ABSTRACT
The relevance of biogeography to the distal gut microbiota has been investigated in both health and inflammatory bowel disease (IBD), however multiple factors, including sample type and methodology, microbiota characterization and interpersonal variability make the construction of a core model of colonic biogeography challenging. In addition, how phylogenetic classification relates to immunogenicity and whether consistent alterations in the microbiota are associated with ulcerative colitis (UC) remain open questions. This addendum seeks to review the human colonic microbiota in health and UC as currently understood, in the broader context of the human microbiome.
KEYWORDS: Biogeography, inflammatory bowel disease, microbiome, microbiota, spatial ecology, ulcerative colitis
Spatial ecology, or biogeography, is the study of how organisms are dispersed throughout their native environment and how these patterns change with time.1 While the field developed initially in relation to macroecology, many of the concepts have been fruitfully applied to microbial communities. With respect to the microbiota, biogeography seeks to establish the community structure of the microbiota across different body habitats and niches within those habitats, elucidating the environmental and biological selection pressures responsible for shaping these communities, quantifying their stability over time and across diverse ethnogeographic populations and ultimately, discovering how disturbances in the overall community structure are associated with disease states. These goals are exemplified by the results of the human microbiome project (HMP), which provides a fundamental reference point for any study of organ-specific biogeography.2,3
The microbiota across body habitats
The HMP, which cataloged the healthy microbiomes of 242 individuals across different body sites at different time points, identified clear clustering based on biogeographical location, with specific skin, oral and vaginal sites as well as stool samples representing the gut microbiota. For stool samples, the degree of α diversity, or within-subject diversity, was high, while the degree of β-diversity, or between-subject diversity, was low with respect to other habitats in the human body.
When investigating the effects of location on the microbiota, it is important to consider how degrees of variation between the spatial sites relate to inter-personal variability. For instance in the HMP, the major variation in community structure was accounted for by differences between body habitats, making it easy to observe separation between the communities of different body sites by aggregating individuals together, findings evident also in another study of the microbiome across body habitats.4 For example, a person's oral microbiota would usually be more similar to another person's oral microbiota than their own skin or stool microbiota. In these examples, differences occur due to explicit driving mechanisms, such as oxygen and environmental factors influencing skin communities and fermentation influencing anaerobic communities in the distal gut. This combination of environmental and biological pressure accounts for the dominant community structure and function demonstrated between the habitats selected in this pivotal study.
However in studies involving changes in the microbiota within single organs or habitats, the variation between individuals can be significantly greater than the variation between different regions within the habitat (Fig. 1). This poses a significant challenge to both characterization and interpretation of different samples from throughout the same body habitat or organ.
Figure 1.
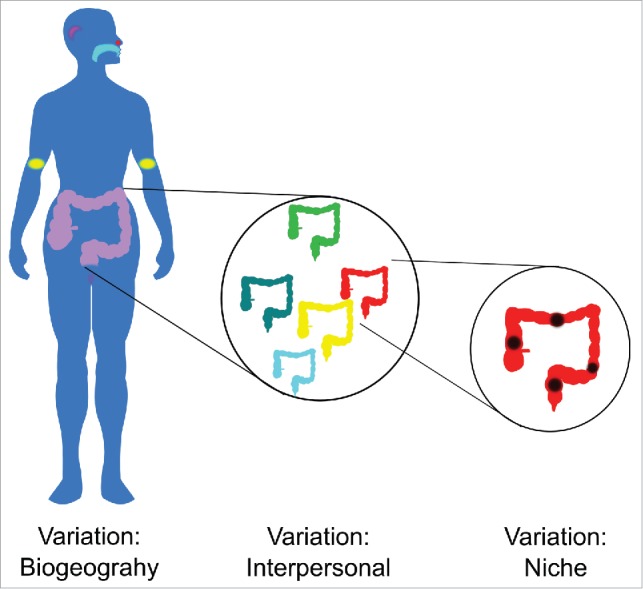
Clear distinctions exist between the microbial communities in different body habitats. However, within the same body habitat, variation between individuals can be much larger than variations between different niches or locations within that habitat.
While it is well established that single body habitats can have “sub-habitats,” such as in the oral cavity, with clear niches in very closely apposed areas, such as the hard palate, the sub-and supra-gingival plaque and saliva5 and similarly clear differences along the gastrointestinal tract as a whole,6 the colon appears to be more homogenous. In assessing the degree of interpersonal variability, Zhou and colleagues applied a model, the Dirichlet-Multinomial Distribution, which was able to characterize variation between individuals by using an over-dispersion parameter to quantify variation of the whole community between individuals for each body habitat. Using this method, stool and oral samples had the lowest interpersonal variability of the body habitats studied in terms of community structure.3,7 Despite this, variation between individuals is still significantly greater than the variation between different colonic regions.8-10 Notably, this variation was preserved in UC (Fig. 2A). Keeping in mind another major finding of the HMP, that large interpersonal variation at a phylogenetic level was countered by marked similarity at a genetic or microbiome level, the relevance of small differences in biogeography within an organ must be considered in a larger perspective.
Figure 2.
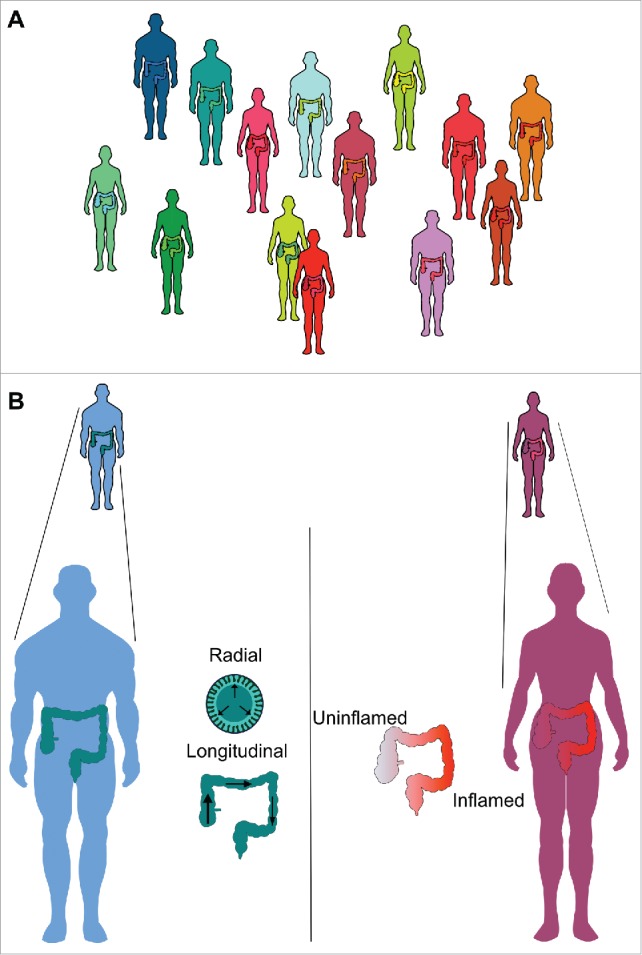
Interpersonal variation is a characteristic feature in health and UC (A), with significant overlap between health (green-blue figures) and UC (yellow-red figures). Within healthy individuals, bigeography may relate to radial or longitdunal distributions, while local inflammation is an additional feature that could influence the biogeography in UC (B).
Spatial structure of the colonic microbiota
Biogeography within the human colon can refer to radial (or cross-sectional) and longitudinal structure (Fig. 2B). Additionally, the effect of inflammation on community structure and biogeography will be considered here. Various potential mechanisms have been put forward to explain purported cross-sectional gradients, including gradients in radial oxygen tension, alterations in pH at the luminal interface or adaptation of butyrate producers to metabolizing liberated glycans from syntrophic mucin degrading organisms, while proposed mechanisms for longitudinal variation include changes in pH, substrate availability and host expression of toll-like receptors.11-13
Radial structure within the colon
Radial structure involves comparing paired luminal and mucosal communities in a region-specific manner. The most abundant luminal bacteria relative to the mucous gel in control volunteers in our study were Bacteroidaceae, Porphyromonadaceae, Alcaligenaceae, Lactobacillaceae and Rikenellaceae, while the most differentially abundant mucous gel bacteria relative to the lumen were Coriobacteriaceae, Planctomycetaceae, Lachnospiraceae, Family XIII Incertae Sedis and Ruminococcaceae. A previous study in mice, which also used laser capture microdissection and 454 pyrosequencing to interrogate the cross-sectional colonic axis, identified gradients similar to those in our study, with increased abundance of Lachnospiraceae and Ruminococcaceae in the inter-fold region of the mucosa, dominated by secreted mucus gel and increased proportion of Bacteroidaceae, Porphyromonodaceae, Prevotellaceae, Rikenellaceae and Lactobacillaceae in the digesta, corresponding to the luminal compartment.14,15 The similarities, both in terms of methodology and findings, suggest overlap in terms of mice and humans and support the hypothesis of the authors in terms of bacterial groups that have evolved to inhabit the mucus gel niche.15,16
The finding of an increase in mucosal associated Lachnospiraceae and Ruminococcaceae has been suggested in another murine models as well,17 while a recent murine study demonstrated increases in Bacteroidetes in the lumen with corresponding increases in clostridia and Lactobacillus in the mucus of the distal colon of mice.18 An in vitro model of lumen and mucosal environments (L-SHIME and M-SHIME, respectively) also found differential enrichment of Firmicutes in the mucosal relative to the luminal model when inoculated with the same donor fecal samples.12 These results support some studies in humans,19,20 particularly the pyrosequencing studies of Willing et al of healthy twins and twins with inflammatory bowel disease and the pyrosequencing study of Hong et al in health.
The findings of differential enrichment of the mucosal environment are however not universal as some studies have demonstrated fecal enrichment of Firmicutes when compared to the mucosal niche microbiota identified in colonic biopsies.21-23 Particularly, bowel preparation may have an effect on biogeography and work using mucosal biopsies from unprepared colons demonstrated increased Firmicutes and Proteobacteria in stool samples.23 It is worth noting that bowel preparation has been shown to reduce stool quantities of Clostridium cluster XIVa, while increasing both Proteobacteria and Clostridium cluster IV.24 While this has not been compared across the luminal and mucosal niches, it is possible that colonic lavage has a greater effect on the luminal and in turn faecal communities. The results also stand in contrast to those of a recently published study examining the biogeography of the Macaque intestine, demonstrating a reversal of this pattern, with a high abundance of Firmicutes in the lumen and an increased abundance of facultative anaerobes and gram negative bacteria associated with the mucosa.25 Notable differences between this study and human studies include the predominance of mucosal ε Proteobacteria families (largely Helicobacter but also Treponema), the lack of a gradient in Bacteroidaceae and the prominence of sampling location over interpersonal variation as the main variable.
Longitudinal gradients
Some studies have demonstrated longitudinal structure, including a tendency for increased γ proteobacteria9 and Lactobacillus levels in distal colon26 and increased proportions of Streptococcus and Enterococcus in the proximal colon9 and it has been suggested that bacterial dispersion in the colon may occur in a non-linear fashion.27 In the murine colon, regional changes in the mucosa-associated microbiota have been linked to host expression of toll-like receptors.13 Other studies however have not demonstrated conserved longitudinal gradients in health or IBD20,21,28-31 and our study did not demonstrate any consistent alterations in the community structure of the colonic microbiota across the long axis of the colon or any regional associations with single organisms.
Community alterations associated with UC
Whether inflammation causes alterations in the microbiota, or the microbiota instigates inflammation in IBD, the 2 processes become inextricably linked.32 As there is no evidence of a “core microbiota” in health,2 there is consequently no core disturbance characteristic of inflammation in IBD. This is particularly true in UC. Certain bacterial families have, however, been more strongly implicated and a consensus regarding key members that are enriched or depleted is beginning to emerge.33-35
Bacteria that have been associated with an increase in UC include Fusobacterium spp,35,36 Peptostreptococcus,8,37 Desulfovibrio spp38 and the Enterobacteriaceae8,35
Ruminococcus gnavus and R. torques, reclassified as Lachnospiraceae, have been shown to be increased in one study39 while another study has demonstrated reductions in multiple members of Lachnospiraceae.40 A notable reduction in the butyrate-producing bacteria Faecalibacterium prausnitzii and Roseburia hominis have been both associated with UC and correlated with the severity of inflammation,34 while overlapping members of Ruminococcaceae and Lachnospiraceae, including Clostridium leptum, Ruminococcus bromii, Roseburia and Eubacterium rectale were reduced in proportional abundance in another study.35 In one of the most important recent papers published, only 9 of 350 clades were altered in UC, with only Odoribacter being reduced in patients with pancolitis. Other observered alterations in UC included increases in Clostridiaceae and reductions in Roseburia, Veillonellaceae and Leuconostocaceae, although these findings were far less dramatic than those demonstrated in ileal Crohn's disease.33 Finally, a number of studies have noted a reduction in Akkermensia spp, a key mucin degrading species in health.8,35,39 Other studies have demonstrated a reduction in Firmicutes:Bacteroidetes ratio.10 It is important to note however that based on a recent population-level microbiome paper, powering studies to detect alterations due to clinical conditions may require significantly larger cohort sizes than have previously been employed.41
Differences associated with local inflammation
A natural extension of sampling the mucosa in IBD is to compare inflamed and uninflamed regions to determine if specific associations are evident (Fig. 2B). A number of studies using different techniques, including culture-based methods,42 TTGE,43 clone libraries44,45 and pyrosequencing8,19 have not demonstrated significant differences between inflamed and uninflamed tissue within individuals with IBD.46,47 In contrast, other studies using T-RFLP and ARISA48 and clone-library analysis10 demonstrated differences, although this could not be attributed to specific bacterial groups, while a study using FISH demonstrated increased clostridia and E. coli in inflamed vs. uninflamed tissue in UC.49 Morgan et al looked at anatomical regions rather than inflamed vs. uninflamed regions and noted a reduction in abundance in Roseburia, Allistipes and Ruminococcaceae in the terminal ileum and right colon with an increase in Enterobacteriaceae and Fusobacteria in these regions.33 While this question remains to be fully answered, no clear correlations between specific bacterial groups and inflamed tissue have thus far been demonstrated.
Taking a radically different approach, colitogenic bacterial species in an inflammasome murine model of colitis were identified by the avidity of IgA binding in a recent study.50 Subsequently, the authors isolated bacteria from human subjects with IBD and created inocula for germ free mice with high and low IgA binding bacterial isolates, demonstrating a dramatic increase in the severity of dextran sodium sulfate colitis in mice inoculated with the colitogenic bacteria. Another remarkable finding of this study was that 2 bacterial isolates of the same species (B. fragilis) had dramatically different pro-inflammatory effects, based on their degree of IgA binding, suggesting that even at the species level, taxonomic classification cannot comprehensively identify the key bacterial mediators in inflammation. While IgA binding identified the bacteria already associated with murine colitis, the message applied to human IBD is clear: alternative approaches will be required to identify colitogenic members of the microbiota based on their functional activity, rather than their taxonomy.
Biopsy vs. stool
Most studies have demonstrated differences between the mucosal and stool microbiota.21,28 Yasuda et al found that stool was highly representative of the rhesus macaque luminal and mucosal communities in the large intestine, being within 2 standard deviation for 95% of detected OTUs, although this dropped to 50% and 66% for small intestinal luminal and mucosal samples, respectively.25 However in the patients with new-onset Crohn's disease, the associated dysbiosis was only weakly detected in stool samples when compared to tissue samples.22 Morgan et al demonstrated that the most important covariates in determining community structure was whether samples were faecal or mucosal, as well as patient age, drug therapy and disease phenotype.33
Summary
What can be said about the findings to date and where do our results fit in with these? Inter-personal variation remains the dominant variable in studies of the colonic microbiota. Our results suggest that this variation is retained even in advanced UC. While no single group or species has consistently been associated with UC, reductions in members of butyrate-producing clostridial species, as well as reductions in Akkermensiaceae with an increase in Enterobacteriaceae seem to be frequently described. However in one of the most comprehensive studies to date, none of these groups were altered in patients with the most severe form colonic IBD, pancolitis.33 It is certainly possible that the enormous diversity in the fecal microbiota hides to some extent the signal of disease-associated microbes, something that has been established in pediatric Crohn's disease, making mucosal sampling an important step in focusing sequencing efforts on the mucosal niche22 and potentially reducing the requirement for large cohort sizes implied by a recent population study based on fecal sampling.41
Specific to our study, we found no evidence of longitudinal changes or changes associated with local inflammation, in keeping with the larger body of evidence in health and IBD. Our results do point to a radial structure, suggesting a degree of segregation between the luminal and mucosal niches. Although we have emphasized differences between health and UC, perhaps more remarkable are the similarities, in terms of the retained inter-personal variation, the lack of regional variation associated with local inflammation and the preservation of radial structure despite profound disruption to the mucosal barrier. Ultimately, constructing a full picture of the colonic microbiota will require the integration of spatial, temporal and functional components, their interaction with the mucosa and the host immune system. While consistent community and biogeographic associations with inflammation have been lacking, targeted mucosal sampling further refined by metagenomic analysis and immunogenicity profiling will advance our understanding of the inflammatory landscape of IBD.
Abbreviations
- ARISA
Automated Ribosomal Intergenic Spacer Analysis
- FISH
Fluorescent In-Situ Hybridization
- HMP
Human Microbiome Project
- IBD
Inflammatory Bowel Disease
- OTU
Operational Taxonomic Unit
- T-RFLP
Terminal Restriction Fragment Length Polymorphism analysis
- TTGE
Temporal Temperature Gradient gel Electrophoresis
- UC
Ulcerative Colitis
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
References
- [1].Fierer N. Microbial biogeography: patterns in microbial diversity across space and time. Washington DC: ASM Press; 2008. [Google Scholar]
- [2].The Human Microbiome Consortium: Structure, function and diversity of the healthy human microbiome. Nature 2012; 486:207-14; PMID:22699609; http://dx.doi.org/ 10.1038/nature11234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Zhou Y, Gao H, Mihindukulasuriya KA, La Rosa PS, Wylie KM, Vishnivetskaya T, Podar M, Warner B, Tarr PI, Nelson DE, et al.. Biogeography of the ecosystems of the healthy human body. Genome Biol 2013; 14:R1; PMID:23316946; http://dx.doi.org/ 10.1186/gb-2013-14-1-r1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Caporaso JG, Lauber CL, Costello EK, Berg-Lyons D, Gonzalez A, Stombaugh J, Knights D, Gajer P, Ravel J, Fierer N, et al.. Moving pictures of the human microbiome. Genome Biol 2011; 12:R50; PMID:21624126; http://dx.doi.org/ 10.1186/gb-2011-12-5-r50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Segata N, Haake SK, Mannon P, Lemon KP, Waldron L, Gevers D, Huttenhower C, Izard J. Composition of the adult digestive tract bacterial microbiome based on seven mouth surfaces, tonsils, throat and stool samples. Genome Biol 2012; 13:R42-R; PMID:22698087; http://dx.doi.org/ 10.1186/gb-2012-13-6-r42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Stearns JC, Lynch MD, Senadheera DB, Tenenbaum HC, Goldberg MB, Cvitkovitch DG, Croitoru K, Moreno-Hagelsieb G, Neufeld JD. Bacterial biogeography of the human digestive tract. Sci Rep 2011; 1:170; PMID:22355685; http://dx.doi.org/ 10.1038/srep00170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Holmes I, Harris K, Quince C. Dirichlet multinomial mixtures: generative models for microbial metagenomics. PLoS ONE 2012; 7:e30126; PMID:22319561; http://dx.doi.org/ 10.1371/journal.pone.0030126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Lavelle A, Lennon G, O'Sullivan O, Docherty N, Balfe A, Maguire A, Mulcahy HE, Doherty G, O'Donoghue D, Hyland J, et al.. Spatial variation of the colonic microbiota in patients with ulcerative colitis and control volunteers. Gut 2015; 64:1553-61; PMID:25596182; http://dx.doi.org/ 10.1136/gutjnl-2014-307873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].de Carcer DA, Cuiv PO, Wang T, Kang S, Worthley D, Whitehall V, Gordon I, McSweeney C, Leggett B, Morrison M. Numerical ecology validates a biogeographical distribution and gender-based effect on mucosa-associated bacteria along the human colon. ISME J 2011; 5:801-9; PMID:21124491; http://dx.doi.org/ 10.1038/ismej.2010.177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Walker AW, Sanderson JD, Churcher C, Parkes GC, Hudspith BN, Rayment N, Brostoff J, Parkhill J, Dougan G, Petrovska L. High-throughput clone library analysis of the mucosa-associated microbiota reveals dysbiosis and differences between inflamed and non-inflamed regions of the intestine in inflammatory bowel disease. BMC Microbiol 2011; 11:7; PMID:21219646; http://dx.doi.org/ 10.1186/1471-2180-11-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Albenberg L, Esipova TV, Judge CP, Bittinger K, Chen J, Laughlin A, Grunberg S, Baldassano RN, Lewis JD, Li H, et al.. Correlation between intraluminal oxygen gradient and radial partitioning of intestinal microbiota. Gastroenterology 2014; 147:1055-63 e8; PMID:25046162; http://dx.doi.org/ 10.1053/j.gastro.2014.07.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Van den Abbeele P, Belzer C, Goossens M, Kleerebezem M, De Vos WM, Thas O, De Weirdt R, Kerckhof F-M, Van de Wiele T. Butyrate-producing Clostridium cluster XIVa species specifically colonize mucins in an in vitro gut model. ISME J 2013; 7:949-61; PMID:23235287; http://dx.doi.org/ 10.1038/ismej.2012.158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Wang Y, Devkota S, Musch MW, Jabri B, Nagler C, Antonopoulos DA, Chervonsky A, Chang EB. Regional mucosa-associated microbiota determine physiological expression of TLR2 and TLR4 in murine colon. PLoS ONE 2010; 5:e13607; PMID:21042588; http://dx.doi.org/ 10.1371/journal.pone.0013607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Nava GM, Friedrichsen HJ, Stappenbeck TS. Spatial organization of intestinal microbiota in the mouse ascending colon. ISME J 2011; 5:627-38; PMID:20981114; http://dx.doi.org/ 10.1038/ismej.2010.161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Nava GM, Stappenbeck TS. Diversity of the autochthonous colonic microbiota. Gut microbes 2011; 2:99-104; PMID:21694499; http://dx.doi.org/ 10.4161/gmic.2.2.15416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Donaldson GP, Lee SM, Mazmanian SK. Gut biogeography of the bacterial microbiota. Nat Rev Micro 2016; 14:20-32; http://dx.doi.org/ 10.1038/nrmicro3552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Hill DA, Hoffmann C, Abt MC, Du Y, Kobuley D, Kirn TJ, Bushman FD, Artis D. Metagenomic analyses reveal antibiotic-induced temporal and spatial changes in intestinal microbiota with associated alterations in immune cell homeostasis. Mucosal immunol 2010; 3:148-58; PMID:19940845; http://dx.doi.org/ 10.1038/mi.2009.132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Jakobsson HE, Rodríguez-Piñeiro AM, Schütte A, Ermund A, Boysen P, Bemark M, Sommer F, Bäckhed F, Hansson GC, Johansson MEV. The composition of the gut microbiota shapes the colon mucus barrier. EMBO reports 2015; 16:164-77; PMID:25525071; http://dx.doi.org/ 10.15252/embr.201439263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Willing BP, Dicksved J, Halfvarson J, Andersson AF, Lucio M, Zheng Z, Järnerot G, Tysk C, Jansson JK, Engstrand L. A pyrosequencing study in twins shows that gastrointestinal microbial profiles vary with inflammatory bowel disease phenotypes. Gastroenterology 2010; 139:1844-54.e1; PMID:20816835; http://dx.doi.org/ 10.1053/j.gastro.2010.08.049 [DOI] [PubMed] [Google Scholar]
- [20].Hong PY, Croix JA, Greenberg E, Gaskins HR, Mackie RI. Pyrosequencing-based analysis of the mucosal microbiota in healthy individuals reveals ubiquitous bacterial groups and micro-heterogeneity. PLoS ONE 2011; 6:e25042; PMID:21966408; http://dx.doi.org/ 10.1371/journal.pone.0025042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Eckburg PB, Bik EM, Bernstein CN, Purdom E, Dethlefsen L, Sargent M, Gill SR, Nelson KE, Relman DA. Diversity of the human intestinal microbial flora. Science 2005; 308:1635; PMID:15831718; http://dx.doi.org/ 10.1126/science.1110591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Gevers D, Kugathasan S, Denson LA, Vázquez-Baeza Y, Van-Treuren W, Ren B, Schwager E, Knights D, Song S-J, Yassour M, et al.. The treatment-naive microbiome in new-onset Crohn's disease. Cell Host Microbe 2014; 15:382-92; PMID:24629344; http://dx.doi.org/ 10.1016/j.chom.2014.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Ringel Y, Maharshak N, Ringel-Kulka T, Wolber EA, Sartor RB, Carroll IM. High throughput sequencing reveals distinct microbial populations within the mucosal and luminal niches in healthy individuals. Gut microbes 2015; 6:173-81; PMID:25915459; http://dx.doi.org/ 10.1080/19490976.2015.1044711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Jalanka J, Salonen A, Salojärvi J, Ritari J, Immonen O, Marciani L, Gowland P, Hoad C, Garsed K, Lam C, et al.. Effects of bowel cleansing on the intestinal microbiota. Gut 2014; 64:1562-8; PMID:25527456 [DOI] [PubMed] [Google Scholar]
- [25].Yasuda K, Oh K, Ren B, Tickle TL, Franzosa EA, Wachtman LM, Miller AD, Westmoreland SV, Mansfield KG, Vallender EJ, et al.. Biogeography of the intestinal mucosal and lumenal microbiome in the rhesus macaque. Cell Host Microbe 2015; 17:385-91; PMID:25732063; http://dx.doi.org/ 10.1016/j.chom.2015.01.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Ahmed S, Macfarlane GT, Fite A, McBain AJ, Gilbert P, Macfarlane S. Mucosa-associated bacterial diversity in relation to human terminal ileum and colonic biopsy samples. Appl Environ Microbiol 2007; 73:7435-42; PMID:17890331; http://dx.doi.org/ 10.1128/AEM.01143-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Zhang Z, Geng J, Tang X, Fan H, Xu J, Wen X, Ma Z, Shi P. Spatial heterogeneity and co-occurrence patterns of human mucosal-associated intestinal microbiota. ISME J 2014; 8:881-93; PMID:24132077; http://dx.doi.org/ 10.1038/ismej.2013.185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Zoetendal E, Von Wright A, Vilpponen-Salmela T, Ben-Amor K, Akkermans A, De Vos W. Mucosa-associated bacteria in the human gastrointestinal tract are uniformly distributed along the colon and differ from the community recovered from feces. Appl Environ Microbiol 2002; 68:3401; PMID:12089021; http://dx.doi.org/ 10.1128/AEM.68.7.3401-3407.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Wang M, Ahrné S, Jeppsson B, Molin G. Comparison of bacterial diversity along the human intestinal tract by direct cloning and sequencing of 16S rRNA genes. FEMS microbiol ecol 2005; 54:219-31; PMID:16332321; http://dx.doi.org/ 10.1016/j.femsec.2005.03.012 [DOI] [PubMed] [Google Scholar]
- [30].Lepage P, Seksik P, Sutren M, de la Cochetiere MF, Jian R, Marteau P, Dore J. Biodiversity of the mucosa-associated microbiota is stable along the distal digestive tract in healthy individuals and patients with IBD. InflammBowel Dis 2005; 11:473-80 [DOI] [PubMed] [Google Scholar]
- [31].Green GL, Brostoff J, Hudspith B, Michael M, Mylonaki M, Rayment N, Staines N, Sanderson J, Rampton DS, Bruce KD. Molecular characterization of the bacteria adherent to human colorectal mucosa. J Appl Microbiol 2006; 100:460-9; PMID:16478485; http://dx.doi.org/ 10.1111/j.1365-2672.2005.02783.x [DOI] [PubMed] [Google Scholar]
- [32].Garrett WS, Lord GM, Punit S, Lugo-Villarino G, Mazmanian SK, Ito S, Glickman JN, Glimcher LH. Communicable ulcerative colitis induced by T-bet deficiency in the innate immune system. Cell 2007; 131:33-45; PMID:17923086; http://dx.doi.org/ 10.1016/j.cell.2007.08.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Morgan XC, Tickle TL, Sokol H, Gevers D, Devaney KL, Ward DV, Reyes JA, Shah SA, LeLeiko N, Snapper SB, et al.. Dysfunction of the intestinal microbiome in inflammatory bowel disease and treatment. Genome Biol 2012; 13:R79; PMID:23013615; http://dx.doi.org/ 10.1186/gb-2012-13-9-r79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Machiels K, Joossens M, Sabino Jo, De Preter V, Arijs I, Eeckhaut V, Ballet V, Claes K, Van Immerseel F, Verbeke K, et al.. A decrease of the butyrate-producing species Roseburia hominis and Faecalibacterium prausnitzii defines dysbiosis in patients with ulcerative colitis. Gut 2014; 63:1275-83; PMID:24021287; http://dx.doi.org/ 10.1136/gutjnl-2013-304833 [DOI] [PubMed] [Google Scholar]
- [35].Rajilic-Stojanovic M, Shanahan F, Guarner F, de Vos WM. Phylogenetic analysis of dysbiosis in ulcerative colitis during remission. Inflamm Bowel Dis 2013; 19:481-8; PMID:23385241; http://dx.doi.org/ 10.1097/MIB.0b013e31827fec6d [DOI] [PubMed] [Google Scholar]
- [36].Ohkusa T, Sato N, Ogihara T, Morita K, Ogawa M, Okayasu I. Fusobacterium varium localized in the colonic mucosa of patients with ulcerative colitis stimulates species-specific antibody. J Gastroenterol Hepatol 2002; 17:849-53; PMID:12164960; http://dx.doi.org/ 10.1046/j.1440-1746.2002.02834.x [DOI] [PubMed] [Google Scholar]
- [37].Macfarlane S, Furrie E, Cummings JH, Macfarlane GT. Chemotaxonomic Analysis of Bacterial Populations Colonizing the Rectal Mucosa in Patients with Ulcerative Colitis. Clin Infect Dis 2004; 38:1690-9; PMID:15227614; http://dx.doi.org/ 10.1086/420823 [DOI] [PubMed] [Google Scholar]
- [38].Rowan F, Docherty NG, Murphy M, Murphy B, Calvin Coffey J, O'Connell PR. Desulfovibrio bacterial species are increased in ulcerative colitis. Dis Colon Rectum 2010; 53:1530; PMID:20940602; http://dx.doi.org/ 10.1007/DCR.0b013e3181f1e620 [DOI] [PubMed] [Google Scholar]
- [39].Png CW, Linden SK, Gilshenan KS, Zoetendal EG, McSweeney CS, Sly LI, McGuckin MA, Florin TH. Mucolytic bacteria with increased prevalence in IBD mucosa augment in vitro utilization of mucin by other bacteria. Am J Gastroenterol 2010; 105:2420-8; PMID:20648002; http://dx.doi.org/ 10.1038/ajg.2010.281 [DOI] [PubMed] [Google Scholar]
- [40].Frank DN, St. Amand AL, Feldman RA, Boedeker EC, Harpaz N, Pace NR. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc Natl Acad Sci USA 2007; 104:13780-5; PMID:17699621; http://dx.doi.org/ 10.1073/pnas.0706625104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Falony G, Joossens M, Vieira-Silva S, Wang J, Darzi Y, Faust K, Kurilshikov A, Bonder MJ, Valles-Colomer M, Vandeputte D, et al.. Population-level analysis of gut microbiome variation. Science 2016; 352:560-4; PMID:27126039; http://dx.doi.org/ 10.1126/science.aad3503 [DOI] [PubMed] [Google Scholar]
- [42].Poxton IR, Brown R, Sawyerr A, Ferguson A. Mucosa-associated bacterial flora of the human colon. J Med Microbiol 1997; 46:85-91; PMID:9003751; http://dx.doi.org/ 10.1099/00222615-46-1-85 [DOI] [PubMed] [Google Scholar]
- [43].Sokol H, Lepage P, Seksik P, Dore J, Marteau P. Molecular comparison of dominant microbiota associated with injured versus healthy mucosa in ulcerative colitis. Gut 2007; 56:152-4; PMID:17172591; http://dx.doi.org/ 10.1136/gut.2006.109686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Gophna U, Sommerfeld K, Gophna S, Doolittle WF, Veldhuyzen van Zanten SJ. Differences between tissue-associated intestinal microfloras of patients with Crohn's disease and ulcerative colitis. J Clin Microbiol 2006; 44:4136-41; PMID:16988016; http://dx.doi.org/ 10.1128/JCM.01004-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Bibiloni R, Mangold M, Madsen KL, Fedorak RN, Tannock GW. The bacteriology of biopsies differs between newly diagnosed, untreated, Crohn's disease and ulcerative colitis patients. J Med Microbiol 2006; 55:1141-9; PMID:16849736; http://dx.doi.org/ 10.1099/jmm.0.46498-0 [DOI] [PubMed] [Google Scholar]
- [46].Seksik P, Lepage P, de la Cochetiere MF, Bourreille A, Sutren M, Galmiche JP, Dore J, Marteau P. Search for localized dysbiosis in Crohn's disease ulcerations by temporal temperature gradient gel electrophoresis of 16S rRNA. J Clin Microbiol 2005; 43:4654-8; PMID:16145122; http://dx.doi.org/ 10.1128/JCM.43.9.4654-4658.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Vasquez N, Mangin I, Lepage P, Seksik P, Duong J-P, Blum S, Schiffrin E, Suau A, Allez M, Vernier G, et al.. Patchy distribution of mucosal lesions in ileal Crohn's disease is not linked to differences in the dominant mucosa-associated bacteria: A study using fluorescence in situ hybridization and temporal temperature gradient gel electrophoresis. Inflamm Bowel Dis 2007; 13:684-92 10.1002/ibd.20084; PMID:17206669; http://dx.doi.org/ 10.1002/ibd.20084 [DOI] [PubMed] [Google Scholar]
- [48].Sepehri S, Kotlowski R, Bernstein CN, Krause DO. Microbial diversity of inflamed and noninflamed gut biopsy tissues in inflammatory bowel disease. Inflamm Bowel Dis 2007; 13:675-83; PMID:17262808; http://dx.doi.org/ 10.1002/ibd.20101 [DOI] [PubMed] [Google Scholar]
- [49].Mylonaki M, Rayment NB, Rampton DS, Hudspith BN, Brostoff J. Molecular characterization of rectal mucosa-associated bacterial flora in inflammatory bowel disease. Inflamm Bowel Dis 2005; 11:481-7; PMID:15867588; http://dx.doi.org/ 10.1097/01.MIB.0000159663.62651.4f [DOI] [PubMed] [Google Scholar]
- [50].Palm NW, de Zoete MR, Cullen TW, Barry NA, Stefanowski J, Hao L, Degnan PH, Hu J, Peter I, Zhang W, et al.. Immunoglobulin A coating identifies colitogenic bacteria in inflammatory bowel disease. Cell 2014; 158:1000-10; PMID:25171403; http://dx.doi.org/ 10.1016/j.cell.2014.08.006 [DOI] [PMC free article] [PubMed] [Google Scholar]


