Abstract
Long noncoding RNAs (lncRNAs) have emerged as a class of pivotal regulators of gene expression. Recent studies have shown that lncRNAs contribute to the initiation, maintenance, and development of acute myeloid leukemia (AML). In this review, we summarize the current knowledge of the lncRNAs that play critical roles in AML. We first briefly describe the characteristics of lncRNAs, and then focus on their regulatory roles in AML, including the modulation of differentiation, proliferation, cell cycle, and apoptosis. We further emphasize the action of lncRNAs during leukemogenesis by describing how they interact with RNA, protein and chromatin DNA to exert their functions. We also highlight an urgent need to investigate the mechanisms by which lncRNAs contribute to the pathogenesis of AML. Finally, we discuss the prognostic value of lncRNAs in AML patients.
Keywords: Long noncoding RNAs, Acute myeloid leukemia, Myelopoiesis
Background
Acute myeloid leukemia (AML) is a group of hematopoietic malignancies with various genetic abnormalities, including chromosomal translocations and/or somatic mutations, which are mainly responsible for the abnormal proliferation, differentiation or survival of myeloid progenitors [1, 2]. Extensive studies have established the regulatory roles of protein-coding genes in the initiation, maintenance, and development of AML, which constitutes our main knowledge of the pathogenesis of AML.
Recently, long noncoding RNAs (lncRNAs) have emerged as a novel class of pivotal regulators of gene expression and has received increasing attention in the field of AML. LncRNAs are operationally defined as RNA larger than 200 base pairs that appear to lack coding potential. They participate in various cellular processes, like inflammatory response [3], neuronal activity [4] and erythropoiesis [5]. Mechanistically, lncRNAs could promote the strength of specific enhancer-promoter looping and thus contribute to gene activation [6–8], regulate protein modifications and activities, sequester microRNAs, and serve as precursors of small RNAs [9–11]. Based on the genomic locations where lncRNAs are transcribed, they can be classified into the following groups: (1) sense, which overlap with at least part of another gene in the same strand; (2) antisense, which overlap with at least part of another gene on the opposite strand; (3) intronic, which originate from the intron of another gene; (4) intergenic, which does not overlap with any gene. In addition to the above four classes, it is possible to characterize an additional fifth category: (5) chimeric, which are the fusion products due to chromosomal rearrangements, based on a study showing the existence of fusion transcripts between protein-coding genes and lncRNAs in AML [12]. Since only two known protein-lncRNA fusions have been identified in AML and their function has not been elucidated [12], the further investigation would be required. In this review, we summarize the recent progress in the knowledge of lncRNAs in AML, including their biological functions, the mechanisms behind their actions, the upstream regulation and the prognostic values in the clinic.
The roles of lncRNAs in myeloid leukemia
The latest studies have demonstrated that lncRNAs contribute to many critical signaling pathways in AML development and therapy. We summarize the reported AML-related lncRNAs and their roles in Table 1.
Table 1.
LncRNAs in AML
| LncRNAs | Classification | Function | Target genes | Reference |
|---|---|---|---|---|
| RUNXOR | Sense | Be involved in chromosomal translocation | RUNX1 | [13] |
| HOTAIRM1 | Antisense | Regulate myeloid differentiation and cell cycle enhance the autophagy pathway regulates chromatin state and architecture | HOXA1, HOXA4, CD11b and CD18 miR-20a/106b and miR-125b | [14–17] |
| HOXA-AS2 | Antisense | Act as an apoptosis repressor | Unknown | [18] |
| PU.1-AS | Antisense | Inhibit the translation of PU.1 | PU.1 | [19] |
| WT1-AS | Antisense | Control WT1 expression | WT1 | [20] |
| EGO | Intronic | Regulate MBP and EDN expression | MBP and EDN | [22] |
| IRAIN | Intronic | Be engaged in long-range intra chromosomal interactions | IGF1R | [23] |
| BGL3 | Intergenic | Sensitize leukemic cells to undergo apoptosis | miR-17, miR-93, miR-20a, miR-20b, miR-106a and miR-106b | [24] |
| CCAT1 | Intergenic | Repress monocytic differentiation and promote cell growth | miR-155 | [25] |
| CCDC26 | Intergenic | Control the growth of AML cells | c-Kit | [26] |
| HOTAIR | Intergenic | Induce cell growth and inhibit apoptosis | miR-193a and c-Kit | [27] |
| NEAT1 | Intergenic | Regulate ATRA-induced myeloid differentiation | Unknown in AML | [28] |
| PVT1 | Intergenic | Regulate proliferation of promyelocytes | MYC | [21] |
| UCA1 | Intergenic | Sustain proliferation of AML cells | p27kip1 | [29] |
| PVT1-NSMCE2 | Fusion | Unknown | Unknown | [12] |
| BF104016-NSMCE2 | Fusion | Unknown | Unknown | [12] |
LncRNAs exert important roles in myeloid differentiation and can respond to differentiation induction therapy. At present, two lncRNAs, HOTAIRM1 and NEAT1 are known to regulate the differentiation of AML cells. HOTAIRM1 is a myeloid-specific long non-coding transcript, which is transcribed from the locus between HOXA1 and HOXA2 genes. HOTAIRM1 regulates myeloid differentiation genes such as CD11b and CD18, and its knockdown impairs all-trans retinoic acid (ATRA)-induced granulocytic differentiation [14]. This work could serve as a paradigm for exploring lncRNAs in AML, from the discovery of the lncRNA candidate to the investigation of the biological function. This work also has an interesting finding—HOTAIRM1 is derived from HOXA clusters, and in turn regulates the nearby genes in HOXA cluster. Whether this regulation is direct or indirect awaits further investigation. Another example is NEAT1, a widespread and abundant long noncoding RNA. Although myeloid differentiation is usually considered to be dominantly controlled by highly myeloid-specific factors, NEAT1 has been reported to be responsive to ATRA and be indispensable for ATRA-mediated myeloid differentiation [28]. This observation indicates that myeloid differentiation also requires commonly expressed long noncoding transcripts. However, since both studies of HOTAIRM1 and NEAT1 mentioned above were conducted in myeloid leukemia cells, it has yet to be determined whether HOTAIRM1 and NEAT1 are required for normal myelopoiesis. Further in vivo studies are needed to evaluate their roles in normal hematopoiesis and the development of AML.
LncRNAs also exert effects on proliferation, cell cycle and apoptosis in AML cells. Such lncRNAs tend to be expressed more widely than those myeloid-specific transcripts that regulate differentiation. A typical example is lncRNA PVT1, which can promote the proliferation of AML cells [21]. The oncogenic activity of PVT1 is closely related to MYC, whose overexpression can lead to hyper proliferation of cancer cells. A gain-of-function of both the MYC gene and the PVT1 lncRNA due to the amplification of 8q24.21 is observed in about 10% of AML patients [30]. On the one hand, PVT1 protects MYC from degradation by direct physical interaction [30]. On the other hand, PVT1 acts as a microRNA precursor for indirect regulation of MYC [31]. The PVT1 locus can produce six annotated oncogenic microRNAs [31, 32], and one of these microRNAs, hsa-miR-1204, which is derived from the exon 1b, has been reported to be able to enhance the expression level of MYC [31]. From the example of PVT1, we should note that lncRNAs may have multifaceted roles and can function through multiple ways. Also, the co-amplification of PVT1 and MYC informs us that it might be interesting to see the association or causal relationship of copy number of lncRNAs with leukemogensis. In addition to PVT1, lncRNA UCA1 has also been reported to have the capability to modulate the proliferation of AML cells [29]. UCA1 silencing by short-hairpin RNA transduction results in a significant reduction of cell proliferation, with an increase in the G1 phase and a decrease in the S phase. Another example is lncRNA CCAT1, which can coordinate the proliferation and differentiation of AML cells [25]. Overexpression and knockdown experiments demonstrate that CCAT1 inhibits the PMA-induced monocytic differentiation as well as promotes the proliferation of AML-derived HL60 cells [25]. As to apoptosis of AML cells, Xing et al. [27] found that HOTAIR knockdown inhibits cell growth and colony formation, and also induces the apoptosis of AML cells [27]. Of note, the oncogenic properties of these lncRNAs are not limited to leukemia, but can also be observed in solid tumors. For example, PVT1 can promote the proliferation of hepatocellular carcinoma and non-small cell lung cancer cells [33, 34]. Similarly, CCAT1 can promote the proliferation and invasion of colon cancer cells, gallbladder cancer cells and hepatocellular carcinoma cells [25, 35, 36]. These observations indicate that such lncRNAs usually share similar functions in leukemia and other types of malignancies. Further experiments are required to investigate the underlying mechanism by which these lncRNAs control the cellular phenotypes—proliferation, cell cycle and apoptosis in AML cells.
Recently, the discovery of a novel class of lncRNAs piques the curiosity of scientists and potentially ignites their efforts to characterize them: fusion transcripts between coding genes and lncRNAs. In an AML patient sample and an AML-derived HL-60 cell line, it is verified that NSMCE2 rearrangement gives rise to two novel chimeric genes, PVT1-NSMCE2 and CCDC26-NSMCE2 [12]. Although their identities have been revealed, their involvement in leukemogenesis is elusive to date. It is likely that PVT1-NSMCE2 and CCDC26-NSMCE2 may contribute to the leukemogenesis through the oncogenic activity of PVT1 and CCDC26 [26]. At present, our understanding of the fusion transcripts between lncRNAs and protein-coding genes is still in its infancy. Further experiments are required to determine the causal relationship between the protein-lncRNA-fusion transcripts and leukemia as well as the mechanism behind their behaviors.
Mechanisms of lncRNA action in acute myeloid leukemia
LncRNAs exert their functions through multiple ways (Fig. 1).
Fig. 1.
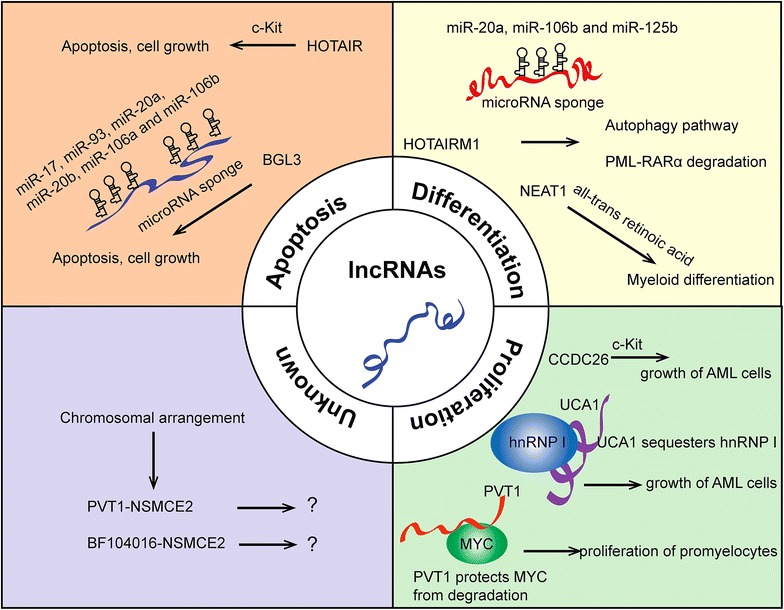
LncRNAs in acute myeloid leukemia. LncRNAs influence apopotosis, differentiation and proliferation of acute myeloid leukemia cells. Selected examples of AML lncRNAs and their molecular partners as well as the associated cellular phenotpyes are shown
First, lncRNAs can interact with other RNA molecules, e.g. microRNAs and mRNAs. The lncRNAs that can act by competing with endogenous RNA to sequester microRNAs include BGL3, CCAT-1, HOTAIR and HOTAIRM1. BGL3 functions as a competitive endogenous RNA for binding miR-17, miR-93, miR-20a, miR-20b, miR-106a and miR-106b to cross-regulate PTEN expression [24]. CCAT-1 exerts its oncogenic activity by sequestering tumor-suppressive miR-155, an inducer of apoptosis and cell differentiation, which is down-regulated in AML [25, 37]. HOTAIR competitively binds to and sequesters miR-193a, thus modulating the expression of c-KIT in AML cells [27]. A recent study showed that HOTAIRM1 regulates the ATRA-induced degradation of PML-RARα by acting as microRNA sponge sequestering miR-20a, miR-106b and miR-125b to target autophagy-associated genes [16]. The lncRNAs can also bind to the protein-coding transcripts and induce the translation inhibition. One such lncRNA is PU.1-AS. It is transcribed from the opposite strand of PU.1, a master transcription factor controlling myeloid differentiation. It has been found that PU.1-AS negatively regulates the expression of PU.1 by stalling the translation of PU.1 mRNA [19]. Furthermore, the preliminary data shows that PU.1-AS interferes with the binding of eEF1A to PU.1 mRNA, impairing the elongating complexes [19], although further studies are required to determine the precise mechanism.
Second, lncRNAs can bind protein partners to form a complex. For instance, a proposed mechanism for UCA1 action in proliferation regulation is to sequester hnRNP I, which is a positive translation regulator of p27 protein. Consistent with this hypothesis, it has been shown that UCA1 has a moderate binding capacity to hnRNP I and that UCA1 knock-down leads to an increased expression of p27 protein in AML cells [29]. Further evidence is required to support the hypothesis that UCA1 functions by sequestering hnRNP I, since the investigation of the biological effects and molecular mechanism underlying the interaction between lncRNAs and proteins is more difficult than the identification of factors interacting with the lncRNA of interest. A possible effect of the lncRNAs’ interaction with proteins is involved in regulating the stability of protein partners, as exemplified by PVT1. In both breast cancer and AML cells, it has been reported that PVT1 protects MYC from phosphorylation by the direct interaction with MYC, stabilizing and enhancing MYC [30].
Currently, orchestrating the chromosomal looping is a seldom-observed yet critical aspect regarding the action of lncRNAs in AML. Two lncRNAs have been reported to mediate the long-range regulation. Interestingly, both lncRNAs are transcribed from the loci that produce critical protein regulators. LncRNA RUNXOR is derived from the locus of transcription factor RUNX1 (AML1). It is 216 kb long and covers the entire locus of RUNX1, an important transcription factor in AML [13]. The 3′ UTR of RUNXOR is capable of binding to the promoter and enhancer of RUNX1, forming an intra-chromosomal looping modulating the expression of RUNX1. Also, RUNXOR participates in the long-range interaction between the chromosomes, and thereby might be involved in chromosomal translocations and leukemia development [13]. Another example is IRAIN, an imprinted lncRNA transcribed from the intron of insulin-like growth factor type I receptor (IGF1R), an oncogene promoting cell growth in AML cells. IRAIN interacts with chromatin DNA and participates in scaffolding the long-distance DNA regions to form an intra chromosomal loop involving the IGF1R promoter and the intronic enhancer [23].
However, apart from the cases mentioned above, the mechanisms of lncRNA action in AML are still largely unknown. Further studies are required to investigate the interactome of lncRNAs, and to determine the causal relationship between the direct physical interaction and the resultant phenotypic changes. For example, although HOTAIRM1 and NEAT1 are found to be required for ATRA-induced myeloid differentiation in AML cells, the mechanism of how they exactly exert their functions is poorly known.
Regulation and dysregulation of lncRNAs in myelopoiesis
Emerging studies have indicated that lncRNAs specifically expressed during myelopoiesis are under the direct control of hematopoietic transcription factors. C/EBPα (CCAAT/enhancer-binding protein-α) is one of key regulators in myelopoiesis. A genome-wide study on investigating C/EBPα-regulated lncRNAs has revealed that 930 lncRNAs, including 600 up-regulated and 330 down-regulated, are regulated by C/EBPα [38]. At the individual gene level, a more in-depth study has shed light on the regulatory role of C/EBPα in the control of lncRNA UCA1 [29]. About 10% of AML cases bear CEBPA mutations [39], which leads to overexpression of C/EBPα-p30. Hughes et al. [29] and his colleagues identified that UCA1 is a novel target of both C/EBPα isoforms, i.e. C/EBPα-p42 and C/EBPα-p30, and the short isoform C/EBPα-p30 induces the expression of UCA1, leading to the abnormal up-regulation of UCA1 in CEBPA-mutated AML patients [29]. The work suggests the complexity of the transcriptional modulation of lncRNAs, as they could be controlled in a distinct regulatory pattern by different isoforms produced from the same gene.
PU.1 is another master regulator in myeloid differentiation. Most recently, we have found that the myeloid differentiation lncRNA HOTAIRM1 is a direct target of PU.1 [40]. The up-regulation of HOTAIRM1 during granulopoiesis depends on PU.1. Furthermore, low HOTAIRM1 expression is observed in APL cells, which is attributed to the reduced PU.1 expression, rather than the direct binding and repression by PML-RARα, the unique oncofusion protein in APL cells. This work identifies HOTAIRM1 as a novel target of PU.1, suggests the role of HOTAIRM1 in PU.1-mediated regulation network during myeloid differentiation and elucidates the mechanism by which HOTAIRM1 is deregulated in APL cells.
In addition to the direct regulation by transcription factors, lncRNA expression is closely associated with recurrent mutations in AML [39], indicating an indirect way of regulation. For example, lncRNA HOXB-AS3, MEIS1-AS2, PVT1, and CCD26 are up-regulated in cytogenetically normal AML patients with mutated NPM1. WT1-AS is associated with FLT3-ITD mutations. IDH1R132 mutated patient samples have up-regulated DLEU2 as well as down-regulated RP11-147N17.1. RUNX1-mutated patients have up-regulated vault RNA 1-1 (VTRNA1-1) [39]. Whether these observations merely represent a co-expression pattern or these lncRNAs indeed have roles in AML with the associated mutations remains to be explored.
Presently, mechanisms behind the dysregulation of lncRNAs in AML are far less clear than their biological functions. For example, previous studies have shown the involvement of PU.1-AS and EGO in myelopoiesis [19, 22], but whether they are dysregulated in AML is unknown, and how they are controlled during hematopoiesis and leukemogenesis is unexplored. Thus, more attention should be paid into the mechanisms behind the abnormal expression of lncRNAs in AML.
Prognostic value and therapeutic promise of lncRNAs in acute myeloid leukemia
LncRNAs are associated with AML clinical features and outcomes. A recent study has shown that lncRNAs can be used to predict treatment response and outcome in older patients with cytogenetically normal AML. Garzon et al. have investigated the associations of lncRNA expression with clinical characteristics, recurrent mutations, and outcome in 148 cytogenetically normal older (age >60 years) AML patients and built a prognostic score based on the expression values of 48 lncRNAs that can been used to for outcome prediction [39]. Future studies are required to investigate whether these lncRNAs are functional in the AML development and whether they are regulated directly or indirectly by biologic changes influenced by specific mutations. Moreover, AML patients expressing a higher level of HOTAIR are associated with a worse clinical outcome in comparison with those with lower expression of HOTAIR [27]. Based on 215 intermediate-risk AML patients, Díaz-Beyá et al. [41] have reported that high HOTAIRM1 expression is independently associated with worse prognosis: a shorter overall survival and a higher cumulative incidence of relapse [41]. Besides, a higher HOTAIRM1 expression level is also associated with worse clinical outcome in AML patients with mutated NPM1 [41]. Overall, only a few published reports of lncRNAs’ prognostic value in AML are available currently, so more work is needed to explore the association between lncRNA and clinical characteristics, mutations and outcome.
LncRNAs are promising candidates for cancer therapy, especially considering some of them are tissue-specific drivers of cancer. Preclinical researches have implicated the efficacy of lncRNA-targeted therapeutics in certain types of cancer. In the MMTV-PyMT breast cancer mouse model, Malat1 exerts oncogenic activity by enhancing cell proliferation and tumor metastasis, and serves as a potential target for treatment [42]. Perturbation of Malat1 by RNA depletion induces differentiation of mammary tumors, increases cell adhesion and decreases metastasis [42]. However, to the best of our knowledge, there is still no therapeutic examples targeting lncRNAs in AML to date. Progress of developing lncRNA-targeted therapeutics is impeded by the limited understanding of their roles in AML and the molecular mechanisms by which they exert their functions in AML.
Conclusions and perspectives
It becomes increasingly clear that lncRNAs play critical regulatory roles in AML. However, there are as yet unidentified lncRNAs and undiscovered machineries involved in AML. The road is still long before we fully understand their disease relevance and biological significance in AML. Genome-wide studies, in parallel with careful and in-depth investigations of individual lncRNAs, will greatly advance our understanding of how lncRNAs are involved in the initiation, maintenance and development of AML. Furthermore, the nature of the interactions between lncRNAs and other molecules, to a large extent, remains a mystery in AML. Resolving such issues requires the implementation of methodologies such as chromatin isolation by RNA purification (ChIRP) [43], capture hybridization analysis of RNA targets (CHART) [44] and RNA immune precipitation (RIP) to identify the binding chromatin DNA or binding proteins of lncRNAs. Finally, an imperative aspect of research is the inquiry at the mechanism under the dysregulation of lncRNAs in AML. In particular, for those functionally characterized lncRNAs that have regulatory roles in AML, it is really worthwhile to investigate how they are modulated in physiological conditions and whether they are dysregulated in the pathogenesis of AML. Understanding the mechanisms underlying lncRNA’ actions and dysregulation will ultimately pave the way to completely understand the pathology and to better treat AML patients.
Authors’ contributions
SYW drafted the manuscript and KKW revised the manuscript. Both authors read and approved the final manuscript.
Acknowledgements
None.
Competing interests
Both authors declare that they have no competing interests.
Availability of data and materials
Data sharing not applicable to this article as no datasets were generated or analyzed during the current study.
Funding
This work was supported in part by the National Key Research and Development Program (2016YFC0902800), National Natural Science Foundation Grants of China (81530003, 81270625 and 91440114), Shanghai Leading Talent Projects (2015008) and the Academic Leader Program of Shanghai Science and Technology Committee (2015137).
Abbreviations
- lncRNA
long non-coding RNA
- AML
acute myeloid leukemia
- APL
acute promyelocytic leukemia
- ATRA
all-trans retinoic acid
Contributor Information
Shuyong Wei, Email: weishuyong@hotmail.com.
Kankan Wang, Phone: 86-21-64370045, Email: kankanwang@shsmu.edu.cn.
References
- 1.Shih AH, Abdel-Wahab O, Patel JP, Levine RL. The role of mutations in epigenetic regulators in myeloid malignancies. Nat Rev Cancer. 2012;12:599–612. doi: 10.1038/nrc3343. [DOI] [PubMed] [Google Scholar]
- 2.Meyer SC, Levine RL. Translational implications of somatic genomics in acute myeloid leukaemia. Lancet Oncol. 2014;15:e382–e394. doi: 10.1016/S1470-2045(14)70008-7. [DOI] [PubMed] [Google Scholar]
- 3.Ilott NE, Heward JA, Roux B, Tsitsiou E, Fenwick PS, Lenzi L, Goodhead I, Hertz-Fowler C, Heger A, Hall N, Donnelly LE. Long non-coding RNAs and enhancer RNAs regulate the lipopolysaccharide-induced inflammatory response in human monocytes. Nat Commun. 2014;5:3979. doi: 10.1038/ncomms5536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kim T-K, Hemberg M, Gray JM, Costa AM, Bear DM, Wu J, Harmin DA, Laptewicz M, Barbara-Haley K, Kuersten S, Markenscoff-Papadimitriou E, Kuhl D, Bito H, et al. Widespread transcription at neuronal activity-regulated enhancers. Nature. 2010;465:182–187. doi: 10.1038/nature09033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cells RED, Alvarez-dominguez JR, Hu W, Yuan B, Shi J, Park SS, Gromatzky AA, Van Oudenaarden A, Lodish HF. Regular article global discovery of erythroid long noncoding RNAs reveals novel regulators of red cell maturation. Blood. 2014;123:570–581. doi: 10.1182/blood-2013-10-530683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lam MTY, Cho H, Lesch HP, Gosselin D, Heinz S, Tanaka-Oishi Y, Benner C, Kaikkonen MU, Kim AS, Kosaka M, Lee CY, Watt A, Grossman TR, et al. Rev-Erbs repress macrophage gene expression by inhibiting enhancer-directed transcription. Nature. 2013;498:511–515. doi: 10.1038/nature12209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hsieh C-L, Fei T, Chen Y, Li T, Gao Y, Wang X, Sun T, Sweeney CJ, Lee G-SM, Chen S, Balk SP, Liu XS, Brown M, et al. Enhancer RNAs participate in androgen receptor-driven looping that selectively enhances gene activation. Proc Natl Acad Sci USA. 2014;111:7319–7324. doi: 10.1073/pnas.1324151111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li W, Notani D, Ma Q, Tanasa B, Nunez E, Chen AY, Merkurjev D, Zhang J, Ohgi K, Song X, Oh S, Kim H-S, Glass CK, et al. Functional roles of enhancer RNAs for oestrogen-dependent transcriptional activation. Nature. 2013;498:516–520. doi: 10.1038/nature12210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huang M, Chen W, Qi F, Xia R, Sun M, Xu T, Yin L, Zhang E, De W, Shu Y. Long non-coding RNA ANRIL is upregulated in hepatocellular carcinoma and regulates cell apoptosis by epigenetic silencing of KLF2. J Hematol Oncol. 2015;8:50. doi: 10.1186/s13045-015-0146-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang P, Xue Y, Han Y, Lin L, Wu C, Xu S, Jiang Z, Xu J, Liu Q, Cao X. The STAT3-binding long noncoding RNA lnc-DC controls human dendritic cell differentiation. Science. 2014;344:310–313. doi: 10.1126/science.1251456. [DOI] [PubMed] [Google Scholar]
- 11.Dhir A, Dhir S, Proudfoot NJ, Jopling CL. Microprocessor mediates transcriptional termination in genes encoding long noncoding microRNAs. Nat Struct Mol Biol. 2015;22:319–327. doi: 10.1038/nsmb.2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chinen Y, Sakamoto N, Nagoshi H, Taki T, Maegawa S, Tatekawa S, Tsukamoto T, Mizutani S, Shimura Y, Yamamoto-Sugitani M, Kobayashi T, Matsumoto Y, Horiike S, et al. 8q24 amplified segments involve novel fusion genes between NSMCE2 and long noncoding RNAs in acute myelogenous leukemia. J Hematol Oncol. 2014;7:68. doi: 10.1186/s13045-014-0068-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang H, Li W, Guo R, Sun J, Cui J, Wang G, Hoffman AR, Hu J-F. An intragenic long noncoding RNA interacts epigenetically with the RUNX1 promoter and enhancer chromatin DNA in hematopoietic malignancies. Int J Cancer. 2014;135(12):2783–2794. doi: 10.1002/ijc.28922. [DOI] [PubMed] [Google Scholar]
- 14.Zhang X, Lian Z, Padden C, Gerstein MB, Rozowsky J, Snyder M, Gingeras TR, Kapranov P, Weissman SM, Newburger PE. A myelopoiesis-associated regulatory intergenic noncoding RNA transcript within the human HOXA cluster. Blood. 2009;113:2526–2534. doi: 10.1182/blood-2008-06-162164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang X, Weissman SM, Newburger PE. Long intergenic non-coding RNA HOTAIRM1 regulates cell cycle progression during myeloid maturation in NB4 human promyelocytic leukemia cells. RNA Biol. 2014;11:777–787. doi: 10.4161/rna.28828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen ZH, Wang WT, Huang W, Fang K, Sun YM, Liu SR, Luo XQ, Chen YQ. The lncRNA HOTAIRM1 regulates the degradation of PML-RARA oncoprotein and myeloid cell differentiation by enhancing the autophagy pathway. Cell Death Differ. 2016 doi: 10.1038/cdd.2016.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang XQD, Dostie J. Reciprocal regulation of chromatin state and architecture by HOTAIRM1 contributes to temporal collinear HOXA gene activation. Nucleic Acids Res. 2016 doi: 10.1093/nar/gkw966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhao H, Zhang X, Frazão JB, Condino-Neto A, Newburger PE. HOX antisense lincRNA HOXA-AS2 is an apoptosis repressor in all Trans retinoic acid treated NB4 promyelocytic leukemia cells. J Cell Biochem. 2013;114:2375–2383. doi: 10.1002/jcb.24586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ebralidze AK, Guibal FC, Steidl U, Zhang P, Lee S, Bartholdy B, Jorda MA, Petkova V, Rosenbauer F, Huang G, Dayaram T, Klupp J, O’Brien KB, et al. PU.1 expression is modulated by the balance of functional sense and antisense RNAs regulated by a shared cis-regulatory element. Genes Dev. 2008;22:2085–2092. doi: 10.1101/gad.1654808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McCarty G, Loeb DM. Hypoxia-sensitive epigenetic regulation of an antisense-oriented lncRNA controls WT1 expression in myeloid leukemia cells. PLoS ONE. 2015;10:1–21. doi: 10.1371/journal.pone.0119837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zeng C, Yu X, Lai J, Yang L, Chen S, Li Y. Overexpression of the long non-coding RNA PVT1 is correlated with leukemic cell proliferation in acute promyelocytic leukemia. J Hematol Oncol. 2015;8:126. doi: 10.1186/s13045-015-0223-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wagner LA, Christensen CJ, Dunn DM, Spangrude GJ, Georgelas A, Kelley L, Esplin MS, Weiss RB, Gleich GJ. EGO, a novel, noncoding RNA gene, regulates eosinophil granule protein transcript expression. Blood. 2007;109:5191–5198. doi: 10.1182/blood-2006-06-027987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sun J, Li W, Sun Y, Yu D, Wen X, Wang H, Cui J, Wang G, Hoffman AR, Hu J-F. A novel antisense long noncoding RNA within the IGF1R gene locus is imprinted in hematopoietic malignancies. Nucleic Acids Res. 2014;42:9588–9601. doi: 10.1093/nar/gku549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guo G, Kang Q, Zhu X, Chen Q, Wang X, Chen Y, Ouyang J, Zhang L, Tan H, Chen R, Huang S, Chen J-L. A long noncoding RNA critically regulates Bcr-Abl-mediated cellular transformation by acting as a competitive endogenous RNA. Oncogene. 2014;34(14):1768–1779. doi: 10.1038/onc.2014.131. [DOI] [PubMed] [Google Scholar]
- 25.Chen L, Wang W, Cao L, Li Z, Wang X. Long non-coding RNA CCAT1 acts as a competing endogenous RNA to regulate cell growth and differentiation in acute myeloid leukemia. Mol Cells. 2016;39:330–336. doi: 10.14348/molcells.2016.2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hirano T, Yoshikawa R, Harada H, Harada Y, Ishida A, Yamazaki T. Long noncoding RNA, CCDC26, controls myeloid leukemia cell growth through regulation of KIT expression. Mol Cancer. 2015;14:90. doi: 10.1186/s12943-015-0364-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xing C, Hu X, Xie F, Yu Z, Li H. Bin-Zhou, Wu J, Tang L, Gao S. Long non-coding RNA HOTAIR modulates c-KIT expression through sponging miR-193a in acute myeloid leukemia. FEBS Lett. 2015;589:1981–1987. doi: 10.1016/j.febslet.2015.04.061. [DOI] [PubMed] [Google Scholar]
- 28.Zeng C, Xu Y, Xu L, Yu X, Cheng J, Yang L, Chen S, Li Y. Inhibition of long non-coding RNA NEAT1 impairs myeloid differentiation in acute promyelocytic leukemia cells. BMC Cancer. 2014;14:693. doi: 10.1186/1471-2407-14-693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hughes JM, Legnini I, Salvatori B, Masciarelli S, Marchioni M, Fazi F, Morlando M, Bozzoni I, Fatica A. C/EBPα-p30 protein induces expression of the oncogenic long non-coding RNA UCA1 in acute myeloid leukemia. Oncotarget. 2015;6:18534–18544. doi: 10.18632/oncotarget.4069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tseng Y-Y, Moriarity BS, Gong W, Akiyama R, Tiwari A, Kawakami H, Ronning P, Reuland B, Guenther K, Beadnell TC, Essig J, Otto GM, O’Sullivan MG, et al. PVT1 dependence in cancer with MYC copy-number increase. Nature. 2014;512:82–86. doi: 10.1038/nature13311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huppi K, Volfovsky N, Runfola T, Jones TL, Mackiewicz M, Martin SE, Mushinski JF, Stephens R, Caplen NJ. The identification of microRNAs in a genomically unstable region of human chromosome 8q24. Mol Cancer Res. 2008;6:212–221. doi: 10.1158/1541-7786.MCR-07-0105. [DOI] [PubMed] [Google Scholar]
- 32.Beck-Engeser GB, Lum AM, Huppi K, Caplen NJ, Wang BB, Wabl M. Pvt1-encoded microRNAs in oncogenesis. Retrovirology. 2008;5:4. doi: 10.1186/1742-4690-5-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wan L, Sun M, Liu G-J, Wei C-C, Zhang E-B, Kong R, Xu T-P, Huang M-D, Wang Z-X. Long noncoding RNA PVT1 promotes non-small cell lung cancer cell proliferation through epigenetically regulating LATS2 expression. Mol Cancer Ther. 2016;15:1082–1094. doi: 10.1158/1535-7163.MCT-15-0707. [DOI] [PubMed] [Google Scholar]
- 34.Wang F, Yuan J-H, Wang S-B, Yang F, Yuan S-X, Ye C, Yang N, Zhou W-P, Li W-L, Sun S-H. Oncofetal long noncoding RNA PVT1 promotes proliferation and stem cell-like property of hepatocellular carcinoma cells by stabilizing NOP2. Hepatology. 2014;60:1278–1290. doi: 10.1002/hep.27239. [DOI] [PubMed] [Google Scholar]
- 35.Zhu H, Zhou X, Chang H, Li H, Liu F, Ma C, Lu J. CCAT1 promotes hepatocellular carcinoma cell proliferation and invasion. Int J Clin Exp Pathol. 2015;8:5427–5434. [PMC free article] [PubMed] [Google Scholar]
- 36.Ma M-Z, Chu B-F, Zhang Y, Weng M-Z, Qin Y-Y, Gong W, Quan Z-W. Long non-coding RNA CCAT1 promotes gallbladder cancer development via negative modulation of miRNA-218-5p. Cell Death Dis. 2015;6:e1583. doi: 10.1038/cddis.2014.541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Palma CA, Al Sheikha D, Lim TK, Bryant A, Vu TT, Jayaswal V, Ma DDF. MicroRNA-155 as an inducer of apoptosis and cell differentiation in acute myeloid leukaemia. Mol Cancer. 2014;13:79. doi: 10.1186/1476-4598-13-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hughes JM, Salvatori B, Giorgi FM, Bozzoni I, Fatica A. CEBPA-regulated lncRNAs, new players in the study of acute myeloid leukemia. J Hematol Oncol. 2014;7:69. doi: 10.1186/s13045-014-0069-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Garzon R, Volinia S, Papaioannou D, Nicolet D, Kohlschmidt J, Yan PS, Mrózek K, Bucci D, Carroll AJ, Baer MR, Wetzler M, Carter TH, Powell BL, et al. Expression and prognostic impact of lncRNAs in acute myeloid leukemia. Proc Natl Acad Sci U S A. 2014;111(52):18679–18684. doi: 10.1073/pnas.1422050112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wei S, Zhao M, Wang X, Li Y, Wang K. PU.1 controls the expression of long noncoding RNA HOTAIRM1 during granulocytic differentiation. J Hematol Oncol. 2016;9:44. doi: 10.1186/s13045-016-0274-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Díaz-Beyá M, Brunet S, Nomdedéu J, Pratcorona M, Cordeiro A, Gallardo D, Escoda L, Tormo M, Heras I, Ribera JM, Duarte R, de Llano MPQ, Bargay J, et al. The lincRNA HOTAIRM1, located in the HOXA genomic region, is expressed in acute myeloid leukemia, impacts prognosis in patients in the intermediate-risk cytogenetic category, and is associated with a distinctive microRNA signature. Oncotarget. 2015;6:31613–31627. doi: 10.18632/oncotarget.5148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Arun G, Diermeier S, Akerman M, Chang K-C, Wilkinson JE, Hearn S, Kim Y, MacLeod AR, Krainer AR, Norton L, Brogi E, Egeblad M, Spector DL. Differentiation of mammary tumors and reduction in metastasis upon Malat1 lncRNA loss. Genes Dev. 2016;30:34–51. doi: 10.1101/gad.270959.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chu C, Qu K, Zhong FL, Artandi SE, Chang HY. Genomic maps of long noncoding RNA occupancy reveal principles of RNA-chromatin interactions. Mol Cell. 2011;44:667–678. doi: 10.1016/j.molcel.2011.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Simon MD, Wang CI, Kharchenko PV, West JA, Chapman BA, Alekseyenko A, Borowsky ML, Kuroda MI, Kingston RE. The genomic binding sites of a noncoding RNA. Proc Natl Acad Sci USA. 2011;108:20497–20502. doi: 10.1073/pnas.1113536108. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data sharing not applicable to this article as no datasets were generated or analyzed during the current study.


