesenchymal progenitor cells. Stem Cells Dev 2009; 18(3): 487–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakopoulou A, Leyhausen G, Volk J et al. Comparative characterization of STRO-1(neg)/CD146(pos) and STRO-1(pos)/CD146(pos) apical papilla stem cells enriched with flow cytometry. Arch Oral Biol 2013; 58(10): 1556–1568. [DOI] [PubMed] [Google Scholar]
- Mikami Y, Ishii Y, Watanabe N et al. CD271/p75(NTR) inhibits the differentiation of mesenchymal stem cells into osteogenic, adipogenic, chondrogenic, and myogenic lineages. Stem Cells Dev 2011; 20(5): 901–913. [DOI] [PubMed] [Google Scholar]
- Shi S, Gronthos S. Perivascular niche of postnatal mesenchymal stem cells in human bone marrow and dental pulp. J Bone Miner Res 2003; 18(4): 696–704. [DOI] [PubMed] [Google Scholar]
- Alvarez-Viejo M, Menendez-Menendez Y, Blanco-Gelaz MA et al. LNGFR (CD271) as a marker to identify mesenchymal stem cells from different human sources: umbilical cord blood, wharton's jelly and bone marrow. Bone Marrow Res 2013; 1(4): 1–6. [Google Scholar]
- Bühring HJ, Battula VL, Treml S et al. Novel markers for the prospective isolation of human MSC. Ann N Y Acad Sci 2007; 1106: 262–271. [DOI] [PubMed] [Google Scholar]
- Quirici N, Soligo D, Bossolasco P et al. Isolation of bone marrow mesenchymal stem cells by anti-nerve growth factor receptor antibodies. Exp Hematol 2002; 30(7): 783–791. [DOI] [PubMed] [Google Scholar]
- Tomellini E, Lagadec C, Polakowska R et al. Role of p75 neurotrophin receptor in stem cell biology: more than just a marker. Cell Mol Life Sci 2014; 71(13): 2467–2481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- About I, Laurent-Maquin D, Lendahl U et al. Nestin expression in embryonic and adult human teeth under normal and pathological conditions. Am J Pathol 2000; 157(1): 287–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki S, Namiki J, Shibata S et al. The neural stem/progenitor cell marker nestin is expressed in proliferative endothelial cells, but not in mature vasculature. J Histochem Cytochem 2010; 58(8): 721–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilder RL. Integrin alpha V beta 3 as a target for treatment of rheumatoid arthritis and related rheumatic diseases. Ann Rheum Dis 2002; 61(Suppl 2): ii96–ii99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yubero N, Jiménez-Marín A, Barbancho M et al. Two cDNAs coding for the porcine CD51 (αv) integrin subunit: cloning, expression analysis, adhesion assays and chromosomal localization. Gene 2011; 481(1): 29–40. [DOI] [PubMed] [Google Scholar]
- Chong JJ, Reinecke H, Iwata M et al. Progenitor cells identified by PDGFR-alpha expression in the developing and diseased human heart. Stem Cells Dev 2013; 22(13): 1932–1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawashima N. Characterisation of dental pulp stem cells: a new horizon for tissue regeneration? Arch Oral Biol 2012; 57(11): 1439–1458. [DOI] [PubMed] [Google Scholar]
- Coura GS, Garcez RC, de Aguiar CB et al. Human periodontal ligament: a niche of neural crest stem cells. J Periodont Res 2008; 43(5): 531–536. [DOI] [PubMed] [Google Scholar]
- Kolf CM, Cho E, Tuan RS. Mesenchymal stromal cells. Biology of adult mesenchymal stem cells: regulation of niche, self-renewal and differentiation. Arthritis Res Ther 2007; 9(1): 204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, He H, Tang C et al. Differentiation potential of STRO-1+ dental pulp stem cells changes during cell passaging. BMC Cell Biol 2010; 11: 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- d'Aquino R, De Rosa A, Laino G et al. Human dental pulp stem cells: from biology to clinical applications. J Exp Zool B Mol Dev Evol 2009; 312B(5): 408–415. [DOI] [PubMed] [Google Scholar]
- Navabazam AR, Sadeghian Nodoshan F, Sheikhha MH et al. Characterization of mesenchymal stem cells from human dental pulp, preapical follicle and periodontal ligament. Iran J Reprod Med 2013; 11(3): 235–242. [PMC free article] [PubMed] [Google Scholar]
- Stevens A, Zuliani T, Olejnik C et al. Human dental pulp stem cells differentiate into neural crest-derived melanocytes and have label-retaining and sphere-forming abilities. Stem Cells Dev 2008; 17(6): 1175–1184. [DOI] [PubMed] [Google Scholar]
- Miura M, Gronthos S, Zhao M et al. SHED: stem cells from human exfoliated deciduous teeth. Proc Natl Acad Sci USA 2003; 100(10): 5807–5812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierret C, Spears K, Maruniak JA et al. Neural crest as the source of adult stem cells. Stem Cells Dev 2006; 15(2): 286–291. [DOI] [PMC free article] [PubMed] [Google Scholar]