Abstract
Objective
We recently identified a locus on chromosome 18q11.2 for high serum triglycerides (TGs) in Mexicans. We hypothesize that the lead GWAS single nucleotide polymorphism (SNP) rs9949617, or its linkage disequilibrium (LD) proxies, regulate one of the 5 genes in the TG-associated region.
Approach and Results
We performed an LD analysis and found 9 additional variants in LD (r2>0.7) with the lead SNP. To prioritize the variants for functional analyses, we annotated the 10 variants using DNase I hypersensitive sites, transcription factor (TF) and chromatin states, and identified rs17259126 as the lead candidate variant for functional in-vitro validation. Using luciferase transcriptional reporter assay in liver HepG2 cells, we found that the G allele exhibits a significantly lower effect on transcription (p<0.05). The electrophoretic mobility shift and ChIPqPCR assays confirmed that the minor G allele of rs17259126 disrupts an HNF4A binding site. To find the regional candidate gene, we performed a local expression quantitative trait locus (cis-eQTL) analysis and found that rs17259126 and its LD proxies alter expression of the regional transmembrane protein 241 (TMEM241) gene in 795 adipose RNAs from the METSIM cohort (p=6.11x10−07–5.80x10−04). These results were replicated in expression profiles of TMEM241 from the MuTHER resource (n=856).
Conclusions
The Mexican GWAS signal for high serum TGs on chromosome 18q11.2 harbors a regulatory SNP, rs17259126, which disrupts normal HNF4A binding and decreases the expression of the regional TMEM241 gene. Our data suggest that decreased transcript levels of TMEM241 contribute to increased TG levels in Mexicans.
Keywords: Genome-wide association study (GWAS), Triglycerides, Mexican, cis-expression quantitative trait locus (cis-eQTL), HNF4A
Introduction
Serum triglyceride (TG) levels are heritable and environmentally modifiable risk factor for cardiovascular disease (CVD)1. Several groups have successfully utilized genome-wide association studies (GWAS) to identify signals for TGs and other lipid traits, including high-density lipoprotein cholesterol (HDL-C), low-density lipoprotein cholesterol (LDL-C), and total cholesterol (TC)2. However, the lead GWAS signals may not themselves be functional rather in linkage disequilibrium (LD) with the actual underlying susceptibility variant. This limitation in GWAS derives from the fact that the human genome is only relatively superficially screened in GWAS using common tag single nucleotide polymorphisms (SNPs). Furthermore, the functional variant often acts through a regional gene. Therefore, GWASs are only a starting point and require subsequent fine mapping and functional validation studies to identify the actual susceptibility variants and genes.
According to a recent survey, both the U.S. Hispanic men and women have higher levels of serum TGs than non-Hispanic whites or blacks3, a result consistently reported for the last two decades4. Recent studies utilizing Latino cohorts have successfully narrowed European lipid loci5. Moreover, due to the higher incidence of metabolic disease in the Amerindian origin populations, the investigation of their admixed genomes provides an opportunity to identify Amerindian-specific susceptibility variants for complex cardiovascular traits6. Despite their high predisposition to dyslipidemias Hispanics remain underinvestigated as the discovery study stage in genomic cardiovascular studies. Previously we identified a locus on chromosome 18q11.2 associated with high serum TGs in Mexicans using GWAS5. However, similarly to other GWAS, the functional variants and the underlying gene(s) through which these variants exert their effects in the TG phenotype remain to be elucidated. To find the actual functional risk variant(s), we systematically annotated the SNPs in the TG-associated LD block with chromatin state marks and transcription factor (TF) binding events which nominated rs17259126 as the top candidate functional variant. Its genomic landscape harbors regulatory sites and is predicted to disrupt an HNF4A binding site. We show that the G allele of rs7259126 reduces expression of the luciferase reporter gene in a human liver cell line. Consistent with this result, the mobility shift and ChIPqPCR assays confirmed that the same allele disrupts an HNF4A binding site. Replicated cis-eQTL analyses also implicate the minor G allele of rs17259126 for reduced expression of transmembrane protein 241 (TMEM241), suggesting TMEM241 as the regional candidate gene. Taken together, we found that the TG locus on chromosome 18q11.2 harbors at least one functional variant, rs17259126, associated with a decreased expression of the regional TMEM241 gene, a novel gene for TGs in the rapidly growing Hispanic population with a high predisposition to dyslipidemias.
Materials and Methods
Materials and Methods are available in the online-only Data Supplement.
Results
Pairwise linkage disequilibrium analysis to identify LD proxies
In our original GWAS5, conditional association analyses at the top 12 genotyped loci did not reveal additional independent SNPs with p≤2.5×10−3. To identify the full set of variants in LD with the lead GWAS SNP rs9949617, we first performed a regional LD analysis in the TG-associated LD block. The LD block was determined in our previous study as the region spanning SNPs in LD of r2≥0.5 with the lead SNP rs99496175. For the LD analysis, we used our genotyped and imputed GWAS data5 and we also verified utilizing the 1000 Genomes Project data that no additional SNP(s) inside or outside this LD block (+/- 500 kb from the block borders) have emerged to be in LD with the lead SNP rs9949617 since our previous study5. We found 3 genotyped and 6 imputed SNPs in LD (r2≥0.7) with the lead SNP rs9949617 (table 1). Two of these 10 SNPs (rs9949617 and rs4800467) were genotyped in stages 1 and 2 of our original GWAS scan5, both resulting in p-values <5x10−8. As any of these 10 SNPs in LD can be the functional variant underlying the TG association on chromosome 18q11.2, we first performed functional annotation followed by hypothesis driven functional assays to uncover the functional variant in the TG-associated LD block. We also tested the candidate variant and its LD proxies for regional effects on gene expression among the five genes in the TG-associated LD block using a cis-eQTL analysis to investigate if the variant changes expression of a particular regional gene.
Table 1.
The lead SNP rs9949617 and its LD proxies are cis-eQTLs for the TMEM241 gene.
| SNP | Minor allele |
r2 | MAF (AMR) |
MAF (EUR) |
MAF (FIN) |
cis-eQTL p-value* (beta) |
|---|---|---|---|---|---|---|
| rs9949617† | T | 1.00 | 0.34 | 0.17 | 0.14 | 5.96×10-06 (−0.129) |
| rs9962573† | T | 0.97 | 0.32 | 0.17 | 0.14 | 5.96×10−06 (−0.129) |
| rs4800467† | G | 0.92 | 0.35 | 0.2 | 0.18 | 6.11×10−07 (−0.131) |
| rs1276322† | G | 0.82 | 0.33 | 0.17 | 0.14 | 1.7×10−04 (−0.109) |
| rs17259126 | G | 0.77 | 0.25 | 0.06 | 0.08 | 1.1×10−04 (−0.149) |
| rs9954334 | G | 1.00 | 0.34 | 0.17 | 0.14 | 5.96×10−06 (−0.128) |
| rs67124903 | G | 0.97 | 0.34 | 0.17 | 0.14 | 5.96×10−06 (−0.128) |
| rs71360517 | A | 0.88 | 0.29 | 0.14 | 0.10 | 5.80×10−04 (−0.112) |
| rs77127070 | A | 0.74 | 0.22 | 0.03 | 0.04 | N/A |
| rs4800154 | A | 0.74 | 0.23 | 0.03 | 0.04 | N/A |
The regional LD analysis uncovered 3 genotyped and 6 imputed variants in LD (r2≥0.7) with the lead SNP, rs9949617 in Mexicans. MAF indicates the minor allele frequency in the 1000 Genomes Project on the admixed American (AMR) individuals, European ancestry (EUR) individuals, and Finns (FIN). NA indicates not available.
The cis-eQTL p-values obtained in the analysis of the Finnish METSIM RNA-seq data (n=795) pass the Bonferroni correction for 50 tests (10 SNPs and 5 regional genes in the TG-associated LD block; p<0.001). The beta is shown for the minor allele.
Genotyped SNPs.
Functional genomics analysis using ENCODE data
Cis-eQTL variants often reside in regulatory elements such as transcription factor binding sites (TFBS) and interrupt TF occupancy, leading to transcriptional changes. However functional variants may also act through multiple other mechanisms making functional validation studies challenging. To facilitate the identification of suitable functional assays, we used the ENCODE data sets to give biological interpretation to the variants, and based on their predicted functionality, we conducted hypothesis driven functional assays. TFBS often coincide with regions of open chromatin, hence we annotated the chromatin state using ENCODE DNase I hypersensitive sites (DHS) and histone marks in disease relevant cell lines and control cell lines. In addition to the ENCODE biochemical annotations, we looked for TF motif disruptions using HaploReg. We hypothesized that variants with the greatest amount of regulatory evidence from experimental data sets and bioinformatic predictions are more likely to be functional. Utilizing this approach, we screened all 10 SNPs (the lead SNP and its 9 LD proxies) and selected rs17259126 as a top candidate for functional validation because it resides in a TFBS and a likely regulatory element defined by the co-occurrence of H3K27ac and H3K4me1. The G allele of rs17259126 is also predicted to disrupt a hepatocyte nuclear factor 4 alpha (HNF4A) regulatory motif (suppl figure I). HNF4A is a known regulator of several metabolic genes7. Based on these annotations, we hypothesized that rs17259126 resides in a TFBS and regulates expression of one of the regional genes on chromosome 18q11.
Functional validation of candidate variants
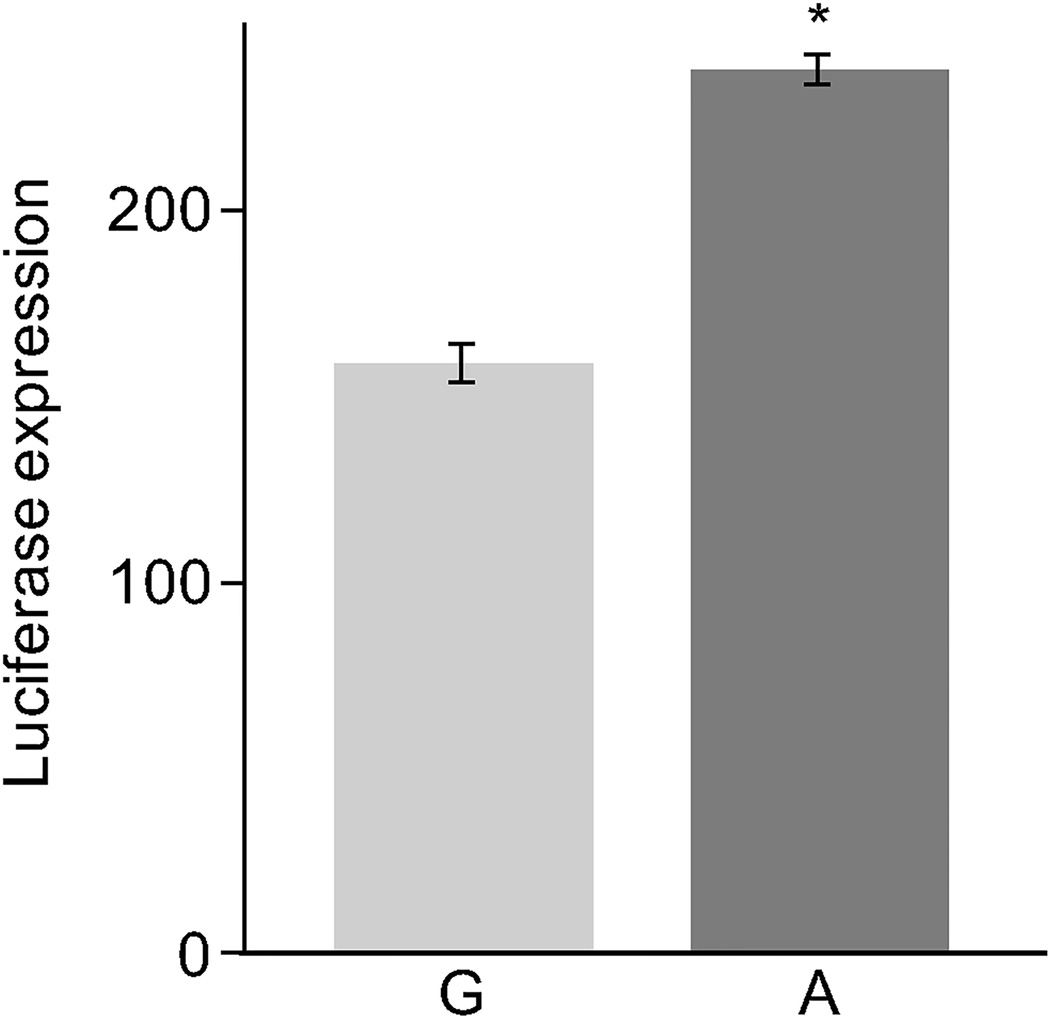
We sought to validate our predicted functional variant rs17259126. We performed luciferase reporter assays using engineered vectors containing a 600-bp sequence around the SNP. At 48 hrs post transfection of HepG2 cells we found that the minor allele G displays a 1.5-fold decreased reporter expression (p<0.05) compared to the major A allele in 3 biological replicates (figure 1). These results are consistent with the observed direction of the cis-eQTL effect (beta=-0.149, table 1) (see below). Similar assays for the lead SNP rs9949617 and rs4800467 did not reveal significant expression changes in the luciferase assay.
Figure 1.
Luciferase expression of rs17259126 alleles in HepG2 cells. The reference allele A exhibits a 1.5-fold increased reporter expression (p= 2.97x10−4) when compared to the minor allele G. Luciferase assays were read at 48 hrs post transfection. Statistical analysis was performed using the t test function in R. The Y axis represents the percent change in reporter expression relative to the empty pGL4.23[luc2/ minP] vector. Error bars indicate the standard errors (SE) for the means of three independent biological replicates each done in triplicate.
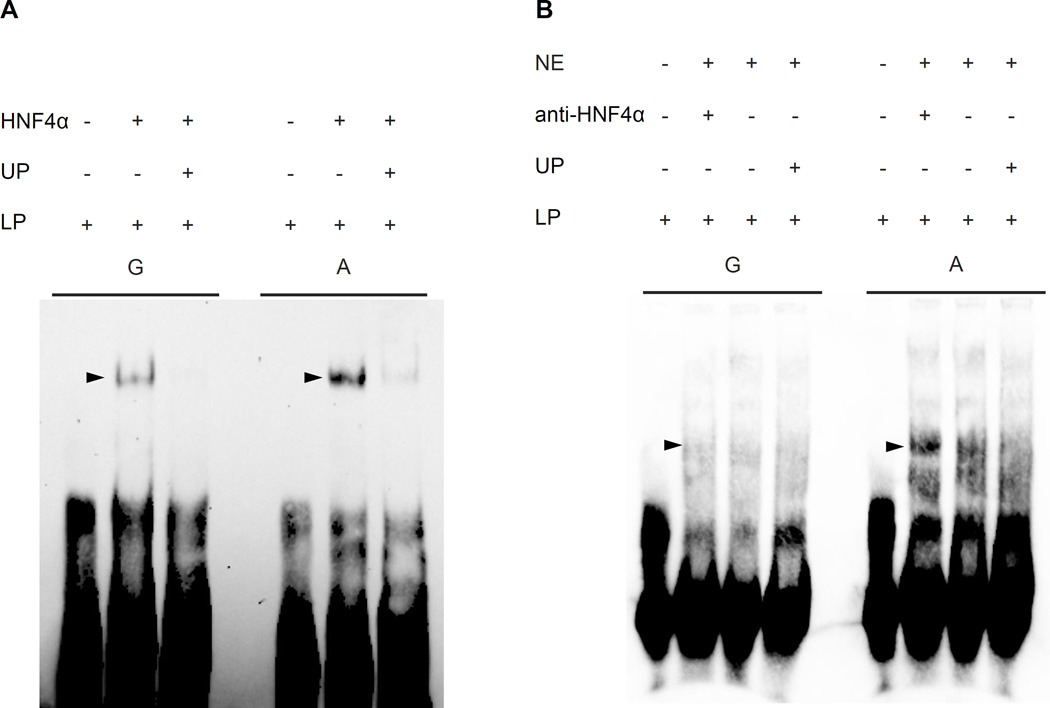
To further investigate whether the variant disrupts an HNF4A motif, we performed EMSA assays using isolated HNF4A protein (figure 2A) or HepG2 cell nuclear extracts (figure 2B) and found evidence that HNF4A preferentially binds the major A allele of rs17259126 in four biological replicates. We also performed EMSAs for the 9 other LD proxies variants. No allele-specific shifts were observed (suppl figure II). Together the luciferase (figure 1) and EMSA (figure 2) assays suggest that HNF4A may regulate expression of a target gene by directly binding to the rs17259126 regulatory site.
Figure 2.
As predicted by motif analysis (see suppl figure I), EMSA assays revealed that HNF4A recombinant protein directly interacts and displays a higher affinity for the reference A allele of the TMEM241 eQTL SNP rs17259126 (figure 2A) although no super shift was obtained when using HepG2 nuclear extract and anti-HNF4A (figure 2B). a) HNF4A isolate. b) HepG2 hepatoma nuclear extract. HNF4A (HNF4A isolated protein), unlabeled probe (UP), labeled probe (LP), nuclear extract (NE), and anti-HNF4A (anti-HNF4A).
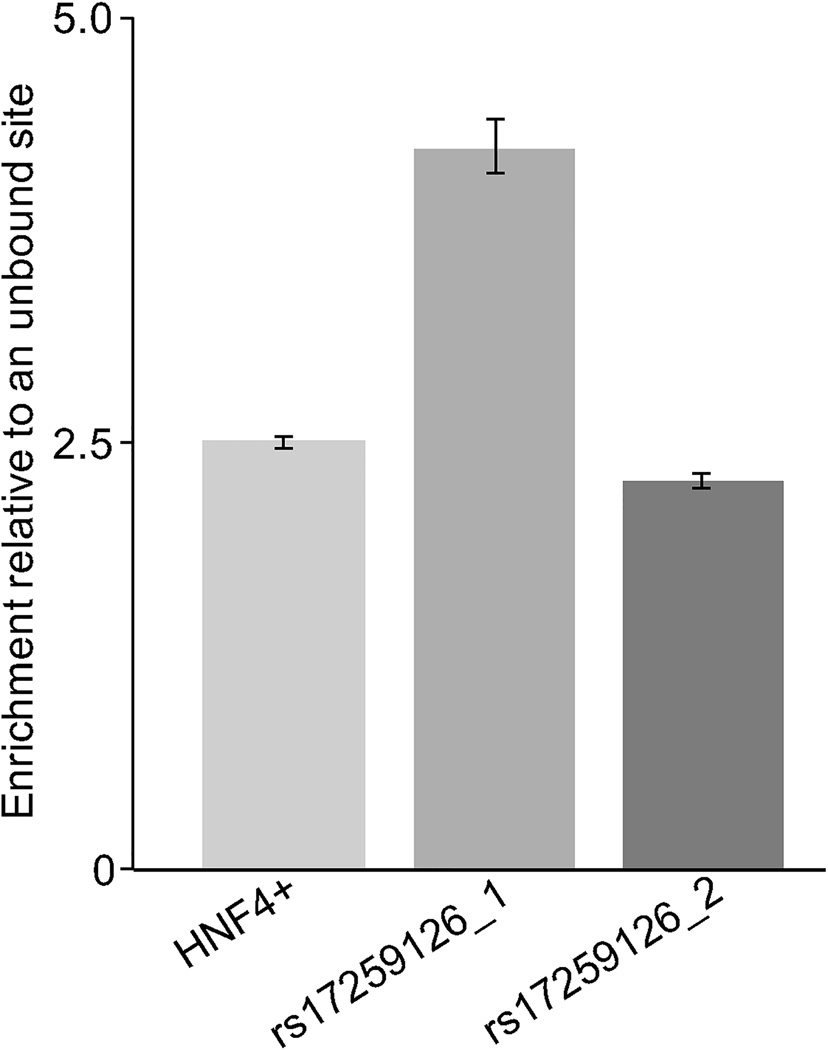
To confirm that HNF4A interacts with the variant site in HepG2 cells, we performed chromatin immunoprecipitation followed by qPCR targeting a 71-bp (site 1) or 151-bp (site 2) sequence surrounding rs17259126 (figure 3). We found an average enrichment of 4.23 and 2.29 for the sequences, respectively, when compared to an unbound control site. Our functional studies provide converging evidence that the sequence underlying rs17259126 is an HNF4A binding site and that the G minor allele significantly inhibits this interaction in vitro.
Figure 3.
ChIP DNA analyzed by real-time PCR showed that HNF4A binds the TMEM241 eQTL SNP rs17259126 site in hepatoma cells. The Y axis represents the enrichment of HNF4A at the rs17259126 site relative to an unbound control site. Error bars represent the standard error for the means of three experimental replicates each done in triplicate. The region surrounding rs17259126 was targeted using two different pairs of oligos, indicated as rs17259126_1 and rs17259126_2. The sequences of the oligos are given in suppl table I.
Cis-expression quantitative trait locus (cis-eQTL) analysis
GWAS variants residing in regulatory elements such as TFBS can lead to gene expression changes and contribute to disease susceptibility. We investigated whether the lead GWAS SNP may affect expression of the regional genes in the ~300-kb region defining the TG-associated window on chromosome 18q11.2 (LD r2>0.5 with the lead SNP)5. We performed a cis-eQTL analysis for the 5 genes within this TG-associated LD block using adipose RNA-seq samples (n=795) from the METSIM cohort and discovered that the lead SNP rs9949617 (i.e. the SNP with the strongest TG association signal4) and its LD proxies are a cis-eQTL, regulating the expression of one regional gene, the transmembrane protein 241 (TMEM241) (p=6.11x10−07–5.80x10−04) (table 1). These results pass the Bonferroni correction for the 50 performed tests (10 SNPs tested for 5 regional genes; p<0.001) (table 1), and the 10 TG-associated SNPs did not regulate expression of any of the 4 other genes within the LD block (Bonferroni corrected p>0.05).
To validate and replicate these regional cis-eQTL results, we utilized expression data from 856 publicly available human adipose, skin, and lymphocyte RNA microarray samples from the MuTHER resource8, and similarly discovered that the lead SNP rs9949617 is a cis-eQTL (suppl figure III), regulating the expression of TMEM241 (p<1x10−5 across all three tissues, beta=-0.107 for adipose). These replication data are consistent, including the direction of the effect, with our cis-eQTL signal in Finns and our luciferase assays in which the minor G allele results in a decreased expression (table 1 and figure 1). We also found comparable cis-eQTL results for the lead SNP rs9949617 in the HapMap3 data sets for the CEU (p=0.0010), CHB (p=0.0019), and JPT (p=6.0x10−4) samples in lymphoblastoid cells. Although there was a trend towards significance, this relationship did not hold for the MEX HapMap sample (p=0.20), perhaps due to the low number of Mexican-American samples (n=45) included in the HapMap project. These results implicate TMEM241 as a likely regional gene underlying the GWAS association because the lead SNP and/or its LD proxies robustly regulate TMEM241 expression through multiple cohorts. Taken together these data suggest that rs17259126 is at least one of the functional SNPs underlying the original TG GWAS signal5 on chromosome 18q11.2 in Amerindian origin populations.
Discussion
We recently identified a locus on chromosome 18q11.2 associated with high serum TGs in Mexicans using GWAS5. However, GWAS typically do not conclusively identify a functional regulatory variant and candidate gene, rather they require statistical and biochemical follow-up studies9, 10. We used statistical fine mapping to first identify variants in the TG-associated LD block. Since all variants represent 3’UTR or non-coding variants, we annotated their biological function using available regulatory datasets and bioinformatic tools, and subsequently validated our recorded annotations using appropriate molecular assays.
Our LD analyses, uncovered 9 variants in LD with the lead GWAS SNP rs9949617. Functional annotations using HaploReg11 found that rs17259126 is predicted to disrupt an HNF4A binding site, the minor G allele exhibiting a lower enrichment score. Furthermore, the ENCODE TF ChIP-seq data in HepG2 showed evidence of HNF4A enrichment around rs17259126. These findings prompted us to nominate rs17259126 as the lead candidate for molecular validation. We performed HNF4A ChIPqPCR targeting the SNP region, and confirmed that HNF4A indeed binds the SNP site. HNF4A is a well-known, central regulator of hepatocyte development, differentiation, and gene expression12, 7 associated with type 2 diabetes (T2D), consistent with the TG association. In line with our bioinformatics prediction, we also show that the G allele of rs17259126 reduces transcription of the luciferase reporter and significantly inhibits HNF4A binding in mobility shift assays. It is worth noting that Amerindian origin populations have >3-fold higher frequency of the minor allele G of rs17259126 when compared to Europeans (MAFs for AMR=0.22, EUR=0.06, AFR=0.08, and ASN=0.20, respectively).
To identify the regional gene, we performed cis-eQTL analyses using expression data from multiple cohorts, tissues, and platforms. We provide replicated evidence that the minor G allele of rs17259126 and its LD proxies are a robust cis-eQTL decreasing expression of the regional TMEM241 gene across many cohorts. Our results suggest that HNF4A binds the A allele of rs17259126 site and increases expression of the TMEM241 gene, one of the five regional genes in the LD block. We hypothesize that individuals with the G allele have decreased TMEM241 expression which affects the normal TG synthesis and/or secretory pathways through an unknown mechanism.
The TMEM241 gene is a yeast VRG4 homolog, a Golgi-localized GDP-mannose transporter. Yeast VRG4 is pleiotropically required for a range of Golgi functions, including N-linked glycosylation, secretion, protein sorting, and the maintenance of a normal endomembrane system13, 14. In the mammalian Golgi, carbohydrate processing is a highly diverse process. Carbohydrate chains may contain galactose, sialic acid, fucose, xylose, N-acetylglucosamine, and N-acetylgalactosamine unlike in the yeast S. cerevisiae, where glycosylation is restricted to mannosylation. Thus, human TMEM241 may function in the transport of other nucleotide sugars required in mammalian systems. In addition to glycoproteins, sphingolipids are also modified in the Golgi and have been implicated in metabolic disease15. TMEM241 is believed to function as a nucleotide sugar transporter, and when defective, may lead to underglycosylation of glycoproteins and sphingolipids, potentially resulting in dysregulation of TG synthesis.
Together, our results provide converging evidence suggesting rs17259126 as one of the functional variants underlying the GWAS association signal on 18q11.25, and TMEM241 as the underlying gene for TGs in Amerindian origin populations. However, because not all individuals of Mexican ancestry share the same composition of Amerindian DNA, additional cohorts may or may not replicate this particular association.
Future studies focusing on characterizing the role of TMEM241 in TG metabolism could include CRISPR/Cas916, an emerging technology for targeted genomic modification. This technology allows a site-specific genetic engineering in disease relevant cell lines to interrogate the function of specific genes and single nucleotide variants in their native chromatin state. Elucidation of the role of TMEM241 in TG metabolism may help guide future research and development of new therapies for effective TG management and prevention of heart disease in the rapidly growing Hispanic populations, currently underinvestigated in genomic cardiovascular studies despite their high predisposition to dyslipidemias.
Supplementary Material
Highlights.
The TG locus on chromosome 18q11.2 harbors at least one functional variant, rs17259126, associated with a decreased expression of the regional TMEM241 gene, a novel gene for TGs in the Hispanic population.
HNF4A may regulate the expression of the TMEM241 gene by directly binding the rs17259126 regulatory site.
Our findings suggest that decreased transcript levels of TMEM241 contribute to increased TG levels in Mexicans.
Acknowledgments
We thank the Mexican and Finnish individuals who participated in this study. We also thank Saúl Cano-Colín for laboratory technical assistance. Michael Boehnke and Francis Collins are thanked for providing the METSIM genotype data.
Funding
This study was funded by the NIH grants HL-095056, HL-28481, and DK093757. AR was supported by the National Science Foundation Graduate Research Fellowship Program NSF grant number DGE-1144087, and AK by the NIH grant F31HL127921. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Genotyping services for the METSIM cohort were supported by NIH grants DK072193, DK093757, DK062370, and Z01HG000024 and provided by the Center for Inherited Disease Research (CIDR). CIDR is fully funded through a federal contract from the National Institutes of Health to The Johns Hopkins University, contract number HHSN268201200008I.
Abbreviations
- TG
Triglycerides
- GWAS
Genome-wide association study
- SNP
Single nucleotide polymorphism
- LD
Linkage disequilibrium
- cis-eQTL
cis-expression quantitative trait locus
- HNF4A
Hepatocyte nuclear factor 4 alpha
- ENCODE
Encyclopedia of DNA elements
Footnotes
Disclosures
The authors have declared that no conflict of interest exists.
References
- 1.Willer CJ, Schmidt EM, Sengupta S, et al. Discovery and refinement of loci associated with lipid levels. Nat Genet. 2013;45:1274–1283. doi: 10.1038/ng.2797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Go AS, Mozaffarian D, Roger VL, et al. Executive Summary: Heart Disease and Stroke Statistics-2014 Update: A Report From the American Heart Association. Circulation. 2014;129:399–410. doi: 10.1161/01.cir.0000442015.53336.12. [DOI] [PubMed] [Google Scholar]
- 3.Aguilar-Salinas CA, Canizales-Quinteros S, Rojas-Martínez R, Mehta R, Villarreal-Molina MT, Arellano-Campos O, Riba L, Gómez-Pérez FJ, Tusié-Luna MT. Hypoalphalipoproteinemia in populations of Native American ancestry: an opportunity to assess the interaction of genes and the environment. Curr Opin Lipidol. 2009;20:92–97. doi: 10.1097/mol.0b013e3283295e96. [DOI] [PubMed] [Google Scholar]
- 4.Aguilar-Salinas CA, Tusie-Luna T, Pajukanta P. Genetic and environmental determinants of the susceptibility of Amerindian derived populations for having hypertriglyceridemia. Metabolism. 2014;63:887–894. doi: 10.1016/j.metabol.2014.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Weissglas-Volkov D, Aguilar-Salinas CA, Nikkola E, et al. Genomic study in Mexicans identifies a new locus for triglycerides and refines European lipid loci. J Med Genet. 2013;50:298–308. doi: 10.1136/jmedgenet-2012-101461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ko A, Cantor RM, Weissglas-Volkov D, et al. Amerindian-specific regions under positive selection harbour new lipid variants in Latinos. Nat Commun. 2014;5:1–12. doi: 10.1038/ncomms4983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hayhurst GP, Lee YH, Lambert G, Ward JM, Gonzalez FJ. Hepatocyte nuclear factor 4alpha (nuclear receptor 2A1) is essential for maintenance of hepatic gene expression and lipid homeostasis. Mol Cell Biol. 2001;21:1393–1403. doi: 10.1128/MCB.21.4.1393-1403.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grundberg E, Small KS, Hedman ÅK, et al. Mapping cis- and trans-regulatory effects across multiple tissues in twins. Nat Genet. 2012;44:1084–1089. doi: 10.1038/ng.2394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schaub MA, Boyle AP, Kundaje A, Batzoglou S, Snyder M. Linking disease associations with regulatory information in the human genome. Genome Res. 2012;22:1748–1759. doi: 10.1101/gr.136127.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lo KS, Vadlamudi S, Fogarty MP, Mohlke KL, Lettre G. Strategies to fine-map genetic associations with lipid levels by combining epigenomic annotations and liver-specific transcription profiles. Genomics. 2014;104:105–112. doi: 10.1016/j.ygeno.2014.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ward LD, Kellis M. HaploReg: a resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Res. 2011;40:D930–D934. doi: 10.1093/nar/gkr917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Parviz F, Matullo C, Garrison WD, Savatski L, Adamson JW, Ning G, Kaestner KH, Rossi JM, Zaret KS, Duncan SA. Hepatocyte nuclear factor 4α controls the development of a hepatic epithelium and liver morphogenesis. Nat Genet. 2003;34:292–296. doi: 10.1038/ng1175. [DOI] [PubMed] [Google Scholar]
- 13.Hansen HG, Schmidt JD, Soltoft CL, Ramming T, Geertz-Hansen HM, Christensen B, Sorensen ES, Juncker AS, Appenzeller-Herzog C, Ellgaard L. Hyperactivity of the Ero1 Oxidase Elicits Endoplasmic Reticulum Stress but No Broad Antioxidant Response. J Biol Chem. 2012;287:39513–39523. doi: 10.1074/jbc.M112.405050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dean N, Zhang YB, Poster JB. The VRG4 Gene Is Required for GDP-mannose Transport into the Lumen of the Golgi in the Yeast, Saccharomyces cerevisiae. J Biol Chem. 1997;272:31908–31914. doi: 10.1074/jbc.272.50.31908. [DOI] [PubMed] [Google Scholar]
- 15.Brice SE, Cowart LA. Sphingolipid metabolism and analysis in metabolic disease. Adv Exp Med Biol. 2011;721:1–17. doi: 10.1007/978-1-4614-0650-1_1. [DOI] [PubMed] [Google Scholar]
- 16.Hsu PD, Lander ES, Zhang F. Development and Applications of CRISPR-Cas9 for Genome Engineering. Cell. 2014;157:1262–1278. doi: 10.1016/j.cell.2014.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.