Abstract
In untargeted metabolomics analysis, several factors (e.g., unwanted experimental & biological variations and technical errors) may hamper the identification of differential metabolic features, which requires the data-driven normalization approaches before feature selection. So far, ≥16 normalization methods have been widely applied for processing the LC/MS based metabolomics data. However, the performance and the sample size dependence of those methods have not yet been exhaustively compared and no online tool for comparatively and comprehensively evaluating the performance of all 16 normalization methods has been provided. In this study, a comprehensive comparison on these methods was conducted. As a result, 16 methods were categorized into three groups based on their normalization performances across various sample sizes. The VSN, the Log Transformation and the PQN were identified as methods of the best normalization performance, while the Contrast consistently underperformed across all sub-datasets of different benchmark data. Moreover, an interactive web tool comprehensively evaluating the performance of 16 methods specifically for normalizing LC/MS based metabolomics data was constructed and hosted at http://server.idrb.cqu.edu.cn/MetaPre/. In summary, this study could serve as a useful guidance to the selection of suitable normalization methods in analyzing the LC/MS based metabolomics data.
Metabolomics aims at characterizing metabolic biomarkers by analytically describing complex biological samples1. At present, the metabolomics based on liquid chromatography mass spectrometry (LC/MS) is capable of simultaneously monitoring thousands of metabolites in bio-fluid, cell and tissue, and is widely applied to various aspects of biomedical research. In particular, metabolomics analysis on LC/MS data can aid the choice of therapy2, provide powerful tools for drug discovery by revealing drug mechanism of actions and potential side effects3, and help to identify biomarkers4,5,6 of various diseases such as hepatocellular carcinoma (HCC)7, colorectal cancer8, insulin resistance9, and so on.
Several factors (e.g., unwanted experimental & biological variations and technical errors) may hamper the identification of differential metabolic profiles and effectiveness of metabolomics analysis (e.g., paired or nested studies)10,11,12,13,14. To remove specific types of unwanted variations, the signal drift correction (when quality control samples are available), the batch effect removal (when internal standards or quality control samples are available), and the scaling (not suitable when the self-averaging property does not hold) are adopted13. These commonly used strategies are generally grouped into two categories: (1) method-driven normalization approaches extrapolating external model that is based upon internal standards or quality control samples and (2) data-driven normalization approaches scaling or transforming metabolomics data15,16,17,18,19,20. As reported in Ejigu’s work, the method-driven strategies may not be practical due to several reasons, especially their unsuitability for treating untargeted metabolomics data, while data-driven ones are better choices for untargeted LC/MS based metabolomics data15. The capacities of 11 data-driven normalization methods (“normalization method” in short for the rest of this paper) for processing nuclear magnetic resonance (NMR) based metabolomics data were systematically compared21. Two methods (the Quantile and the Cubic Splines) were identified as the “best” performed normalization methods, while other two methods (the Contrast and the Li-Wong) could “hardly” reduce bias at all and could not improve the comparability between samples21. For gas chromatography mass spectrometry (GC/MS) based metabolomics, a comparative research on the performances of 8 normalization methods discovered two (the Auto Scaling and the Range Scaling) of “overall best performance”12. Similar to NMR and GC/MS, the LC/MS is one of the most popular sources of current metabolomics data, and it is of great importance to analyze the differential influence of those methods on LC/MS based data. Ejigu et al. measured the performance of 6 methods according to their “average metabolite specific coefficient of variation (CV)”15. The CV showed that the Cyclic Loess and the Cubic Splines performed “slightly better” than other methods, but no statistical difference among CVs of those methods was observed15.
For the past decade, no less than 16 methods have been developed for normalizing the LC/MS based metabolomics data13,22,23, some of which (e.g., the VSN24, the Quantile25, the Cyclic Loess26) are directly adopted from those previously used for processing transcriptomics data. Both metabolomics data and transcriptomics data are high-dimensional. However, the dimension of transcriptomics data can reach 10 thousands, while that of metabolomics data is about a few thousands. Moreover, unlike transcriptomics, correlation among metabolites identified from metabolomics data may not indicate a common biological function27. Apart from the above differences, there are significant similarities between two OMICs data: (1) right-skewed distribution23, (2) great data sparsity28, (3) substantial amount of noise29,30 and (4) significantly varied sample sizes31,32. Due to these similarities, it is feasible to apply some of the normalization methods used in transcriptomics data analysis to the metabolomics one.
Those 16 methods specifically normalizing LC/MS based metabolomics data can be classified into two groups21. Methods in group one (including the Contrast Normalization33, the Cubic Splines34, the Cyclic Loess35, the Linear Baseline Scaling25, the MSTUS22, the Non-Linear Baseline Normalization36, the Probabilistic Quotient Normalization37 and the Quantile Normalization25) aim at removing the unwanted sample-to-sample variations, while methods of the second group (including the Auto Scaling38, the Level Scaling12, the Log Transformation39, the Pareto Scaling40, the Power Scaling41, the Range Scaling42, the VSN43,44 and the Vast Scaling45) adjust biases among various metabolites to reduce heteroscedasticity. However, the performance and the sample size dependence of those methods widely adopted in current metabolomics studies (e.g., the Pareto Scaling and the VSN)28,46 have not yet been exhaustively compared in the context of LC/MS metabolomics data analysis.
Moreover, several comprehensive metabolomics pipelines are currently available online, where various normalization algorithms are integrated in as one step in their corresponding analysis chain. These online pipelines include the MetaboAnalyst28, the Metabolomics Workbench47, the MetaDB48, the MetDAT49, the MSPrep50, the Workflow4Metabolomics51 and the XCMS online52. Based on a comprehensive review, the number of normalization algorithms provided by the above pipelines varies significantly from 2 (the Workflow4Metabolomics) to 13 (the MetaboAnalyst). 6 out of those 7 pipelines only provide <50% of those 16 methods analyzed in this study. The MetaboAnalyst is the only pipeline offering 13 methods, but some methods reported as “well-performed” in LC/MS based metabolomics analysis (e.g., the VSN and the PQN)28,37,46 are not provided. The inadequate coverage of these methods may weaken the applicability range of those pipelines. Moreover, the suitability of a normalization method was reported to be greatly dependent on the nature of the analyzed data53, a comparative performance evaluation among methods is therefore essential to the determination of the most appropriate method for professional/inexperienced researchers. However, no comparative evaluation among those normalization methods was conducted in the above pipelines. So far, the Normalyzer53 is the only online tool offering comparative evaluation of 12 different normalization methods treating high-throughput OMICs data53. In particular, this tool accepted a variety of data types including metabolomics, proteomics, DNA microarray and the real-time polymerase chain reaction data53. However, since the Normalyzer was designed to process a wide range of OMICs, it did not cover 8 of those 16 methods specifically for LC/MS based metabolomics studies. Thus, it is in urgent need to construct a publicly available tool for comparatively and comprehensively evaluating the performances of methods used specifically for normalizing LC/MS based metabolomics data.
In this study, a comprehensive comparison on the normalization capacities of 16 methods was conducted. Firstly, the differential metabolic features selected based on each method were validated by a benchmark spike-in dataset and by experimentally validated markers. To further understand the influence of sample size on the method performance, 10 sub-datasets of various sample size were generated to evaluate the variation of normalization performance among 16 methods, and to categorize these methods into 3 groups (superior, good and poor performance group). Finally, a web-based tool used to comprehensively evaluate the performance of all 16 methods was constructed. In sum, this study could serve as valuable guidance to the selection of suitable normalization methods in analyzing the LC/MS based metabolomics data.
Materials and Methods
Benchmark datasets collection and sub-datasets generation
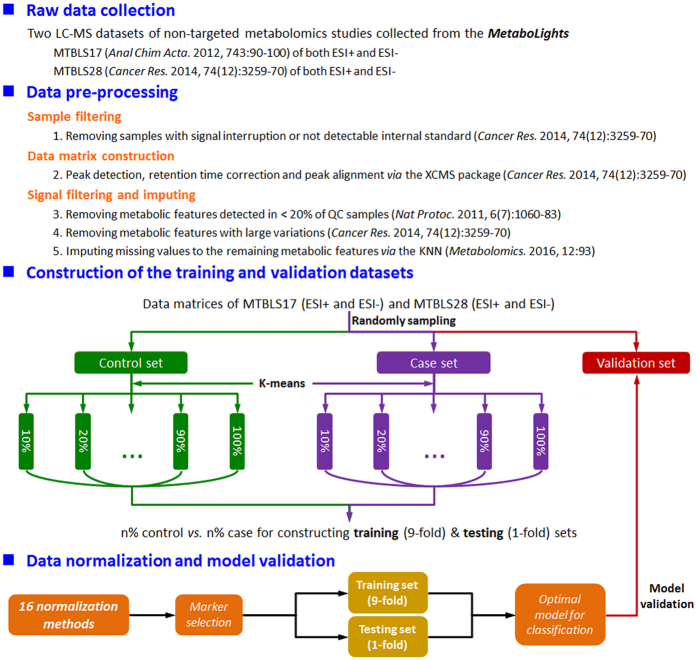
Five criteria were used to select datasets from the MetaboLights (http://www.ebi.ac.uk/metabolights/)32 in this study, which include: (1) data type set as “study”; (2) technology set as “mass spectrometry”; (3) organism set as “homo sapiens”; (4) study validation set as “fully validated”; (5) untargeted LC/MS based metabolomics data with >100 samples selected by manual literature and dataset reviews. Based on the above criteria, 4 benchmark datasets were collected for analysis, which include the positive (ESI+) and negative (ESI−) ionization modes of both MTBLS2854 and MTBLS1755. For MTBLS17, only the dataset of experiment 1 with >100 studied samples was included. For the remaining text of this paper, MTBLS17 was used to stand for the dataset of experiment 1 in Ressom’s work55. Both ESI+ and ESI− of MTBLS28 provided LC/MS based metabolomics profiles of 1,005 samples (469 lung cancer patients and 536 healthy individuals)54, and MTBLS17 ESI+ and ESI− gave profiles of 189 samples (60 HCC patients and 129 people with cirrhosis) and 185 samples (59 HCC patients and 126 people with cirrhosis), respectively55.
To construct training and validation datasets and sub-datasets of various sample size, random sampling and k-means clustering were applied. Taking MTBLS28 ESI+ as an example, 1,005 samples were divided into training dataset (400 lung cancer patients and 500 healthy individuals) and validation dataset (105 samples) by random sampling. Moreover, to generate the sub-datasets from training dataset, the k-means clustering56 was used to sample 10 sub-datasets of various sample size. In particular, the number of lung cancer patients versus that of healthy individuals were 50 vs. 40, 100 vs. 80, 150 vs. 120, 200 vs. 160, 250 vs. 200, 300 vs. 240, 350 vs. 280, 400 vs. 320, 450 vs. 360, and 500 vs. 400 for 10%, 20%, 30%, 40%, 50%, 60%, 70%, 80%, 90%, 100% of the samples in the training group, respectively.
LC/MS based metabolomics data pre-processing
Biological variance and technical error are two key factors introducing biases to the metabolomics data. Biological variance arises from the spread of metabolic signals detected from various biological samples57, while technical error results from machine drift58. In particular, biological variances (e.g., varying concentration levels of bio-fluid, different cell sizes, varying sample measurements) are commonly encountered in metabolomics data13, while technical errors (e.g., a sudden drop in peak intensities or measurements on different instruments) are the major issues in large-scale metabolomics studies58. Apart from those above methods widely adopted to remove biological variances22, quality-control (QC) samples were used to significantly reduce technical errors58.
Moreover, sparsity is the nature of metabolomics data, which can be represented by a substantial amount of missing values (10~40%), which can affect up to 80% of all metabolic features59. The direct assignment of zero to the missing values could be useful for cluster analysis, but it may lead to poor performance or even malfunction if normalization method is applied50, especially for those methods based on the logarithm (e.g., the Log Transformation)50,53. Several missing value imputation methods are currently available, among which the KNN algorithm60 was reported as the most robust one for analyzing mass spectrometry based metabolomics data60. Therefore, the KNN algorithm was adopted in this work to impute the missing signals of the metabolic features.
In this study, a widely adopted data pre-processing procedure54,60,61 was applied, which included sample filtering, data matrix construction and signal filtering & imputing (Fig. 1). In particular, (1) samples with signal interruption or not detectable internal standard were removed based on Mathé’s work54; (2) peak detection, retention time correction and peak alignment54 were applied to the UHPLC/Q-TOF-MS raw data (in CDF format) using the xcmsSet, the group and the rector functions in the XCMS package62 with both the full width at half-maximum (fwhm) and the retention time window (bw) set as 10; (3) metabolic features detected in <20% of QC samples61 or with large variations54 were removed based on Mathé’s work, and missing signals of the remaining metabolic features were imputed by the KNN algorithm60. The detailed workflow of data pre-processing used in this study was illustrated in Fig. 1.
Figure 1. The overall research design and flowchart of this study.
Normalization methods analyzed in this study
16 methods were analyzed in this work, which include the Auto Scaling (unit variance scaling, UV)38, the Contrast Normalization33, the Cubic Splines34, the Cyclic Locally Weighted Regression (Cyclic Loess)35, the Level Scaling12, the Linear Baseline Scaling25, the Log Transformation39, the MS Total Useful Signal (MSTUS)22, the Non-Linear Baseline Normalization (Li-Wong)36, the Pareto Scaling40, the Power Scaling41, the Probabilistic Quotient Normalization (PQN)37, the Quantile Normalization25, the Range Scaling42, the Variance Stabilization Normalization (VSN)43,44 and the Vast Scaling45.
Auto Scaling (unit variance scaling, UV) is one of the simplest methods adjusting metabolic variances21, which scales metabolic signals based on the standard deviation of metabolomics data. This method makes all metabolites of equal importance, but analytical errors may be amplified due to dilution effects21. Auto scaling has been used to improve the diagnosis of bladder cancer using gas sensor arrays63 and to identify urinary nucleoside markers from urogenital cancer patients64.
Contrast Normalization is originated from the integration of MA-plots and logged Bland-Altman plots, which assumes the presence of non-linear biases21. The use of a log function in this method may impede the processing of zeros and negative numbers, which requires the conversion of non-positive numbers to an extremely small value21. The contrast method has been employed to reveal the role of polychlorinated biphenyls in non-alcoholic fatty liver disease by metabolomics analysis65.
Cubic Splines is one of the non-linear baseline methods assuming the existence of non-linear relationships between baseline and individual spectra21. Cubic splines has been adopted to reduce variability in DNA microarray experiments by normalizing all signal channels to a target array34. Moreover, this method has been performed to evaluate differential effects of clinical and biological variables in breast cancer patients66.
Similar to contrast normalization, Cyclic Locally Weighted Regression (Cyclic Loess) comes also from the combination of MA-plot and logged Bland-Altman plot by assuming the existence of non-linear bias21. However, cyclic loess is the most time-consuming one among those studied normalization methods, and the amount of time grows exponentially as the number of sample increases67. This method has been used to discover microRNA candidates regulating human osteosarcoma68.
Level Scaling transforms metabolic signal variation into variation relative to the average metabolic signal by scaling according to the mean signal12. This method is especially suitable for the circumstances when huge relative variations are of great interest (e.g., studying the stress responses, identifying relatively abundant biomarkers)12. Level Scaling has been used to identify urinary nucleoside markers from urogenital cancer patients64.
Linear Baseline Scaling maps each sample spectrum to the baseline based on the assumption of a constant linear relationship21. However, this assumption of a linear correlation among sample spectra may be oversimplified21. This method has been conducted to identify differential metabolomics profiles among the banana’s 5 different senescence stages69. Moreover, linear baseline scaling has been performed to discover the toxicity profiling of capecitabine in patients with inoperable colorectal cancer70.
Log Transformation converts skewed metabolomics data to symmetric via the non-linear transformation, which is usually used to adjust heteroscedasticity and transform metabolites’ relations from multiplication to addition12. In metabolomics, relations among metabolites may not always be additive, this method is thus needed to identify multiplicative relation with linear techniques12. This method has been used to delineate potential role of sarcosine in prostate cancer progression71.
MS Total Useful Signal (MSTUS) utilizes the total signals of metabolites that are shared by all samples by assuming that the number of increased and decreased metabolic signals is relatively equivalent22,72. However, the validity of this hypothesis is questionable since an increase in the concentration of one metabolite may not necessarily be accompanied by a decrease in that of another metabolite72,73. MSTUS has been reported as among the best choices for overcoming sample variability in urinary metabolomics73 and used to identify diagnostic and prognostic markers for lung cancer patients54.
Non-Linear Baseline Normalization (Li-Wong) is one of the normalization methods aiming at removing unwanted sample-to-sample variations21. This method is first used to analyze oligonucleotide arrays based on a multiplicative parametrization36,74, and currently adopted to improve NMR-based metabolomics analysis21. This method has already been successfully integrated into the dChip74.
Different from the auto scaling, Pareto Scaling uses the square root of the standard deviation of the data as scaling factor40. Therefore, comparing to the auto scaling, this method is able to reduce more significantly the weights of large fold changes in metabolite signals, but the dominant weight of extremely large fold changes may still be unchanged21. Pareto scaling has been performed for improving the pattern recognition for targeted75 and untargeted76 metabolomics data.
Power Scaling aims at correcting for the pseudo scaling and the heteroscedasticity12. Different from the log transformation, the method is able to handle and zero values12. Power scaling has been used to study the serum amino acid profiles and their variations in colorectal cancer patients77.
Probabilistic Quotient Normalization (PQN) transforms the metabolomics spectra according to an overall estimation on the most probable dilution37. This algorithm has been reported to be significantly robust and accurate comparing to the integral and the vector length normalizations37. PQN has been used to discover potential diagnostic technique for ovarian and breast cancers from urine metabolites78.
Quantile Normalization aims at achieving the same distribution of metabolic feature intensities across all samples, and the quantile-quantile plot in this method is used to visualize the distribution similarity21. Quantile normalization has been used to probe differential molecular profiling between pancreatic adenocarcinoma and chronic pancreatitis79, and currently adopted to improve NMR-based metabolomics analysis21.
Range Scaling scales the metabolic signals by the variation of biological responses63. A disadvantage of this method lies in a limited number (usually only 2) of values used to describe the variation unlike other scaling methods taking all measurements into account using the standard deviation, which makes this algorithm relatively sensitive to outliers12. Because all variation levels of the metabolites are treated equally by the range scaling, it has been used to fuse mass spectrometry-based metabolomics data42.
Variance Stabilization Normalization (VSN) is one of the non-linear methods aiming at remain variances unchanged across the whole data range21. The method is reported to be a preferred approach for exploratory analysis such as the principal component analysis80. VSN was originally developed for normalizing single and two-channel microarray data81, and currently used to determine metabolic profiles of liver tissue during early cancer development82.
As an extension of the auto scaling, Vast Scaling scales the metabolic signals based on the coefficient of variation12. Vast scaling has been used to identify prognostic factors for breast cancer patients from the magnetic resonance based metabolomics83.
Detailed descriptions on these methods could be found in Supplementary Note S1, and their source codes programed in this study could be found in Supplementary Note S2.
Assessment of the normalization performance by classification algorithm
Firstly, the differential metabolic features were identified by VIP value (>1) of the partial least squares discriminant analysis (PLS-DA)84 in R package ropls85 together with p-value (<0.05) of Student t-test71. All computational assessments were conducted in R (http://www.r-project.org) version 3.2.4 running on 64-bit Mac OS X EI Capitan (v10.11.5) platform. Source codes of related programs designed in this study could be found in Supplementary Note S2.
Secondly, classification algorithm was applied to assess the performance of each normalization method based on the identified differential metabolic features. Several classification algorithms were adopted to evaluate the performance of normalization methods, which include the Support Vector Machine (SVM)21, the k-Nearest Neighbors (k-NN)86, the Gaussian Mixture Model (GMM)87, and so on. As illustrated in Fig. 1, the SVM algorithm in the R package e1071 (http://cran.r-project.org/web/packages/e1071) was selected to assess normalization performance in this study. In the process of training the classification models, 10-fold cross validation was used to optimize parameters, and the validation dataset was then used to assess the classification performance of the selected differential features by the receiver operating characteristic (ROC) plots generated by R package ROCR88. Source codes of the classification algorithm programed in this study could be found in Supplementary Note S2.
Identification of the performance relationship among normalization methods
The hierarchical clustering56,89,90 was adopted to identify the relationship of sample size dependent performance among 16 methods. Firstly, the area under the curve values (AUCs) of a specific method among 10 sub-datasets of various sample size were used to generate a 10 dimensional vector. Secondly, hierarchical clustering was adopted to investigate the relationship among vectors, and therefore among corresponding methods. As an assessment of consistency between different distance metrics, two metrics (the Manhattan and the Euclidean) were applied:
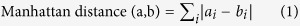 |
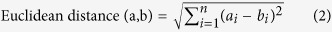 |
In Eq. (1) and Eq. (2), i refers to each AUC of method a and b. Clustering approach adopted is the Ward’s minimum variance method91, which is used to reduce the total within-cluster variance to the maximum extent. In this work, Ward’s minimum variance module in R package was used92. Source codes of the hierarchical clustering algorithm programed in this study could be found in Supplementary Note S2.
Construction of web-based tool for evaluating performance of 16 normalization method
A web-based tool named as MetaPre for comprehensively evaluating the normalization performance of all 16 methods was constructed and hosted at http://server.idrb.cqu.edu.cn/MetaPre/. MetaPre was developed in R environment, and further extended using HTML, CSS and JavaScript. The R package Shiny (http://shiny.rstudio.com/) was used to construct web application (comprised of a front end and a back end). R package DiffCorr93 and vsn from Bioconductor Project94 were utilized to support background processes. MetaPre server was deployed at Apache HTTP web server v2.2.15 (http://httpd.apache.org).
Results and Discussion
Validation of the differential metabolic features selected based on 16 normalization methods
Supplementary Table S1 showed the number of differential metabolic features identified by PLS-DA based on 16 normalization methods. As demonstrated, the numbers of features selected based on some methods were the same as each other, while the numbers identified by some others varied significantly. SVM classifier based on those features was used in this work, the validity of these features were therefore crucial for assessing performances of 16 methods. In this study, two lines of evidence were provided for this assessment. First, a benchmark spike-in dataset from Franceschi’s work95 was analyzed. As shown in Supplementary Table S2, the performances on identifying spike-in compounds based on 16 methods were equivalent to that of Franceschi’s work, which indirectly reflected the reliability of strategy applied in this study. Secondly, 2 markers (creatine riboside and 561.3432) from positive and other 2 markers (cortisol sulfate and N-acetylneuraminic acid) from negative ionization mode were experimentally validated in Mathé’s work54. Supplementary Table S3 listed the number of experimentally validated markers identified by this work from the same datasets as that in Mathé’s work (MTBLS28 ESI+ and ESI−). For all methods of various sample sizes, the absolute majority (91.6%) identified all experimentally validated markers, which could server as another line of evidence for the validity of metabolic features selected by this study.
Variation of normalization performances among 16 methods based on benchmark datasets
Table 1 demonstrated the prediction accuracy (ACC) of each method trained by 10 sub-datasets based on MTBLS28 (ESI+ and ESI−). For the training set of 900 samples from MTBLS28 ESI+, the ACC values of 11 methods fell in the range from 0.6095 (the Level Scaling) to 0.6952 (the Log Transformation, the Power Scaling and the Range Scaling). The ACC values of 4 methods (the VSN, the PQN, the Cyclic Loess and the Cubic Splines) exceeded 0.7, while that of another method (the Contrast) was only 0.5143. For training set of 900 samples from MTBLS28 ESI−, the ACC values of 14 methods fell in the range from 0.6095 (the Level Scaling) to 0.6857 (the Cyclic Loess and the VSN). The ACC value of only one method (the Quantile) exceeded 0.7, while that of another method (the Contrast) was only 0.3333. Moreover, Supplementary Table S4 showed the ACC values of each method trained by 10 sub-datasets based on MTBLS17 (ESI+ and ESI−). For the training set of 170 samples from MTBLS17, the Contrast method always underperformed comparing to other methods, which was similar to that of MTBLS28. However, the top-ranked normalization methods for each ionization mode of each dataset vary significantly, which is in accordance with Chawade’s conclusion that the effectiveness of a method in normalizing data relied on the nature of the analyzed data53. Thus, this significant variation reminded us that it is essential to take various sample size into account, if one try to compare the performance among normalization methods.
Table 1. Performance evaluation of 16 normalization methods across 10 sub-datasets based on the benchmark data MTBLS28 (ESI+ and ESI−).
| Normalization method | MetaboLights ID (ionization mode) | Sample size of 10 various sub-datasets used in the training set |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 90 | 180 | 270 | 360 | 450 | 540 | 630 | 720 | 810 | 900 | ||
| Auto Scaling | MTBLS28 (ESI+) | 0.5905 | 0.6286 | 0.6381 | 0.6952 | 0.5810 | 0.6286 | 0.6381 | 0.6381 | 0.6476 | 0.6667 |
| MTBLS28 (ESI−) | 0.5524 | 0.5333 | 0.5810 | 0.6190 | 0.6000 | 0.6190 | 0.6571 | 0.6476 | 0.6381 | 0.6667 | |
| Contrast | MTBLS28 (ESI+) | 0.4762 | 0.5238 | 0.4095 | 0.4667 | 0.5238 | 0.5619 | 0.5429 | 0.5619 | 0.4762 | 0.5143 |
| MTBLS28 (ESI−) | 0.4381 | 0.3429 | 0.4667 | 0.3524 | 0.3524 | 0.3714 | 0.4000 | 0.3619 | 0.3524 | 0.3333 | |
| Cubic Splines | MTBLS28 (ESI+) | 0.6095 | 0.6095 | 0.6762 | 0.6952 | 0.6095 | 0.7143 | 0.6476 | 0.7048 | 0.6762 | 0.7048 |
| MTBLS28 (ESI−) | 0.6667 | 0.6095 | 0.6381 | 0.6000 | 0.6476 | 0.6952 | 0.6952 | 0.6762 | 0.7238 | 0.6667 | |
| Cyclic Loess | MTBLS28 (ESI+) | 0.6000 | 0.6190 | 0.6667 | 0.6381 | 0.6476 | 0.6476 | 0.6381 | 0.6762 | 0.7524 | 0.7238 |
| MTBLS28 (ESI−) | 0.5905 | 0.6667 | 0.6571 | 0.6190 | 0.6476 | 0.6667 | 0.6857 | 0.6762 | 0.6857 | 0.6857 | |
| Level Scaling | MTBLS28 (ESI+) | 0.5905 | 0.6190 | 0.6381 | 0.6476 | 0.6286 | 0.6476 | 0.6286 | 0.6381 | 0.6286 | 0.6095 |
| MTBLS28 (ESI−) | 0.5714 | 0.5619 | 0.6095 | 0.6190 | 0.6000 | 0.6095 | 0.6667 | 0.6476 | 0.6381 | 0.6095 | |
| Li-Wong | MTBLS28 (ESI+) | 0.5524 | 0.6095 | 0.5524 | 0.6000 | 0.5619 | 0.5714 | 0.6190 | 0.6286 | 0.6667 | 0.6762 |
| MTBLS28 (ESI−) | 0.5048 | 0.6286 | 0.5429 | 0.6571 | 0.6476 | 0.6762 | 0.6381 | 0.5905 | 0.6476 | 0.6286 | |
| Linear Baseline | MTBLS28 (ESI+) | 0.6095 | 0.6095 | 0.6476 | 0.7238 | 0.6095 | 0.6286 | 0.6667 | 0.6762 | 0.6952 | 0.6286 |
| MTBLS28 (ESI−) | 0.5619 | 0.5619 | 0.5905 | 0.6095 | 0.6190 | 0.6000 | 0.6762 | 0.6571 | 0.6286 | 0.6476 | |
| Log Transformation | MTBLS28 (ESI+) | 0.6476 | 0.6857 | 0.6952 | 0.6476 | 0.7048 | 0.6667 | 0.6857 | 0.6952 | 0.6762 | 0.6952 |
| MTBLS28 (ESI−) | 0.6095 | 0.6286 | 0.6190 | 0.6762 | 0.6095 | 0.6571 | 0.6857 | 0.6190 | 0.6952 | 0.6476 | |
| MSTUS | MTBLS28 (ESI+) | 0.5905 | 0.6095 | 0.6286 | 0.6476 | 0.6000 | 0.6571 | 0.6667 | 0.6762 | 0.6667 | 0.6762 |
| MTBLS28 (ESI−) | 0.5619 | 0.5714 | 0.5619 | 0.6000 | 0.6095 | 0.6286 | 0.6762 | 0.6762 | 0.6667 | 0.6476 | |
| Pareto Scaling | MTBLS28 (ESI+) | 0.6190 | 0.6571 | 0.6190 | 0.5905 | 0.6476 | 0.6667 | 0.6476 | 0.6952 | 0.6381 | 0.6667 |
| MTBLS28 (ESI−) | 0.5714 | 0.5619 | 0.5905 | 0.6095 | 0.6000 | 0.6190 | 0.6667 | 0.6381 | 0.6381 | 0.6476 | |
| Power Scaling | MTBLS28 (ESI+) | 0.6000 | 0.6571 | 0.5905 | 0.6190 | 0.6000 | 0.6286 | 0.6952 | 0.6476 | 0.7048 | 0.6952 |
| MTBLS28 (ESI−) | 0.5810 | 0.6381 | 0.5905 | 0.5714 | 0.5810 | 0.6095 | 0.6476 | 0.6095 | 0.6286 | 0.6476 | |
| PQN | MTBLS28 (ESI+) | 0.5619 | 0.6381 | 0.6476 | 0.6286 | 0.6381 | 0.6667 | 0.6762 | 0.6952 | 0.7524 | 0.7333 |
| MTBLS28 (ESI−) | 0.6000 | 0.6095 | 0.6476 | 0.6381 | 0.6571 | 0.6857 | 0.6667 | 0.7238 | 0.7429 | 0.6667 | |
| Quantile | MTBLS28 (ESI+) | 0.6095 | 0.6190 | 0.6476 | 0.6952 | 0.6095 | 0.6476 | 0.6762 | 0.7143 | 0.7048 | 0.6857 |
| MTBLS28 (ESI−) | 0.6667 | 0.6190 | 0.6381 | 0.6095 | 0.6286 | 0.6667 | 0.6857 | 0.6571 | 0.6381 | 0.7238 | |
| Range Scaling | MTBLS28 (ESI+) | 0.5714 | 0.6190 | 0.6190 | 0.5905 | 0.6381 | 0.6000 | 0.6190 | 0.6476 | 0.6381 | 0.6952 |
| MTBLS28 (ESI−) | 0.5810 | 0.5429 | 0.5714 | 0.5905 | 0.6000 | 0.6095 | 0.6190 | 0.6476 | 0.6667 | 0.6571 | |
| Vast Scaling | MTBLS28 (ESI+) | 0.5524 | 0.6190 | 0.6286 | 0.5905 | 0.6381 | 0.5905 | 0.6286 | 0.6571 | 0.6476 | 0.6381 |
| MTBLS28 (ESI−) | 0.5333 | 0.5714 | 0.6095 | 0.6095 | 0.6095 | 0.5810 | 0.6571 | 0.6190 | 0.6667 | 0.6190 | |
| VSN | MTBLS28 (ESI+) | 0.6381 | 0.6381 | 0.6286 | 0.7048 | 0.6476 | 0.7048 | 0.7048 | 0.6571 | 0.7524 | 0.7429 |
| MTBLS28 (ESI−) | 0.6571 | 0.6762 | 0.6476 | 0.6762 | 0.6476 | 0.6667 | 0.6762 | 0.7048 | 0.6857 | 0.6857 | |
The performance was evaluated by the prediction accuracies (ACCs) on the validation set. The ACC equals to (true positive + true negative)/(true positive + false positive + true negative + false negative).
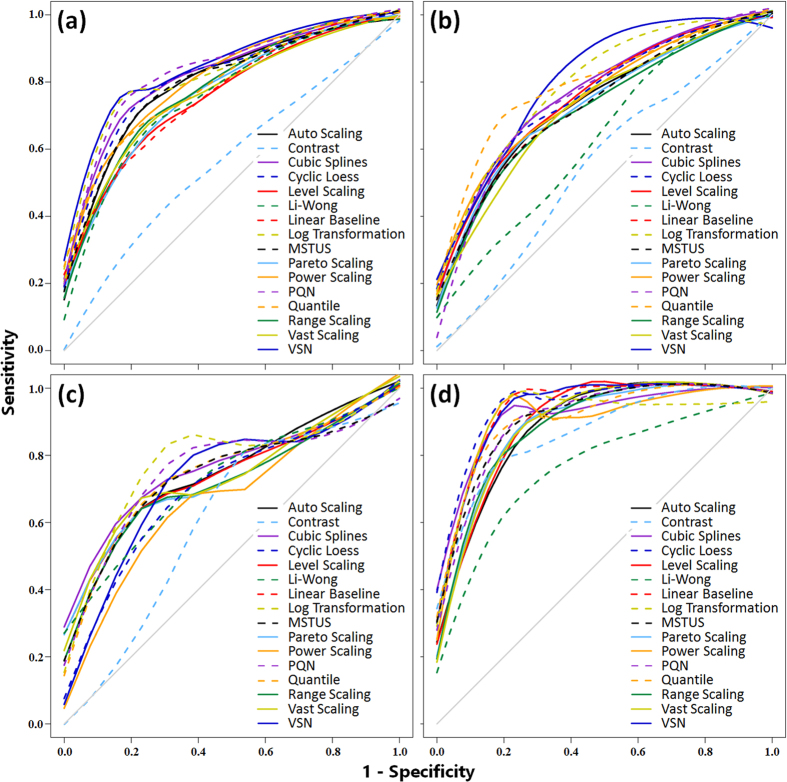
The receiver operating characteristic (ROC) curves and the area under the curve values (ACCs) were used to illustrate the performances of 16 methods in Fig. 2 and Supplementary Table S5. Figure 2a–d illustrated ROC curves of MTBLS28 ESI+, MTBLS28 ESI−, MTBLS17 ESI+ and MTBLS17 ESI−, respectively. The training dataset of Fig. 2a and b consisted of 900 samples (400 lung cancer patients and 500 healthy individuals), and that of Fig. 2c and d consisted of 170 samples (50 HCC patients and 120 people with cirrhosis). The grey diagonal represented an invalid model with the corresponding AUC value equaled to 0.5. As illustrated in Fig. 2a–d, the Contrast method showed a poor normalization performance in all 4 datasets, while the VSN and the Log Transformation outperformed consistently. However, performance rank of the remaining methods fluctuated dramatically, which also requested a collective assessment of normalization performance based on various sample size.
Figure 2.
Normalization performance of 16 methods measured by receiver operating characteristic (ROC) curves based on four benchmark datasets: (a) MTBLS28 ESI+, (b) MTBLS28 ESI−, (c) MTBLS17 ESI+ and (d) MTBLS17 ESI−. The training dataset of (a) and (b) composed of 900 samples (400 lung cancer patients and 500 healthy individuals), and that of (c) and (d) consisted of 170 samples (50 HCC patients and 120 people with cirrhosis). The grey diagonal represented an invalid model with the corresponding area under the curve (AUC) value equaled to 0.5. All lines were generated by the LOESS regression.
Categorization of 16 methods based on their normalization performances
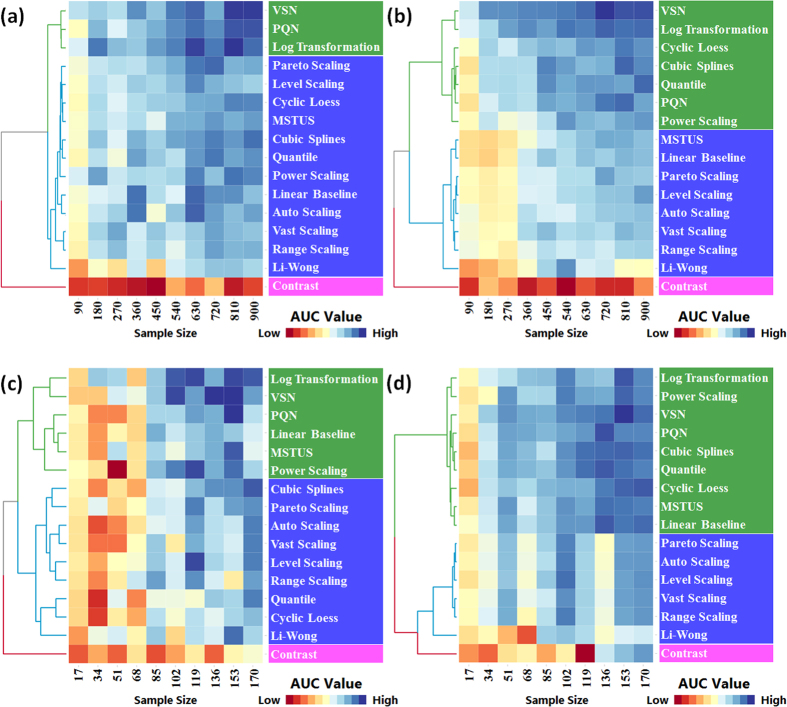
AUCs of a specific method among 10 sub-datasets were calculated to construct a 10 dimensional vector. The resulting 16 vectors were then hierarchically clustered based on two popular distance metrics (the Manhattan in Fig. 3 and the Euclidean in Supplementary Figure S1). Cluster analysis of 16 methods was conducted based on 4 benchmark datasets: (a) MTBLS28 ESI+, (b) MTBLS28 ESI−, (c) MTBLS17 ESI+ and (d) MTBLS17 ESI−. As shown in Fig. 3a–d, 16 methods were divided by the corresponding dendrogram on the left side of each figure into three areas: top, middle and bottom areas colored by green, blue and magenta, respectively. Clearly, 3 methods (the VSN, the Log Transformation and the PQN) were consistently ranked into the top area of all 4 figures, while one method (the Contrast) always stayed in the bottom area. Therefore, 16 normalization methods could be categorized into 3 groups (A, B and C) by comprehensively considering their performances across all 4 benchmark datasets.
Figure 3.
Cluster analysis of 16 normalization methods according to their AUC values (across 10 various sample sizes) calculated based on four benchmark datasets: (a) MTBLS28 ESI+, (b) MTBLS28 ESI−, (c) MTBLS17 ESI+ and (d) MTBLS17 ESI−. The data were presented in matrix format in which columns represent specific training dataset of various sample size and rows represent each normalization method. Each cell in heat map represents AUC value of a normalization method trained on one specific training sample. The cell of the highest AUC value was set as exact blue with those lower AUC values gradually fading towards red (the lowest AUC value). Hierarchical clustering analyses were conducted using Manhattan metric and Ward’s minimum variance algorithm.
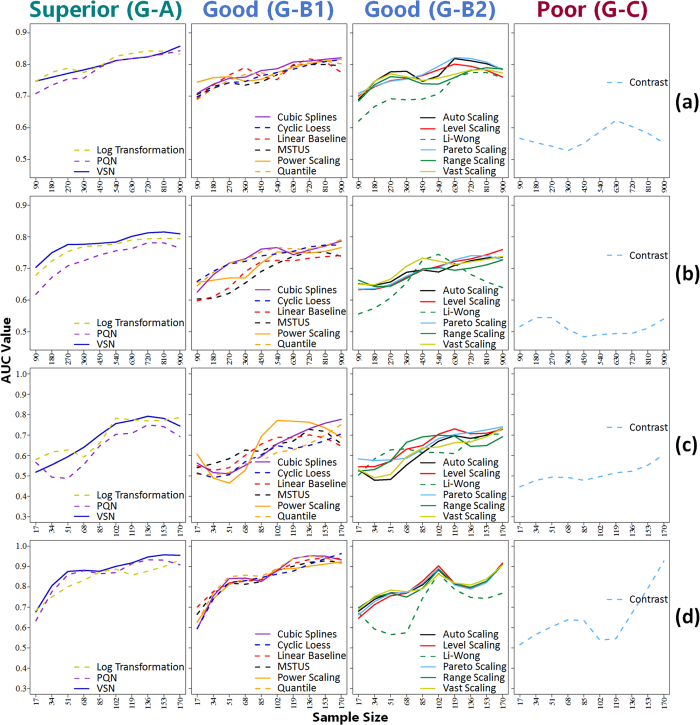
As illustrated by Fig. 4, normalization methods in group A (the VSN, the Log Transformation and the PQN) demonstrated the best performance among all 16 methods, which made group A (G-A) the Superior Performance Group. The VSN and the PQN had been discovered as robust and well-performed methods in metabolomics for various dilutions of biological samples37,96. The Log Transformation was reported to be a powerful tool for making skewed distributions symmetric12, it was therefore a very suitable method for treating metabolomics data (the distribution of which was right-skewed)23. Moreover, some methods (e.g., the VSN) in G-A was also found to be the most capable one in reducing variation between technical replicates in proteomics, and consistently well-performed in identifying differential expression profiles97. The Contrast was the only one method in group C (G-C, the Poor Performance Group), the performance of which was consistently the worst across 10 sub-datasets among all 16 methods. As reported by Kohl et al.21, the Contrast hardly reduced bias at all and could not improve comparability among samples21.
Figure 4.
Method groups categorized according to the normalization performances across various sample sizes based on four benchmark datasets: (a) MTBLS28 ESI+, (b) MTBLS28 ESI−, (c) MTBLS17 ESI+ and (d) MTBLS17 ESI−. (G–A) superior performance group; (G-B1) good performance group including methods occasionally classified to top green area of Fig. 3; (G-B2) good performance group including methods consistently staying in middle blue area of Fig. 3; (C) poor performance group. All lines were generated by the LOESS regression.
Moreover, the remaining 12 methods in group B (Good Performance Group) could be further divided into G-B1 (including 6 methods occasionally classified to the top area of Fig. 3) and G-B2 (including 6 methods consistently staying in the middle area of Fig. 3). As illustrated in Fig. 4, although slightly underperformed comparing to G-A, methods in G-B1 showed good normalization performances across 10 sub-datasets of various sample size. Furthermore, the majority of the methods in G-B2 followed a similar fluctuation trends across various sample sizes, with the Li-Wong distinguished as an outlier. The Li-Wong performed the worst among other assessed methods in reducing within- and between-group variations96, and could hardly reduce the biases among samples at all21.
Similar to the Manhattan metric (Fig. 3), 16 methods could also be re-categorized with the Euclidean metric. As illustrated in Supplementary Figure S1, the categorization generated based on the Euclidean metric identified 3 groups with exactly the same methods in each group as that of the Manhattan metric, which reflected the independent nature of method categorization on different distance metrics. Moreover, in Supplementary Figure S1d, the Li-Wong was clustered into the bottom area (magenta) together with the Contrast, which again reflected its unsuitability in analyzing LC/MS based metabolomics data21,96.
Online interactive analysis tool for normalizing LC/MS based metabolomics data
With R package Shiny (http://shiny.rstudio.com/), an interactive web tool, named MetaPre, was developed in this study and hosted at http://server.idrb.cqu.edu.cn/MetaPre/. The MetaPre constructed to normalize LC/MS based metabolomics data could be easily accessed by modern web browsers such as Chrome, Foxfire, IE, Safari, and so on. Meanwhile, the local version of MetaPre was freely provide in this study and could also be readily downloaded from https://github.com/libcell/MetaPre in Github. The procedure for using online version of the MetaPre was provided in Fig. 5, which included 4 steps: (1) uploading the dataset; (2) data pre-processing; (3) data normalization; (4) performance evaluation.
Figure 5. General operational procedure for using MetaPre.
Uploading the dataset provided the option to upload data with or without QC samples. In large-scale metabolomics study (especially the LC/MS based one), not all samples can be analyzed in the same experimental batch61. To cope with these difficulties, QC samples were frequently applied58,61. In the MetaPre, batch correction based on QC samples was provided, which made this tool one of the few currently available online servers51,98 offering such kind of function.
Data pre-processing offered the function to correct metabolic features and impute missing signals. For data with QC samples, the MetaPre firstly applied within-block signal correction61 to correct metabolic features. Then, multiple popular imputing algorithms were provided to fill missing signals. For data without QC samples, only the process of missing signal imputing was implemented.
Data normalization integrated 16 normalization methods discussed in this study to remove the unwanted biological variations. After selecting any of these methods, the normalized data matrix was displayed on the web page and a corresponding csv file could be downloaded directly. Moreover, two box plots used to visualizing the distributions of data before and after normalization were illustrated on the web page.
Performance evaluation was quantified based on AUC values of the constructed SVM models. Firstly, the differential metabolic features were identified by VIP value (>1) of PLS-DA model. Then, SVM models were constructed based on these identified differential features. After k-folds cross validation, ROC curve together with its AUC value were calculated and displayed on the web page.
MetaPre is valuable online tool to select suitable methods for normalizing LC/MS based metabolomics data, and is a useful complement to the currently available tools in modern metabolomics analysis.
Conclusion
Based on the 4 datasets tested in this work, 16 methods for normalizing LC/MS based metabolomics data were categorized into three groups based on their normalization performances across various sample sizes, which included the superior (3 methods), good (12 methods) and poor (1 method) performance groups. The VSN, the Log Transformation and the PQN were identified as methods of the best normalization performance, while the Contrast consistently underperformed across all sub-datasets of different benchmark data among those 16 methods. Moreover, an interactive web tool comprehensively evaluating the performance of all 16 methods for normalizing LC/MS based metabolomics data was constructed and hosted at http://server.idrb.cqu.edu.cn/MetaPre/. In sum, this study could serve as guidance to the selection of suitable normalization methods in analyzing the LC/MS based metabolomics data.
Additional Information
How to cite this article: Li, B. et al. Performance Evaluation and Online Realization of Data-driven Normalization Methods Used in LC/MS based Untargeted Metabolomics Analysis. Sci. Rep. 6, 38881; doi: 10.1038/srep38881 (2016).
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Material
Acknowledgments
This work was funded by the research support of National Natural Science Foundation of China (81202459, 21505009 and 21302102); by Innovation Project on Industrial Generic Key Technologies of Chongqing (cstc2015zdcy-ztzx120003); by the Chongqing Graduate Student Research Innovation Project (CYB14027); by the Fundamental Research Funds for the Central Universities (CDJZR14468801, CDJKXB14011, 2015CDJXY).
Footnotes
Author Contributions F.Z. designed research. B.L., J.T., Q.Y., X.C. and S.C. performed research and developed the web tool. B.L., W.X., N.C., S.L. and Q.C wrote the scripts and prepared the example data. B.L. and F.Z. wrote the manuscript. All authors reviewed the manuscript.
References
- Xia J., Broadhurst D. I., Wilson M. & Wishart D. S. Translational biomarker discovery in clinical metabolomics: an introductory tutorial. Metabolomics 9, 280–299 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss R. H. & Kim K. Metabolomics in the study of kidney diseases. Nat. Rev. Nephrol. 8, 22–33 (2012). [DOI] [PubMed] [Google Scholar]
- Kaddurah-Daouk R. & Krishnan K. R. Metabolomics: a global biochemical approach to the study of central nervous system diseases. Neuropsychopharmacology 34, 173–186 (2009). [DOI] [PubMed] [Google Scholar]
- Yang H. et al. Therapeutic target database update 2016: enriched resource for bench to clinical drug target and targeted pathway information. Nucleic Acids Res. 44, D1069–1074 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu F. et al. Therapeutic target database update 2012: a resource for facilitating target-oriented drug discovery. Nucleic Acids Res. 40, D1128–1136 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J. et al. Comparison of FDA approved kinase targets to clinical trial ones: insights from their system profiles and drug-target interaction networks. Biomed Res. Int. 2016, 2509385 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leng D. et al. Meta-analysis of genetic programs between idiopathic pulmonary fibrosis and sarcoidosis. PLoS One 8, e71059 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. et al. Meta-analysis of differentially expressed genes in osteosarcoma based on gene expression data. BMC Med. Genet. 15, 80 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milburn M. V. & Lawton K. A. Application of metabolomics to diagnosis of insulin resistance. Annu. Rev. Med. 64, 291–305 (2013). [DOI] [PubMed] [Google Scholar]
- Matthan N. R. et al. Plasma phospholipid fatty acid biomarkers of dietary fat quality and endogenous metabolism predict coronary heart disease risk: a nested case-control study within the Women’s Health Initiative observational study. J. Am. Heart Assoc. 3, e000764 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weckwerth W. Metabolomics in systems biology. Annu. Rev. Plant Biol. 54, 669–689 (2003). [DOI] [PubMed] [Google Scholar]
- van den Berg R. A., Hoefsloot H. C. J., Westerhuis J. A., Smilde A. K. & van der Werf M. J. Centering, scaling, and transformations: improving the biological information content of metabolomics data. BMC Genomics 7, 142 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Livera A. M. et al. Statistical methods for handling unwanted variation in metabolomics data. Anal. Chem. 87, 3606–3615 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu F. et al. Clustered patterns of species origins of nature-derived drugs and clues for future bioprospecting. Proc. Natl. Acad. Sci. USA 108, 12943–12948 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ejigu B. A. et al. Evaluation of normalization methods to pave the way towards large-scale LC-MS-based metabolomics profiling experiments. OMICS 17, 473–485 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nezami Ranjbar M. R., Zhao Y., Tadesse M. G., Wang Y. & Ressom H. W. Gaussian process regression model for normalization of LC-MS data using scan-level information. Proteome Sci. 11, S13 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranjbar M. R., Tadesse M. G., Wang Y. & Ressom H. W. Bayesian normalization model for label-free quantitative analysis by LC-MS. IEEE/ACM Trans. Comput. Biol. Bioinform. 12, 914–927 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranjbar M. R., Di Poto C., Wang Y. & Ressom H. W. SIMAT: GC-SIM-MS data analysis tool. BMC Bioinformatics 16, 259 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Befekadu G. K., Tadesse M. G. & Ressom H. W. A Bayesian based functional mixed-effects model for analysis of LC-MS data. Conf. Proc. IEEE Eng. Med. Biol. Soc. 2009, 6743–6746 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranjbar M. R. N., Tadesse M. G., Wang Y. & Ressom H. W. Normalization of LC-MS data using Gaussian process. in 2012 IEEE International Workshop on Genomic Signal Processing and Statistics (Gensips) 187–190 (2012). [Google Scholar]
- Kohl S. M. et al. State-of-the art data normalization methods improve NMR-based metabolomic analysis. Metabolomics 8, 146–160 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warrack B. M. et al. Normalization strategies for metabonomic analysis of urine samples. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 877, 547–552 (2009). [DOI] [PubMed] [Google Scholar]
- De Livera A. M. et al. Normalizing and integrating metabolomics data. Anal. Chem. 84, 10768–10776 (2012). [DOI] [PubMed] [Google Scholar]
- Lin S. M., Du P., Huber W. & Kibbe W. A. Model-based variance-stabilizing transformation for Illumina microarray data. Nucleic Acids Res. 36, e11 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolstad B. M., Irizarry R. A., Astrand M. & Speed T. P. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics 19, 185–193 (2003). [DOI] [PubMed] [Google Scholar]
- Edwards D. Non-linear normalization and background correction in one-channel cDNA microarray studies. Bioinformatics 19, 825–833 (2003). [DOI] [PubMed] [Google Scholar]
- Fukushima A., Kusano M., Redestig H., Arita M. & Saito K. Integrated omics approaches in plant systems biology. Curr. Opin. Chem. Biol. 13, 532–538 (2009). [DOI] [PubMed] [Google Scholar]
- Xia J. G. & Wishart D. S. Web-based inference of biological patterns, functions and pathways from metabolomic data using MetaboAnalyst. Nat. Protoc. 6, 743–760 (2011). [DOI] [PubMed] [Google Scholar]
- Mak T. D., Laiakis E. C., Goudarzi M. & Fornace A. J. Jr. Selective paired ion contrast analysis: a novel algorithm for analyzing postprocessed LC-MS metabolomics data possessing high experimental noise. Anal. Chem. 87, 3177–3186 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grun D., Kester L. & van Oudenaarden A. Validation of noise models for single-cell transcriptomics. Nat. Methods 11, 637–640 (2014). [DOI] [PubMed] [Google Scholar]
- Barrett T. et al. NCBI GEO: archive for functional genomics data sets--update. Nucleic Acids Res. 41, D991–995 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haug K. et al. MetaboLights–an open-access general-purpose repository for metabolomics studies and associated meta-data. Nucleic Acids Res. 41, D781–786 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Astrand M. Contrast normalization of oligonucleotide arrays. J. Comput. Biol. 10, 95–102 (2003). [DOI] [PubMed] [Google Scholar]
- Workman C. et al. A new non-linear normalization method for reducing variability in DNA microarray experiments. Genome Biol. 3, research0048 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudoit S., Yang Y. H., Callow M. J. & Speed T. P. Statistical methods for identifying differentially expressed genes in replicated cDNA microarray experiments. Stat. Sin. 12, 111–139 (2002). [Google Scholar]
- Li C. & Wong W. H. Model-based analysis of oligonucleotide arrays: expression index computation and outlier detection. Proc. Natl. Acad. Sci. USA 98, 31–36 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dieterle F., Ross A., Schlotterbeck G. & Senn H. Probabilistic quotient normalization as robust method to account for dilution of complex biological mixtures. Application in 1H NMR metabonomics. Anal. Chem. 78, 4281–4290 (2006). [DOI] [PubMed] [Google Scholar]
- Hu C. X. & Xu G. W. Mass-spectrometry-based metabolomics analysis for foodomics. Trac-Trend Anal. Chem. 52, 36–46 (2013). [Google Scholar]
- Purohit P. V., Rocke D. M., Viant M. R. & Woodruff D. L. Discrimination models using variance-stabilizing transformation of metabolomic NMR data. OMICS 8, 118–130 (2004). [DOI] [PubMed] [Google Scholar]
- Eriksson L. et al. Using chemometrics for navigating in the large data sets of genomics, proteomics, and metabonomics (gpm). Anal. Bioanal. Chem. 380, 419–429 (2004). [DOI] [PubMed] [Google Scholar]
- Brodsky L., Moussaieff A., Shahaf N., Aharoni A. & Rogachev I. Evaluation of peak picking quality in LC-MS metabolomics data. Anal. Chem. 82, 9177–9187 (2010). [DOI] [PubMed] [Google Scholar]
- Smilde A. K., van der Werf M. J., Bijlsma S., van der Werff-van der Vat B. J. & Jellema R. H. Fusion of mass spectrometry-based metabolomics data. Anal. Chem. 77, 6729–6736 (2005). [DOI] [PubMed] [Google Scholar]
- Huber W., von Heydebreck A., Sultmann H., Poustka A. & Vingron M. Variance stabilization applied to microarray data calibration and to the quantification of differential expression. Bioinformatics 18 Suppl 1, S96–104 (2002). [DOI] [PubMed] [Google Scholar]
- Karp N. A. et al. Addressing accuracy and precision issues in iTRAQ quantitation. Mol. Cell Proteomics 9, 1885–1897 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keun H. C. et al. Improved analysis of multivariate data by variable stability scaling: application to NMR-based metabolic profiling. Anal. Chim. Acta 490, 265–276 (2003). [Google Scholar]
- Theodoridis G., Gika H. G. & Wilson I. D. LC-MS-based methodology for global metabolite profiling in metabonomics/metabolomics. Trac-Trend Anal. Chem. 27, 251–260 (2008). [Google Scholar]
- Sud M. et al. Metabolomics Workbench: An international repository for metabolomics data and metadata, metabolite standards, protocols, tutorials and training, and analysis tools. Nucleic Acids Res. 44, D463–470 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franceschi P. et al. Metadb a data processing workflow in untargeted MS-based metabolomics experiments. Front. Bioeng. Biotechnol. 2, 72 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswas A. et al. MetDAT: a modular and workflow-based free online pipeline for mass spectrometry data processing, analysis and interpretation. Bioinformatics 26, 2639–2640 (2010). [DOI] [PubMed] [Google Scholar]
- Hughes G. et al. MSPrep–summarization, normalization and diagnostics for processing of mass spectrometry-based metabolomic data. Bioinformatics 30, 133–134 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giacomoni F. et al. Workflow4Metabolomics: a collaborative research infrastructure for computational metabolomics. Bioinformatics 31, 1493–1495 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gowda H. et al. Interactive XCMS Online: simplifying advanced metabolomic data processing and subsequent statistical analyses. Anal. Chem. 86, 6931–6939 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chawade A., Alexandersson E. & Levander F. Normalyzer: a tool for rapid evaluation of normalization methods for omics data sets. J. Proteome Res. 13, 3114–3120 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathe E. A. et al. Noninvasive urinary metabolomic profiling identifies diagnostic and prognostic arkers in lung cancer. Cancer Res. 74, 3259–3270 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ressom H. W. et al. Utilization of metabolomics to identify serum biomarkers for hepatocellular carcinoma in patients with liver cirrhosis. Anal. Chim. Acta. 743, 90–100 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libbrecht M. W. & Noble W. S. Machine learning applications in genetics and genomics. Nat. Rev. Genet. 16, 321–332 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moseley H. N. Error analysis and propagation in metabolomics data analysis. Comput. Struct. Biotechnol. J. 4, e201301006 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Kloet F. M., Bobeldijk I., Verheij E. R. & Jellema R. H. Analytical error reduction using single point calibration for accurate and precise metabolomic phenotyping. J. Proteome Res. 8, 5132–5141 (2009). [DOI] [PubMed] [Google Scholar]
- Xia J., Psychogios N., Young N. & Wishart D. S. MetaboAnalyst: a web server for metabolomic data analysis and interpretation. Nucleic Acids Res. 37, W652–660 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Guida R. et al. Non-targeted UHPLC-MS metabolomic data processing methods: a comparative investigation of normalisation, missing value imputation, transformation and scaling. Metabolomics 12, 93 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn W. B. et al. Procedures for large-scale metabolic profiling of serum and plasma using gas chromatography and liquid chromatography coupled to mass spectrometry. Nat. Protoc. 6, 1060–1083 (2011). [DOI] [PubMed] [Google Scholar]
- Smith C. A., Want E. J., O’Maille G., Abagyan R. & Siuzdak G. XCMS: processing mass spectrometry data for metabolite profiling using nonlinear peak alignment, matching, and identification. Anal. Chem. 78, 779–787 (2006). [DOI] [PubMed] [Google Scholar]
- Weber C. M. et al. Evaluation of a gas sensor array and pattern recognition for the identification of bladder cancer from urine headspace. Analyst 136, 359–364 (2011). [DOI] [PubMed] [Google Scholar]
- Struck W. et al. Liquid chromatography tandem mass spectrometry study of urinary nucleosides as potential cancer markers. J. Chromatogr. A 1283, 122–131 (2013). [DOI] [PubMed] [Google Scholar]
- Shi X. et al. Metabolomic analysis of the effects of polychlorinated biphenyls in nonalcoholic fatty liver disease. J. Proteome Res. 11, 3805–3815 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boracchi P., Biganzoli E. & Marubini E. Joint modelling of cause-specific hazard functions with cubic splines: an application to a large series of breast cancer patients. Comput. Stat. Data An. 42, 243–262 (2003). [Google Scholar]
- Ballman K. V., Grill D. E., Oberg A. L. & Therneau T. M. Faster cyclic loess: normalizing RNA arrays via linear models. Bioinformatics 20, 2778–2786 (2004). [DOI] [PubMed] [Google Scholar]
- Duan Z. et al. MicroRNA-199a-3p is downregulated in human osteosarcoma and regulates cell proliferation and migration. Mol. Cancer Ther. 10, 1337–1345 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Y. et al. Metabolomic analyses of banana during postharvest senescence by 1H-high resolution-NMR. Food Chem. 218, 406–412 (2017). [DOI] [PubMed] [Google Scholar]
- Backshall A., Sharma R., Clarke S. J. & Keun H. C. Pharmacometabonomic profiling as a predictor of toxicity in patients with inoperable colorectal cancer treated with capecitabine. Clin. Cancer Res. 17, 3019–3028 (2011). [DOI] [PubMed] [Google Scholar]
- Sreekumar A. et al. Metabolomic profiles delineate potential role for sarcosine in prostate cancer progression. Nature 457, 910–914 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- Jacob C. C., Dervilly-Pinel G., Biancotto G. & Le Bizec B. Evaluation of specific gravity as normalization strategy for cattle urinary metabolome analysis. Metabolomics 10, 627–637 (2014). [Google Scholar]
- Chen Y. et al. Combination of injection volume calibration by creatinine and MS signals’ normalization to overcome urine variability in LC-MS-based metabolomics studies. Anal. Chem. 85, 7659–7665 (2013). [DOI] [PubMed] [Google Scholar]
- Chu T. M., Weir B. S. & Wolfinger R. D. Comparison of Li-Wong and loglinear mixed models for the statistical analysis of oligonucleotide arrays. Bioinformatics 20, 500–506 (2004). [DOI] [PubMed] [Google Scholar]
- Yan Z. & Yan R. Tailored sensitivity reduction improves pattern recognition and information recovery with a higher tolerance to varied sample concentration for targeted urinary metabolomics. J. Chromatogr. A 1443, 101–110 (2016). [DOI] [PubMed] [Google Scholar]
- Yang J., Zhao X., Lu X., Lin X. & Xu G. A data preprocessing strategy for metabolomics to reduce the mask effect in data analysis. Front. Mol. Biosci. 2, 4 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leichtle A. B. et al. Serum amino acid profiles and their alterations in colorectal cancer. Metabolomics 8, 643–653 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slupsky C. M. et al. Urine metabolite analysis offers potential early diagnosis of ovarian and breast cancers. Clin. Cancer Res. 16, 5835–5841 (2010). [DOI] [PubMed] [Google Scholar]
- Logsdon C. D. et al. Molecular profiling of pancreatic adenocarcinoma and chronic pancreatitis identifies multiple genes differentially regulated in pancreatic cancer. Cancer Res. 63, 2649–2657 (2003). [PubMed] [Google Scholar]
- Zhang S. et al. Interdependence of signal processing and analysis of urine 1H NMR spectra for metabolic profiling. Anal. Chem. 81, 6080–6088 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kultima K. et al. Development and evaluation of normalization methods for label-free relative quantification of endogenous peptides. Mol. Cell Proteomics 8, 2285–2295 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibarra R. et al. Metabolomic analysis of liver tissue from the VX2 rabbit model of secondary liver tumors. HPB Surg. 2014, 310372 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giskeodegard G. F. et al. Multivariate modeling and prediction of breast cancer prognostic factors using MR metabolomics. J. Proteome Res. 9, 972–979 (2010). [DOI] [PubMed] [Google Scholar]
- Ballabio D. & Consonni V. Classification tools in chemistry. Part 1: linear models. PLS-DA. Anal. Methods 5, 3790–3798 (2013). [Google Scholar]
- Thévenot E. A., Roux A., Xu Y., Ezan E. & Junot C. Analysis of the human adult urinary metabolome variations with age, body mass index, and gender by implementing a comprehensive workflow for univariate and OPLS statistical analyses. J. Proteome Res. 14, 3322–3335 (2015). [DOI] [PubMed] [Google Scholar]
- Wu W., Xing E. P., Myers C., Mian I. S. & Bissell M. J. Evaluation of normalization methods for cDNA microarray data by k-NN classification. BMC Bioinformatics 6, 191 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brahim A., Ramirez J., Gorriz J. M., Khedher L. & Salas-Gonzalez D. Comparison between different intensity normalization methods in 123I-Ioflupane imaging for the automatic detection of Parkinsonism. PLoS One 10, (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sing T., Sander O., Beerenwinkel N. & Lengauer T. ROCR: visualizing classifier performance in R. Bioinformatics 21, 3940–3941 (2005). [DOI] [PubMed] [Google Scholar]
- Zheng G. et al. Exploring the inhibitory mechanism of approved selective norepinephrine reuptake inhibitors and reboxetine enantiomers by molecular dynamics study. Sci. Rep. 6, 26883 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue W. et al. Identification of the inhibitory mechanism of FDA approved selective serotonin reuptake inhibitors: an insight from molecular dynamics simulation study. Phys. Chem. Chem. Phys. 18, 3260–71 (2016). [DOI] [PubMed] [Google Scholar]
- Kim J. et al. Somatic ERCC2 mutations are associated with a distinct genomic signature in urothelial tumors. Nat. Genet. 48, 600–606 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tippmann S. Programming tools: Adventures with R. Nature 517, 109–110 (2015). [DOI] [PubMed] [Google Scholar]
- Fukushima A. DiffCorr: an R package to analyze and visualize differential correlations in biological networks. Gene 518, 209–214 (2013). [DOI] [PubMed] [Google Scholar]
- Huber W. et al. Orchestrating high-throughput genomic analysis with Bioconductor. Nat. Methods 12, 115–121 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franceschi P., Masuero D., Vrhovsek U., Mattivi F. & Wehrens R. A benchmark spike-in data set for biomarker identification in metabolomics. J. Chemom. 26, 16–24 (2012). [Google Scholar]
- Hochrein J. et al. Data normalization of 1H NMR metabolite fingerprinting data sets in the presence of unbalanced metabolite regulation. J. Proteome Res. 14, 3217–3228 (2015). [DOI] [PubMed] [Google Scholar]
- Valikangas T., Suomi T. & Elo L. L. A systematic evaluation of normalization methods in quantitative label-free proteomics. Brief. Bioinform. bbw095 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia J., Sinelnikov I. V., Han B. & Wishart D. S. MetaboAnalyst 3.0--making metabolomics more meaningful. Nucleic Acids Res. 43, W251–257 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.