Abstract
Circadian clocks are fundamental properties of all eukaryotic organisms and at least some prokaryotic organisms. Recent studies in our laboratory have shown that the gastrointestinal system contains a circadian clock that controls many, if not all, aspects of gastrointestinal function. We now report that at least one species of intestinal bacteria, Enterobacter aerogenes, responds to the pineal and gastrointestinal hormone melatonin by an increase in swarming activity. This swarming behavior is expressed rhythmically, with a period of approximately 24 hrs. Transformation of E. aerogenes to express luciferase with a MotA promoter reveals circadian patterns of bioluminescence that are synchronized by melatonin and whose periods are temperature compensated from 26°C to 40°C. Bioinformatics suggest similarities between the E. aerogenes and cyanobacterial clocks, suggesting the circadian clock may have evolved very early in the evolution of life. They also point to a coordination of host circadian clocks with those residing in the microbiota themselves.
KEYWORDS: Circadian, microbiome, melatonin, gastrointestinal, swarming
The bacterial microbiome is an important modulator of gastrointestinal function. The circadian biological clock figures to play a central role in that process, both from the perspective of circadian regulation of the microbiome by the host's circadian clock and from the perspective that microbiota may influence gastrointestinal (GI) clocks. There is extensive evidence from rodent studies in our laboratory and others1-4 that the gastrointestinal system contains a self-sustaining circadian clock capable of regulating circadian patterns of motility, secretion and gene expression.1-3 This clock is coordinated with the light: dark cycle and the circadian clock within the brain's suprachiasmatic nucleus (SCN) via the sympathetic nervous system.4 Further, the GI clock can independently entrain to the timing of the presence or absence of food.1,4
Microbial signaling affects both homeostatic maintenance of intestinal function as well as circadian control of gastrointestinal function.5,6 Disruption of the circadian clock, either via dietary restriction or phase shifting (e.g. jet-lag) affects temporal distribution of the gut microbiome constituents.7-9 While it is clear from these studies that commensal bacteria and gut tissues do communicate, it has not been clear how the GI tract communicates with the microbiome or how the microbiome exploits to sustain its own homeostasis.
A potential human signal that may affect gastrointestinal microbiota is the secretion of the hormone melatonin into the lumen of the gut. Although melatonin is widely regarded as a pineal and retinal neuromodulator of circadian and photoperiodic function,10,11 it is present throughout the gastrointestinal system,10-12 in part from pineal melatonin secretion,16,17 but there is evidence for melatonin biosynthetic enzymes in the GI tract itself.14,15 In addition, many foods contain melatonin.14-16 Interestingly, GI-synthesized melatonin has never been shown to enter the systemic circulatory system in any species.
To address the hypothesis that gut bacteria have adapted to recognize and respond to melatonin signaling from the gut, we identified from metagenomics data in GenBank several enteric bacteria that expressed sequences with 24–42% identity to known melatonin binding sites in the human genome. Among these were sequences in the Gram-negative, indole-negative motile Enterobacter aerogenes. Colonies formed by clinical isolates of this bacterium proliferated – with respect to overall growth area – on semi-solid Agar significantly more rapidly in the presence of melatonin than in its absence in a specific, dose-dependent fashion. This phenomenon was dose-dependent, melatonin-specific, and not observed in clinical strains of Klebsiella pneumoniae and E. coli, nor in a lab strain of E. coli. Lab strains of E. aerogenes have not been tested.
The more widely-spread cultures of E. aerogenes in the presence of melatonin exhibited patterns of swarming within the cultures, evidenced by stereotypical, concentric rings of colonies. These patterns were less apparent in the smaller, control cultures of E. aerogenes in melatonin's absence. Remarkably, the number of rings consistently coincided with the number of incubation days. Calculation of banding periodicity – the number of bands visually observed divided by the number of hours of incubation – revealed a period of much greater than 24 hours in control-treated cultures. In contrast, in 1nM melatonin's presence, the period of swarming behavior was 25.1 ± 1.4 (SD) hours.
We therefore hypothesized that the swarming rhythms might represent the output of a circadian clock. To test this, cultures of E. aerogenes were transformed to express luciferase using a luxCDABE construct driven by the motA promoter. Bioluminescence from these cultures measured in a Lumicycle photomultiplier system indicated robust circadian patterns when cultures were maintained in temperatures ranging from ambient 26°C to those corresponding to human body temperatures (TB) to 40°C. The circadian periods of these bioluminescence rhythms were temperature compensated with a Q10 = 0.96 from 26°C to 40°C. While there was no effect of melatonin on circadian period, there was a significant effect of melatonin on the phase of peak bioluminescence. In the absence of melatonin, the circadian phases of multiple replicates were highly variable. However, in the presence of 1 nM melatonin the phases of these rhythms were synchronized, especially at 34–37°C.
This is the first demonstration of a bona fide circadian clock in a prokaryote outside Phylum Cyanobacteria. In Cyanobacteria, such as Synechococcus elongatus, the only species in which a circadian clock has been definitively demonstrated, circadian rhythmicity results from the rhythmic phosphorylation and dephosphorylation of the Kai protein complex, due in part by the rhythmic autokinase activity of Kai C, which is in turn rhythmically modulated by Kai A and Kai B.17 In vivo, this oscillator responds to light, temperature and metabolic state through the CikA, LdpA and Pex pathways, each of which can entrain the Kai oscillator to environmental cues.18-20 This relatively simple oscillator in turn regulates a wide array of processes through transcriptional regulation and other downstream processes. Cladistic analyses places several E. aerogenes sequences closely nested within Kai protein phylograms, suggesting, albeit not demonstrating yet, that the cyanobacterial and E. aerogenes clocks share common evolutionary ancestors.
These observations have at least 2 significant consequences. First, should our future work demonstrate that the molecular mechanisms underlying the circadian rhythm in E. aerogenes are phylogenetically related to the cyanobacterial clock, then, the evolution of circadian organization likely predates the emergence of oxygen generating photosynthesis some 3.5 billion years ago. This has far-reaching importance for the understanding of biological evolution on early Earth. Secondly, the data point to the view that overall vertebrate circadian organization is an orchestration of multiple circadian pacemakers organized in an hierarchical symphony of clocks. Light-sensitive circadian pacemakers within the SCN synchronize peripheral oscillators throughout the body. Among these, the GI tract contains its own circadian clock, coordinated to the light: dark cycle via sympathetic afferents from the SCN and to the timing of a meal directly. The GI clock in turn synchronizes at least one circadian clock within the enteric microbiome (Fig. 1), which, in turn, may affect other components of the microbiome. The biological and clinical significance of this grand scale of circadian coordination will be important to discover in the very near future.
Figure 1.
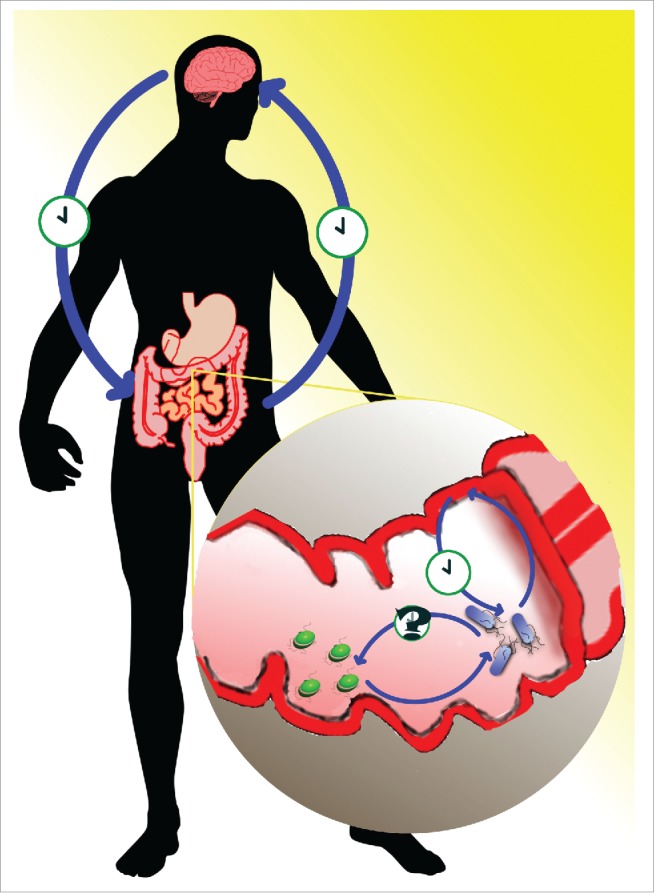
The discovery of an enteric bacterium with an endogenous clock adds to the hierarchical organization of human circadian rhythms. Circadian rhythms are generated in both the suprachiasmatic nucleus (SCN) of the brain as well as in the gut. These rhythms are independent of, but feed back onto, each other. Within the gut, previous studies have shown that bacterial signaling to the gut via TLRs may influence circadian rhythms. The discovery of an enteric bacterium that expresses circadian rhythms and is sensitive to melatonin (as depicted by blue rod-shaped bacteria) adds a novel signaling modality, and suggests that the rhythmic gut environment acts as a zeitgeber, or time-giver, to the microbiome. Whether or not this signal is propagated to other, melatonin-insensitive bacteria (represented by green coccoid bacteria), remains to be determined.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Funding
This research was funded by NIH R01 AG045833 to VMC.
References
- [1].Hoogerwerf WA, Hellmich HL, Cornélissen G, Halberg F, Shahinian VB, Bostwick J, Savidge TC, Cassone VM. Clock gene expression in the murine gastrointestinal tract: endogenous rhythmicity and effects of a feeding regimen. Gastroenterology 2007; 133(4):1250-60; PMID:17919497; http://dx.doi.org/ 10.1053/j.gastro.2007.07.009 [DOI] [PubMed] [Google Scholar]
- [2].Hoogerwerf WA, Sinha M, Conesa A, Luxon BA, Shahinian VB, Cornélissen G, Halberg F, Bostwick J, Timm J, Cassone VM. Transcriptional profiling of mRNA expression in the mouse distal colon. Gastroenterology 2008; 135(6):2019-29; PMID:18848557; http://dx.doi.org/ 10.1053/j.gastro.2008.08.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Hoogerwerf WA, Shahinian VB, Cornélissen G, Halberg F, Bostwick J, Timm J, Bartell PA, Cassone VM. Rhythmic changes in colonic motility are regulated by period genes. Am J Physiol Gastrointest Liver Physiol 2010; 298(2):G143-50; PMID:19926812; http://dx.doi.org/ 10.1152/ajpgi.00402.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Malloy JN, Paulose JK, Li Y, Cassone VM. Circadian rhythms of gastrointestinal function are regulated by both central and peripheral oscillators. Am J Physiol Gastrointest Liver Physiol 2012; 303(4):G461-73; PMID: 22723262; http://dx.doi.org/ 10.1152/ajpgi.00369.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Mukherji A, Kobiita A, Ye T, Chambon P. Homeostasis in intestinal epithelium is orchestrated by the circadian clock and microbiota cues transduced by TLRs. Cell [Internet] 2013. [cited 2014September3]; 153:812-27. Available from: http://www.ncbi.nlm.nih.gov/pubmed/23663780; PMID:23663780; http://dx.doi.org/ 10.1016/j.cell.2013.04.020 [DOI] [PubMed] [Google Scholar]
- [6].de Kivit S, Tobin MC, Forsyth CB, Keshavarzian A, Landay AL. Regulation of Intestinal Immune Responses through TLR Activation: Implications for Pro- and Prebiotics. Front Immunol [Internet] 2014. [cited 2015January29]; 5:60. Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3927311&tool=pmcentrez&rendertype=abstract; PMID:24600450; http://dx.doi.org/ 10.3389/fimmu.2014.00060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Voigt RM, Forsyth CB, Green SJ, Mutlu E, Engen P, Vitaterna MH, Turek FW, Keshavarzian A. Circadian disorganization alters intestinal microbiota. PLoS One [Internet] 2014. [cited 2015April27]; 9:e97500. Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=4029760&tool=pmcentrez&rendertype=abstract; PMID:24848969; http://dx.doi.org/ 10.1371/journal.pone.0097500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Zarrinpar A, Chaix A, Yooseph S, Panda S. Diet and feeding pattern affect the diurnal dynamics of the gut microbiome. Cell Metab [Internet] 2014. [cited 2014December3]; 20:1006-17. Available from: http://www.ncbi.nlm.nih.gov/pubmed/25470548; PMID:25470548; http://dx.doi.org/ 10.1016/j.cmet.2014.11.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Thaiss CA, Zeevi D, Levy M, Zilberman-Schapira G, Suez J, Tengeler AC, Abramson L, Katz MN, Korem T, Zmora N, et al.. Transkingdom control of microbiota diurnal oscillations promotes metabolic homeostasis. Cell [Internet] 2014. [cited 2014October16]; 159:514-29. Available from: http://www.ncbi.nlm.nih.gov/pubmed/25417104; PMID:25417104; http://dx.doi.org/ 10.1016/j.cell.2014.09.048 [DOI] [PubMed] [Google Scholar]
- [10].Acuña-Castroviejo D, Escames G, Venegas C, Díaz-Casado ME, Lima-Cabello E, López LC, Rosales-Corral S, Tan D-X, Reiter RJ. Extrapineal melatonin: sources, regulation, and potential functions. Cell Mol Life Sci [Internet] 2014. [cited 2014November10]; 71:2997-3025. Available from: http://www.ncbi.nlm.nih.gov/pubmed/24554058; PMID:24554058; http://dx.doi.org/ 10.1007/s00018-014-1579-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Brzozowski T, Jaworek J. Basic and clinical aspects of melatonin in the gastrointestinal tract. New advancements and future perspectives. Curr Pharm Des [Internet] 2014. [cited 2015January29]; 20:4785-7. Available from: http://www.ncbi.nlm.nih.gov/pubmed/24251669; PMID:24251669; http://dx.doi.org/ 10.2174/1381612819666131119111201 [DOI] [PubMed] [Google Scholar]
- [12].Bubenik GA. Gastrointestinal melatonin: localization, function, and clinical relevance. Dig Dis Sci [Internet] 2002. [cited 2015January29]; 47:2336-48. Available from: http://www.ncbi.nlm.nih.gov/pubmed/12395907; PMID:12395907; http://dx.doi.org/ 10.1023/A:1020107915919 [DOI] [PubMed] [Google Scholar]
- [13].Bubenik GA, Brown GM. Pinealectomy reduces melatonin levels in the serum but not in the gastrointestinal tract of rats. Biol Signals [Internet] 1997. [cited 2015January29]; 6:40-4. Available from: http://www.ncbi.nlm.nih.gov/pubmed/9098522; PMID:9098522; http://dx.doi.org/ 10.1159/000109107 [DOI] [PubMed] [Google Scholar]
- [14].Reiter RJ, Rosales-Corral S, Coto-Montes A, Boga JA, Tan D-X, Davis JM, Konturek PC, Konturek SJ, Brzozowski T. The photoperiod, circadian regulation and chronodisruption: the requisite interplay between the suprachiasmatic nuclei and the pineal and gut melatonin. J Physiol Pharmacol [Internet] 2011. [cited 2015January29]; 62:269-74; PMID:21893686; Available from: http://www.ncbi.nlm.nih.gov/pubmed/21893686 [PubMed] [Google Scholar]
- [15].Chen C-Q, Fichna J, Bashashati M, Li Y-Y, Storr M. Distribution, function and physiological role of melatonin in the lower gut. World J Gastroenterol [Internet] 2011. [cited 2015January29]; 17:3888-98. Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3198018&tool=pmcentrez&rendertype=abstract; PMID:22025877; http://dx.doi.org/ 10.3748/wjg.v17.i34.3888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Venegas C, García JA, Escames G, Ortiz F, López A, Doerrier C, García-Corzo L, López LC, Reiter RJ, Acuña-Castroviejo D. Extrapineal melatonin: analysis of its subcellular distribution and daily fluctuations. J Pineal Res [Internet] 2012. [cited 2016January14]; 52:217-27. Available from: http://www.ncbi.nlm.nih.gov/pubmed/21884551; PMID:21884551; http://dx.doi.org/ 10.1111/j.1600-079X.2011.00931.x [DOI] [PubMed] [Google Scholar]
- [17].Kitayama Y, Iwasaki H, Nishiwaki T, Kondo T. KaiB functions as an attenuator of KaiC phosphorylation in the cyanobacterial circadian clock system. EMBO J [Internet] 2003. [cited 2015January8]; 22:2127-34. Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=156084&tool=pmcentrez&rendertype=abstract; PMID:12727879; http://dx.doi.org/ 10.1093/emboj/cdg212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Johnson CH, Mori T, Xu Y. A cyanobacterial circadian clockwork. Curr Biol [Internet] 2008. [cited 2015January29]; 18:R816-25. Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=2585598&tool=pmcentrez&rendertype=abstract; PMID:18786387; http://dx.doi.org/ 10.1016/j.cub.2008.07.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Mackey SR, Golden SS, Ditty JL. The itty-bitty time machine genetics of the cyanobacterial circadian clock. Adv Genet [Internet] 2011. [cited 2015January29]; 74:13-53. Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3319097&tool=pmcentrez&rendertype=abstract; PMID:21924974; http://dx.doi.org/ 10.1016/B978-0-12-387690-4.00002-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Kondo T. A cyanobacterial circadian clock based on the Kai oscillator. Cold Spring Harb Symp Quant Biol [Internet] 2007. [cited 2015January29]; 72:47-55. Available from: http://www.ncbi.nlm.nih.gov/pubmed/18419262; PMID:18419262; http://dx.doi.org/ 10.1101/sqb.2007.72.029 [DOI] [PubMed] [Google Scholar]


