Abstract
The von Hippel-Lindau (VHL) tumor suppressor functions as a ubiquitin ligase that mediates proteolytic inactivation of hydroxylated α subunits of hypoxia-inducible factor (HIF). Although studies of VHL-defective renal carcinoma cells suggest the existence of other VHL tumor suppressor pathways, dysregulation of the HIF transcriptional cascade has extensive effects that make it difficult to distinguish whether, and to what extent, observed abnormalities in these cells represent effects on pathways that are distinct from HIF. Here, we report on a genetic analysis of HIF-dependent and -independent effects of VHL inactivation by studying gene expression patterns in Caenorhabditis elegans. We show tight conservation of the HIF-1/VHL-1/EGL-9 hydroxylase pathway. However, persisting differential gene expression in hif-1 versus hif-1; vhl-1 double mutant worms clearly distinguished HIF-1–independent effects of VHL-1 inactivation. Genomic clustering, predicted functional similarities, and a common pattern of dysregulation in both vhl-1 worms and a set of mutants (dpy-18, let-268, gon-1, mig-17, and unc-6), with different defects in extracellular matrix formation, suggest that dysregulation of these genes reflects a discrete HIF-1–independent function of VHL-1 that is connected with extracellular matrix function.
Besides its known function of inactivating hypoxia-inducible factor (HIF), genetically engineered worms clearly demonstrate that there exist HIF-independent effects of the von Hippel- Lindau tumor suppressor
Introduction
The von Hippel-Lindau (VHL) gene is a tumor suppressor that is mutated in the majority of both hereditary and sporadic, clear-cell renal carcinomas (Kaelin 2002). In hereditary VHL disease affected individuals are also predisposed to pheochromocytomas and retinal/central nervous system hemangioblastomas and develop multiple benign lesions in the kidney and other organs. Despite more than a decade of intensive investigation following identification of the defective gene in 1993 (Latif et al. 1993), the nature of the VHL tumor suppressor mechanism and how it relates to the physiological function of VHL remains unclear (Kaelin 2002).
To date, the best-understood function of VHL is as a ubiquitin ligase that affects oxygen-dependent proteolytic targeting of the α subunits of hypoxia-inducible factor (HIF) (Maxwell et al. 1999; Ohh et al. 2000). Oxygen-dependent hydroxylation of two HIF-α prolyl residues by HIF prolyl hydroxylases (Epstein et al. 2001; Ivan et al. 2001; Jaakkola et al. 2001) promotes interaction with VHL and targets HIF-α for degradation by the ubiquitin-proteasome pathway. In VHL-defective cells HIF-α subunits are stabilized and HIF is constitutively activated, resulting in the upregulation of HIF target genes (Maxwell et al. 1999). Whether this, or other putative VHL pathways, accounts for the tumor suppressor action is the subject of active investigation (Kondo et al. 2002, 2003; Maranchie et al. 2002). For instance, a number of different VHL-dependent cellular phenotypes have been defined by contrasting VHL-defective cells with transfectants re-expressing wild-type VHL (Kaelin 2002). These have highlighted effects of VHL on invasiveness, branching morphogenesis, and matrix assembly (Ohh et al. 1998; Koochekpour et al. 1999; Davidowitz et al. 2001; Kamada et al. 2001; Esteban-Barragan et al. 2002). However, mechanistic links to VHL function have not yet been defined and it is unclear whether or not these effects are secondary to dysregulation of HIF. This has led to attempts to define the existence, or otherwise, of non-HIF, VHL-regulated pathways by comparing patterns of gene expression induced by VHL inactivation with those induced by hypoxia (Wykoff et al. 2000; Zatyka et al. 2002; Y. Jiang et al. 2003). The observed patterns are not fully concordant, suggesting that there may be non-HIF, VHL-regulated pathways. However, these studies leave important uncertainties since HIF dysregulation might have secondary effects on pathways that are not themselves responsive to hypoxia and VHL might target hypoxia pathways other than HIF.
To address this we have used a genetic approach in Caenorhabditis elegans. Whereas mammalian cells possess three HIF-α isoforms that are targeted by VHL, C. elegans has a single HIF-α homolog (HIF-1) and a single VHL homolog (VHL-1), simplifying the genetic approach (Epstein et al. 2001; H. Jiang et al. 2001). Since homozygous vhl-1 and hif-1 loss-of-function worms are viable, we created hif-1; vhl-1 worms and compared the effects of vhl-1 inactivation on gene expression in wild-type and HIF-1–defective backgrounds. Our results clearly demonstrate the existence of both HIF-dependent and HIF-independent pathways of VHL-dependent gene expression. HIF-1–dependent effects of vhl-1 inactivation on gene expression were also produced by inactivation of the HIF prolyl hydroxylase homolog EGL-9. In contrast, the HIF-1–independent effects of vhl-1 inactivation were not observed in egl-9 loss-of-function worms but were seen in a panel of mutant worms (dpy-18, let-268, gon-1, mig-17, and unc-6) bearing defects in genes involved in extracellular matrix function, supporting the existence of a conserved non-HIF pathway connecting VHL with an as yet unknown extracellular matrix function.
Results
Effect of VHL-1 Inactivation on Gene Expression in C. elegans
As a first step in defining VHL-1–dependent pathways in C. elegans, a whole-genome microarray was probed to compare transcript patterns in vhl-1 versus wild-type worms (n = 1). From this array a set of genes (selected for amplitude of differential expression, signal intensity, quality of array signal, and putative function) was assayed quantitatively by ribonuclease (RNase) protection (Table 1). Of the 14 genes analyzed, six (F22B5.4, unknown function; nhr-57, predicted nuclear hormone receptor; fmo-12, predicted flavin monooxygenase; egl-9, HIF-1 prolyl hydroxylase [Epstein et al. 2001]; phy-2, procollagen prolyl 4-hydroxylase α subunit [Friedman et al. 2000]; and cah-4, predicted carbonic anhydrase) were strikingly downregulated by VHL-1 (Figure 1A; Table 2, column B). Further analysis in synchronized worm populations indicated that the VHL-1–dependent effects were observed in all stages (Figure 1B and unpublished data).
Table 1. Top 30 Upregulated Genes in the vhl-1 versus Wild-Type Microarray Comparison and Confirmation of Selected Genes by RNase Protection Assays.
Confirmation of selected genes: Y, reproducible upregulation of gene in vhl-1/wild-type worms as confirmed by RNase protection assays; N, no reproducible upregulation of gene as tested by RNase protection assays; —, not determined. Gene name refers to the three-letter gene name where available and open reading frame name (WormBase); details of name changes and primers in arrays are from http://worm-chip.stanford.edu/pcr.all_primers.plus_gels.4–10-02.txt. Description is the predicted protein, annotated by Proteome/Incyte; —, protein of unknown function. Microarrays and RNase protection assays were performed using worms cultured under normoxic conditions
Figure 1. HIF-1–Dependent Effects of VHL-1 Inactivation.
Representative RNase protection assays of genes that were differentially expressed in the vhl-1 versus wild-type microarray in (A) mixed-stage and (B) synchronized populations of worm. All genes are regulated by the VHL-1/HIF-1/EGL-9 pathway.
(C) Regulation of nhr-57 mRNA in egl-9; vhl-1 worms and by egl-9 RNAi and DIP in vhl-1 worms. For RNAi experiments controls were L4440 vector alone (−) and C17G10.1, an irrelevant putative dioxygenase.
F21C3.5 is a constitutively expressed gene used to control for RNA integrity. RNase protection assays were performed using worms cultured under normoxic conditions, unless otherwise indicated.
Table 2. Differential Expression of VHL-1–Regulated Genes in Mutants Affecting the HIF-1/VHL-1/EGL-9 Pathway.
Column A, data from the microarray comparison of vhl-1 versus wild-type worms. Columns B to I, data from RNase protection assays. The figures represent the (fold) differences in expression averaged for the indicated number (n) of independent comparisons. Statistical analysis of differential expression was performed where n ≥ 3; *, p < 0.05. Statistical analysis was performed for differences between vhl-1 versus wild-type (column B) and egl-9 versus wild-type (column C); †, p < 0.05. N, normoxia; H, hypoxia (0.1% oxygen)
Analysis of the EGL-9/HIF-1 Pathway
To determine the extent to which disruption of the conserved EGL-9/HIF-1 pathway mediates these effects we studied wild-type, hif-1, vhl-1, and egl-9 single mutant worms and hif-1; vhl-1 and egl-9; hif-1 double mutant worms. Apart from the mild phenotype of the vhl-1 worms (slightly uncoordinated, slow growth, and reduced brood size) and the egg-laying defective phenotype of egl-9, none of the worm strains showed obvious phenotypic abnormalities. Interestingly, hif-1 corrected the phenotype of egl-9.
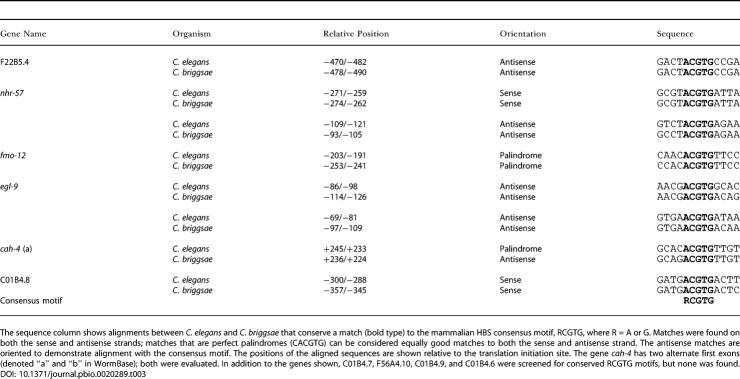
The findings indicate that all six genes are strongly regulated by the EGL-9/HIF-1 pathway (Figure 1A; Table 2). All six genes were inducible by hypoxia in wild-type worms (Table 2, column D) and strikingly upregulated in egl-9 worms (Table 2, column C). hif-1 inactivation abrogated the upregulation by the egl-9 mutation (Table 2, columns C and F) and strikingly reduced induction by hypoxia (Table 2, columns D and G). Computational analysis revealed that five (F22B5.4, nhr-57, fmo-12, egl-9, and cah-4) of the six genes contained a potential HIF-1 binding core motif (RCGTG) within an arbitrarily defined region (−1,000 to +250 nucleotides) that was conserved in Caenorhabditis briggsae (Table 3), suggesting that these genes are direct HIF-1 transcriptional targets.
Table 3. Evolutionarily Conserved HBS Consensus Sequences.
The sequence column shows alignments between C. elegans and C. briggsae that conserve a match (bold type) to the mammalian HBS consensus motif, RCGTG, where R = A or G. Matches were found on both the sense and antisense strands; matches that are perfect palindromes (CACGTG) can be considered equally good matches to both the sense and antisense strand. The antisense matches are oriented to demonstrate alignment with the consensus motif. The positions of the aligned sequences are shown relative to the translation initiation site. The gene cah-4 has two alternate first exons (denoted “a” and “b” in WormBase); both were evaluated. In addition to the genes shown, C01B4.7, F56A4.10, C01B4.9, and C01B4.6 were screened for conserved RCGTG motifs, but none was found
Though the six genes all conformed to the above patterns to demonstrate regulation by the EGL-9/HIF-1 pathway (Figure 1A; Table 2), there were differences. First, for some genes (F22B5.4, nhr-57, and fmo-12) expression in normoxia was entirely dependent on HIF-1, whereas other genes retained substantial normoxic expression in hif-1 worms (Figure 1A). Second, three genes (nhr-57, egl-9, and cah-4) showed modest upregulation, and one gene (phy-2) showed modest downregulation, by hypoxia that was independent of HIF-1, VHL-1, and EGL-9 (Table 2, columns G–I). Finally, for certain genes, upregulation was clearly greater in egl-9 than vhl-1 worms, results being particularly striking for nhr-57 (Table 2, columns C and B). To pursue this, we created egl-9; vhl-1 double mutants and also exposed vhl-1 worms to egl-9 RNAi. Both procedures increased nhr-57 expression, indicating that EGL-9 has non–VHL-1–mediated effects on this pathway (Figure 1C). Interestingly, the effects of genetic inactivation of egl-9 in the vhl-1 background were not mimicked by the dioxygenase inhibitor 2,2′-dipyridyl (DIP), suggesting that the VHL-1–independent repressive effects on nhr-57 may be nonenzymatic.
Evidence for a VHL-1–Dependent, HIF-1–Independent Pathway
To address directly whether HIF-1–independent, VHL-1–mediated pathways exist, we performed further microarray comparisons of RNA from hif-1; vhl-1 and hif-1 worms (n = 3). Fewer genes showed differential expression than in the vhl-1 versus wild-type array; however, persisting differential expression did suggest the existence of VHL-1 pathways that are independent of HIF-1 (Table 4). To test this, a number of genes were selected for further validation by RNase protection assay on the basis of amplitude of differential expression, p value, signal intensity, and quality of array signal. Of the 25 genes analyzed by RNase protection assay (Table 4), six (C01B4.7, F56A4.10, C01B4.9, and C01B4.8, all predicted transmembrane proteins belonging to the major facilitator superfamily [InterPro: IPR007114 and IPR005828]; F56A4.2, a predicted C-type lectin [InterPro: IPR001304]; and C01B4.6, a predicted aldose epimerase [InterPro: IPR008183]) showed clear downregulation by VHL-1 in a HIF-1–independent manner (Figure 2A; Table 5, column C). These effects were observed across essentially all developmental stages of the worm (Figure 2B). Computational analysis revealed that only one (C01B4.8) of the five HIF-1–independent, VHL-1–dependent genes tested (C01B4.7, F56A4.10, C01B4.9, C01B4.8, and C01B4.6; no single ortholog of F56A4.2 could be identified in C. briggsae) contained a potential HIF-1 binding site (HBS) within an arbitrarily defined region that was conserved in C. briggsae (see Table 3). This contrasts with the HIF-1–dependent, VHL-1–dependent genes validated by RNase protection assay (see Figure 1A), for which potential HBSs could be defined for five of the six genes tested (see Table 3).
Table 4. Upregulated Genes in the hif-1; vhl-1 versus hif-1 Microarray Comparisons and Confirmation of Selected Genes by RNase Protection Assays.
Confirmation of selected genes: Y, reproducible upregulation of gene in hif-1; vhl-1/hif-1 worms as confirmed by RNase protection assays; N, no reproducible upregulation of gene as tested by RNase protection assays; asterisk, not assayed, riboprobe could not be constructed; NS, no signal by RNase protection assay; —, not determined. Microarrays and RNase protection assays were performed using worms cultured under normoxic conditions. C35B8.1, C46A5.3, R03D7.5, T11F9.8, and ZK1010.7 were also tested by RNase protection assay based on microarray data; these genes did not show reproducible upregulation by RNase protection assays
Figure 2. HIF-1–Independent Effects of VHL-1 Inactivation.
RNase protection assays of genes that were differentially expressed in the hif-1; vhl-1 versus hif-1 microarrays in (A) mixed-stage and (B) synchronized populations of worm. The results confirm the existence of VHL-1–dependent, HIF-1–independent effects on gene expression.
Table 5. Differential Expression of HIF-1–Independent, VHL-1–Regulated Genes in Mutants Affecting the HIF-1/VHL-1/EGL-9 Pathway and Procollagen Hydroxylases.
Columns A and B, data from microarray comparisons; columns C to K, data from RNase protection assays. The figures represent the (fold) differences in expression averaged for the indicated number (n) of independent comparisons. Statistical analysis of differential expression was performed where n ≥ 3; *, p < 0.05. N, normoxia; H, hypoxia (0.1% oxygen)
Interestingly, all six genes validated by RNase protection assay to be negatively regulated by VHL-1 in a HIF-1–independent manner localize within 45 kb on Chromosome V (although they were not situated in physical proximity on the array). We applied single-linkage clustering (nearest-neighbor method) (Sneath 1957; Dillon and Goldstein 1984; Roy et al. 2002) to identify spatial clusters of genes considered to be negatively regulated by VHL-1 in a HIF-1–independent manner from the microarray data (Table 4) and random sampling to evaluate the significance of such clusters. Using a clustering threshold of 96,985 bp (see Materials and Methods), one cluster of ten genes and four clusters of two genes were identified (Figure 3A). On 100,000 simulated datasets of 57 randomly selected genes (equal number to that of VHL-1–dependent, HIF-1–independent genes; Table 4), the frequency of observed cluster sizes was as follows: one gene, 5,043,442; two genes, 298,425; three genes, 18,198; four genes, 1,190; five genes, 66; six genes, 4. No clusters of more than six genes were observed. Therefore, the cluster of ten VHL-1–regulated (HIF-1–independent) genes, which extends over 110 kb to include F56A4.9, Y45G12C.9, Y45G12C.12, and Y45G12C.2 in addition to the six genes validated by RNase protection assays, can be considered statistically significant to p ≪ 10−5 (Figure 3B). Recent C. elegans genomic assemblies (for example, WS120) have shown that the entire 110-kb region containing the coregulated gene cluster is arranged in tandem with a second nearly identical segmental duplication of the locus (>99.9% identical in alignment). At this level of identity, our microarray and RNase protection analyses cannot discriminate between the two copies of each gene, so for all of our analyses we have only used the names of the distal copy and genes from the proximal copy were excluded from computational analyses.
Table 6. C. elegans Strains and Alleles.

a Note that eDf18 carries a weak gon-1 mutation that renders the CB4504 strain temperature sensitive for the Gon phenotype
Figure 3. Chromosomal Clustering of VHL-1–Dependent (HIF-1–Independent) Genes.
(A) Chromosomal localization of VHL-1–dependent, HIF-1–independent genes. The positions of the genes from Table 4 are indicated by vertical ticks along the C. elegans chromosomes (shown to scale). Where two such genes are too close to be clearly resolved, the tick is marked by an asterisk. The single significant spatial clustering of VHL-1–dependent, HIF-1–independent genes is indicated by a red rectangle. The histogram under each chromosome shows the gene density (deeper bar, greater density) calculated as a sliding window of 100,000 bp moving with 10,000-bp increments along each chromosome. Dark blue indicates total annotated gene density, and light blue indicates the density of genes from the microarray that passed preliminary quality control.
(B) Organization of the VHL-1–regulated (HIF-1–independent) gene cluster from Chromosome V. The relative positions and sizes of gene transcription units are shown to scale, with genes transcribed left to right above the horizontal line and right to left below the line. Names in black indicate genes that passed all selection criteria to be considered upregulated in hif-1; vhl-1 versus hif-1 worms (see Table 4). Genes with a mean >2.0-fold upregulation are indicated by green boxes, 1.5- to 2-fold are yellow, and <1.5-fold are red. Genes for which no data were obtained are shown as light grey.
Extracellular Matrix Link to Novel VHL-1 Pathway
Since ubiquitin ligases commonly recognize more than one substrate, we considered whether these HIF-1–independent genes might be regulated by prolyl hydroxylation of another VHL-1 substrate by EGL-9. However, this was not supported by any differential expression in egl-9; hif-1 versus hif-1 worms (Figure 4A; Table 5, column E). Nevertheless, two genes, C01B4.7 and C01B4.8, were upregulated in hif-1 worms by hypoxia and the 2-oxoglutarate dioxygenase inhibitors, DIP and dimethyloxalylglycine (DMOG) (Figure 4; Table 5, column F), suggesting that another enzyme in this class might be involved. The procollagen prolyl hydroxylases DPY-18, PHY-2, and PHY-3 (Friedman et al. 2000; Riihimaa et al. 2002) and the procollagen lysyl hydroxylase LET-268 (Norman and Moerman 2000) were tested as potential candidates. A clear pattern was observed. All six VHL-1–regulated, HIF-1–independent genes were reproducibly downregulated by DPY-18 and LET-268 but not by PHY-2 or PHY-3 (Figure 5A; Table 5, columns H–K). The strain carrying the heterozygous let-268 mutation is heterozygous for unc-4, dpy-10, and unc-52; however, the VHL-1–dependent, HIF-1–independent genes were not differentially expressed in unc-4, dpy-10, or unc-52 worms, indicating that the effects were due to LET-268 (unpublished data). Further experiments on dpy-18; hif-1 double mutant worms clearly indicated that the effects of DPY-18 on this group of genes were (like the effects of VHL-1) HIF-1 independent (Figure 5C and unpublished data).
Figure 4. Responses of VHL-1–Dependent, HIF-1–Independent Genes to egl-9 Inactivation, Hypoxia, and 2-Oxoglutarate Dioxygenase Inhibitors.
RNase protection assays showing regulation of VHL-1–dependent, HIF-1–independent genes by (A) EGL-9 and hypoxia and (B) pharmacological inhibitors of 2-oxoglutarate dioxygenases: DIP and DMOG. None of the genes is regulated by EGL-9, but two genes (C01B4.7 and C01B4.8) show modest induction by hypoxia, DIP, and DMOG.
Figure 5. Sensitivity of VHL-1–Regulated Genes to Defects in Extracellular Matrix-Associated Proteins.
RNase protection assays showing altered expression of VHL-1–regulated genes that are HIF-1 independent (upper six panels) and HIF-1 dependent (F22B5.4) in worms bearing mutations affecting (A) procollagen prolyl and lysyl hydroxylases and (B) other extracellular matrix-associated proteins. A common pattern of upregulation is observed in hif-1; vhl-1, vhl-1, dpy-18, let-268, gon-1, mig-17, and unc-6 worms but not other mutants. This contrasts with the HIF-1–dependent gene F22B5.4, which is upregulated in vhl-1 worms but none of the other mutants.
(C) RNase protection assay for C01B4.9 illustrating DPY-18–mediated changes in expression that are independent of HIF-1.
Downregulation by DPY-18 and LET-268 is consistent with the positive effects of hypoxia, DIP, and DMOG, since all these stimuli inhibit DPY-18 and LET-268. However, the involvement of a lysyl, as well as a prolyl, hydroxylase suggests that the effects were unlikely to arise from failure of hydroxylation of a second prolyl hydroxylation substrate recognized by VHL-1 and were more likely to be related to a common function of DPY-18 and LET-268, such as a function in extracellular matrix formation. To pursue this, we tested the effects of defects in proteins involved in other aspects of extracellular matrix formation (either in the cuticle or basement membrane) that are distinct from protein hydroxylation. These experiments indicated that the six genes were, to varying extents, upregulated in the basement membrane-associated gon-1 (heterozygote), mig-17, and unc-6 mutant worms but not in the cuticle-associated dpy-11, bli-4, or sqt-3 mutant worms (Figure 5B and unpublished data). In contrast, none of the HIF-1–dependent genes was upregulated in these mutants (Figure 5B and unpublished data). GON-1 and MIG-17 encode secreted metalloproteases and UNC-6 encodes a netrin; all are thought to be involved in basement membrane remodeling/cell migration during gonadal morphogenesis (Hedgecock et al. 1990; Blelloch and Kimble 1999; Nishiwaki et al. 2000). Conversely, DPY-11 (a thioredoxin) and BLI-4 (a serine endoprotease) are both involved in collagen formation in the worm cuticle (Thein et al. 2003) and SQT-3 encodes a cuticular collagen. These results therefore extend the characterization of the VHL-1–dependent, HIF-1–independent pathway and support a connection with extracellular matrix/basement membrane function.
Discussion
By comparing the effects of vhl-1 inactivation in different genetic backgrounds, these data clearly distinguish HIF-1–dependent and –independent effects of VHL-1 on gene expression. Somewhat surprisingly, all of the VHL-regulated genes analyzed fell into one of two patterns: independent of HIF-1 and EGL-9 and dependent on DPY-18, LET-268, GON-1, MIG-17, and UNC-6, or the reverse, suggesting that they reflect perturbation of two discrete aspects of VHL-1 function.
The HIF-1–dependent expression pattern of all six genes chosen for detailed analysis from the vhl-1 versus wild-type array underlines the importance of the HIF-1 pathway in VHL-1 function. Computational analysis revealed that five of these genes (F22B5.4, nhr-57, fmo-12, egl-9, and cah-4) have at least one HIF-1 binding core motif (RCGTG) that is conserved in C. briggsae within an arbitrarily defined (−1,000 to +250 nucleotides) promoter region, suggesting that they are direct HIF-1 transcriptional targets. Several genes (egl-9, HIF prolyl hydroxylase; phy-2, procollagen prolyl 4-hydroxylase α subunit; and cah-4, carbonic anhydrase) have mammalian homologs that are HIF targets (Ivanov et al. 1998; Takahashi et al. 2000; Epstein et al. 2001), emphasizing the extent of conservation of the pathway. Others, such as flavin monooxygenase fmo-12 and the nuclear hormone receptor nhr-57, are apparently novel HIF-1 target genes. Interestingly, some of these HIF-1–dependent genes were partly downregulated by EGL-9 in a VHL-1– and iron-independent manner, suggesting that, in addition to the HIF-1/VHL-1 pathway, EGL-9 regulates HIF-1 transcriptional activity via a novel pathway.
Remarkably, among the candidate genes tested from the hif-1; vhl-1 versus hif-1 screens, all six that showed reproducible (HIF-1–independent) regulation by VHL-1 were located within 45 kb on Chromosome V. Analysis of the microarray data revealed that there was indeed a single, highly significant (p < 10−5) chromosomal cluster of genes negatively regulated by VHL-1 in a HIF-1–independent manner and that in total this cluster extended over 110 kb to include F56A4.9, Y45G12C.9, Y45G12C.12, and Y45G12C.2 in addition to the six genes validated by RNase protection assay. The chromosomal localization of genes in C. elegans is not random, with functionally related genes located close to one another (Roy et al. 2002) or even organized into operons (Blumenthal and Gleason 2003). Even though, based on the absence of spliced leader SL2 sequences (Blumenthal et al. 2002) and the presence of inverse transcriptional orientations, the genes in this cluster do not appear to be within the same operon, there may be a functional relevance to their physical proximity. Four of the six genes validated by RNase protection assay (C01B4.7, F56A4.10, C01B4.9, and C01B4.8) encode membrane transporters of the major facilitator superfamily, a family of transporters involved in passive transport of small solutes. C01B4.9 clusters phylogenetically with monocarboxylate transporters, and C01B4.7, F56A4.10, and C01B4.8 cluster with sodium phosphate transporters (unpublished data). Both gene families have been subject to rounds of gene duplication in the vertebrate and nematode lineages. As such, it is not possible to define one-to-one orthologous relationships for these genes between C. elegans and Homo sapiens. Nevertheless, the genomic clustering, predicted functional similarities, and common pattern of perturbed expression across an extensive set of mutant worms suggest that the upregulation of the genes in vhl-1 worms reflects the disturbance of a specific function of VHL-1.
The common effects of inactivating mutations in vhl-1 and in genes that manifest functional overlap in extracellular matrix formation—dpy-18 and let-268 (encoding procollagen hydroxylases) (Friedman et al. 2000; Norman and Moerman 2000), gon-1 and mig-17 (encoding secreted metalloproteases) (Blelloch and Kimble 1999; Nishiwaki et al. 2000), and unc-6 (encoding the extracellular guidance protein, netrin) (Hedgecock et al. 1990)—suggest a related function for this HIF-1–independent VHL pathway. Interestingly, VHL-defective renal carcinoma cells demonstrate a variety of matrix-related abnormalities, including abnormal fibronectin assembly, defective formation of fibrillar adhesions, and changes in branching morphogenesis and migration (Ohh et al. 1998; Koochekpour et al. 1999; Davidowitz et al. 2001; Kamada et al. 2001; Esteban-Barragan et al. 2002). These abnormalities can be corrected by transfection of renal carcinoma cells with wild-type vhl, indicating that they are attributable, either directly or indirectly, to VHL loss of function. Furthermore, immunoprecipitation studies using renal carcinoma cell extracts have indicated that VHL binds to fibronectin (Ohh et al. 1998). Most tumor-associated VHL mutants, when transfected into VHL-defective renal carcinoma cells, are defective in both complementing HIF dysregulation and fibronectin binding (Kaelin 2002). However, mutations associated with type 2C (predisposition to pheochromocytoma only) VHL disease complement defective HIF regulation but bind fibronectin with lower affinity than wild-type VHL (Hoffman et al. 2001). Though the precise link to abnormal matrix assembly remains unclear, this has suggested a HIF-independent function of VHL. The present study supports the existence of a HIF-independent pathway connected with extracellular matrix function and suggests that this may be a highly conserved function of VHL that is potentially amenable to genetic analysis in model organisms.
Materials and Methods
Strains and culturing conditions.
Worms were studied as mixed-stage populations or as synchronized populations following brief exposure to sodium hypochlorite. Exposure to hypoxia (2% or 0.1% oxygen), DIP (200 μM), and DMOG (1 mM) was for 18 h (Epstein et al. 2001). RNA interference (RNAi) was performed by feeding worms Escherichia coli strain HT115(DE3) expressing double-stranded (ds) RNA on Nematode Growth Medium containing 1 mM isopropyl-β-D-thiogalactopyranoside (ITPG) and 50 μg/ml ampicillin for 72 h. Plasmids for ds RNA production were derivatives of the L4440 vector and were obtained from J. Ahringer (Cambridge, United Kingdom); ds RNA sequences are available on WormBase (http://www.wormbase.org). Wild-type worms were Bristol strain (N2); mutant strains were obtained from the Caenorhabditis Genetics Center (Table 6). Strains were maintained at room temperature except for the temperature-sensitive gon-1 worms (maintained at 18 °C). The double mutants hif-1; vhl-1, egl-9; hif-1, and dpy-18; hif-1 were constructed using either fog-2 or unc-51 to mark hif-1(+); the double mutant egl-9; vhl-1 was constructed using unc-42 to mark egl-9(+); PCR was used to confirm homozygosity.
Microarray screening.
Microarray comparisons of wild-type versus vhl-1 worms and hif-1 versus hif-1; vhl-1 worms were performed on independent samples of RNA (n = 1 and 3, respectively), using near full-genome C. elegans DNA microarrays (M. Jiang et al. 2001). Total RNA was extracted from mixed-stage populations of worm cultured under normoxic conditions using Tri-reagent (Sigma, Poole, Dorset, United Kingdom) and mRNA purified using oligo-dT beads (Qiagen, Crawley, West Sussex, United Kingdom). cDNA synthesis and microarray hybridization and scanning were performed as described previously (M. Jiang et al. 2001). Cy5-dUTP was used to label cDNA from wild-type and hif-1 worms and Cy3-dUTP was used to label cDNA from vhl-1 and hif-1; vhl-1 worms. The arrays were computer normalized by the default procedure in the Stanford Microarray Database (SMD); primary array data are available on the SMD (http://genome-www.stanford.edu/microarray) and are also shown in Tables S1 through S4. Fold change was calculated as the ratio of the means of Cy3-dUTP intensity to normalized Cy5-dUTP intensity (normalized to correct for signal differences between Cy3-dUTP and Cy5-dUTP intensities across the whole array) with median background intensities subtracted from both signal intensities to correct for the background (see SMD). Genes with background-corrected signal intensities below zero or with array spots that were flagged in the SMD as being unreliable were discarded as a preliminary quality control. For the hif-1 versus hif-1; vhl-1 microarray comparisons (n = 3) the log2 fold change was calculated as the mean of the three log2 transformed fold changes. To test for significant upregulation, the mean log2 fold change was compared with zero using a Student's t test. The genes were ranked by amplitude of fold upregulation and a subset of genes was selected for potential validation by RNase protection assays (see Tables 1 and 4) based on the following criteria: (a) t test, p < 0.10 (for the hif-1 versus hif-1; vhl-1 microarray comparisons, n = 3); (b) mean Cy3-dUTP and Cy5-dUTP background-corrected signal intensities exceeding 300 and 100 U, respectively (lower intensities than these were difficult to detect by RNase protection assay); and (c) high spot quality as judged by manual inspection. For the hif-1 versus hif-1; vhl-1 microarray comparisons (n = 3), genes (which had been filtered as described above) were considered to be differentially expressed if the mean fold change was greater than 2.0 (see Table 4).
RNase protection assays.
Assays were performed on total RNA from mixed-stage populations of worm cultured under normoxic conditions, unless otherwise indicated. Details of riboprobe templates are provided in Table 7; details of genes tested are shown in Tables 1 and 4. Quantification was performed using a phosphorimager (Molecular Dynamics, Sunnyvale, California, United States) and related to an internal control assay for the constitutively expressed F21C3.5 (protein with similarity to mouse prefoldin subunit 6). Where n ≥ 3, the log2 fold change was calculated from the mean of the log2 transformed fold changes and statistical significance was calculated by comparing the mean log2 fold change with zero using a Student's t test.
Table 7. Sequence and Length of Riboprobes.

a Note that the protected region of the egl-9 transcript does not overlap the sa307 deletion in the JT307 egl-9 strain
Computational analyses
(1) Identification of potential HBSs (see Table 3). Orthologs of C. elegans genes were identified in the C. briggsae genome assembly (cb25) as reciprocal best matches by BLASTN, initiated with the C. elegans gene coding sequence (a single ortholog of F56A4.2 could not be defined). Translation initiation sites (well-annotated surrogates for transcriptional start sites; none of these genes are annotated as having spliced 5′ UTRs) were inferred in both C. briggsae and C. elegans through alignment with the C. elegans coding sequence (WormBase, WS117). Sequences encompassing the 1,000 nucleotides upstream to 250 nucleotides downstream of the translation start sites for orthologous genes were aligned using DNA Block Aligner (Jareborg et al. 1999) with the following options: gap = 0.001 and blockopen = 0.005. Sequence alignments were searched with the HBS motif RCGTG (Camenisch et al. 2001), identifying cases where HBS-like motifs were conserved between both C. elegans and C. briggsae.
(2) Single-linkage analysis to determine spatial clusters of VHL-1–dependent, HIF-1–independent genes (see Figure 3). A maximum distance for the linking of two clusters was determined by ranking the distance between 10,000 randomly selected pairs of genes from the same chromosome (but sampled over all six nuclear chromosomes) and selecting as a threshold the first percentile of the distribution, 96,985 bp. Intergene distances were calculated from the closest point between the annotated coding sequence of each gene; genes on separate chromosomes were considered to have an infinite intergene distance. Simulations were performed using genes selected at random from genes that were represented on the microarray and that passed preliminary quality control criteria. All genomic coordinates were based on genomic assembly WS120 and the associated WormBase annotation of genes obtained from the University of California, Santa Cruz Genome Browser (http://genome.ucsc.edu/). The software used for simulations and clustering was implemented in Perl and is available on request.
Supporting Information
Primary microarray data can be viewed at http://genome-www.stanford.edu/microarray.
Primary microarray data for the vhl-1 (green, channel 1) versus wild-type (red, channel 2) comparison.
(6.5 MB XLS).
Primary microarray data for the three independent hif-1; vhl-1 (green, channel 1) versus hif-1 (red, channel 2) microarray comparisons. Continued in Tables S3 and S4.
(6.6 MB XLS).
Continuation of Table S2.
(6.6 MB XLS).
Continuation of Tables S2 and S3.
(6.6 MB XLS).
Accession Numbers
Primary array data have been deposited in ArrayExpress (http://www.ebi.ac.uk/arrayexpress/) under accession number E-SMDB-23.
The H. sapiens VHL gene discussed in this paper can be found in Online Mendelian Inheritance in Man (OMIM) under accession number 608537 (http://www.ncbi.nlm.nih.gov:80/entrez/dispomim.cgi?id=608537).
The C. elegans genes discussed in this paper (bli-4, C01B4.6, C01B4.7, C01B4.8, C01B4.9, cah-4, dpy-10, dpy-11, dpy-18, egl-9, F21C3.5, F22B5.4, F56A4.2, F56A4.9, F56A4.10, fmo-12, fog-2, gon-1, hif-1, let-268, mig-17, nhr-57, phy-2, phy-3, sqt-3, unc-4, unc-6, unc-42, unc-51, unc-52, vhl-1, Y45G12C.2, Y45G12C.9, and Y45G12C.12) can be found in the WormBase database by including the name of the gene at the end of the URL (e.g., for bli-4, http://wormbase.org/db/gene/gene?name=bli-4).
Table 4. Continued.
Acknowledgments
The authors would like to thank Richard Mott and James Lund for helpful discussions. This work was supported by the Wellcome Trust, the British Heart Foundation, and the Medical Research Council (United Kingdom); the Agency for Science, Technology and Research (Singapore); and the National Center for Research Resources (United States of America).
Abbreviations
- DIP
2,2′-dipyridyl
- DMOG
dimethyloxalylglycine
- HBS
HIF-1 binding site
- HIF
hypoxia-inducible factor
- RNase
ribonuclease
- SMD
Stanford Microarray Database
- VHL
von Hippel-Lindau
Conflicts of interest. The authors have declared that no conflicts of interest exist.
Author contributions. TB, KWL, ACRE, CWP, JH, and PJR conceived and designed the experiments. TB, KWL, ACRE, MJ, and DOR performed the experiments. TB, KWL, CWP, JMG, MST, JH, and PJR analyzed the data. MST designed and implemented the computational analyses. SKK, JH, and PJR contributed reagents/materials/analysis tools. TB and PJR wrote the paper.
Academic Editor: Christopher Kemp, Fred Hutchinson Cancer Research Center
Citation: Bishop T, Lau KW, Epstein ACR, Kim SK, Jiang M, et al. (2004) Genetic analysis of pathways regulated by the von Hippel-Lindau tumor suppressor in Caenorhabditis elegans. PLoS Biol 2(10): e289.
References
- Blelloch R, Kimble J. Control of organ shape by a secreted metalloprotease in the nematode Caenorhabditis elegans . Nature. 1999;399:586–590. doi: 10.1038/21196. [DOI] [PubMed] [Google Scholar]
- Blumenthal T, Gleason KS. Caenorhabditis elegans operons: Form and function. Nat Rev Genet. 2003;4:112–120. doi: 10.1038/nrg995. [DOI] [PubMed] [Google Scholar]
- Blumenthal T, Evans D, Link CD, Guffanti A, Lawson D, et al. A global analysis of Caenorhabditis elegans operons. Nature. 2002;417:851–854. doi: 10.1038/nature00831. [DOI] [PubMed] [Google Scholar]
- Camenisch G, Stroka DM, Gassmann M, Wenger RH. Attenuation of HIF-1 DNA-binding activity limits hypoxia-inducible endothelin-1 expression. Pflugers Arch. 2001;443:240–249. doi: 10.1007/s004240100679. [DOI] [PubMed] [Google Scholar]
- Davidowitz EJ, Schoenfeld AR, Burk RD. VHL induces renal cell differentiation and growth arrest through integration of cell-cell and cell-extracellular matrix signaling. Mol Cell Biol. 2001;21:865–874. doi: 10.1128/MCB.21.3.865-874.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dillon WR, Goldstein M. New York: John Wiley & Sons; 1984. Multivariate analysis: Methods and applications; 608 pp. [Google Scholar]
- Epstein AC, Gleadle JM, McNeill LA, Hewitson KS, O'Rourke J, et al. C. elegans EGL-9 and mammalian homologs define a family of dioxygenases that regulate HIF by prolyl hydroxylation. Cell. 2001;107:43–54. doi: 10.1016/s0092-8674(01)00507-4. [DOI] [PubMed] [Google Scholar]
- Esteban-Barragan MA, Avila P, Alvarez-Tejado M, Gutierrez MD, Garcia-Pardo A, et al. Role of the von Hippel-Lindau tumor suppressor gene in the formation of beta1-integrin fibrillar adhesions. Cancer Res. 2002;62:2929–2936. [PubMed] [Google Scholar]
- Friedman L, Higgin JJ, Moulder G, Barstead R, Raines RT, et al. Prolyl 4-hydroxylase is required for viability and morphogenesis in Caenorhabditis elegans . Proc Natl Acad Sci U S A. 2000;97:4736–4741. doi: 10.1073/pnas.97.9.4736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedgecock EM, Culotti JG, Hall DH. The unc-5, unc-6 and unc-40 genes guide circumferential migrations of pioneer axons and mesodermal cells on the epidermis in C. elegans . Neuron. 1990;4:61–85. doi: 10.1016/0896-6273(90)90444-k. [DOI] [PubMed] [Google Scholar]
- Hoffman MA, Ohh M, Yang H, Klco JM, Ivan M, et al. von Hippel-Lindau protein mutants linked to type 2C VHL disease preserve the ability to downregulate HIF. Hum Mol Genet. 2001;10:1019–1027. doi: 10.1093/hmg/10.10.1019. [DOI] [PubMed] [Google Scholar]
- Ivan M, Kondo K, Yang H, Kim W, Valiando J, et al. HIFalpha targeted for VHL-mediated destruction by proline hydroxylation: Implications for O2 sensing. Science. 2001;292:464–468. doi: 10.1126/science.1059817. [DOI] [PubMed] [Google Scholar]
- Ivanov SV, Kuzmin I, Wei MH, Pack S, Geil L, et al. Down-regulation of transmembrane carbonic anhydrases in renal cell carcinoma cell lines by wild-type von Hippel-Lindau transgenes. Proc Natl Acad Sci U S A. 1998;95:12596–12601. doi: 10.1073/pnas.95.21.12596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaakkola P, Mole DR, Tian YM, Wilson MI, Gielbert J, et al. Targeting of HIF-alpha to the von Hippel-Lindau ubiquitylation complex by O2-regulated prolyl hydroxylation. Science. 2001;292:468–472. doi: 10.1126/science.1059796. [DOI] [PubMed] [Google Scholar]
- Jareborg N, Birney E, Durbin R. Comparative analysis of noncoding regions of 77 orthologous mouse and human gene pairs. Genome Res. 1999;9:815–824. doi: 10.1101/gr.9.9.815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H, Guo R, Powell-Coffman JA. The Caenorhabditis elegans hif-1 gene encodes a bHLH-PAS protein that is required for adaptation to hypoxia. Proc Natl Acad Sci U S A. 2001;98:7916–7921. doi: 10.1073/pnas.141234698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang M, Ryu J, Kiraly M, Duke K, Reinke V, et al. Genome-wide analysis of developmental and sex-regulated gene expression profiles in Caenorhabditis elegans . Proc Natl Acad Sci U S A. 2001;98:218–223. doi: 10.1073/pnas.011520898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y, Zhang W, Kondo K, Klco JM, St Martin TB, et al. Gene expression profiling in a renal cell carcinoma cell line: Dissecting VHL and hypoxia-dependent pathways. Mol Cancer Res. 2003;1:453–462. [PubMed] [Google Scholar]
- Kaelin WG Jr. Molecular basis of the VHL hereditary cancer syndrome. Nat Rev Cancer. 2002;2:673–682. doi: 10.1038/nrc885. [DOI] [PubMed] [Google Scholar]
- Kamada M, Suzuki K, Kato Y, Okuda H, Shuin T. von Hippel-Lindau protein promotes the assembly of actin and vinculin and inhibits cell motility. Cancer Res. 2001;61:4184–4189. [PubMed] [Google Scholar]
- Kondo K, Klco J, Nakamura E, Lechpammer M, Kaelin WG. Inhibition of HIF is necessary for tumor suppression by the von Hippel-Lindau protein. Cancer Cell. 2002;1:237–246. doi: 10.1016/s1535-6108(02)00043-0. [DOI] [PubMed] [Google Scholar]
- Kondo K, Kim WY, Lechpammer M, Kaelin WG. Inhibition of HIF2alpha is sufficient to suppress pVHL-defective tumor growth. PLoS Biol. 2003;1:e83. doi: 10.1371/journal.pbio.0000083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koochekpour S, Jeffers M, Wang PH, Gong C, Taylor GA, et al. The von Hippel-Lindau tumor suppressor gene inhibits hepatocyte growth factor/scatter factor-induced invasion and branching morphogenesis in renal carcinoma cells. Mol Cell Biol. 1999;19:5902–5912. doi: 10.1128/mcb.19.9.5902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latif F, Tory K, Gnarra J, Yao M, Duh FM, et al. Identification of the von Hippel-Lindau disease tumor suppressor gene. Science. 1993;260:1317–1320. doi: 10.1126/science.8493574. [DOI] [PubMed] [Google Scholar]
- Maranchie JK, Vasselli JR, Riss J, Bonifacino JS, Linehan WM, et al. The contribution of VHL substrate binding and HIF1-alpha to the phenotype of VHL loss in renal cell carcinoma. Cancer Cell. 2002;1:247–255. doi: 10.1016/s1535-6108(02)00044-2. [DOI] [PubMed] [Google Scholar]
- Maxwell PH, Wiesener MS, Chang GW, Clifford SC, Vaux EC, et al. The tumour suppressor protein VHL targets hypoxia-inducible factors for oxygen-dependent proteolysis. Nature. 1999;399:271–275. doi: 10.1038/20459. [DOI] [PubMed] [Google Scholar]
- Nishiwaki K, Hisamoto N, Matsumoto K. A metalloprotease disintegrin that controls cell migration in Caenorhabditis elegans . Science. 2000;288:2205–2208. doi: 10.1126/science.288.5474.2205. [DOI] [PubMed] [Google Scholar]
- Norman KR, Moerman DG. The let-268 locus of Caenorhabditis elegans encodes a procollagen lysyl hydroxylase that is essential for type IV collagen secretion. Dev Biol. 2000;227:690–705. doi: 10.1006/dbio.2000.9897. [DOI] [PubMed] [Google Scholar]
- Ohh M, Yauch RL, Lonergan KM, Whaley JM, Stemmer-Rachamimov AO, et al. The von Hippel-Lindau tumor suppressor protein is required for proper assembly of an extracellular fibronectin matrix. Mol Cell. 1998;1:959–968. doi: 10.1016/s1097-2765(00)80096-9. [DOI] [PubMed] [Google Scholar]
- Ohh M, Park CW, Ivan M, Hoffman MA, Kim TY, et al. Ubiquitination of hypoxia-inducible factor requires direct binding to the beta-domain of the von Hippel-Lindau protein. Nat Cell Biol. 2000;2:423–427. doi: 10.1038/35017054. [DOI] [PubMed] [Google Scholar]
- Riihimaa P, Nissi R, Page AP, Winter AD, Keskiaho K, et al. Egg shell collagen formation in Caenorhabditis elegans involves a novel prolyl 4-hydroxylase expressed in spermatheca and embryos and possessing many unique properties. J Biol Chem. 2002;277:18238–18243. doi: 10.1074/jbc.M200895200. [DOI] [PubMed] [Google Scholar]
- Roy PJ, Stuart JM, Lund J, Kim SK. Chromosomal clustering of muscle-expressed genes in Caenorhabditis elegans . Nature. 2002;418:975–979. doi: 10.1038/nature01012. [DOI] [PubMed] [Google Scholar]
- Sneath PH. The application of computers to taxonomy. J Gen Microbiol. 1957;17:201–226. doi: 10.1099/00221287-17-1-201. [DOI] [PubMed] [Google Scholar]
- Takahashi Y, Takahashi S, Shiga Y, Yoshimi T, Miura T. Hypoxic induction of prolyl 4-hydroxylase alpha (I) in cultured cells. J Biol Chem. 2000;275:14139–14146. doi: 10.1074/jbc.275.19.14139. [DOI] [PubMed] [Google Scholar]
- Thein MC, McCormack G, Winter AD, Johnstone IL, Shoemaker CB, et al. Caenorhabditis elegans exoskeleton collagen COL-19: An adult-specific marker for collagen modification and assembly, and the analysis of organismal morphology. Dev Dyn. 2003;226:523–539. doi: 10.1002/dvdy.10259. [DOI] [PubMed] [Google Scholar]
- Wykoff CC, Pugh CW, Maxwell PH, Harris AL, Ratcliffe PJ. Identification of novel hypoxia dependent and independent target genes of the von Hippel-Lindau (VHL) tumour suppressor by mRNA differential expression profiling. Oncogene. 2000;19:6297–6305. doi: 10.1038/sj.onc.1204012. [DOI] [PubMed] [Google Scholar]
- Zatyka M, da Silva NF, Clifford SC, Morris MR, Wiesener MS, et al. Identification of cyclin D1 and other novel targets for the von Hippel-Lindau tumor suppressor gene by expression array analysis and investigation of cyclin D1 genotype as a modifier in von Hippel-Lindau disease. Cancer Res. 2002;62:3803–3811. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Primary microarray data for the vhl-1 (green, channel 1) versus wild-type (red, channel 2) comparison.
(6.5 MB XLS).
Primary microarray data for the three independent hif-1; vhl-1 (green, channel 1) versus hif-1 (red, channel 2) microarray comparisons. Continued in Tables S3 and S4.
(6.6 MB XLS).
Continuation of Table S2.
(6.6 MB XLS).
Continuation of Tables S2 and S3.
(6.6 MB XLS).