Abstract
Cervical cancer (CC) is one of the most common malignancies in women. Paclitaxel is the front-line chemotherapeutic agent for treating CC. However, its therapeutic efficacy is limited because of chemoresistance, the mechanism of which remains poorly understood. Here, we used microRNA (miRNA) arrays to compare miRNA expression levels in the CC cell lines, HeLa and CaSki, with their paclitaxel resistance counterparts, HeLa/PR and CaSki/PR. We demonstrate that miR-125a was one of most significantly downregulated miRNAs in paclitaxel-resistant cells, which also acquired cisplatin resistance. And that the upregulation of miR-125a sensitized HeLa/PR and CaSki/PR cells to paclitaxel both in vitro and in vivo and to cisplatin in vitro. Moreover, we determined that miR-125a increased paclitaxel and cisplatin sensitivity by downregulating STAT3. MiR-125a enhanced paclitaxel and cisplatin sensitivity by promoting chemotherapy-induced apoptosis. Clinically, miR-125a expression was associated with an increased responsiveness to paclitaxel combined with cisplatin and a more favorable outcome. These data indicate that miR-125a may be a useful method to enable treatment of chemoresistant CC and may also provide a biomarker for predicting paclitaxel and cisplatin responsiveness in CC.
Introduction
Cervical cancer (CC) is a common gynecological malignancy that is a leading cause of cancer-related mortality among women worldwide.1, 2, 3 Paclitaxel is a front-line chemotherapeutic agent for treating CC, usually in combination with other chemotherapeutic agents.4, 5 However, the therapeutic efficacy of paclitaxel is limited, with response rates between 29–63% because of chemoresistance.4, 5, 6 Paclitaxel resistance is caused by several mechanisms, including overexpression of P-glycoprotein or other drug efflux pumps,7, 8 alterations to microtubules involved in drug-binding or altered expression of tubulin isotypes and microtubule-associated proteins,9, 10, 11 alterations to cell cycle and cell survival pathways12, 13, 14 and the induction of treatment-related autophagy.15, 16, 17 However, the molecular mechanisms by which resistance to paclitaxel occurs are not fully understood and further investigation is required.
MicroRNAs (miRNAs) are a class of endogenous short noncoding RNAs that inhibit post-transcriptional gene expression by binding to target mRNA at their 3′-untranslated region (UTR).18 Aberrant expression of miRNA has been associated with cancer chemoresistance, including resistance to paclitaxel.16, 17, 19 MiR-125a is an anti-oncogene that has a key role in tumorigenesis in multiple cancers20 and is crucial for paclitaxel sensitivity in colon cancer21 and cisplatin sensitivity in nasopharyngeal carcinoma.22 However, its importance in enabling sensitivity of CC to paclitaxel has not been explored.
In this study, we found miR-125a was significantly downregulated in paclitaxel-resistant CC cells. Overexpressing miR-125a in paclitaxel-resistant cells increases the cell sensitivity not only to paclitaxel both in vitro and in vivo but also to cisplatin in vitro by enabling apoptosis via suppressing STAT3 expression. High expression of miR-125a in CC patients was associated with a favorable response to paclitaxel combined with cisplatin treatment and prognosis. Therefore, upregulating miR-125a may be a novel way to treat chemoresistant CC and miR-125a may be a useful biomarker for predicting the response of CC to paclitaxel and cisplatin.
Results
miRNA profiles in paclitaxel-sensitive and -resistant CC cells
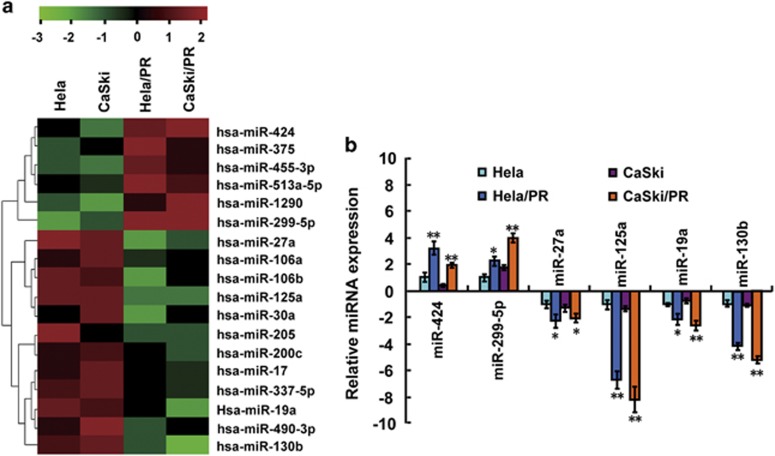
To screen critical miRNAs associated with paclitaxel resistance in CC, we simultaneously analyzed miRNA expression in two CC cell lines (HeLa and CaSki) and their paclitaxel-resistant counterparts (HeLa/PR and CaSki/PR cells; Supplementary Figures 1A and 1B) using miRNA array chips that covered a total 2549 miRNAs. A total of 18 differentially expressed miRNAs were detected in paclitaxel-resistant cells compared with paclitaxel-sensitive cells, including six upregulated miRNAs and 12 downregulated miRNAs (Figure 1a). To further validate the miRNA array chip results, we randomly selected six miRNAs (two upregulated miRNAs, miR-424 and miR-229-5p, and four downregulated miRNAs, miR-27a, miR-125a, miR-19a and miR-130b) to determine their expression by reverse transcription–polymerase chain reaction (RT–PCR). The results were consistent with the miRNA array experiments (Figure 1b). MiR-125a was the most differentially expressed miRNA detected, with more than sixfold lower expression in HeLa/PR and CaSki/PR cells when compared with HeLa and CaSki cells (Figure 1b). As miR-125a expression was associated with paclitaxel-sensitive cells, we hypothesized that miR-125a has a prominent role in paclitaxel resistance of CC.
Figure 1.
Different expression of miRNAs in paclitaxel-sensitive and -resistant CC cells. (a) A heat map showing the miRNA expression profiles and supervised hierarchical clustering analysis for paclitaxel-sensitive (HeLa and CaSki) and -resistant (HeLa/PR and CaSki/PR) CC cell lines. Significantly differentially expressed miRNAs matched the threshold (differential ⩾2-fold, chip signal value >500) and statistical analysis standard were selected (P<0.05). Each column represents a cell line and each row shows the relative expression level for individual miRNAs. The red and green colors indicate high of low expression, respectively. (b) Expression of six miRNAs (miR-424, miR-229-5p, miR-27a, miR-125a, miR-19a and miR-130b) in the CC cell lines were validated by RT–PCR. U6 small nuclear RNA was used as an internal control. All values are the mean±s.d. of triplicate measurements, and experiments were repeated three times. *P<0.05, **P<0.01 compared with matched CC cells.
Paclitaxel and cisplatin sensitivities are modulated by changes in miR-125a expression in vitro
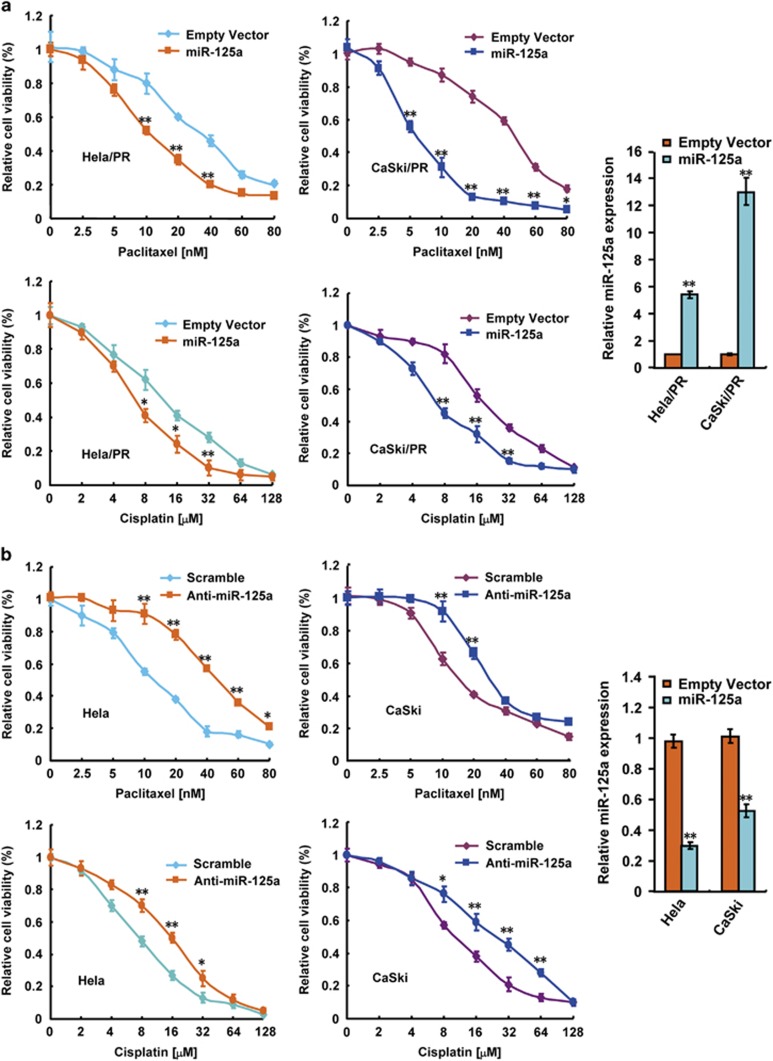
To investigate the biological functions of miR-125a in paclitaxel sensitivity of CC cells, we analyzed the relationship between miR-125a and the IC50 of paclitaxel. The IC50 values derived from paclitaxel-treated HeLa, CaSki, HeLa/PR and CaSki/PR were 7.76±0.55, 14.1±1.1, 39.89±2.87 and 41.67±3.88 nM, respectively (Supplementary Figure 2A). miR-125a expression was negatively correlated with the IC50 values of paclitaxel in all four cell lines (P=0.0484, r=−0.9372; Supplementary Figure 2B). To confirm the association between paclitaxel resistance and miR-125a expression, miR-125a was transfected into HeLa/PR and CaSki/PR cells, which were then treated with increasing concentrations of paclitaxel. In this viability assay, miR-125a overexpression increased the sensitivity of HeLa/PR and CaSki/PR cells to paclitaxel (Figure 2a). In addition, suppression of miR-125a with specific miR-125a inhibitor in HeLa and CaSki cells increased the paclitaxel resistance (Figure 2b). Recently, Chen et al.22 reported that miR-125a correlated with cisplatin and cisplatin is another important agent for treating CC. Consistent with the results reported by Chen et al. in CC, we repeated the experiments as paclitaxel using increasing concentrations of cisplatin. As expected, paclitaxel-resistant cells also acquired cisplatin resistance (Supplementary Figures 1A and 1B). Moreover, similar effects were observed that miR-125a increased cisplatin resistance as paclitaxel (Figures 2a and b). These results suggest that miR-125a expression levels are positively correlated with the sensitivity of CC cells to paclitaxel and cisplatin.
Figure 2.
Altered miR-125a expression modulates sensitivity to paclitaxel in vitro. (a) Overexpression of miR-125a in paclitaxel-resistant cervical cells (HeLa/PR and CaSki/PR) were treated with increasing concentrations of paclitaxel as indicated. After 24 h, cell viability assays were performed using a CCK-8 kit. The group not treated with paclitaxel was presented as 100% viable cells and was used as an internal control for comparison. Histograms on the right show the level of miR-125a overexpression. (b) A cell viability assay on paclitaxel-sensitive cells (HeLa and CaSki) following knockdown of miR-125a expression. The cells were treated as in a. Histograms on the right show the level of miR-125a knockdown. All values shown are the mean±s.d. of triplicate samples. Experiments were repeated three times (*P<0.05, **P<0.01).
miR-125a inhibits STAT3 expression by binding to its 3′-UTR
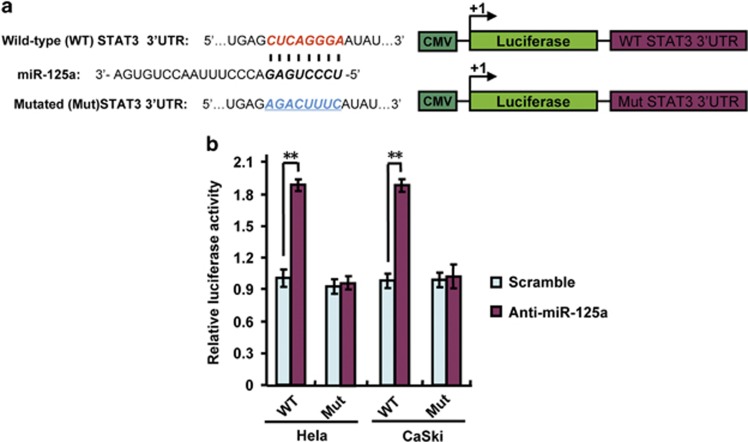
Previous studies have indicated that miR-125a directly targets STAT3.23, 24 To confirm this, we analyzed the binding ability of anti-miR-125a to wild-type or mutant STAT3 3′-UTR using the luciferase reporter assays. Our results indicated that miR-125a suppression increased wild-type STAT3 3′-UTR reporter activity in HeLa and CaSki cells but did not alter luciferase activity in cells with mutations in the binding sites for miR-125a (Figures 3a and b), which further confirmed previous study results in CC cells. These results indicate that miR-125a inhibits STAT3 expression by directly binding to its 3′-UTR in CC cells.
Figure 3.
Anti-miR-125a activates STAT3 expression by targeting its 3′-UTR. (a) A schematic diagram of the STAT3 3′-UTR luciferase reporter constructs. Bold and italicized fonts indicate putative miR-125a-binding sites in the human STAT3 3′-UTR. Underlining indicates mutations introduced into the STAT3 3′-UTR. (b) A miRNA luciferase reporter assay using HeLa and CaSki cells transfected with wild-type or mutated STAT3 reporters plus anti-miR-125a. All values shown are the mean±s.d. of triplicate samples. Experiments were repeated three times (**P<0.01).
miR-125a increased sensitivity of CC to paclitaxel by altering apoptosis because of the downregulation of STAT3
STAT3 has been shown to inhibit apoptosis, the mechanism of which contributes to Lee et al.25 Thus, we analyzed the expression of STAT3, STAT3 phosphorylated tyrosine 705 (p-STAT3 (Tyr705)), two key apoptosis inhibitors Bcl-2 and Bcl-xL using western blot assays in paclitaxel-sensitive and -resistant CC cell lines. The results indicated that the expression of STAT3 and p-STAT3 (Tyr705) were increased in paclitaxel-resistant cells (HeLa/PR and CaSki/PR) when compared with paclitaxel-sensitive cells (HeLa and CaSki). Similar trends were observed for the expression of Bcl-2 and Bcl-xL, which were also downstream effectors of STAT3 (Supplementary Figure 3).
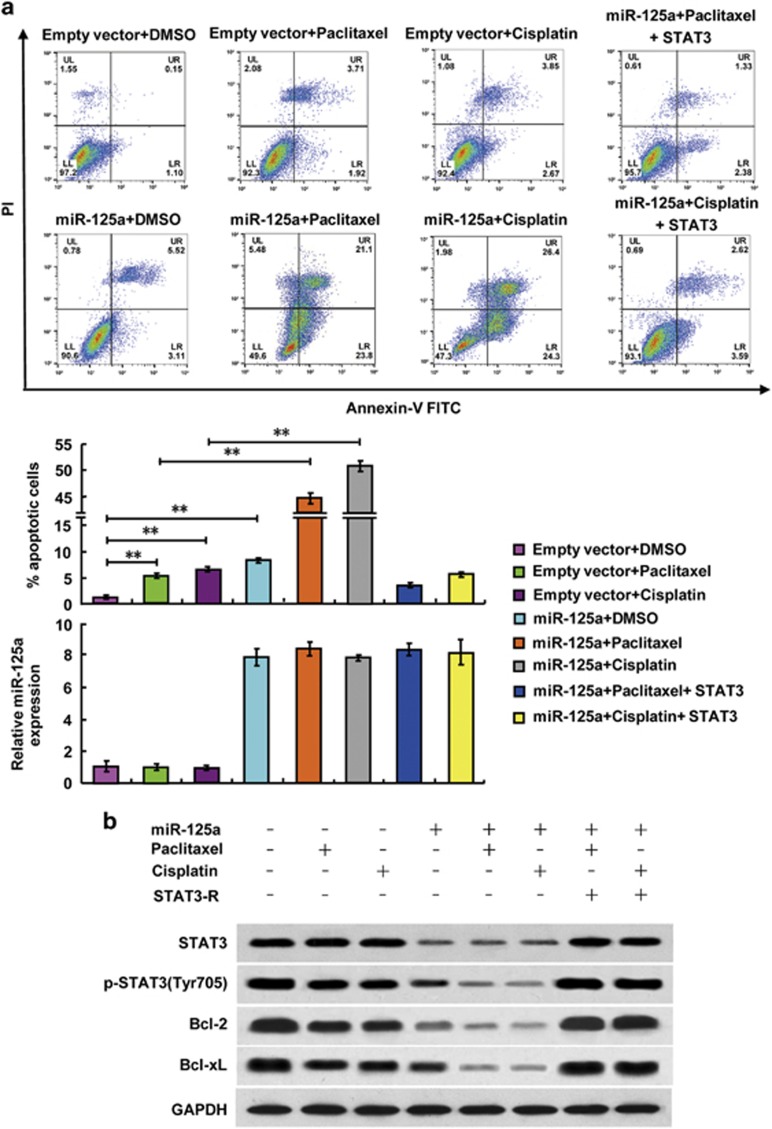
As miR-125a directly targeted STAT3 and increased paclitaxel and cisplatin sensitivity, we hypothesized that miR-125a increased paclitaxel and cisplatin sensitivity in CC by enabling apoptosis through downregulation of STAT3. To confirm this hypothesis, we examined the effects of miR-125a and chemotherapeutic agents on the apoptosis of HeLa/PR cells by flow cytometry. The overexpression of miR-125a, paclitaxel treatment and cisplatin treatment, individually, increased the proportion of apoptotic HeLa/PR cells to 8.63%, 5.63% and 6.52%, respectively, compared with that of the control cells (1.25% apoptotic; Figure 4a). Moreover, HeLa/PR cells that overexpressed miR-125a and were treated with paclitaxel or cisplatin had a significantly higher proportion of apoptotic cells (44.9% and 50.7%, respectively; Figure 4a). Similar results were obtained using CaSki/PR cells (Supplementary Figure 4A). Furthermore, treatment with paclitaxel, cisplatin or overexpression of miR-125a inhibited the expression of p-STAT3 (Tyr705), Bcl-2 and Bcl-xL. However, STAT3 expression was suppressed only by miR-125a overexpression. The simultaneous treatment with paclitaxel or cisplatin plus overexpression of miR-125a induced the greatest inhibition of p-STAT3 (Tyr705), Bcl-2 and Bcl-xL (Figure 4b and Supplementary Figure 4B). Inducing the re-expression of STAT3 in the CC cells reversed the effects of miR-125a overexpression and chemotherapeutic agents treatment-induced apoptosis (Figures 4a and b). These data indicate that, miR-125a enables CC cell paclitaxel and cisplatin sensitivity by inducing apoptosis pathway via downregulation of STAT3.
Figure 4.
miR-125a suppresses paclitaxel resistance in HeLa/PR cells by downregulating STAT3 resulting in altered expression of apoptosis-related genes. (a) Representative flow cytometry analysis of annexin V (1:1000) and propidium iodide (1:1000) staining in HeLa/PR cells transfected with miR-125a or miR-125a plus STAT3 and treated with or without paclitaxel (20 nM) or cisplatin (10 μM) for 24 h. The proportion of apoptotic cells is shown as the mean±s.d. from three independent experiments (**P<0.01). Histograms show the miR-125a expression levels. (b) Representative western blots using the indicated antibodies in HeLa/PR cells transfected and treated as in a.
Role of miR-125a in modulating paclitaxel resistance in vivo
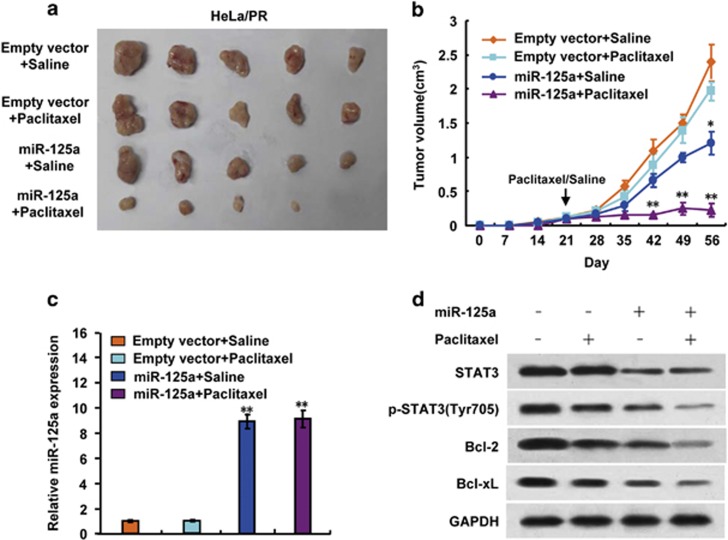
After determining that miR-125a mediates paclitaxel resistance in different CC cell lines in vitro, we investigated the phenotype of cells overexpressing miR-125a in vivo. MiR-125a-overexpressing HeLa/PR cells or control HeLa/PR cells were subcutaneously injected into the backs of BALB/c nu/nu mice. Once the mice developed palpable tumors (>5 mm in diameter within 3 weeks), they were randomly assigned into paclitaxel or saline treatment groups. There was no significant difference in initial tumor volumes (within 3 weeks) between the four groups. Paclitaxel (15 mg/kg) or the same volumes of saline were intraperitoneally injected once a week for 5 weeks. Paclitaxel induced a slight inhibition of tumor growth, however, miR-125a overexpression in paclitaxel-resistant HeLa cells induced an inhibition of tumor growth. The combination of miR-125a overexpression and paclitaxel treatment induced a significant reduction of tumor growth (Figures 5a and b). RT–PCR and immunoblot analysis confirmed that the expression level of miR-125a, STAT3, p-STAT3 (Tyr705), Bcl-2 and Bcl-xL in HeLa/PR cells from the representative tumor mass (Figures 5c and d). These findings indicate that miR-125a upregulation increases paclitaxel sensitivity in paclitaxel-resistant CC cells.
Figure 5.
miR-125a mediates paclitaxel sensitivity in vivo. HeLa/PR and HeLa/PR cells overexpressing miR-125a were injected into nude mice. After 3 weeks, paclitaxel (15 mg/kg) or saline was intraperitoneally injected once a week for 5 weeks. At the times indicated, tumors were measured with Vernier Calipers (mean±s.d.; n=5) (a, b). Arrow represents the beginning treatment with paclitaxel or saline. (c) RT–PCR analysis of miR-125a expression from representative excised tumors. (d) Immunoblot analysis of representative excised tumor mass from a (*P<0.05, **P<0.01).
miR-125a expression correlates with PFS and OS in CC patients who received paclitaxel-based chemotherapy
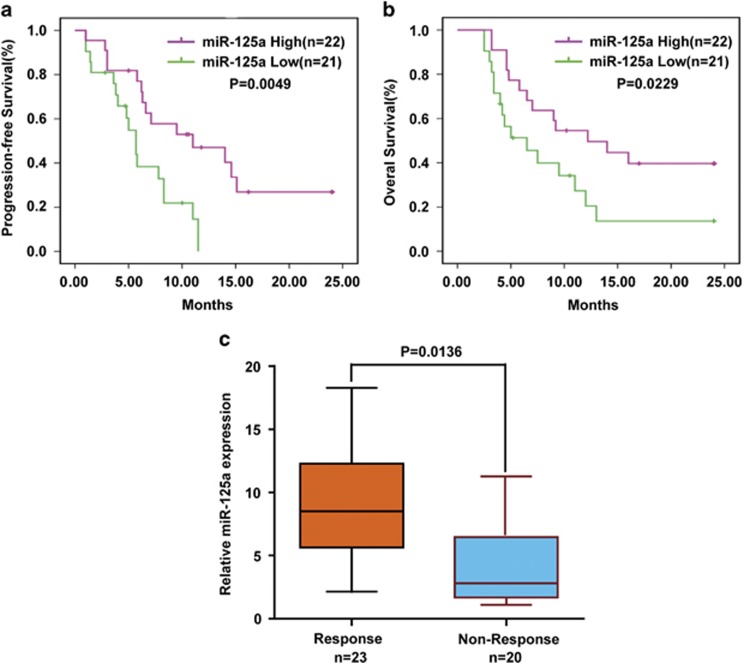
To study the relationship between miR-125a and paclitaxel resistance in CC, we selected 43 patients who had received paclitaxel combined with cisplatin chemotherapy as the first-line treatment. We stratified patients into high miR-125a expression and low miR-125a expression according to miR-125a expression levels in their CC cells. The effect of chemotherapy on the patient's progression-free survival (PFS) and overall survival (OS) was evaluated according to the Response Evaluation Criteria in Solid Tumors. Patients in the low miR-125a expression group had poorer PFS (P=0.0049; Figure 6a), OS (P=0.0229; Figure 6b) and response rate (P=0.015; Table 1) than those in the high miR-125a expression group. Furthermore, compared with patients who responsed to chemotherapy, miR-125a expression was significantly downregulated in non-responsed patients (P=0.0136; Figure 6c). These data indicate important roles for miR-125a in CC paclitaxel resistance.
Figure 6.
Expression of miR-125a correlates with PFS and OS in paclitaxel treated CC patients. (a, b) CC patients who received paclitaxel-based chemotherapy were separated into groups based on low or high miR-125a expression levels. Kaplan–Meier survival curves and log-rank tests were used to compare the (a) PFS and (b) OS between the two groups. (c) Expression of miR-125a in patients responded to paclitaxel (n=18) and no-responsed (n=20) to paclitaxel was compared using the two-tailed Student's t-test. U6 small nuclear RNA was used as an internal control.
Table 1. Clinical correlations of miR-125a expression in cervical carcinoma.
| Variables |
miR-125a expression in tumor tissues (T) |
P-value | |
|---|---|---|---|
| High expression | Low expression | ||
| N=22 (51.2%) | N=21 (48.8%) | ||
| Age (mean±s.d.), years | 53.6±9.78 | 55.9±10.15 | 0.911 |
| Tumor size | |||
| <4 cm | 10 (23.3%) | 3 (7.0%) | 0.045a |
| ⩾4 cm | 12 (27.9%) | 18 (41.9%) | |
| SCC-Ag | |||
| <1.5 ng/ml | 13 (30.2%) | 10 (23.3%) | 0.547 |
| ⩾1.5 ng/ml | 9 (20.9%) | 11 (25.6%) | |
| Histology | |||
| Adenocarcinoma | 2 (4.7%) | 3 (7.0%) | 0.664 |
| Squamous | 20 (46.5%) | 18 (41.9%) | |
| FIGO stage | |||
| I/II | 15 (34.9%) | 6 (14.0%) | 0.015a |
| III/IV | 7 (16.3%) | 15 (34.9%) | |
| Treat with paclitaxel | |||
| Response | 16 (37.2%) | 7 (16.3%) | 0.015a |
| No response | 6 (14.0%) | 14 (34.9%) | |
Abbreviations: FIGO, International Federation of Gynecology and Obstetrics; SCC-Ag, squamous cell carcinoma antigen. P-values of age were calculated by the t-test, others by Pearson's chi-square test.
Statistically significant.
Discussion
Chemoresistance in cancers is the main cause of treatment failure.26, 27 Although recent data indicate that aberrant miRNA expression is closely linked to chemoresistance by targeting genes related to chemosensitivity28, 29 or chemoresistance,16, 17, 21 the specific chemoresistance-related miRNAs are largely unknown. More research is needed to identify miRNAs associated with chemoresistance and the mechanisms by which they induce chemoresistance. In this study, we profiled miRNA expression in CC cells by miRNA microarray and compared miRNA expression between paclitaxel-resistant and -sensitive cells. In total, 18 miRNAs had altered expression between the cell lines, several of which have been reported to associate with paclitaxel resistance in CC28 and other cancers,17, 21 however, miR-125a was first reported in this paper to associate with paclitaxel resistance in CC. miR-125a was the most significantly downregulated miRNA in the resistance cells, and therefore miR-125a may have an essential role in modulating paclitaxel resistance in CC. Based on this, the function of miR-125a in paclitaxel resistance was further analyzed.
In solid tumors, the studies identified miR-125a as an anti-oncogene that inhibits tumorigenesis and cancer progression.20, 23 In breast cancer, miR-125a has been demonstrated to inhibit cancer growth and migration by suppressing ERBB2 and ERBB3.30 The antitumor functions also confirmed in gastric cancer through ERBB2/miR-125a loop31 and by regulating angiogenesis through VEGF-A.32 Moreover, Ninio-Many et al.33 indicated miR-125a modulates molecular pathway of motility and migration via Fyn expression in prostate cancer cells. In addition, miR-125a has been associated with many diseases, including autoimmune diseases,20 cardiovascular diseases,34, 35 microbial infection36, 37 and hematological malignancies.20, 30 Recent data indicate that miR-125a upregulation sensitized paclitaxel-resistant colon cancer cells to paclitaxel.21 However, the association between miR-125a and paclitaxel resistance in CC is unknown. Consistent with the function of miR-125a in colon cancer, we demonstrate that miR-125a expression is suppressed in paclitaxel-resistant CC cells. As expected, overexpressing miR-125a increased the sensitivity of resistant CC cells to paclitaxel and miR-125a knockdown in paclitaxel-sensitive CC cells resulted in resistance to paclitaxel. Meanwhile, we found that miR-125a sensitizes acquired cisplatin resistance of paclitaxel-resistant CC cells. Therefore, a negative correlation between miR-125a expression and chemoresistance was identified in CC.
STAT3 is a well-characterized transcription factor that has been demonstrated to contribute to tumorigenesis and chemoresistance by regulating apoptosis by promoting Bcl-2 and Bcl-xL expression.25 In this study, we determined that miR-125a can directly bind to the STAT3 3′-UTR and suppress its expression. Moreover, the expression of STAT3, Bcl-2 and Bcl-xL were significantly increased in paclitaxel-resistant CC cells. Therefore, it is possible that miR-125a regulates chemoresistance by inhibiting STAT3 expression. We determined that enforced overexpression of miR-125a enhanced cell apoptosis, suppressed apoptosis-related proteins. Furthermore, re-expression of STAT3 reversed the function of miR-125a. These data suggest that miR-125a may be a novel molecular mechanism contributing to chemoresistance by inhibiting STAT3 expression in CC. Therefore, miR-125a and STAT3 regulation may be beneficial for the treatment of paclitaxel and cisplatin resistant CC.
Treatment of CC cells with paclitaxel or cisplatin for 24 h did not change the expression of miR-125a or STAT3, but it did decrease p-STAT3 (Tyr705) protein levels. This suggests that paclitaxel and cisplatin inhibits STAT3 not through changing miR-125a/STAT3 pathway but through other pathways, such as the IL-6/Jak2 pathway.38 There are several explanations for why miR-125a is downregulated in CC. In this study, we found that overexpressing miR-125a in CC cells and treatment with paclitaxel significantly increased the apoptosis rate than either treatment individually. Thus, sub-populations of cancer cells that have low miR-125a expression may be a potential explanation for resistance against paclitaxel and acquired cisplatin resistance, and combining miR-125a reactivation with paclitaxel and cisplatin treatment may be a useful therapeutic intervention against chemoresistant CC.
In our study, we indicate for the first time that miR-125a upregulation can sensitize HeLa/PR cells to paclitaxel treatment in mice. Tumor volume was more significantly reduced after miR-125a overexpression and paclitaxel treatment than in the other three groups (control group, miR-125a overexpression or paclitaxel treatment groups). We also demonstrated that CC patients with low miR-125a expression had shorter chemotherapy-induced PFS and OS. Moreover, miR-125a expression was significantly downregulated in patients no-responsed to chemotherapy. These data confirm the function of miR-125a in maintaining sensitivity of CC cells to paclitaxel in vivo and paclitaxel combined with cisplatin in the clinic.
In conclusion, paclitaxel-resistant CCs have reduced miR-125a expression. MiR-125a negatively regulates paclitaxel and cisplatin resistance in CC by reducing STAT3 expression, which promotes apoptosis and microtubule stabilization. Upregulating miR-125a or inhibiting STAT3 may be useful in combination with paclitaxel and cisplatin for treating chemoresistant CC for these two agents.
Materials and methods
Patients and tumor tissues
A total of 43 human CC samples were obtained from the Chinese PLA 309th Hospital and General Hospital with the informed consent of patients and approval for experimentation from the Chinese PLA 309th Hospital and PLA General Hospital. Diagnoses were based on pathological evidence. Patients had not undergone immunotherapy, chemotherapy, hormone therapy or radiotherapy before specimen collection. The clinical stage and histological grades were based on the International Federation of Gynecology and Obstetrics staging. Tissue samples were snap frozen in liquid nitrogen and stored at −80 °C until RNA extraction. All patients underwent intravenous neo-adjuvant chemotherapy (paclitaxel 135 mg/m2 and cisplatin 75 mg/m2 at 3-week intervals for 5 weeks) after surgery. The effect of chemotherapy was evaluated by following response rate, PFS and OS according to the Response Evaluation Criteria in Solid Tumors.
Cell culture and transfection
CC cell lines, HeLa and CaSki, were obtained from the American Type Culture Collection (Manassas, VA, USA) and tested for mycoplasma contamination. Paclitaxel-resistant HeLa/PR and CaSki/PR cells were developed from HeLa and CaSki cell lines by treatment with gradually increasing concentrations of paclitaxel in cell culture medium. Briefly, cells were seeded in six-well plates and reached about 80% confluency in fresh medium before treating with paclitaxel. The dose of paclitaxel range from 0.1 to 20 nM and it was increased by a dose gradient that was 25–50% of the previous dose. The next dose was given until the cells were stable in proliferation without significant death.
Stable cell lines overexpressing miR-125a were established by lentiviral transduction using a pCDH plasmid (System Biosciences, Mountain View, CA, USA) carrying miR-125a. All cells were cultured at 37 °C in a humidified atmosphere with 5% CO2 in Dulbecco's modified Eagle's medium or RPMI-1640 medium (Life Technologies, Carlsbad, CA, USA) supplemented with 10% fetal bovine serum (Atlanta Biologicals, Lawrenceville, GA, USA) and 1% penicillin/streptomycin (Life Technologies). For transfection,39 cells were seeded in 24-well or 6-well plates and then transfected with the indicated plasmids using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) according to the manufacturer's protocol.
Plasmid construction and reagents
The expression vector for the miR-125a precursor sequence was produced by cloning the PCR product into a pcDNA3.1 vector (Invitrogen) or pCDH plasmid (System Biosciences) with the primers as follows: 5′-CGGGATCCTCTTTCTGTCTCTGGCTCTCAGAA-3′ (forward) and 5′-CGGAATTCAGTGGTCTGGGGTCAGAGGTCA-3′ (reverse). Wild-type and mutated STAT3 3′-UTRs were cloned into a dual-luciferase reporter vector (Promega, Madison, WI, USA) as described previously39 using the primers displayed as follows: 5′-CGGAATTCAGGAATCCTGGTCTCAGGACCTC-3′ (forward) and 5′-GCTCTAGATCATACGAGGGCAGACTCAAGT-3′ (reverse) (wild-type STAT3 3′-UTR); 5′-TGGGGCCCCAGCGACGTGTCTGGTTGAGAGACTTTCA-3′ (forward) and 5′-GAAAGTCTCTCAACCAGACACGTCGCTGGGGCCCCA-3′ (reverse) (mutant STAT3 3′UTR). The expression vector for STAT3 was its CDS sequence, which lacks 3′UTR sequence with miR-125a binding sites. The antisense miR-125a oligonucleotide (anti-hsa-miR-125a) and antisense miRNA control were purchased from Qiagen (Valencia, CA, USA).
Anti-Bcl-2 (cat. #sc-429), anti-Bcl-xL (cat. #sc-7195) and anti-GAPDH (cat. #sc-25778) antibodies were purchased from Santa Cruz Biotechnology (Dallas, TX, USA). Anti-STAT3 (cat. #ab126834) and anti-STAT3 (Tyr705) (cat. #ab76315) antibodies were purchased from Abcam (Cambridge, MA, USA). Paclitaxel and cisplatin were obtained from Sigma-Aldrich (St Louis, MO, USA).
Luciferase reporter assay
Cells were seeded in 24-well plates at a density of 1 × 105 cells per well. The cells were co-transfected with luciferase reporters, either wild-type or mutant STAT3 3′-UTR, in combination with anti-miR-125a or a scramble using Lipofectamine 2000. Forty-eight hours later, cells were harvested and analyzed for luciferase activity using a luciferase assay kit (Promega) according to the manufacturer's protocol.
miRNA microarray analysis
Total RNA was extracted from HeLa and CaSki cells and their paclitaxel-resistant counterparts using a Qiagen miRNeasy mini kit and following the manufacturer protocol. Total RNA was sent to CapitalBio Corporation (Beijing, China) for miRNA labeling, quality control, chip hybridization and microarray analysis. Briefly, total RNA was labeled with Hy3 and Hy5 fluorescent dyes. Pairs of labeled samples were hybridized to miRCURY LNA miRNA array slides with 2549 human miRNAs. Normalization was performed using a LOWESS filter (Locally Weighted Regression) method to remove system-related variations. An analysis of variance was first applied to produce a miRNA expression profile overview across all samples and then t-tests were performed to identify significantly differentiated miRNA expression among all interested combinations of paired groups.
miRNA extraction and quantitative RT–PCR
Total RNA, including miRNA, was extracted from cultured cells or tissues samples with a miRNeasy Mini kit (Qiagen). Target miRNA was reverse transcribed to complementary DNA using a specific miRNA primer and miScript Reverse Transcription Kit (Qiagen). miRNA expression was measured with a miScript SYBR Green PCR Kit (Qiagen) using the ABI7500 Real-Time PCR System (Applied Biosystems, Foster City, CA, USA). Primers for miRNAs and the endogenous control, U6 gene were displayed in Table 2. The relative fold expression of the target was calculated by the comparative Ct method and was normalized to control.
Table 2. The real-time PCR primers for miRNAs and U6.
| Gene | Forward primer | Reverse primer |
|---|---|---|
| hsa-miR-424 | 5′-AGCGGCAGCAGCAATTCATG-3′ | 5′-GTGCAGGGTCCGAGGT-3′ |
| hsa-miR-299-5p | 5′-AGCCGTGGTTTACCGTCCCA-3′ | 5′-GTGCAGGGTCCGAGGT-3′ |
| hsa-miR-27a | 5′-CGCGCTTCACAGTGGCTAAG-3′ | 5′-GTGCAGGGTCCGAGGT-3′ |
| hsa-miR-125a | 5′-CGGCGTCCCTGAGACCCTTT-3′ | 5′-GTGCAGGGTCCGAGGT-3′ |
| hsa-miR-19a | 5′-CGCGGAGTTTTGCATAGTTG-3′ | 5′-GTGCAGGGTCCGAGGT-3′ |
| hsa-miR-130b | 5′-AGCCGACTCTTTCCCTGTTG-3′ | 5′-GTGCAGGGTCCGAGGT-3′ |
| U6snRNA | 5′-TGCGGTGGGTGTCATCAAA-3′ | 5′-AACGCTTCACGAATTTGCGT-3′ |
Abbreviation: miRNA, microRNA.
Cell viability assay
Cell viability was measured using a CCK-8 Kit (Dojindo, Kumamoto, Japan) according to the manufacturer's protocol. To analyze the effects of miR-125a in combination with paclitaxel or cisplatin, cells transfected with either miR-125a or anti-miR-125a were treated at concentrations of 0, 2.5, 5, 10, 20, 40 and 80 nM with paclitaxel or at concentrations of 0, 2, 4, 8, 16, 32, 64, 128 μM with cisplatin for 24 h. The IC50 value was calculated as the concentration of paclitaxel that reduced cell viability by 50%.
Apoptosis and flow cytometry analysis
Stable miR-125a-overexpressing and miR-125a plus STAT3-overexpressing or empty vector control paclitaxel-resistant cells (1 × 106 cells) were cultured in 60 mm dishes and treated with paclitaxel (20 nM) or cisplatin (10 μM) for 24 h before harvesting. The cells were labeled with propidium iodide and annexin V according to the manufacturer's instructions (BD Biosciences, San Jose, CA, USA). A minimum of 10 000 events for each sample were collected and analyzed using a FACScalibur Flow Cytometer (Becton Dickinson, BD Biosciences).
In vivo cervical tumor xenograft model
All animal experiments were undertaken in accordance with the National Institute of Health Guide for the Care and Use of Laboratory Animals, with the approval of the Scientific Investigation Board of PLA General Hospital, Beijing. Female 6-week-old BALB/c nu/nu mice were purchased from Vital River Inc. (Beijing, China). For the tumor growth model, HeLa/PR cells (2 × 107 cells) stably transfected with the pCDH control vector or pCDH-miR-125a were injected subcutaneously into the backs of BALB/c nu/nu mice (n=10), which divided into two groups (n=5 based on minimal 30% decrease from 1 g tumors with 250 μg s.d., α error of 0.05 and β error of 0.8) using random number method. After 3 weeks, if the tumor diameter was >5 mm, either paclitaxel (15 mg/kg) or saline was injected intraperitoneally once a week for 5 weeks with no blinding. Tumor sizes were measured at the indicated times using calipers. Tumor volumes were estimated according to the following formula: volume=(longest diameter × shortest diameter2)/2.
Statistical analysis
All in vitro experiments were performed in triplicate and repeated three times. Differences between variables were assessed by a χ2 test or two-tailed Student's t-test. The survival rates in relation to miR-125a expression were estimated using the Kaplan–Meier method and the difference in survival curves was analyzed with a log-rank test. The relationship between miR-125a and the IC50 of paclitaxel was examined using the Spearman's rank correlation. The SPSS 17.0 statistical software package (SPSS Inc, Chicago, IL, USA) was used to perform all statistical analyses. Data are presented as the means±s.d. P<0.05 was considered statistically significant.
Acknowledgments
This work was supported by the General Program of 309th Hospital of PLA (2015MS-010), the National Natural Science Foundation (81472589 and 31100604) and Beijing Nova Program (Z141102001814055).
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on the Oncogenesis website (http://www.nature.com/oncsis).
Supplementary Material
References
- Jemal A, Siegel R, Xu J, Ward E. Cancer statistics, 2010. CA Cancer J Clin 2010; 60: 277–300. [DOI] [PubMed] [Google Scholar]
- Ferlay J, Shin HR, Bray F, Forman D, Mathers C, Parkin DM. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer 2010; 127: 2893–2917. [DOI] [PubMed] [Google Scholar]
- Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin 2011; 61: 69–90. [DOI] [PubMed] [Google Scholar]
- Moore KN, Herzog TJ, Lewin S, Giuntoli RL, Armstrong DK, Rocconi RP et al. A comparison of cisplatin/paclitaxel and carboplatin/paclitaxel in stage IVB, recurrent or persistent cervical cancer. Gynecol Oncol 2007; 105: 299–303. [DOI] [PubMed] [Google Scholar]
- Kitagawa R, Katsumata N, Shibata T, Kamura T, Kasamatsu T, Nakanishi T et al. Paclitaxel plus carboplatin versus paclitaxel plus cisplatin in metastatic or recurrent cervical cancer: the open-label randomized phase III trial JCOG0505. J Clin Oncol 2015; 33: 2129–2135. [DOI] [PubMed] [Google Scholar]
- Kudelka AP, Winn R, Edwards CL, Downey G, Greenberg H, Dakhil SR et al. An update of a phase II study of paclitaxel in advanced or recurrent squamous cell cancer of the cervix. Anticancer Drugs 1997; 8: 657–661. [DOI] [PubMed] [Google Scholar]
- Zheng H, Liu Z, Liu T, Cai Y, Wang Y, Lin S et al. Fas signaling promotes chemoresistance in gastrointestinal cancer by up-regulating P-glycoprotein. Oncotarget 2014; 5: 10763–10777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vargas JR, Stanzl EG, Teng NN, Wender PA. Cell-penetrating, guanidinium-rich molecular transporters for overcoming efflux-mediated multidrug resistance. Mol Pharm 2014; 11: 2553–2565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goncalves A, Braguer D, Kamath K, Martello L, Briand C, Horwitz S et al. Resistance to Taxol in lung cancer cells associated with increased microtubule dynamics. Proc Natl Acad Sci USA 2001; 98: 11737–11742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He W, Zhang D, Jiang J, Liu P, Wu C. The relationships between the chemosensitivity of human gastric cancer to paclitaxel and the expressions of class III β-tubulin, MAPT, and survivin. Med Oncol 2014; 31: 950. [DOI] [PubMed] [Google Scholar]
- Kavallaris M. Microtubules and resistance to tubulin-binding agents. Nat Rev Cancer 2010; 10: 194–204. [DOI] [PubMed] [Google Scholar]
- Ingemarsdotter CK, Tookman LA, Browne A, Pirlo K, Cutts R, Chelela C et al. Paclitaxel resistance increases oncolytic adenovirus efficacy via upregulated CAR expression and dysfunctional cell cycle control. Mol Oncol 2015; 9: 791–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi JH, Sheu JJ, Guan B, Jinawath N, Markowski P, Wang TL et al. Functional analysis of 11q13.5 amplicon identifies Rsf-1 (HBXAP) as a gene involved in paclitaxelresistance in ovarian cancer. Cancer Res 2009; 69: 1407–1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le XF, Bast RC Jr. Src family kinases and paclitaxel sensitivity. Cancer Biol Ther 2011; 12: 260–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng X, Gong F, Chen Y, Jiang Y, Liu J, Yu M et al. Autophagy promotes paclitaxel resistance of cervical cancer cells: involvement of Warburg effect activated hypoxia-induced factor 1-α-mediated signaling. Cell Death Dis 2014; 5: e1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee A, Chattopadhyay D, Chakrabarti G. MiR-16 targets Bcl-2 in paclitaxel-resistant lung cancer cells and overexpression of miR-16 along with miR-17 causes unprecedented sensitivity by simultaneously modulating autophagy and apoptosis. Cell Signal 2015; 27: 189–203. [DOI] [PubMed] [Google Scholar]
- Chatterjee A, Chattopadhyay D, Chakrabarti G. miR-17-5p downregulation contributes to paclitaxel resistance of lung cancer cells through altering beclin1 expression. PLoS ONE 2014; 9: e95716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths-Jones S, Grocock RJ, van Dongen S, Bateman A, Enright AJ. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res 2006; 34: (Database issue) D140–D144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanakkanthara A, Miller JH. MicroRNAs: novel mediators of resistance to microtubule-targeting agents. Cancer Treat Rev 2013; 39: 161–170. [DOI] [PubMed] [Google Scholar]
- Sun YM, Lin KY, Chen YQ. Diverse functions of miR-125 family in different cell contexts. J Hematol Oncol 2013; 6: 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Chen Y, Chen Z. MiR-125a/b regulates the activation of cancer stem cells in paclitaxel-resistant colon cancer. Cancer Invest 2013; 31: 17–23. [DOI] [PubMed] [Google Scholar]
- Chen JJ, Liu SX, Chen MZ, Zhao ZY. Has-miR-125a and 125b are induced by treatment with cisplatin in nasopharyngeal carcinoma and inhibit apoptosis in a p53-dependent manner by targeting p53 mRNA. Mol Med Rep 2015; 12: 3569–3574. [DOI] [PubMed] [Google Scholar]
- Pan W, Zhu S, Dai D, Liu Z, Li D, Li B et al. MiR-125a targets effector programs to stabilize Treg-mediated immune homeostasis. Nat Commun 2015; 6: 7096. [DOI] [PubMed] [Google Scholar]
- Wang C, Li D, Zhang S, Xing Y, Gao Y, Wu J. MicroRNA-125a-5p induces mouse granulosa cell apoptosis by targeting signal transducer and activator of transcription 3. Menopause 2015; 23: 100–107. [DOI] [PubMed] [Google Scholar]
- Lee JH, Kim C, Sethi G, Ahn KS. Brassinin inhibits STAT3 signaling pathway through modulation of PIAS-3 and SOCS-3 expression and sensitizes human lung cancer xenograft in nude mice to paclitaxel. Oncotarget 2015; 6: 6386–6405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Dimassi S, Abou-Antoun T, El-Sibai M. Cancer cell resistance mechanisms: a mini review. Clin Transl Oncol 2014; 16: 511–516. [DOI] [PubMed] [Google Scholar]
- Periti P, Mini E. Drug resistance in cancer: an overview of the clinical aspects. J Chemother 1989; 1: 5–9. [DOI] [PubMed] [Google Scholar]
- Shen Y, Wang P, Li Y, Ye F, Wang F, Wan X et al. miR-375 is upregulated in acquired paclitaxel resistance in cervical cancer. Br J Cancer 2013; 109: 92–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Ren S, Li X, Wang Y, Garfield D, Zhou S et al. MiR-21 overexpression is associated with acquired resistance of EGFR-TKI in non-small cell lung cancer. Lung Cancer 2014; 83: 146–153. [DOI] [PubMed] [Google Scholar]
- Scott GK, Goga A, Bhaumik D, Berger CE, Sullivan CS, Benz CC. Coordinate suppression of ERBB2 and ERBB3 by enforced expression of micro-RNA miR-125a or miR-125b. J Biol Chem 2007; 282: 1479–1486. [DOI] [PubMed] [Google Scholar]
- Fassan M, Pizzi M, Realdon S, Balistreri M, Guzzardo V, Zagonel V et al. The HER2-miR125a5p/miR125b loop in gastric and esophageal carcinogenesis. Hum Pathol 2013; 44: 1804–1810. [DOI] [PubMed] [Google Scholar]
- Dai J, Wang J, Yang L, Xiao Y, Ruan Q. miR-125a regulates angiogenesis of gastric cancer by targeting vascular endothelial growth factor A. Int J Oncol 2015; 47: 1801–1810. [DOI] [PubMed] [Google Scholar]
- Ninio-Many L, Grossman H, Levi M, Zilber S, Tsarfaty I, Shomron N et al. MicroRNA miR-125a-3p modulates molecular pathway of motility and migration in prostate cancer cells. Oncoscience 2014; 1: 250–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimitrakopoulou K, Vrahatis AG, Bezerianos A. Integromics network meta-analysis on cardiac aging offers robust multi-layer modular signatures and reveals micronome synergism. BMC Genomics 2015; 16: 147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong LL, Armugam A, Sepramaniam S, Karolina DS, Lim KY, Lim JY et al. Circulating microRNAs in heart failure with reduced and preserved left ventricular ejection fraction. Eur J Heart Fail 2015; 17: 393–404. [DOI] [PubMed] [Google Scholar]
- Squillace N, Bresciani E, Torsello A, Bandera A, Sabbatini F, Giovannetti C et al. Changes in subcutaneous adipose tissue microRNA expression in HIV-infected patients. J Antimicrob Chemother 2014; 69: 3067–3075. [DOI] [PubMed] [Google Scholar]
- Inchley CS, Sonerud T, Fjærli HO, Nakstad B. Nasal mucosal microRNA expression in children with respiratory syncytial virus infection. BMC Infect Dis 2015; 15: 150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang WL, Yeh HH, Lin CC, Lai WW, Chang JY, Chang WT et al. Signal transducer and activator of transcription 3 activation up-regulates interleukin-6 autocrine production: a biochemical and genetic study of established cancer cell lines and clinical isolated human cancer cells. Mol Cancer 2010; 9: 309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu J, Xu X, Kang L, Zhou L, Wang S, Lu J et al. miR-30a suppresses breast cancer cell proliferation and migration by targeting Eya2. Biochem Biophys Res Commun 2014; 445: 314–319. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.