Abstract
The non-conventional yeast Pichia pastoris is a popular host for recombinant protein production in scientific research and industry. Typically, the expression cassette is integrated into the genome via homologous recombination. Due to unknown integration events, a large clonal variability is often encountered consisting of clones with different productivities as well as aberrant morphological or growth characteristics. In this study, we analysed several clones with abnormal colony morphology and discovered unpredicted integration events via whole genome sequencing. These include (i) the relocation of the locus targeted for replacement to another chromosome (ii) co-integration of DNA from the E. coli plasmid host and (iii) the disruption of untargeted genes affecting colony morphology. Most of these events have not been reported so far in literature and present challenges for genetic engineering approaches in this yeast. Especially, the presence and independent activity of E. coli DNA elements in P. pastoris is of concern. In our study, we provide a deeper insight into these events and their potential origins. Steps preventing or reducing the risk for these phenomena are proposed and will help scientists working on genetic engineering of P. pastoris or similar non-conventional yeast to better understand and control clonal variability.
The non-conventional yeast Pichia pastoris is a popular host for recombinant protein production, due to a highly efficient secretion mechanism and the possibility of reaching high product titres for simpler enzymes such as phytase and complex proteins containing multiple post-translational modifications, e.g. monoclonal antibodies1,2,3,4,5. Research by Kurtzman et al.6,7 resulted in the reclassification of the P. pastoris genus as Komagatella, including the subspecies K. phaffii and K. pastoris. However, they are still commonly referred to as P. pastoris. In recent years, the genetic toolbox for P. pastoris has been markedly expanded with several newly discovered native promoters, synthetic promoters and other regulatory elements8,9,10,11. The construction of optimized strains and vectors enabled new applications, e.g. the production of metabolites or expression of proteins lacking yeast specific hypermannosylation patterns12,13. Additionally, in a very recent publication Weninger et al.14 reported the first CRISPR/Cas9 system for P. pastoris opening up new possibilities for genetic engineering approaches.
Nevertheless, the most frequently used approach for introducing the target gene in P. pastoris is still the integration of an expression cassette into the genome via homologous recombination. The most popular target for integration is the AOX1 (alcohol oxidase 1) locus that represents the stronger expressed of the two alcohol oxidases in P. pastoris. This approach usually involves the utilization of the AOX1 promoter (pAOX1) as homologous sequence and as promoter of the target gene, because it offers very high expression levels and tight regulation15. After a successful integration, the gene expression can be induced with methanol. However, a clone with an intact AOX1 can metabolize methanol at a higher rate, designated as “methanol utilization plus” (Mut+), complicating the maintenance of a constant induction16,17. Therefore, the application of expression cassettes with two homologous sequences targeted for mediating the replacement of AOX1 is one possible technique. A knock-out mutation of AOX1 leads to the phenotype “methanol utilization slow” (MutS) easing process control and allowing to select for correct integration based on the phenotype.
Despite using comparatively long homologous sequences (ca. 1000 bp), a high variance in targeting efficiency is observed, indicating the prevalence of the non-homologous end joining (NHEJ) pathway in P. pastoris18,19,20. As a result, a high clonal variability is found after transformation, necessitating a time and labour-intensive screening process for the clone with the desired characteristics21,22. Concerning productivity characteristics, the different expression levels of clones typically originate from varying gene copy numbers23,24. The clonal variability is regarded as an inherent property of P. pastoris that is a by-product of the available and established transformation techniques in combination with the strong NHEJ pathway in this yeast species. Many other yeasts, filamentous fungi and higher eukaryotes that are used in biotechnological applications display similar or more pronounced clonal variabilities due to a predominant NHEJ pathway25,26,27, while in the model yeast Saccharomyces cerevisiae homologous recombination is dominant over the NHEJ pathway28. Different techniques to reduce the clonal variability in P. pastoris have been proposed14,18,20,29. However, disadvantages like high complexity, lower strain fitness or not yet fully realized methods for donor cassette integration have kept these techniques from replacing the established ones for the efficient construction of producer strains. Therefore, successive genetic manipulation steps, e.g. for the construction of biosynthetic pathways, have been comparatively challenging in P. pastoris and only a few applications have been reported so far30,31,32,33,34. To date, the humanisation of the N-glycosylation pathway in P. pastoris via multiple consecutive cloning steps has been the most sophisticated and successful genetic engineering endeavour13,35. Similar observations have been made for other non-conventional yeast like Hansenula polymorpha, Kluyveromyces lactis and Yarrowia lipolytica25,36,37.
In our previous study24, we concentrated on analysing the clonal variability found in 845 P. pastoris strains transformed with a GFP expression cassette targeted for AOX1 replacement. By screening all clones for their Mut-phenotype, GFP productivity and gene copy number 31 clones with interesting features were selected for genome sequencing. The combination of experimental and genome data allowed the discovery of novel insights into the correlation between productivity and integration event. In the majority of cases variations in the productivity could be traced back to gene copy number related effects. No off-target integrations were found in clones with a high productivity, suggesting that the impact of such events on productivity was low.
During the Mut-phenotype assay, strains were discovered that displayed an abnormal colony morphology on plate. Some of these clones were also selected for genome sequencing. Here, we discuss the results for these strains, elucidating a variety of as yet unreported off-target integration events. Besides the disruption of random genes, the co-integration of DNA elements from the plasmid host E. coli KRX as well as the relocation of the AOX1 locus to another chromosome were discovered. The detrimental effect of these events on the genetic integrity and strain morphology presents an additional burden for the screening process. These observations are especially relevant for metabolic engineering or knock-out experiments in P. pastoris, while the integration events detailed in our previous study24 are more pertinent to experiments targeting the creation of high producer strains.
In recent years, metabolic engineering of non-conventional yeast in general and P. pastoris in particular has gained more interest11,38. Therefore, the discussed integration events and the proposed theories explaining their origin will enable scientists working in this field to modify existing transformation and mutagenesis protocols in order to avoid these events. Based on this strategy, clonal diversity can be reduced and genetic engineering processes of higher complexity can be realized.
Results
Identification of clones with abnormal colony morphology
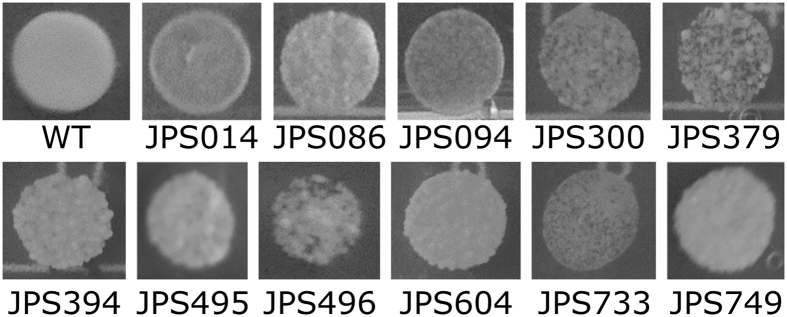
During the plating assay for determination of the Mut phenotype multiple clones with a crenulated colony morphology were found (Fig. 1). The change in colony morphology indicated growth deficiencies, similar to the ones reported for OCH1 knock-out strains in P. pastoris and other non-conventional yeast20,39,40. OCH1 codes for a mannosyltransferase and its loss leads to a lower cell wall integrity, negatively affecting growth and temperature sensitivity as well as an increased flocculation13. It is also a popular knock-out target for humanizing the glycosylation pattern in P. pastoris40. Therefore, it was of interest to discover a potential genetic cause affecting the growth behaviour of clones with abnormal colony morphology. In total, 26 (=3%) of 845 analysed P. pastoris strains displayed a crenulated colony morphology and were divided into the group “Abnormal morphology”. Most of the clones of this group also belonged to other (productivity related) groups, mentioned in our previous report24. Table 1 summarizes the characteristics for all 26 clones with abnormal morphology. Except for the two strains JPS277 and JPS315, all clones had the Mut+-phenotype. Furthermore, only JPS094 and JPS280 displayed clearly elevated GFP expression levels of 2.6 and 3.5, respectively. Since most clones with an abnormal colony morphology had a gene copy number and GFP expression level of approximately 1, they would likely not be selected if looking for high producer strains. However, from a strain engineering perspective the integration events that lead to their change in morphology could be relevant and the strains were therefore further investigated. Eleven strains belonging to this group were sequenced and a variety of different integration events were discovered.
Figure 1. Colony morphology of wildtype P. pastoris CBS 7435 (WT) and irregular clones with crenulated morphology, grown on MD plates for 3 days at 28 °C.
All mutant strains shown here were selected for genome sequencing.
Table 1. Characteristics of P. pastoris strains displaying an abnormal colony morphology, determined as detailed in Schwarzhans et al. 24.
| Strain | Mut-Phenotype | GCN | ExpLvl |
|---|---|---|---|
| JPS014 | Mut+ | 0.7 | 0.3 |
| JPS086 | Mut+ | 0.4 | 1.1 |
| JPS094 | Mut+ | 2.5 | 2.6 |
| JPS165 | Mut+ | 0.9 | 1.0 |
| JPS247 | Mut+ | 0.9 | 0.6 |
| JPS277 | MutS | 0.9 | 0.9 |
| JPS280 | Mut+ | 2.4 | 3.5 |
| JPS300 | Mut+ | 0.5 | 0.5 |
| JPS315 | MutS | 0.9 | 0.8 |
| JPS336 | Mut+ | 1.0 | 0.1 |
| JPS347 | Mut+ | 0.3 | 1.1 |
| JPS379 | Mut+ | 0.1 | 0.0 |
| JPS386 | Mut+ | 5.2 | 1.2 |
| JPS394 | Mut+ | 0.6 | 0.8 |
| JPS495 | Mut+ | 0.1 | 1.7 |
| JPS496 | Mut+ | 0.5 | 0.5 |
| JPS581 | Mut+ | 0.8 | 1.4 |
| JPS603 | Mut+ | 3.1 | 0.6 |
| JPS604 | Mut+ | 0.2 | 0.0 |
| JPS626 | Mut+ | 0.6 | 0.7 |
| JPS636 | Mut+ | 0.8 | 1.0 |
| JPS733 | Mut+ | 0.6 | 0.3 |
| JPS749 | Mut+ | 0.7 | 0.4 |
| JPS768 | Mut+ | 0.6 | 0.5 |
| JPS804 | Mut+ | 0.7 | 0.6 |
| JPS835 | Mut+ | 0.8 | 0.8 |
A gene copy number (GCN) <1 can result from e.g. a reduced PCR efficiency during qPCR, but is still indicative of a single copy of the GFP cassette. The GFP expression level (ExpLvl) was normalized against a reference MutS strain with one copy of the cassette. Underlined strains were selected for genome sequencing.
Gene disruptions
In some cases, a gene disruption is the likely cause of the morphology change. For example, JPS496 (EMBL FBTT01000000) contained a disruption in the MXR1 gene on chromosome 4, which codes for a methionine sulfoxide reductase (not to be confused with the similarly abbreviated methanol expression regulator 1 (MXR1)41). The gene product of MXR1 in S. cerevisiae is part of the oxidative stress response and involved in the repair of damaged proteins by reducing methionine sulfoxide to methionine. Upon inactivation of MXR1, a change in colony morphology was noted in S. cerevisiae42. A disrupted MXR1 in clone JPS496 could lead to the accumulation of damaged proteins in the cell and disrupt metabolic pathways, potentially explaining the observed morphology change. Interpretation of sequencing results for this clone were complicated by the fact that downstream of MXR1 the paralogous gene yahK (alcohol dehydrogenase GroES domain protein) is located and also present on chromosome 3. Furthermore, MXR1 itself has a paralogous copy on chromosome 3, although this copy is a fragment and only 567 bp long (MXR1 on chromosome 4 is 669 bp long) and contains multiple mutations (>90 mismatches). In consequence, multiple possibilities existed to assemble the contigs related to the expression cassette and the MXR1 locus in JPS496. To obtain a clearer picture, a PCR targeting the MXR1 locus on chromosome 4 was performed, the primers are described in Table 2. The PCR assay indicated the integration of an >6 kb fragment in the MXR1 locus on chromosome 4. Figure 2(A) shows the proposed integration event in JPS496. Accordingly, the integration occurred between the base pairs 40 and 41 of the MXR1 gene. In frame transcription of the MXR1 gene would thus continue in the newly integrated pAOX1 and end after adding 10 amino acids (AA) at the stop codon TGA. Additionally, due to the integration between base pairs 40 and 41, a p. Lys14Arg mutation occurs. The resulting 23 AA gene product of the MXR1-pAOX1-hybrid is of different composition and misses the catalytic domain compared to the full length MXR1 gene product (222 AA) (Fig. S1). It is therefore probably inactive. No significant homologies between the integrated DNA and MXR1 exist, indicating that the insertion was likely the result of the NHEJ pathway. The effect of a MXR1 deletion on the morphology of P. pastoris has not been reported yet.
Table 2. Primers used for PCR assays and qRT-PCR.
| Primer name | Sequence | Purpose |
|---|---|---|
| MXR1-FW | GAGAAGGATAGGATCACGGTGGCC | MXR1 locus amplification |
| MXR1-RV | GCGCCAGAGACATTCTACAGGGAG | |
| ADH-FW | CCCTTTATCACCAACTTGAACTCTCACATTCCCCC | Amplification of Adhesin AidA precursor ychA found in JPS094 |
| ADH-RV | GGCCTGGTGCAACAGGATGGATTAATATTTTTAATGG | |
| FIM-FW | CTCAAATATCTTTAATAGCACCAATAACCAGCCAGGG | Amplification of Adhesin fragment fimH and Fimbrial protein found in JPS086 |
| FIM-RV | CATCTGCCTAAATCTACCGTCTTATCAATATCCGC | |
| ACT1-FW | GGTGTGGTGCCAGATCTTTT | qRT-PCR; Housekeeping gene ACT1 in P. pastoris; Amplicon = 181 bp |
| ACT1-RV | AGTGTTCCCATCGGTCGTAG | |
| sfmF-FW | TACTGAACGCTGGCGATACC | qRT-PCR; sfmF in JPS086 and E. coli KRX, Amplicon = 163 bp |
| sfmF-RV | AATTCGATGGCGACGGTTTG | |
| ychA-FW | GCAGCGTGCCATTATTACCG | qRT-PCR; ychA in JPS094 and E. coli KRX, Amplicon = 189 bp |
| ychA-RV | TTGGGCAATCTGATGCACCT | |
| ybaA-FW | TCGGAACGGAACGGATAAGAC | qRT-PCR; ybaA in JPS300 and E. coli KRX, Amplicon = 160 bp |
| ybaA-RV | TGCAAGTGTGAAAGCTCAGCA | |
| finO-FW | AATGTCACCACGCCACCAAA | qRT-PCR; finO in JPS496 and E. coli KRX, Amplicon = 159 bp |
| finO-RV | CTTCAGGGTGTTCACGGCAT |
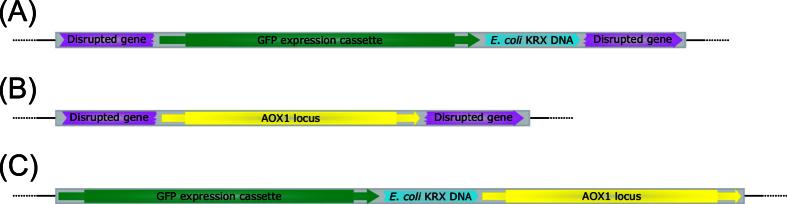
Figure 2. The different kinds of integration events discovered in sequenced P. pastoris clones with abnormal colony morphology.
(A) Disruption of an untargeted gene by a GFP expression cassette fused to E. coli KRX DNA (B) Disruption of an untargeted gene by a re-integrated AOX1 locus. (C) Co-integration of E. coli KRX DNA in fusion with an expression cassette at the AOX1 locus.
A different kind of gene disruption was found in JPS014 (EMBL FBTG01000000). In this strain the AOX1 locus relocated from its native site on chromosome 4 to chromosome 2 between base 2,062,817 and 2,062,818. As a result, gene PP7435_Chr2-1130 was disrupted (Fig. 2(B)). PP7435_Chr2-1130 is annotated as a hypothetical protein in P. pastoris CBS 743543, but shows 100% identity to PAS_chr2-1_0178 in P. pastoris GS11544, which codes for a polyphosphatidylinositol (PtdIns) phosphatase. In JPS014 the in frame transcription of PP7435_Chr2-1130 would continue after the 781st AA in the newly inserted pAOX1, adding 7 AA and end at a TGA stop codon. Furthermore, the insertion results in a c.G2346A silent mutation (p.Lys782Lys). In comparison to the full-length PP7435_Chr2-1130 protein (1069 AA), the gene product in JPS014 (788AA) is distinctly shorter and 79 AA of the predicted catalytic centre of the PtdIns phosphatase are mutated or missing (Fig. S2.). It is therefore likely, that the mutant protein in JPS014 is impaired in its activity. Due to the absence of sequence similarities between the disrupted gene and AOX1, the integration was likely caused by the NHEJ pathway. No knock-out study of this gene in P. pastoris was available at the time of writing the manuscript. However, it was shown that PtdIns phosphatases in S. cerevisiae are membrane bound proteins involved in cytoskeletal organization, signal transduction and membrane trafficking45. Stolz et al.46 could also observe morphological changes, namely increased cell wall thickness, of S. cerevisiae clones by performing knock-out experiments with different PtdIns phosphatases. Therefore, it is likely that the observed growth deficiency of strain JPS014 was caused by the disrupted PP7435_Chr2-1130 gene. PP7435_Chr2-1130 was disrupted by the AOX1 region targeted for replacement by the GFP cassette, implying that P. pastoris used the excised AOX1 locus (including the AOX1 promoter and terminator) as a substrate for DNA repair. JPS014 displayed the Mut+-phenotype indicating the full functionality of the re-integrated AOX1 locus.
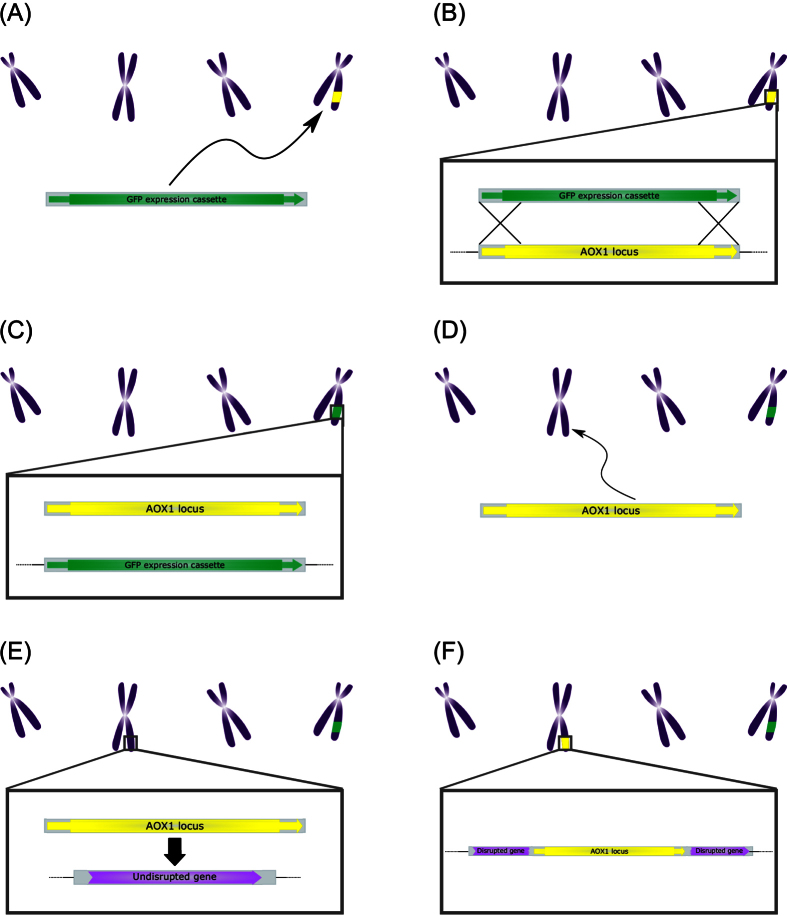
Based on the assembled genome data, it was deduced, which chain of events could have led to the abnormal colony morphology observed for JPS014 (Fig. 3). Firstly, the native AOX1 locus on chromosome 4 was replaced with the GFP expression cassette via the homologous double crossing over event, as intended. Secondly, the excised linear AOX1 locus was not degraded, but instead managed to reach chromosome 2. Potentially, a double-strand break (DSB) in PP7435_Chr2-1130 induced the DSB repair pathway. The NHEJ pathway accepted the linear AOX1 locus as substrate for the DSB repair, disrupting PP7435_Chr2-1130 in the process. In consequence, a P. pastoris clone was created with a single copy of the expression cassette at the desired locus, a functioning AOX1 locus on chromosome 2 and a disrupted PP7435_Chr2-1130, likely causing the observed morphological changes.
Figure 3. Proposed mechanism facilitating the relocation of the AOX1 locus as found in clone JPS014.
(A) After transformation the linear GFP expression cassette from pAHBgl-GFP moved to chromosome 4, where the AOX1 locus is situated. (B) Two distal homologous sequences of the cassette with the chromosomal AOX1 locus mediate a double cross-over recombination event (C) The cassette has replaced the AOX1 locus on chromosome 4 (D) The excised linear AOX1 locus moved to chromosome 2 (E) Potentially due to a DSB initiating the NHEJ pathway, the AOX1 locus is integrated into a random gene on chromosome 2 (F) The untargeted gene PP7435_Chr2-1130 was disrupted. While the AOX1 locus is still functional, the disruption of the untargeted gene likely led to the observed abnormal colony morphology in the case of JPS014.
It seems that even the successful replacement of the AOX1 locus with an expression cassette can result in unforeseen and detrimental off-target effects. It is difficult to estimate, how frequently the excised AOX1 locus was re-integrated, because in case of JPS014 it was only discovered due to morphological changes. Multiple other re-insertions of the AOX1 locus with no or other gene disruptions could have occurred, but bypassed the selection process. If screening for deletion mutants in P. pastoris, it seems possible to miss successful recombination events due to the re-integration of the knock-out target at a different locus. Depending on the aim of the experiment, clones with interesting properties might be overlooked this way. It seems advisable to check for reintegration of the knock-out target at a different locus in knock-out experiments, in addition to confirming the removal of the target from its original site. Additionally, transformation strategies with only one homology sequence for integration should prevent the described event and ought to reduce the clonal variation caused by relocating knock-out targets.
Co-integration of E. coli DNA
In four clones with irregular colony morphology (JPS086, JPS094, JPS300 and JPS496; EMBL FBTK01000000, FBTE01000000, FBTO01000000 and FBTT01000000, respectively) genome sequencing revealed the integration of E. coli KRX DNA elements directly adjacent to the GFP cassette (Fig. 2(C)). E. coli KRX was used as the plasmid host in all experiments. For a better understanding of the exact origin of these elements, we sequenced the E. coli KRX genome (unpublished). Using the genome data, the KRX DNA origin could be traced back to the F plasmid and the chromosome of its original host. In one P. pastoris clone a fusion of chromosomal and F plasmid DNA was discovered. Elements of E. coli KRX DNA present in strain JPS086, JPS094 and JPS300 contained multiple genes coding for proteins that are membrane associated in E. coli, identified via BLASTn. If not mentioned otherwise, KRX DNA elements in P. pastoris showed 100% identity to the chromosomal or F plasmid sequence of E. coli KRX. The ca. 4.6 kb of chromosomal E. coli KRX DNA in JPS086 contained genes coding for fimbria (sfmF) or that are involved in their expression and assembly (sfmZ and smfH). Adhesin associated genes (e.g. ychA) were encoded on the approx. 9.3 kb of F plasmid DNA integrated in JPS094. KRX F plasmid DNA (ca. 4.4 kb) found in clone JPS300 contained multiple membrane proteins or hypothetical proteins with assumed membrane association (ybdA, yuaD, ybbA and ybaA). In JPS496, a fusion of 553 bp chromosomal DNA and 947 bp F plasmid DNA from KRX was found. On the chromosomal portion, two sequences associated with transposases and on the F plasmid portion finO (fertility Inhibition) as well as a fragment of traX (acetylation of F-pilus) were identified. No genes coding for KRX proteins with (hypothetical) membrane association were detected in JPS496.
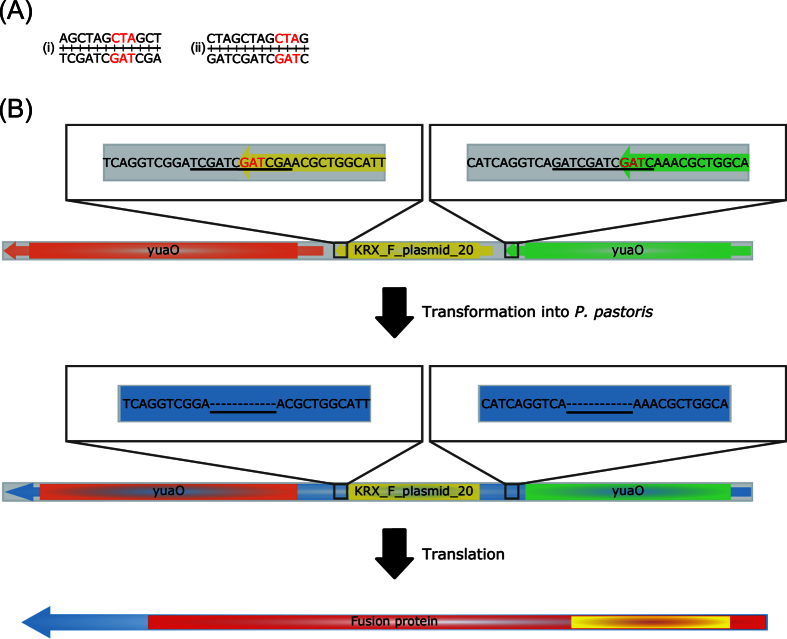
Interestingly, the putative F plasmid DNA in clone JPS094 contained two 12 bp long deletions in comparison to the reference (Fig. 4). Both missing 12 bp sequences are palindromic, which indicates a common cause for their deletion. Potentially, the DNA repair mechanism that facilitated the integration of the pAHBgl-GFP and F plasmid hybrid in P. pastoris caused the deletions. It cannot be excluded that the excision already occurred in E. coli KRX. However, the presence of both 12 bp sequences in the KRX genome data contraindicates this theory. DNA repair mechanisms in yeast have problems with (long) palindromic sequences. In some cases, this phenomenon can lead to DSBs or inhibition of mismatch repair47,48. This fact suggests that the deletion occurred post-transformational in P. pastoris during or after integration of the foreign DNA. The deleted palindromic sequences lead to the excision of two stop-codons (of yuaO and KRX_F_plasmid_20) and the creation of a ca. 157 kDa large fusion protein. Even more surprisingly, the putative ORF ends 4 bp outside the integrated F plasmid DNA. In the adjacent E. coli backbone of pAHBgl-GFP, a TGA base-triplet serves as a presumed stop-codon. The DNA sequence of the resulting fusion protein shows similarity with ycbB (98% identity, BLASTn) of the E. coli ECC-1470 plasmid pECC-1470_100 (GenBank: CP010345) and the encoded protein has a high similarity (99% identity, BLASTp) to an outer membrane auto transporter barrel domain protein from E. coli OK1357 (GenBank: EFZ69416). In comparison to the auto transporter from E. coli OK1357, the putative fusion protein in JPS094 is 131 AA shorter. Using the NCBI Conserved Domain Database, two conserved domains associated with membrane anchoring and transport function (GenBank: PRK14849 and TIGR01414; BLASTx e-values 5.85 × 10−33 and 6.49 × 10−23, respectively) were found in the fusion protein49.
Figure 4. Observed modification of E. coli KRX DNA in P. pastoris clone JPS094.
(A) Sequence of the two 12 bp long palindromic repeats found at the ends of (i) KRX_F_plasmid_20 and (ii) yuaO, respectively. The stop codons have been highlighted in red. (B) In its native form on the F plasmid of E. coli KRX both palindromic repeats (underlined) are present and contain a stop codon. After transformation into P. pastoris a deletion of both palindromic repeats was found in the genome assembly of JPS094. The loss of the stop codons indicated the formation of a ca. 1500 AA long fusion protein (blue), containing two conserved domains (red and yellow) after translation. The larger domain is associated with lipoproteins and autotransporters and the smaller one with outer membrane autotransporters.
With the exception of strain JPS496, a high abundance of genes coding for proteins that interact with the cell membrane were found on the integrated E. coli KRX DNA. This suggests that the observed morphological change have been the result of these proteins compromising the cellular integrity of P. pastoris. Since the irregular morphology was observed on both MD and MM plates, the expression of the E. coli proteins seemed to be independent of the methanol induction. In order to confirm the transcription of the E. coli fragments in the P. pastoris mutant strains, four representative targets were chosen for a qRT-PCR experiment. The targets were sfmF in JPS086, ychA in JPS094, ybaA in JPS300 and finO in JPS496. All four transcripts could be detected during the exponential growth phase in MD medium in their respective strains (Fig. 5). sfmF exhibited the highest and ychA the lowest relative transcript level compared to the endogenous control ACT1, with 0.38 and 0.16, respectively. At the same time, no expression of all four targets could be detected in P. pastoris CBS 7435 wild type. Interestingly, for most targets the Cq-value suggests that the transcript level was similar or higher in the mutant strains than in E. coli KRX (Fig. S3). It remains unclear how the transcription of the E. coli fragments is regulated in P.pastoris, but the results strongly suggest their activity independent of the methanol induction. Although the aberrant phenotypes indicate that the transcripts are also translated, further analysis via e.g. LC-MS would be needed to confirm the presence of the translated E. coli proteins in the mutant strains.
Figure 5. Relative transcriptional levels of E. coli KRX genes found in P. pastoris strains.
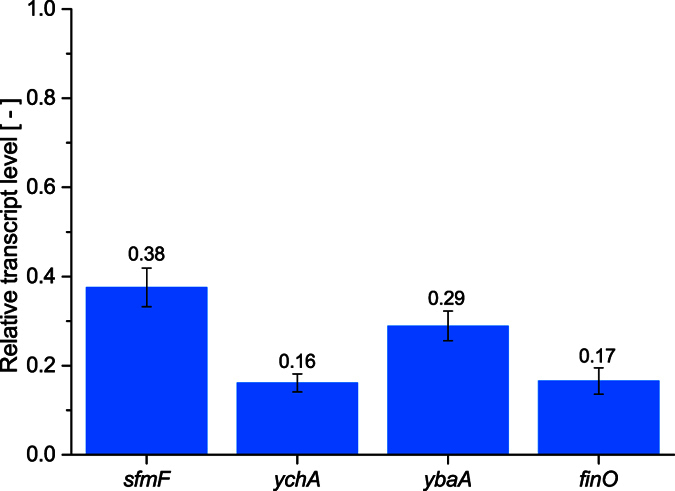
Sfmf (fimbrial protein) in JPS086, ychA (adhesin AidA precursor) in JPS094, ybaA (signalling protein) in JPS300 and finO (fertility inhibitor) in JPS496. Samples for qRT-PCR experiments were taken in the exponential growth phase from cells grown in MD-medium. ACT1 was used as housekeeping gene and values were normalized relative to the expression level of the ACT1 gene in each sample. Error bars represent the standard deviation with n = 3.
The morphological change of JPS496 was likely due to the disruption of MXR1, mentioned above. Other clones with irregular morphology were tested via PCR for the presence of the adhesin and fimbria genes seen in JPS094 and JPS086 ychA and fimH, respectively. It was found that three of them contained adhesin genes and six were positive for fimbria synthesis genes. Considering that these particular clones were only found to contain E. coli KRX DNA due to their abnormal colony morphology, the yet undiscovered presence of different elements from KRX in other strains appears possible. Taken together with the activity of these elements without methanol induction this could present multiple challenges for the biotechnological application of P. pastoris. Integration via the NHEJ pathway could lead to the disruption of untargeted genes. Gene products that interfere with the metabolism of P. pastoris could lead to decreased product yields or negatively affect growth behaviour. Further analysis on the activity of E. coli operons in P. pastoris is needed to clarify this theory. It has to be noted that no separate integration of KRX DNA was observed, it only occurred in fusion with an expression cassette. Therefore it seems, that P. pastoris efficiently degraded most foreign DNA and only the combination of KRX DNA and expression cassette facilitated an integration of E. coli DNA into the genome of P. pastoris.
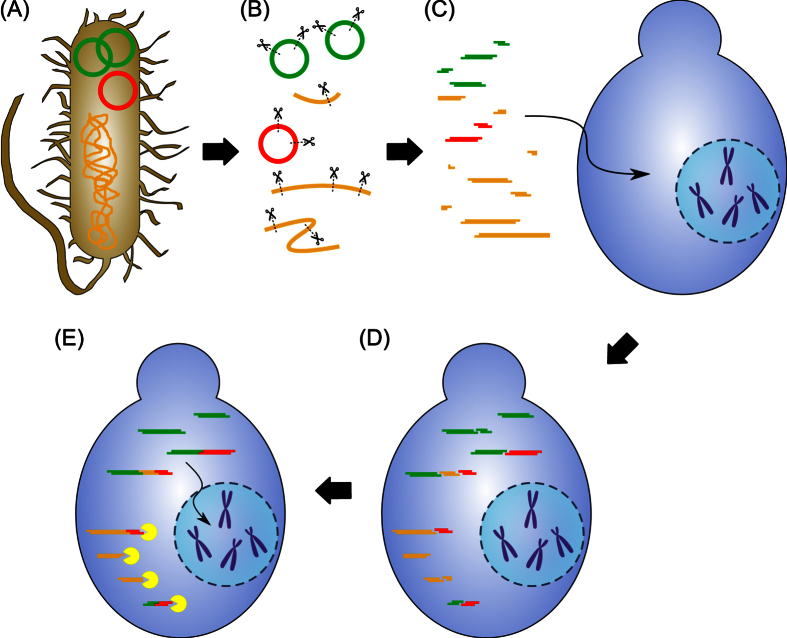
Integrated E. coli KRX DNA was always directly adjacent to at least one BglII site on the chromosome or F plasmid of its original host. BglII was used for linearization of the expression cassette from pAHBgl-GFP, prior to transformation. Based on these results a theory regarding the mechanism underlying the observed phenomenon was formulated (Fig. 6). Presumably F plasmids and fragmented chromosomal DNA were co-extracted with the pAHBgl-GFP vector from E. coli KRX during plasmid preparation. Via BglII digestion the DNA was linearized, compatible sticky-ends were introduced and enabled the in vivo ligation of various fragment combinations in P. pastoris after transformation. In vivo ligation of linear DNA fragments in yeast has been reported before50,51. For P. pastoris it has been suspected that this mechanism plays a role in the generation of multi-copy clones21. The different combinations were then integrated into the genome of P. pastoris leading to the observed morphological changes. Combinations of ligated foreign DNA without homology sequence were likely degraded rather than integrated. While integration of such elements via the NHEJ pathway is possible, no integration events of that kind were found in any of the sequenced clones. Since the unwanted integration and activity of E. coli host DNA is not considered in commonly used P. pastoris transformation protocols, prevention mechanisms should be employed. E. coli strains containing F plasmids (like JM109, TOP10F’, XL10-Gold or DH5alphaF’) are often used for plasmid propagation in studies with P. pastoris52,53,54,55, but should be avoided. In order to prevent insertion of chromosomal DNA, either gel purification or PCR amplification of the expression cassette seems advisable. It has to be noted that in some cases the integrated E. coli host DNA fragments were of similar size as the expression cassette, reducing the potential effectiveness of gel purification. Furthermore, the use of blunt-end or rare cutters for expression cassette linearization ought to reduce the likelihood of hybrid formation.
Figure 6. Schematic depiction of the proposed mechanism facilitating integration of E. coli host DNA in fusion with the expression cassette into the genome of P. pastoris.
(A) E. coli cells containing chromosomal (orange) and F plasmid (red) DNA are used to produce the expression vector (green) (B) During plasmid isolation F plasmids and fragmented chromosomal DNA are co-extracted with the expression vector. Subsequent enzymatic digestion creates compatible sticky ends (C) Restricted DNA is transferred into P. pastoris cells via e.g. electroporation (D) in vivo various fragment combinations are ligated and form new hybrids (E) Expression cassettes and hybrids containing expression cassettes with homologies to chromosomal loci of P. pastoris are integrated into the genome, while non-homologous DNA is degraded.
Discussion
Several non-canonical integration events were discovered in P. pastoris clones displaying a change in colony morphology. The disruption of genes analogous to MXR1 and a PtdIns phosphatse from S. cerevisiae was shown to be the likely cause for growth deficiencies in two of the sequenced strains. In the case of the PtdIns phosphatase disruption, a relocation of the AOX1 locus from chromosome 4 to chromosome 2 was observed. This event raises the question of how efficiently P. pastoris can re-integrate knock-out targets. The full functionality of the relocated AOX1 locus urges scientists interested in efficient knock-out studies in P. pastoris (and other non-conventional yeast) to take similar events into consideration for the design of their experiments. Successful knock-outs are complicated, if the locus simply moves to a new site and clonal diversity is increased in case untargeted genes are disrupted by the relocated knock-out target. Novel techniques like the recently presented CRISPR/Cas9 system could help in knock-out studies14.
In multiple other strains morphological changes were possibly caused by the insertion and activity of DNA elements coding for membrane associated proteins from the plasmid propagation strain E. coli KRX. The methanol induction independent transcription of these E. coli elements in P. pastoris could be confirmed in a qRT-PCR assay. These findings strongly suggest that more care is needed, if transforming P. pastoris. Potentially the integration of other E. coli sequences remained undetected. Interestingly, in one case a presumed post-transformational modification of the KRX DNA was observed resulting in the excision of two stop codons and the creation of a triple-fusion protein with two membrane domains. The unwanted insertion of E. coli elements could cause multiple problems, e.g. in the generation of producer strains. Therefore, E. coli F- strains should be used for studies involving P. pastoris transformation. Amplifying the expression cassette via PCR instead of using plasmid isolations should remediate the co-integration problem more effectively than gel purification. While gel purification of the expression cassette is a commonly used technique, we observed the co-integration of E. coli DNA elements of various sizes ranging from 1.5 to 9.3 kb. In some cases, these elements have a similar length in comparison to the expression cassette and thereby could by-pass a gel purification step. Nevertheless, PCR amplification could prove unsuitable in certain cases. Using blunt-end or rare cutters for plasmid linearization should also impede inadvertent co-integrations in these situations. Newly developed plasmids were recently published that offer sites for blunt-end linearization of an expression cassette targeting the AOX1 locus for replacement56. While evolutionary horizontal gene transfer from bacteria to yeast (e.g. the acquisition of URA157) was described previously, no co-integration of E. coli host DNA during genetic manipulation of P. pastoris has been reported so far.
Some of the sequenced strains (JPS379, JPS394, JPS495, JPS604 and JPS733) contained integration events that could not be related to the phenotype. They include the integration of a truncated expression cassette and mutations in AOX1. However, no gene disruptions were observed. While these events in some cases explained the low productivity, no clear correlation to the colony morphology could be deduced. In all analysed clones, no SNPs (single-nucleotide polymorphism) were detected that would explain the change in colony morphology.
In summary, the results elucidate the reason behind some of the causes for the clonal variability encountered in transformation experiments with P. pastoris. By focusing on strains with abnormal colony morphology, a group of clones undesirable for biotechnological applications can be better understood. Their change in morphology or growth behaviour is detrimental for production processes and in similar cases like ΔOCH1 strains, special process strategies had to be developed to accommodate the changed physiology58. Additionally, the documented integration events and the proposed methods of preventing them enable scientists working with P. pastoris to devise optimized strategies for genetic engineering and mutagenesis of strains.
Methods
Strains and vector
Plasmids were constructed and propagated in E. coli KRX (Promega, Madison, WI; Genotype: [F´, traD36, ΔompP, proA + B+, lacIq, Δ(lacZ)M15] ΔompT, endA1, recA1, gyrA96 (Nalr), thi-1, hsdR17 (rk–, mk+), e14– (McrA–), relA1, supE44, Δ(lac-proAB), Δ(rhaBAD)::T7 RNA polymerase). Yeast experiments involved P. pastoris CBS 7435 (ΔHIS4) (Austrian Center of Industrial Biotechnology, Graz, Austria) and the wild type CBS 7435 (identical to NRRL Y-11430 and ATCC76273), which was obtained from the Spanish Type Culture Collection (Valencia, Spain) under the strain number CECT 11047. P. pastoris CBS 7435 (ΔHIS4) was transformed with BglII linearized expression cassette from pAHBgl-GFP according to Wu and Letchworth59. The composition and construction of pAHBgl-GFP was described previously24. In brief, a linearized GFP expression cassette (purified with Wizard® Plus SV Minipreps DNA Purification System, Promega, Madison, WI) was transformed into P. pastoris and targeted the AOX1 locus on chromosome 4 for replacement. Before transformation the linearized DNA was purified with the Wizard® SV Gel and PCR Clean-Up System (Promega, Madison, WI).
Cultivation conditions
E. coli KRX was cultivated in shake flasks at 37 °C with 120 rpm using Lysogeny Broth (LB) medium supplemented with 100 μg/mL Ampicillin.
P. pastoris was cultivated either in shake flasks, in 96-deep-well plates (Eppendorf, Germany) or on agar plates. Shake flask cultivations were performed with (Buffered) Minimal Dextrose ((B)MD, Invitrogen60), or Yeast Peptone Dextrose (YPD) medium and supplemented with 4 mg/L L-histidine when necessary. Deep-well plate experiments were performed according to Weis et al.61 and Hartner et al.9, with BMD and Buffered Minimal Methanol medium with 1 or 5 g/L methanol (BMM2 and BMM10, respectively). Agar plates contained Minimal Dextrose (MD) or Minimal Methanol (MM) medium60. All cultivations were carried out at 28 °C, with shake flasks being agitated at 120 rpm and deep-well plates at 340 rpm.
Identification of Mut-phenotype and colony morphology
The Mut-phenotype was assayed based on the procedure described in the EasySelect™ Pichia Expression Kit60. Briefly, cells were grown in BMD medium for 60 hours, washed twice and then 5 μL aliquots of the resuspended cells were plated onto both MD and MM plates. After 2–3 days of incubation at 28 °C, the Mut-phenotype could be determined based on the growth behaviour. Additionally, the plating test allowed the visual identification of clones with irregular colony morphology.
Isolation of genomic DNA and PCR assays
For the extraction of genomic DNA (gDNA) from P. pastoris the MasterPure™ Yeast DNA Purification Kit from Epicentre (Madison, WI) was used according to the manufacturer’s protocol.
gDNA from relevant clones was used for different PCR assays. The sequence of the primers used in these assays can be found in Table 2.
Genome sequencing and bioinformatic analysis of P. pastoris strains
The performed procedures for P. pastoris clones selected for genome sequencing were recently reported24. In brief, the gDNA quantity and quality was assayed via the Quant-iT PicoGreen dsDNA kit (Invitrogen, Waltham, MA) and gel-electrophoresis, respectively. gDNA of sufficient quality was sequenced on an Illumina MiSeq system using a paired-end library prepared with the TruSeq sample preparation kit (Illumina, San Diego, CA). The raw data was assembled de novo in the GS De Novo Assembler (Version 2.8, Roche, Basel, Switzerland) with default settings. All assembled genomes can be found under the study id PRJEB12220 in the EBI database.
Bioinformatic analysis involved similarity analysis using the BLASTn algorithm62 and a local database including the pAHBgl-GFP vector sequence. Hits with an e-value >1 × 10−20 and a sequence identity of 100% were analysed in more detail. Gaps in the vector sequence were closed in an in silico approach applying CONSED43,63,64. This approach also allowed the identification of insertion sites of the expression cassette into the genome of P. pastoris.
Genome sequencing, annotation and sequence comparison of the E. coli KRX genome
DNA was extracted using the Wizard® Genomic DNA Purification KIT (Promega, Madison, WI). E. coli KRX genome sequencing and bioinformatic analysis was performed analogous to the method described for P. pastoris, resulting in the draft sequence of the chromosome and the F plasmid (unpublished). E. coli KRX contigs were compared to the P. pastoris draft genomes sequences using BLASTn62. Hits with an e-value >1 × 10−20 and a sequence identity of 95% were analysed in detail to detect horizontal gene transfer between the P. pastoris strains and E. coli KRX chromosomal or F plasmid DNA.
qRT-PCR experiments
E. coli KRX, P. pastoris CBS 7435 and the mutant strains JPS086, JPS094, JPS300 and JPS496 were grown as described above in shake flasks containing LB-Medium or MD-Medium, respectively. Samples taken in the exponential growth phase were employed for RNA isolation using the RNeasy Mini Kit (Qiagen, Hilden, Germany), with both an on- and off-column gDNA digestion step. RNA quantity and the absence of gDNA and other contaminants was assayed with the DropSense16 system (Trinean, Ghent, Belgium). Four different primer sets targeting the various E. coli fragments found in the sequenced P. pastoris mutant strains were designed using the NCBI Primer-BLAST tool, making sure no off-target activity on either the E. coli or P. pastoris genome was found. Additionally, the primer pair for quantification of the housekeeping gene ACT1 (β-actin) in P. pastoris was taken from literature65. All primers exhibited similar TM-values (59 ± 2 °C) and amplicon sizes (170 ± 19 bp). The sequences and amplicon sizes of all employed primer pairs can be found in Table 2. Per reaction 50 ng RNA free of gDNA was used as template. Employing the SensiFASTTM SYBR® No-ROX One-Step Kit by Bioline (London, UK), samples were measured on the LightCycler® 96 system (Roche, Basel, Switzerland) in biological triplicates with technical duplicates. For P. pastoris samples the transcript level of E. coli genes was normalized against the ACT1 level of the same strain via the 2−ΔΔCt method66. After 40 cycles the specificity of the amplicons was confirmed via a melting curve.
Additional Information
How to cite this article: Schwarzhans, J.-P. et al. Non-canonical integration events in Pichia pastoris encountered during standard transformation analysed with genome sequencing. Sci. Rep. 6, 38952; doi: 10.1038/srep38952 (2016).
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Material
Acknowledgments
We acknowledge support for the Article Processing Charge by the Deutsche Forschungsgemeinschaft and the Open Access Publication Fund of Bielefeld University. Grants from the Federal State of North Rhine-Westphalia for the CLIB-Graduate Cluster Industrial Biotechnology are gratefully acknowledged. The bioinformatics support of the BMBF-funded project “Bielefeld-Gießen Center for Microbial Bioinformatics – BiGi (Grant number 031A533)’’ within the German Network for Bioinformatics Infrastructure (de. NBI) is gratefully acknowledged. We would like to thank Timo Wolf for support with the qRT-PCR experiments.
Footnotes
The authors declare no competing financial interests.
Author Contributions J.P.S., J.K. and K.F. designed, analysed and interpreted wet lab experiments. J.P.S. and T.L. performed wet lab experiments. A.W. performed genome sequencing work. D.W. analysed and interpreted sequencing data. J.P.S. and D.W. wrote the manuscript. J.K. and K.F. revised the manuscript. J.P.S., J.K. and K.F. conceived the study. J.K. and K.F. supervised the research. All authors reviewed the final manuscript.
References
- Rodriguez E., Mullaney E. J. & Lei X. G. Expression of the Aspergillus fumigatus phytase gene in Pichia pastoris and characterization of the recombinant enzyme. Biochem. Biophys. Res. Commun. 268, 373–378 (2000). [DOI] [PubMed] [Google Scholar]
- Kim S. et al. Real-time monitoring of glycerol and methanol to enhance antibody production in industrial Pichia pastoris bioprocesses. Biochem. Eng. J. 94, 115–124 (2015). [Google Scholar]
- Gasser B. et al. Pichia pastoris : protein production host and model organism for biomedical research. Future Microbiol. 8, 191–208 (2013). [DOI] [PubMed] [Google Scholar]
- Ahmad M., Hirz M., Pichler H. & Schwab H. Protein expression in Pichia pastoris : recent achievements and perspectives for heterologous protein production. Appl. Microbiol. Biotechnol. 98, 5301–5317 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cregg J. M. et al. Expression in the yeast Pichia pastoris. Methods in enzymology 463, (Elsevier Inc., 2009). [DOI] [PubMed] [Google Scholar]
- Kurtzman C. P. Description of Komagataella phaffii sp. nov. and the transfer of Pichia pseudopastoris to the methylotrophic yeast genus. Komagataella. Int. J. Syst. Evol. Microbiol. 55, 973–976 (2005). [DOI] [PubMed] [Google Scholar]
- Kurtzman C. P. Biotechnological strains of Komagataella (Pichia) pastoris are Komagataella phaffii as determined from multigene sequence analysis. J. Ind. Microbiol. Biotechnol. 36, 1435–1438 (2009). [DOI] [PubMed] [Google Scholar]
- Qin X. et al. GAP promoter library for fine-tuning of gene expression in Pichia pastoris. Appl. Environ. Microbiol. 77, 3600–8 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartner F. S. et al. Promoter library designed for fine-tuned gene expression in Pichia pastoris. Nucleic Acids Res. 36, e76 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prielhofer R. et al. Induction without methanol: novel regulated promoters enable high-level expression in Pichia pastoris. Microb. Cell Fact. 12, 5 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogl T. et al. A toolbox of diverse promoters related to methanol utilization – functionally verified parts for heterologous pathway expression in Pichia pastoris. ACS Synth. Biol. 5, 172–186 (2016). [DOI] [PubMed] [Google Scholar]
- Geier M., Fauland P., Vogl T. & Glieder A. Compact multi-enzyme pathways in P. pastoris. Chem. Commun. 51, 1643–1646 (2015). [DOI] [PubMed] [Google Scholar]
- Choi B. et al. Use of Combinatorial Genetic Libraries to Humanize N-Linked Glycosylation in the Yeast Pichia pastoris. Proc. Natl. Acad. Sci. USA. 100, 5022–5027 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weninger A., Hatzl A.-M., Schmid C., Vogl T. & Glieder A. Combinatorial optimization of CRISPR/Cas9 expression enables precision genome engineering in the methylotrophic yeast Pichia pastoris. J. Biotechnol., doi: 10.1016/j.jbiotec.2016.03.027 (2016). [DOI] [PubMed] [Google Scholar]
- Lin Cereghino G. P. et al. New selectable marker/auxotrophic host strain combinations for molecular genetic manipulation of Pichia pastoris. Gene 263, 159–169 (2001). [DOI] [PubMed] [Google Scholar]
- Romanos M. A., Scorer C. A. & Clare J. J. Foreign gene expression in yeast: a review. Yeast 8, 423–488 (1992). [DOI] [PubMed] [Google Scholar]
- Krainer F. W. et al. Recombinant protein expression in Pichia pastoris strains with an engineered methanol utilization pathway. Microb. Cell Fact. 11, 22 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Näätsaari L. et al. Deletion of the Pichia pastoris KU70 homologue facilitates platform strain generation for gene expression and synthetic biology. PLoS One 7, e39720 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nett J. H., Hodel N., Rausch S. & Wildt S. Cloning and disruption of the Pichia pastoris ARG1, ARG2, ARG3, HIS1, HIS2, HIS5, HIS6 genes and their use as auxotrophic markers. Yeast 22, 295–304 (2005). [DOI] [PubMed] [Google Scholar]
- Chen Z. et al. Enhancement of the gene targeting efficiency of non-conventional yeasts by increasing genetic redundancy. PLoS One 8, e57952 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clare J., Rayment F., Sreekrishna K. & Romanos M. A. High-level expression of tetanus toxin fragment C in Pichia pastoris strains containing multiple tandem integrations of the gene. Nat. Biotechnol. 9, 455–460 (1991). [DOI] [PubMed] [Google Scholar]
- Looser V. et al. Cultivation strategies to enhance productivity of Pichia pastoris: A review. Biotechnol. Adv. 33, 1177–1193 (2015). [DOI] [PubMed] [Google Scholar]
- Aw R. & Polizzi K. M. Can too many copies spoil the broth? Microb. Cell Fact. 12, 128 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarzhans J.-P. et al. Integration event induced changes in recombinant protein productivity in Pichia pastoris discovered by whole genome sequencing and derived vector optimization. Microb. Cell Fact. 15, 84 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verbeke J., Beopoulos A. & Nicaud J. M. Efficient homologous recombination with short length flanking fragments in Ku70 deficient Yarrowia lipolytica strains. Biotechnol. Lett. 35, 571–576 (2013). [DOI] [PubMed] [Google Scholar]
- Meyer V. et al. Highly efficient gene targeting in the Aspergillus niger kusA mutant. J. Biotechnol. 128, 770–775 (2007). [DOI] [PubMed] [Google Scholar]
- Guirouilh-Barbat J. et al. Impact of the KU80 pathway on NHEJ-induced genome rearrangements in mammalian cells. Mol. Cell 14, 611–623 (2004). [DOI] [PubMed] [Google Scholar]
- Daley J. M., Palmbos P. L., Wu D. & Wilson T. E. Nonhomologous end joining in yeast. Annu. Rev. Genet. 39, 431–451 (2005). [DOI] [PubMed] [Google Scholar]
- Heiss S., Maurer M., Hahn R., Mattanovich D. & Gasser B. Identification and deletion of the major secreted protein of Pichia pastoris. Appl. Microbiol. Biotechnol. 97, 1241–9 (2013). [DOI] [PubMed] [Google Scholar]
- Marx H., Mattanovich D. & Sauer M. Overexpression of the riboflavin biosynthetic pathway in Pichia pastoris. Microb. Cell Fact. 7, 23 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araya-Garay J. M., Feijoo-Siota L., Rosa-Dos-Santos F., Veiga-Crespo P. & Villa T. G. Construction of new Pichia pastoris X-33 strains for production of lycopene and β-carotene. Appl. Microbiol. Biotechnol. 93, 2483–2492 (2012). [DOI] [PubMed] [Google Scholar]
- Wriessnegger T. et al. Production of the sesquiterpenoid (+)-nootkatone by metabolic engineering of Pichia pastoris. Metab. Eng. 24, 18–29 (2014). [DOI] [PubMed] [Google Scholar]
- Liu X. B. et al. Metabolic engineering of Pichia pastoris for the production of dammarenediol-II. J. Biotechnol. 216, 47–55 (2015). [DOI] [PubMed] [Google Scholar]
- Nocon J. et al. Model based engineering of Pichia pastoris central metabolism enhances recombinant protein production. Metab. Eng. 24, 129–38 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton S. R. et al. Humanization of Yeast to Produce Complex Terminally Sialylated Glycoproteins. Science (80−). 313, 1441–1443 (2006). [DOI] [PubMed] [Google Scholar]
- Kegel A., Martinez P., Carter S. D. & Åström S. U. Genome wide distribution of illegitimate recombination events in Kluyveromyces lactis. Nucleic Acids Res. 34, 1633–1645 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saraya R. et al. Novel genetic tools for Hansenula polymorpha. FEMS Yeast Res. 12, 271–278 (2012). [DOI] [PubMed] [Google Scholar]
- Liu L., Redden H. & Alper H. S. Frontiers of yeast metabolic engineering: Diversifying beyond ethanol and Saccharomyces. Curr. Opin. Biotechnol. 24, 1023–1030 (2013). [DOI] [PubMed] [Google Scholar]
- Nakayama K., Nagasu T., Shimma Y., Kuromitsu J. & Jigami Y. OCH1 encodes a novel membrane bound mannosyltransferase: outer chain elongation of asparagine-linked oligosaccharides. EMBO J. 11, 2511–2519 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krainer F. W. et al. Knockout of an endogenous mannosyltransferase increases the homogeneity of glycoproteins produced in Pichia pastoris. Sci. Rep. 3, 3279 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin-Cereghino G. P. et al. Mxr1p, a Key Regulator of the Methanol Utilization Pathway and Peroxisomal Genes in Pichia pastoris. Mol. Cell. Biol. 26, 883–897 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen J. Inactivation of MXR1 abolishes formation of dimethyl sulfide from dimethyl sulfoxide in Saccharomyces cerevisiae. Appl. Environ. Microbiol. 65, 3915–3919 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Küberl A. et al. High-quality genome sequence of Pichia pastoris CBS7435. J. Biotechnol. 154, 312–20 (2011). [DOI] [PubMed] [Google Scholar]
- De Schutter K. et al. Genome sequence of the recombinant protein production host Pichia pastoris. Nat. Biotechnol. 27, 561–566 (2009). [DOI] [PubMed] [Google Scholar]
- Nguyen P. H. et al. Interaction of Pik1p and Sjl proteins in membrane trafficking. FEMS Yeast Res. 5, 363–371 (2005). [DOI] [PubMed] [Google Scholar]
- Stolz L. E., Huynh C. V., Thorner J. & York J. D. Identification and characterization of an essential family of inositol polyphosphate 5-phosphatases (INP51, INP52 and INP53 gene products) in the yeast Saccharomyces cerevisiae. Genetics 148, 1715–1729 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nag D. K., White M. A. & Petes T. D. Palindromic sequences in heteroduplex DNA inhibit mismatch repair in yeast. Nature 340, 318–320 (1989). [DOI] [PubMed] [Google Scholar]
- Nasar F., Jankowski C. & Nag D. K. Long palindromic sequences induce double-strand breaks during meiosis in yeast. Mol. Cell. Biol. 20, 3449–3458 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchler-Bauer A. et al. CDD: A Conserved Domain Database for the functional annotation of proteins. Nucleic Acids Res. 39, 225–229 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunes S., Botstein D. & Fox M. S. Transformation of yeast with linearized plasmid DNA. Formation of inverted dimers and recombinant plasmid products. J. Mol. Biol. 184, 375–387 (1985). [DOI] [PubMed] [Google Scholar]
- Suzuki K., Imai Y. & Yamashita I. In vivo ligation of linear DNA molecules to circular forms in the yeast Saccharomyces cerevisiae. J. Bacteriol. 155, 747–754 (1983). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong Y., Zhou H., Tu X., Li J. & Xiao Y. Cloning of a laccase gene from a novel basidiomycete Trametes sp. 420 and its heterologous expression in Pichia pastoris. Curr. Microbiol. 54, 260–5 (2007). [DOI] [PubMed] [Google Scholar]
- Liu M. Q. & Liu G. F. Expression of recombinant Bacillus licheniformis xylanase A in Pichia pastoris and xylooligosaccharides released from xylans by it. Protein Expr. Purif. 57, 101–107 (2008). [DOI] [PubMed] [Google Scholar]
- Hu F. et al. A visual method for direct selection of high-producing Pichia pastoris clones. BMC Biotechnol. 11, 23 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rumjantsev A., Bondareva O., Padkina M. & Sambuk E. Effect of nitrogen source and inorganic phosphate concentration on methanol utilization and PEX genes expression in Pichia pastoris. Sci. World J. 2014, 9 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogl T., Ahmad M., Krainer F. W., Schwab H. & Glieder A. Restriction site free cloning (RSFC) plasmid family for seamless, sequence independent cloning in Pichia pastoris. Microb. Cell Fact. 14, 103 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall C., Brachat S. & Dietrich F. S. Contribution of Horizontal Gene Transfer to the Evolution of Saccharomyces cerevisiae. Eukaryot. Cell 4, 1102–1115 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gmeiner C. et al. Development of a fed-batch process for a recombinant Pichia pastoris Δoch1 strain expressing a plant peroxidase. Microb. Cell Fact. 14, 1 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S. & Letchworth G. J. High efficiency transformation by electroporation of Pichia pastoris pretreated with lithium acetate and dithiothreitol. Biotechniques 36, 152–154 (2004). [DOI] [PubMed] [Google Scholar]
- Invitrogen. Pichia Expression Kit. 31, 79 (2010).
- Weis R. et al. Reliable high-throughput screening with Pichia pastoris by limiting yeast cell death phenomena. FEMS Yeast Res. 5, 179–189 (2004). [DOI] [PubMed] [Google Scholar]
- Altschul S. F. et al. Gapped BLAST and PSI- BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon D., Abajian C. & Green P. Consed : A Graphical Tool for Sequence Finishing. Genome Res. 8, 195–202 (1998). [DOI] [PubMed] [Google Scholar]
- Wibberg D. et al. Complete genome sequencing of Agrobacterium sp. H13-3, the former Rhizobium lupini H13-3, reveals a tripartite genome consisting of a circular and a linear chromosome and an accessory plasmid but lacking a tumor-inducing Ti-plasmid. J. Biotechnol. 155, 50–62 (2011). [DOI] [PubMed] [Google Scholar]
- Zhu T., Guo M., Zhuang Y., Chu J. & Zhang S. Understanding the effect of foreign gene dosage on the physiology of Pichia pastoris by transcriptional analysis of key genes. Appl. Microbiol. Biotechnol. 89, 1127–1135 (2011). [DOI] [PubMed] [Google Scholar]
- Livak K. J. & Schmittgen T. D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25, 402–408 (2001). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.