Abstract
Mycobacteria include both environmental species and many pathogenic species such as Mycobacterium tuberculosis, an intracellular pathogen that is the causative agent of tuberculosis in humans. Inducible gene expression is a powerful tool for examining gene function and essentiality, both in in vitro culture and in host cell infections. The theophylline-inducible artificial riboswitch has recently emerged as an alternative to protein repressor-based systems. The riboswitch is translationally regulated and is combined with a mycobacterial promoter that provides transcriptional control. We here provide methods used by our laboratory to characterize the riboswitch response to theophylline in reporter strains, recombinant organisms containing riboswitch-regulated endogenous genes, and in host cell infections. These protocols should facilitate the application of both existing and novel artificial riboswitches to the exploration of gene function in mycobacteria.
Keywords: mycobacteria, theophylline, inducible, GFP, β-galactosidase, infection, homologous recombination, flow cytometry
1. Introduction
Experimental control over gene expression is a critical tool in molecular genetics. Inducible gene regulation by small molecules offers time- and dose-dependent control. Inducible systems are commonly used to express proteins for purification and biochemical characterization or to silence genes by expressing anti-sense mRNA or directly controlling target gene transcription. For protein expression, the dosing control afforded by inducible regulation can help ameliorate the harmful effects of toxic proteins or overexpression. For gene silencing, time-dependent control can be used to test how gene function changes as experimental conditions vary. In pathogens, the essentiality of a gene may evolve over the course of infection depending on the needs and metabolic state of the pathogen. Inducible systems have been used to show, for example, that the proteasome and the gluconeogenic enzyme phosphoenolpyruvate carboxykinase are necessary for survival of the human pathogen Mycobacterium tuberculosis during both the acute and chronic stages of infection in mice (Gandotra, Schnappinger, Monteleone, Hillen & Ehrt, 2007;Marrero, Rhee, Schnappinger, Pethe & Ehrt, 2010).
In general, inducible gene regulation tools for Gram-positive bacteria are not as numerous or diverse as those available for their Gram-negative brethren (see also “Conditional control of gene expression by artificial riboelements in Streptomyces coelicolor” in this volume). For Mycobacteria, which include numerous pathogenic species such as M. tuberculosis, gene regulatory systems have been validated for induction by acetamide, pristinamycin, thiostrepton, nitriles, and tetracycline derivatives (Parish, Mahenthiralingam, Draper, Davis & Colston, 1997;Carroll, Muttucumaru & Parish, 2005;Ehrt, Guo, Hickey, Ryou, Monteleone, Riley et al, 2005;Hernandez-Abanto, Woolwine, Jain & Bishai, 2006;Sassetti, 2008;Forti, Crosta & Ghisotti, 2009;Klotzsche, Ehrt & Schnappinger, 2009;Pandey, Raman, Proff, Joshi, Kang, Rubin et al, 2009). The Tn10-based Tet repressor and its variants have become some of the most widely used, and the only systems that have been validated in an animal model of M. tuberculosis infection (Gandotra et al, 2007). All of these systems rely on allosteric control of a protein repressor that must be expressed in tandem with a target gene controlled by the corresponding promoter. Not all genes respond identically to a given promoter/repressor pair, and empirical testing can be necessary to balance repressor and target expression and achieve the desired level of control.
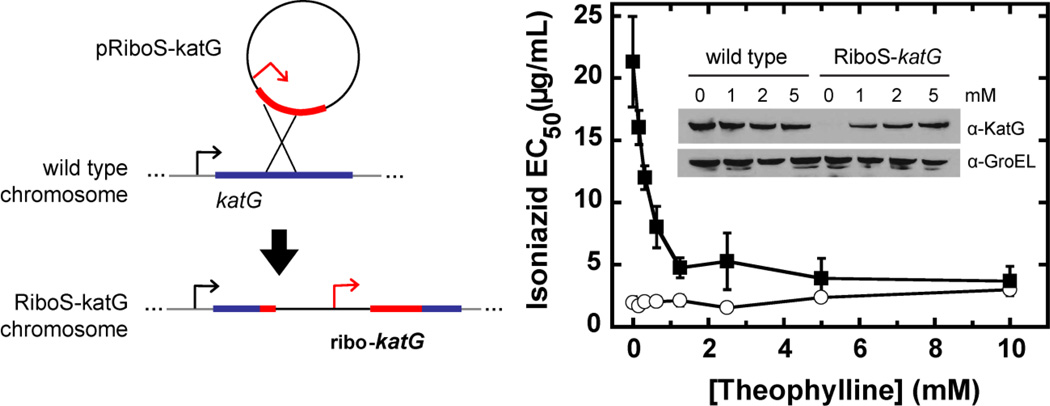
As an alternative to these protein-based systems, we recently reported the application of a theophylline-inducible synthetic riboswitch to gene regulation in mycobacteria such as M. tuberculosis (Topp, Reynoso, Seeliger, Goldlust, Desai, Murat et al, 2010;Seeliger, Topp, Sogi, Previti, Gallivan & Bertozzi, 2012). This system combined transcription driven by the strong constitutive promoter (from the hsp60 gene of the vaccine strain M. bovis BCG) (Stover, de la Cruz, Fuerst, Burlein, Benson, Bennett et al, 1991) with translational control by an optimized synthetic theophylline riboswitch (the E* variant) (Topp et al, 2010). All the regulatory elements are encoded within the 5′UTR of the mRNA and no additional factors are required. Using fluorescence, enzyme activity, immunoblot and phenotypic assays, we demonstrated similar responses to millimolar theophylline for two reporter genes and one endogenous mycobacterial gene (Seeliger et al, 2012). In this last application, we confirmed that the catalase-peroxidase KatG confers sensitivity to the front-line tuberculosis prodrug isoniazid. In the absence of theophylline, KatG was repressed and the half-maximum effective concentration (EC50) for isoniazid was ten-fold higher. Theophylline induction restored both protein expression and antibiotic sensitivity in a dose-dependent manner (Figure 5). We have made similar observations of theophylline riboswitch function for the overexpression of several mycobacterial proteins in both M. smegmatis and M. tuberculosis, with dose-dependent responses and no expression detected by immunoblot in the absence of theophylline (unpublished results). Induction is reversible upon the removal of theophylline. The riboswitch also functions to control GFP expression in M. tuberculosis in an infected macrophage-like cell line. An activation ratio of ~70 was observed in vitro, affording a moderate dynamic range over 0–2 mM theophylline.
Figure 5. An endogenous riboswitch-controlled gene can be obtained by single crossover.
(left) The suicide plasmid pRiboS-katG recombines homologously with the M. smegmatis genome to generate a merodiploid strain with one truncated copy and one full-length riboswitch-controlled copy of katG. (right) Theophylline-dependent expression of KatG was confirmed by immunoblot (inset) and phenotypic resistance to isoniazid. Figure adapted from Seeliger et al, 2012.
Here we provide protocols for assaying riboswitch function and implementing riboswitch-controlled gene expression in mycobacteria. We also report results from a panel of mycobacterial promoters paired with the theophylline riboswitch. Guidelines and representative data follow below for (1) cloning promoters into a theophylline-riboswitch reporter system, (2) assaying promoter-riboswitch reporter constructs by fluorescence or enzyme activity assays, (3) engineering strains by single-crossover homologous recombination for riboswitch-regulated control of endogenous genes, and (4) inducing riboswitch-controlled bacterial genes in infected host cells.
2. Riboswitch Reporter Assays
Because the theophylline riboswitch control translation, it must be combined with a promoter that drives transcription. The choice of promoter can alter the response of the riboswitch by influencing the number of mRNA transcripts and introducing additional regulatory elements. Based on our goals of achieving generality and high dynamic range in mycobacteria, we paired the riboswitch with the strong constitutive promoter Phsp60, but users may want to use different promoters depending on their requirements.
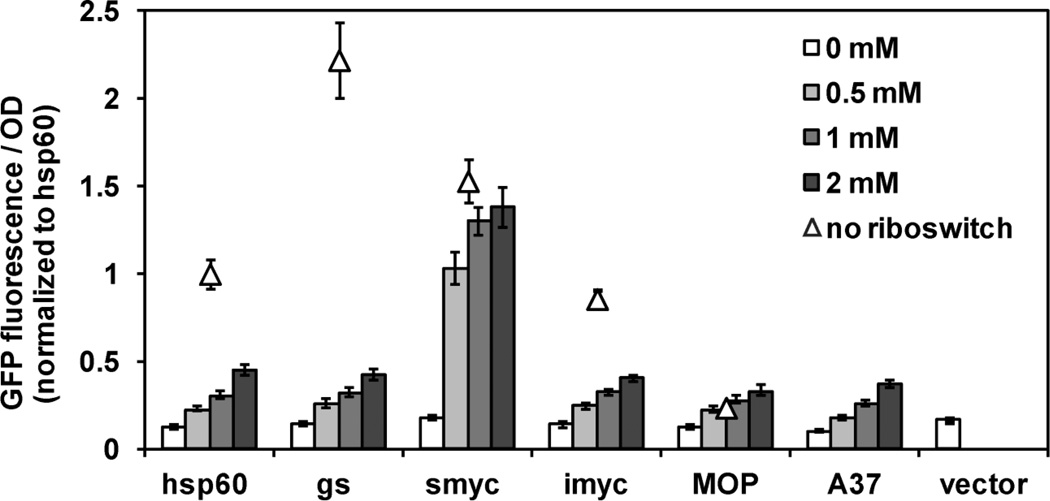
Based on our experiences, we recommend that new promoter-riboswitch combinations be tested in one or more reporter gene systems to confirm and characterize the dose-dependent response to theophylline. Thus far, in addition to Phsp60, we have paired four constitutive mycobacterial promoters, both naturally occurring (smyc, imyc) (Ehrt et al, 2005) and synthetic (MOP, A37) (Hickey, Arain, Shawar, Humble, Langhorne, Morgenroth et al, 1996;Agarwal & Tyagi, 2006), with the E* riboswitch variant (Topp et al, 2010). We found that the strength of the promoter does not correlate with the behavior of the corresponding promoter-riboswitch combination (Figure 1). When driven by the original promoters, GFP expression varied over a half an order of magnitude. When the promoters were combined with the riboswitch, however, the theophylline-dependent GFP response was nearly identical for all constructs, with the exception of Psmyc-ribo. This construct showed higher maximum expression and higher sensitivity to theophylline, but also a small but reproducible signal above background that indicates incomplete repression in the absence of theophylline.
Figure 1. The Psmyc promoter in combination with the riboswitch has the greatest dynamic range and sensitivity to theophylline.
M. smegmatis transformed with promoter-gfp (triangles) and promoter-ribo-gfp (bars) reporter constructs were assayed for GFP fluorescence in response to theophylline over 6–8 hours. For comparison all data were normalized to the GFP fluorescence from Phsp60-gfp.
Factors that could influence theophylline-induced behavior when compared to the native promoter include differences that affect translational strength (such as the Shine-Dalgarno (SD) sequence and the distance between the SD and the start codon) or riboswitch function (such the sequence of the 5’UTR upstream of the riboswitch). However, we did not find any apparent correlation between these factors and the behavior of the promoter-riboswitch constructs (Table 1). Psmyc-ribo is intriguing for its greater dynamic range and as an alternative to Phsp60-ribo for applications in which stringent repression is not required. Efforts are also underway to generate variants with improved repression.
Table 1. Comparison of promoter and promoter-riboswitch properties.
For comparison the approximate promoter strengths and promoter-ribo responses (as measured by GFP fluorescence) are normalized to Phsp60 and Phsp60-ribo, respectively (summarized from Figure 1). For the promoter-ribo constructs, the UNAFold server was used to calculate the predicted RNA secondary structures and calculated free energies for sequences from the transcriptional start through the start codon. The calculated free energy of the riboswitch alone was then subtracted to obtain ΔΔG values as a metric for the secondary structure introduced by the promoter-dependent 5′UTR.
| hsp60 | gs | smyc | imyc | MOP | A37 | ribo | |
|---|---|---|---|---|---|---|---|
| Promoter strength | 1 | 2.5 | 2 | 1 | 0.1 | ||
| Maximum response of promoter-ribo |
1 | 1 | 4 | 1 | 0.6 | 1 | |
| Shine-Dalgarno (deduced) |
GGAGGAA | AAAGGAG | AAGGAGA | AAGGAG | AGGAG | AAGGAGG | AAGGAGG |
| # of bases between SD and ATG |
10 | 6 | 6 | 7 | 9 | 7 | 7 |
| # of bases between transcriptional start and SD (promoter) |
166 | 32 | 91 | * | 37 | 22 | |
| # of bases between transcriptional start and riboswitch (promoter-ribo) |
16 | 33 | 77 | * | 59 | 22 | |
| ΔΔG (kcal/mol) | −7.9 | −13.8 | −30.0 | -- | −22.2 | −10.9 | |
| Constant sequence | Y | Y | N | Y | Y | Y |
The sequence of the 5'UTR is unknown because the transcriptional start site of Pimyc has not been mapped.
Of the two most commonly used reporters, β-galactosidase and GFP, we favor GFP as the primary reporter system. The β -galactosidase assay requires lysing cells and adding a chromogenic substrate. In contrast, GFP fluorescence can be monitored continuously in whole cells and is therefore conducive to screening in multi-well plate format, measuring induction kinetics, and analyzing by flow cytometry. The ability to monitor in whole cells is especially advantageous when working with pathogenic strains, as it minimizes handling and exposure. A membrane-permeable fluorogenic β -galactosidase substrate 5-acetylaminofluorescein di-β-D-galactopyranoside (C2FDG) has been used in whole M. tuberculosis cells (Ehrt et al, 2005). At time of writing, only the more lipophilic alternative (C12FDG) is commercially available and is still significantly more costly than the chromogenic substrate 2-nitrophenyl β-D-galactopyranoside. Independent of the method of detection, β -galactosidase is an important secondary reporter gene for confirming that the theophylline-induced response is not strongly dependent on the downstream coding sequence and for testing the stringency of repression, since the accumulated signal from substrate turnover makes enzyme activity the more sensitive assay.
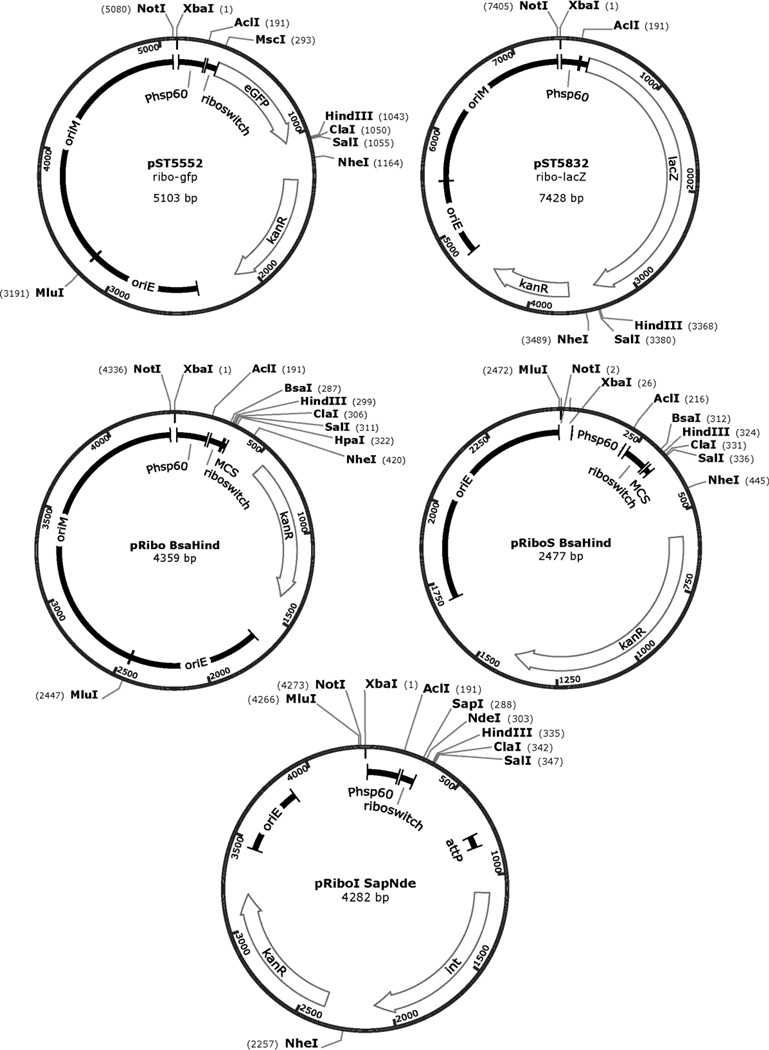
The plasmids pST5552 (ribo-gfp) and pST5832 (ribo-lacZ) are reporter constructs for Phsp60-ribo (Seeliger et al, 2012). When testing novel promoter-riboswitch combinations, these plasmids can be used for comparison and as positive controls for theophylline-induced expression (Figure 2). Below we provide guidance for creating promoter-riboswitch reporter plasmids and detailed protocols for the GFP and β-galactosidase assays. The responses of Phsp60-ribo in non-pathogenic M. smegmatis and pathogenic M. tuberculosis are similar (Seeliger et al, 2012). Therefore, unless the target promoter is known to be species-specific, we recommend using M. smegmatis as the faster, easier and safer choice for reporter assays. Our protocols assume the use of M. smegmatis unless otherwise noted. Additional references should be consulted for information on the handling and manipulation of M. smegmatis and M. tuberculosis (Larsen, Biermann & Jacobs, 2007;Larsen, Biermann, Tandberg, Hsu & Jacobs, 2007;Singh & Reyrat, 2009).
Figure 2. Maps for reporter and cloning plasmids containing the Phsp60-ribo regulatory element.
The restriction sites most relevant to cloning are indicated. Plasmids and partial sequences are available from the AddGene depository. Full maps and sequences are available from the authors upon request.
2.1. Construction of promoter-riboswitch GFP or β-galactosidase reporter plasmids
Because pST5552 and pST5832 are both based on pMV261, an episomal low-copy number shuttle vector, cells containing these constructs or derivatives thereof must be grown in the presence of kanamycin (25 µg/mL in mycobacteria; 50 µg/mL in E. coli) to maintain the plasmid (Stover et al, 1991).
To insert a promoter 5’ of the E* riboswitch, digest the parent vector with XbaI and AclI to excise the Phsp60 promoter (~180 bp). Isolate the vector backbone by agarose gel purification and ligate with the appropriate PCR product containing the desired promoter sequence. This method preserves 14 bases of 5’UTR from the hsp60 gene and 15 bases of a constant sequence that is recommended for poorly characterized promoters (Topp et al, 2010). If removal of the entire 5’UTR and constant sequence is desired, a single digest with KpnI will remove a segment from Phsp60 up to the aptamer (Figure 3).
Figure 3. Sequence of Phsp60-ribo regulatory element.
The 5’UTR of Phsp60 is truncated as described (Topp et al, 2010). Restriction sites KpnI or XbaI/AclI can be used to replace Phsp60 with a different promoter.
General cloning vectors are available to express other proteins as reporters for riboswitch inducible expression. These include the episomal vector pRibo BsaHind (Seeliger et al, 2012) and the integrating vector pRiboI SapNde (unpublished work; Figure 2). The Type IIs restriction endonucleases BsaI and SapI cut outside their recognition sites and allow seamless cloning of target genes. To clone using traditional Type II endonucleases, use pRibo EcoHind. All plasmids described above are available from the AddGene depository.
2.2. GFP fluorescence endpoint assay
For greatest accuracy, grow the bacteria in culture tubes followed by aliquoting into a 96-well plate to measure GFP fluorescence and cell density (OD600). The protocol below is appropriate for all mycobacteria, except for any modifications to the culturing conditions necessary to account for differences in growth rate and pathogenicity. The doubling time for M. smegmatis in standard 7H9 medium is approximately 2.5–3 hours. The time to maximum induction of GFP in fully aerated culture is approximately two doubling times after additional of theophylline.
Due to its low solubility in aqueous solution, theophylline should be dissolved directly in growth medium such as 7H9 and sterilized by 0.2 µm filtration. A 20 mM stock solution may be obtained with sonication and gentle warming at 30–50 °C. At this concentration, the stock is stable at room temperature; avoid storing at 4 °C, as the theophylline tends to crystallize out of solution over time.
Inoculate starter cultures of 3 mL 7H9 medium [4.7 g dehydrated 7H9 (BD Company), 0.5% glycerol, 10% ADC supplement (BD Company), 0.05% Tween 80 per liter) per 14-mL culture tube so that cells will reach late log phase (OD600 of 0.8–1) by the time of subculturing and induction. Inoculate strains containing the appropriate positive control (promoter without riboswitch), negative control (cells carrying empty vector or vector with gfp, but no promoter) and riboswitch (promoter-ribo-gfp) vectors. Incubate with vigorous shaking (250 rpm) at 37 °C.
The next day (after ~12–16 hours), measure the OD600. Aliquot the appropriate volume of overnight culture to obtain OD600 of 0.2 in 2 mL per sample. Pellet cells for 5 min. at 12,000 × g and aspirate the supernatant. Resuspend cells in fresh media containing 0, 0.5, 1 or 2 mM theophylline and incubate for 6 hours at 37 °C to induce target gene expression.
Aliquot 200 uL of each culture into a black, clear-bottom 96-well plate. Also aliquot 200 µL of growth medium as a blank. The excitation and emission should be set by either monochromator or by the choice of filters near 488 nm and 508 nm, respectively. For example, we have used Molecular Devices SpectraMax Gemini XPS (450 nm excitation, 510 nm emission with 495 nm cutoff) and F5 (485/10 nm excitation filter, 535/13 nm emission filter) plate readers. Measure the OD600 for each sample and subtract the buffer blank. Calculate the normalized fluorescence Fnorm for each sample by dividing by the OD600 and subtracting the negative control. Calculate the activation ratio as Fnorm(2 mM)/Fnorm(0 mM). While the absolute fluorescence values will depend on the individual plate reader, relative trends should be comparable and reproducible.
Note on culturing bacteria in multi-well plates
M. smegmatis cultured in a 96-well plate grows more ~40% more slowly (with a doubling time of ~3.5 vs. ~2.5 hours) and the induction kinetics are correspondingly slower. Nevertheless, we have found that the relative behavior of riboswitches can be accurately compared in a multi-well plate, and this format is preferable when appropriate, as it greatly simplifies the procedure and reduces the volume of growth medium required.
If using a 96-well plate, inoculate strains at at OD 0.2 in 200 µL 7H9 (with 0–2 mM theophylline) per well in a black, clear-bottom plate. Cover with a breathable membrane (e.g., Nunc 241205 sealing tape). Shake during incubation and do not cover with a lid, as this will introduce aeration-dependent artifacts in growth and GFP expression. Without humidification, approximately 10–15% of the total volume will evaporate over 6–8 hours, so this format is recommended only for fast-growing non-pathogenic mycobacteria.
2.3. GFP flow cytometry
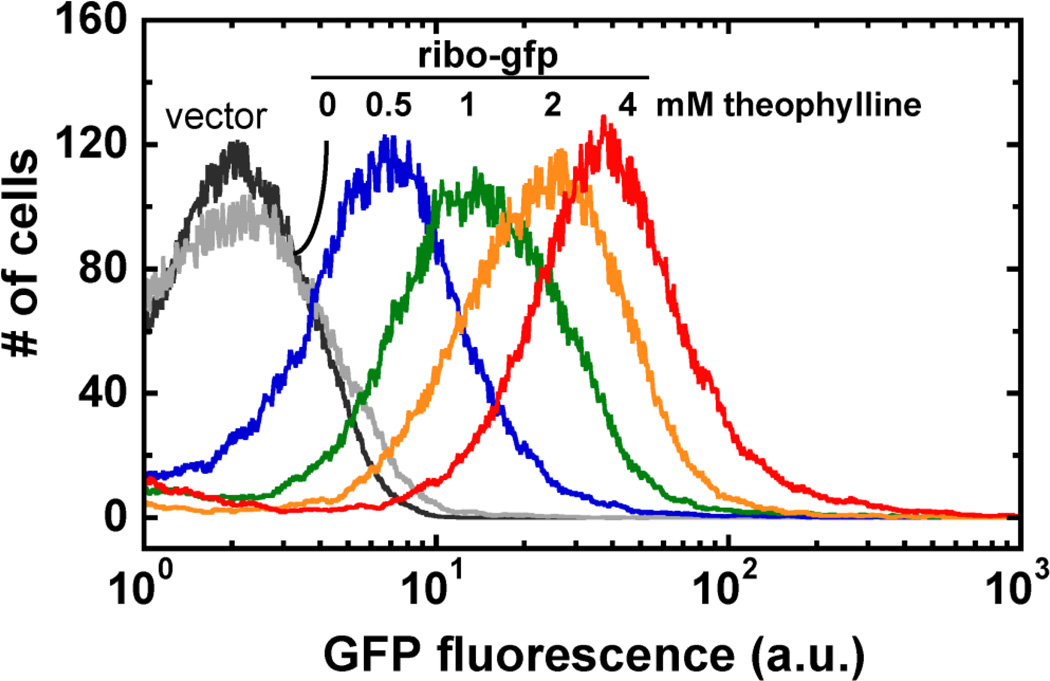
As shown by flow cytometry of M. smegmatis, the theophylline riboswitch is titratable in response to inducer (Figure 4). Due to the tendency of mycobacteria to aggregate, especially in the absence of detergent, cell clumps must be dispersed to obtain single-cell suspensions prior to flow cytometry analysis. A single-setting, low-power sonicator was used for this protocol. The use of sonicators with variable power settings may require optimization to obtain the optimal balance between dispersing and lysing cells. In contrast to the non-pathogeneic M. smegmatis, flow cytometry of pathogenic strains should be performed only when an instrument dedicated to an appropriate Biosafety Level 3 facility is available, due to the generation of aerosols.
Grow overnight starter cultures and induce cells with theophylline as described above in Section 2.2, Steps 1–2. To afford sufficient sample for analysis, a culture volume of 3 mL per sample is recommended for theophylline induction. After the desired period of induction, pellet 1–3×108 cells (based on OD600 of 1 = 3×108 cells/mL) for each strain and wash twice with 1 mL of PBS. Resuspend the final pellet in 1 mL 10% formalin.
After sonication in an ice water bath for 2 minutes, pellet cell clumps by centrifugation for 10 min at 200 × g. Reserve 900 µL supernatant for flow cytometry analysis.
Use the negative control sample (empty vector or no-GFP control strain) to set a gate based on forward and side scatter channels and select against cell debris and any remaining cell clumps. Record flow cytometry data for at least 1 × 104 cells per sample. Calculate the activation ratio as above using the mean fluorescence intensity (MFI) for each sample and first subtracting the negative control. Activation ratio = MFI(2mM)/MFI(0 mM).
Figure 4. Flow cytometry of theophylline-dependent GFP expression shows that the E* riboswitch is titratable.
Figure adapted from Seeliger et al, 2012.
2.4. β-galactosidase activity endpoint assay
For 1 mL of cells at OD600 of ~1, the cell debris after lysis does not scatter significantly at 420 nm, so in this protocol lysates are not cleared after stopping the reaction. However, if lysates are turbid, debris should be removed by centrifugation and the supernatant transferred to a fresh plate for OD420 measurement.
Grow overnight starter culture and induce cells with theophylline as described above. Measure the OD600 for each sample.
Pellet 1 mL (or ~1 × 108 cells) of cells for 5 min. at 12,000 × g and aspirate supernatant. Resuspend cells in 1 mL Z buffer (60 mM Na2HPO4, 40 mM NaH2PO4, 10 mM KCl, 1 mM MgSO4, 50 mM β-mercaptoethanol, pH 7.0). Chill cells on ice.
Lyse cell suspension with a tip sonicator with two pulses of 20 s at power level 0.5. Aliquot 200 µL of uncleared lysate per sample into a clear 96-well plate. Aliquot Z buffer as a negative control.
At t = 0 min, add 50 µL 2-nitrophenyl β-D-galactopyranoside (4 mg/mL in Z buffer) to each well Incubate at 30° C until yellow color is visible (~10 min.). Record the time and stop the reaction with 125 µL 1 M sodium bicarbonate.
Measure the absorbance at 420 nm. Subtract the Z buffer control to account for nonspecific hydrolysis and calculate substrate turnover in Miller units: (OD420 × 1000) / (OD600 × reaction time in min). Calculate the activation ratio as above in the GFP endpoint assay.
3. Construction of Recombinant Strains with Riboswitch-Regulated Genes
The Phsp60-ribo element can be used to control the expression of endogenous genes if inserted directly upstream of the target gene in the chromosome. Below we provide a protocol for generating such a recombinant strain by a single-crossover event, as we have done previously for katG in M. smegmatis (Seeliger et al, 2012). This strategy relies on homologous recombination of a suicide plasmid (pRiboS; Figure 2) to generate one functional copy of the gene controlled by Phsp60-ribo and one non-functional copy controlled by the native promoter (Figure 5). The major advantage of this method is its relative ease; the disadvantages are the low frequency of legitimate homologous recombination in mycobacteria and the retention of a residual, truncated gene copy.
Other methods that are possible, but have not yet been tested, are to introduce an antibiotic resistance marker and Phsp60-ribo upstream of the target gene by homologous recombination using (1) specialized phage transduction (Bardarov, Bardarov, Pavelka, Sambandamurthy, Larsen, Tufariello et al, 2002;Larsen et al, 2007), (2) recombineering (van Kessel & Hatfull, 2007), or (3) selection and counter-selection to obtain double-crossover strains (Parish & Stoker, 2000).
Digest pRiboS with BsaI and ligate with appropriate digested PCR product encoding an N-terminal fragment of the target gene with an in-frame stop codon.
After sequence confirmation, aliquot 1–2 µg of plasmid DNA and treat with 100 mJ cm−2 UV light to promote recombination in the next step (Hinds, Mahenthiralingam, Kempsell, Duncan, Stokes, Parish et al, 1999).
Electroporate DNA into electrocompetent cells (Goude & Parish, 2009). Spread on selective medium and incubate at 37 °C until colonies are visible.
Select single clones and confirm recombination by PCR and sequencing.
4. Induction of Mycobacterial Genes in Infected Host Cells
Mycobacteria residing in infected macrophage cells respond to theophylline in the culture medium (Seeliger et al, 2012). The suggested concentration and period of induction are detailed in the protocol below, although these parameters may need to be optimized depending on host cell type used and experimental needs. Theophylline is moderately toxic to mammalian cells at millimolar concentrations. We therefore recommend treating the selected host cells with theophylline and checking for cell death before performing the infection/induction experiment. The protocol below uses incubation times appropriate for infection with Mycobacterium tuberculosis
Seed mammalian cells (e.g., primary bone marrow-derived macrophages, or cell lines such as RAW264.7, J774.1 or PMA-differentiated THP-1) on 22 × 22 mm sterile glass coverslips in 6-well plates at 3 × 105 cells per well and culture for one day. Synchronize the growth of wild type and riboswitch-containing mycobacteria to reach late log phase by the time the seeded cells have recovered.
Pellet a 10-mL aliquot of bacteria and spin at 500 × g for 5 min. to remove cell clumps. Transfer the supernatant by pipette to a fresh tube and spin at 3500 × g for 5 min. Wash the cell pellet twice with an equal volume of PBS.
Measure OD600 of final cell suspension. Serial dilute in culture medium with 10% horse serum (to promote infectivity) to obtain the concentration necessary for a multiplicity of infection (MOI) of 5 bacteria per host cell at 1 mL/well.
Aspirate medium off mammalian cell monolayers and cover with bacterial cell suspension to initiate infection. Incubate at 37 °C for 4 h.
Aspirate medium and wash cell monolayers twice with an equal volume of PBS. Add 1 mL culture medium without theophylline and allow infected macrophages to recover for 24 hours at 37 °C.
Aspirate medium and replace with fresh medium containing 0 or 0.5 mM theophylline. Return to incubator.
After an additional 24 hours, wash monolayers with PBS and fix in phosphate-buffered 10% formalin for 1 h. Mount coverslips on glass slides with anti-fade and stain nuclei with DAPI (e.g., Vectorlabs Vectashield Mounting Medium with DAPI). Image cells by phase contrast and by GFP and DAPI fluorescence.
References
- Agarwal N, Tyagi AK. Mycobacterial transcriptional signals: requirements for recognition by RNA polymerase and optimal transcriptional activity. Nucleic Acids Res. 2006;34:4245–4257. doi: 10.1093/nar/gkl521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardarov S, et al. Specialized transduction: an efficient method for generating marked and unmarked targeted gene disruptions in Mycobacterium tuberculosis, M. bovis BCG and M. smegmatis. Microbiology. 2002;148:3007–3017. doi: 10.1099/00221287-148-10-3007. [DOI] [PubMed] [Google Scholar]
- Carroll P, Muttucumaru DGN, Parish T. Use of a Tetracycline-Inducible System for Conditional Expression in Mycobacterium tuberculosis and Mycobacterium smegmatis. Appl. Environ. Microbiol. 2005;71:3077–3084. doi: 10.1128/AEM.71.6.3077-3084.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrt S, et al. Controlling gene expression in mycobacteria with anhydrotetracycline and Tet repressor. Nucl. Acids Res. 2005;33:e21. doi: 10.1093/nar/gni013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forti F, Crosta A, Ghisotti D. Pristinamycin-inducible gene regulation in mycobacteria. J. Biotechnol. 2009;140:270–277. doi: 10.1016/j.jbiotec.2009.02.001. [DOI] [PubMed] [Google Scholar]
- Gandotra S, Schnappinger D, Monteleone M, Hillen W, Ehrt S. In vivo gene silencing identifies the Mycobacterium tuberculosis proteasome as essential for the bacteria to persist in mice. Nat. Med. 2007;13:1515–1520. doi: 10.1038/nm1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goude R, Parish T. Electroporation of mycobacteria. Methods Mol. Biol. 2009;465:203–215. doi: 10.1007/978-1-59745-207-6_13. [DOI] [PubMed] [Google Scholar]
- Hernandez-Abanto S, Woolwine S, Jain S, Bishai W. Tetracycline-inducible gene expression in mycobacteria within an animal host using modified Streptomyces tcp830 regulatory elements. Arch. Microbiol. 2006;186:459–464. doi: 10.1007/s00203-006-0160-2. [DOI] [PubMed] [Google Scholar]
- Hickey MJ, et al. Luciferase in vivo expression technology: use of recombinant mycobacterial reporter strains to evaluate antimycobacterial activity in mice. Antimicrob. Agents Chemother. 1996;40:400–407. doi: 10.1128/aac.40.2.400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinds J, et al. Enhanced gene replacement in mycobacteria. Microbiology. 1999;145:519–527. doi: 10.1099/13500872-145-3-519. [DOI] [PubMed] [Google Scholar]
- Klotzsche M, Ehrt S, Schnappinger D. Improved tetracycline repressors for gene silencing in mycobacteria. Nucl. Acids Res. 2009;37:1778–1788. doi: 10.1093/nar/gkp015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen MH, Biermann K, Jacobs WR., Jr Laboratory maintenance of Mycobacterium tuberculosis. Curr Protoc Microbiol. 2007;Chapter 10(Unit 10A):11. doi: 10.1002/9780471729259.mc10a01s6. [DOI] [PubMed] [Google Scholar]
- Larsen MH, Biermann K, Tandberg S, Hsu T, Jacobs WR., Jr Genetic Manipulation of Mycobacterium tuberculosis. Curr Protoc Microbiol. 2007;Chapter 10(Unit 10A):12. doi: 10.1002/9780471729259.mc10a02s6. [DOI] [PubMed] [Google Scholar]
- Marrero J, Rhee KY, Schnappinger D, Pethe K, Ehrt S. Gluconeogenic carbon flow of tricarboxylic acid cycle intermediates is critical for Mycobacterium tuberculosis to establish and maintain infection. Proc. Natl. Acad. Sci. U. S. A. 2010;107:9819–9824. doi: 10.1073/pnas.1000715107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey AK, et al. Nitrile-inducible gene expression in mycobacteria. Tuberculosis. 2009;89:12–16. doi: 10.1016/j.tube.2008.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parish T, Mahenthiralingam E, Draper P, Davis EO, Colston EO. Regulation of the inducible acetamidase gene of Mycobacterium smegmatis. Microbiology. 1997;143:2267–2276. doi: 10.1099/00221287-143-7-2267. [DOI] [PubMed] [Google Scholar]
- Parish T, Stoker NG. Use of a flexible cassette method to generate a double unmarked Mycobacterium tuberculosis tlyA plcABC mutant by gene replacement. Microbiology. 2000;146:1969–1975. doi: 10.1099/00221287-146-8-1969. [DOI] [PubMed] [Google Scholar]
- Sassetti CM. Inducible Expression Systems for Mycobacteria. In: Parish T, Brown AC, editors. Mycobacteria Protocols. Totowa, NJ: Humana Press; 2008. pp. 255–264. [Google Scholar]
- Seeliger JC, et al. A Riboswitch-Based Inducible Gene Expression System for Mycobacteria. PLoS ONE. 2012;7:e29266. doi: 10.1371/journal.pone.0029266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh AK, Reyrat JM. Laboratory maintenance of Mycobacterium smegmatis. Curr Protoc Microbiol. 2009;10(Unit10C):11. doi: 10.1002/9780471729259.mc10c01s14. [DOI] [PubMed] [Google Scholar]
- Stover CK, et al. New use of BCG for recombinant vaccines. Nature. 1991;351:456–460. doi: 10.1038/351456a0. [DOI] [PubMed] [Google Scholar]
- Topp S, et al. Synthetic Riboswitches that Induce Gene Expression in Diverse Bacterial Species. Appl. Environ. Microbiol. 2010;76:7881–7884. doi: 10.1128/AEM.01537-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Kessel JC, Hatfull GF. Recombineering in Mycobacterium tuberculosis. Nat. Meth. 2007;4:147–152. doi: 10.1038/nmeth996. [DOI] [PubMed] [Google Scholar]