Abstract
Recent improvements in experimental and computational techniques used to study the transcriptome have enabled an unprecedented view of RNA processing, revealing many previously unknown non-canonical splicing events. This includes cryptic events located far from the currently annotated exons, and unconventional splicing mechanisms that have important roles in regulating gene expression. These non-canonical splicing events are a major source of newly emerging transcripts during evolution, especially when they involve sequences derived from transposable elements. They are therefore under precise regulation and quality control, which minimises their potential to disrupt gene expression. While non-canonical splicing can lead to aberrant transcripts that cause many diseases, we also explain how it can be exploited for new therapeutic strategies.
Introduction
The vast majority of human genes contain more than one exon, and therefore introns need to be spliced from the nascent transcript and exons joined to form an mRNA that can be translated into a protein. Alternative splicing allows these exons to be joined in different variations to form alternative transcripts, which greatly increases the diversity of proteins encoded by a limited number of genes1. This alternative splicing can be either enhanced or repressed by transacting factors, which are directed to the precursor mRNA by cis-acting regulatory elements1,2. Genes producing long non-coding RNAs (lncRNAs) also typically contain multiple exons, and often display evidence for alternative splicing using similar mechanisms to those used for protein-coding mRNAs3
The mechanisms of splicing regulation and its perturbation in disease have been reviewed elsewhere4,5. Here, our emphasis is on non-canonical splicing: this includes the cryptic splice sites that are located far from the currently annotated exons, and unconventional mechanisms that deviate from the well-defined rules of splicing. New methods to sequence the transcriptome, together with dedicated analysis pipelines (Box 1), have revealed a broad prevalence of non-canonical splicing events that can generate cryptic exons6–10, microexons11–13, recursive splicing14,15, circular RNAs (circRNAs)16–20, retained introns21–25 and exonic introns (exitrons)25,26, among others. In this Review, we discuss the known mechanisms, functions and evolutionary potential of these events. We describe how mutations can cause disease by disrupting transcriptome integrity via non-canonical splicing, and how the cellular quality control systems defend the transcriptome from such perturbations. Finally, we explain how the unconventional splicing mechanisms can be targeted or exploited for new therapeutic strategies.
Box 1. Identification of non-canonical splicing events.
High-throughput methodologies, and in particular RNA-seq, have created opportunities for transcriptome-wide annotation of rare, cell-type-specific transcripts and non-canonical splicing events in our transcriptomes12,14,15. Whilst cDNAs generated from poly(A)-purified RNAs in mRNA-seq primarily detect fully spliced mRNAs that have passed the cellular quality control, cDNAs generated with random-primers in total RNA-seq also identified the intermediate steps of splicing reactions14,15,48,189. To study non-canonical splicing, several alignment algorithms have been tailored for the discovery of novel splice junctions with RNA-seq data11,29,30. A particularly great diversity of transcripts was found in the brain, in agreement with the great variety of cell types and functions that exist in this organ10,14,29,30. Even though many of these are rare and non-functional, some are functionally important non-canonical splicing events. One way to distinguish those that may have a function is to focus on novel junctions that contact conserved sequences that bear features of splicing elements, such as proximally spaced 3' and 5' splice sites (i.e. within typical exon size limits), branch points, exonic enhancers and other regulatory elements10,14,26,31.
More specialised methods for preparing and analysing RNA-seq data have led to discovery of new types of exons and RNAs. For example, several commonly used alignment algorithms require a minimum length of the seed sequence for the alignment, which generally limits detection of exons to those longer than 30 nt, thereby excluding microexons. To overcome this limitation, alignment algorithms were modified to use shorter seeds and to allow longer reads to be mapped in multiple parts11. If much shorter parts of a long read are mapped to two exons in a way that leaves an unmapped intervening sequence, this sequence can then be mapped back to the intronic sequence present between the two exons, with priority given to conserved sites flanked by proximally spaced 3' and 5' splice sites consistent with a <30nt microexon (Figure 1B)11,13,44. Alternatively, custom alignment files incorporating all putative cryptic exons with flanking splice sites can be used for mapping12.
Information on splicing efficiency can also be gained by analysis of intronic reads. For example, intron retention can be examined by the ratio of exon-intron junction reads relative to junction spanning reads, or by comparing read coverage across the intron to the flanking exons21,23,24. Moreover, co-transcriptional splicing patterns can be visualized across introns in total RNA-seq data as ‘saw-tooth’ patterns14,15,31,48. Specifically, the RNA abundance at the start of a long intron is higher than at its end owing to the presence of nascent transcripts in various stages of transcription, and because splicing can’t proceed until transcription of the 3' splice site47,48. Novel junctions that overlap clear troughs in the co-transcriptional splicing patterns often identify recursive splice sites (RS sites)14,15 (Figure 2A).
Dedicated computational approaches also facilitated discovery of circular RNAs (circRNAs) and chimeric transcripts66,67,190,191. In the simplest method for discovery of circRNAs, unaligned reads are split into two parts before being remapped to exons. If the second part maps to an exon upstream of the first part, these are then considered as circRNA candidates17 (Figure 4B). This local reordering of the alignments distinguishes circRNAs from chimeric transcripts that can also be identified by discordant alignments192,193 (Figure 4B-D). Experimentally enriching the sample preparation for non-linear RNAs before cDNA library preparation using the exoribonuclease RNase R can further enhance circRNA discovery65,67.
Types of non-canonical splicing
The fidelity of splicing is achieved by combinatorial recognition of specific sequences within precursor mRNA at many steps during the splicing process4,27. The first, and possibly the most important aspect of combinatorial recognition is described by the exon-definition model, which was proposed to explain how exons are recognised as functional units in metazoan organisms that contain long introns27,28. This process involves interactions between factors bound to the flanking splice sites (e.g. U1 and U2 snRNPs, U2AF complex) and SR proteins bound to the exonic enhancer sequences. We will first discuss the cryptic exons, microexons and recursive splice sites (RS sites), which often require unconventional exon definition mechanisms. Next, we will discuss non-canonical splicing mechanisms that result from lower or higher splicing efficiency than normal (retained introns, exitrons), changes in the usual order of splicing (circRNAs, chimeric RNAs) or changes in the consensus sequence (atypical splice sites).
Cryptic splice sites and exons
Introns of ENSEMBL-annotated genes constitute around 23% of the human genome, and within such a vast sequence space it is inevitable that many sequences similar to the consensus motifs of canonical splice sites will be present by chance. Such sequences are known as cryptic splice sites. To prevent uncontrolled splicing at cryptic sites, exon definition mechanism has evolved to maintain splicing fidelity, which explains why most individual cryptic splice sites do not efficiently initiate splicing27,28. Nevertheless, over half a million non-annotated splicing events have been discovered through the analysis of mouse and human RNA-seq data10,14,29–31. Even though many of these events may be splicing ‘mistakes’ that are tolerated by the cell and have no function, targeted genome editing experiments are beginning to uncover functions of specific cryptic splice sites31. Moreover, cryptic splice sites can be present in a manner that can define non-annotated, or ‘cryptic exons’32. These exons often introduce premature termination codons (PTCs) into the resulting transcripts, which can target them for nonsense-mediated decay (NMD) in the cytoplasm33–35 (Figure 1A). In some cases, abnormal splicing also leads to transcription-coupled surveillance mechanisms that can decrease the expression of resulting transcripts36. These quality control pathways often decrease the expression of transcripts containing cryptic exons, which makes these exons more difficult to detect and annotate during transcriptome sequencing analysis6–10.
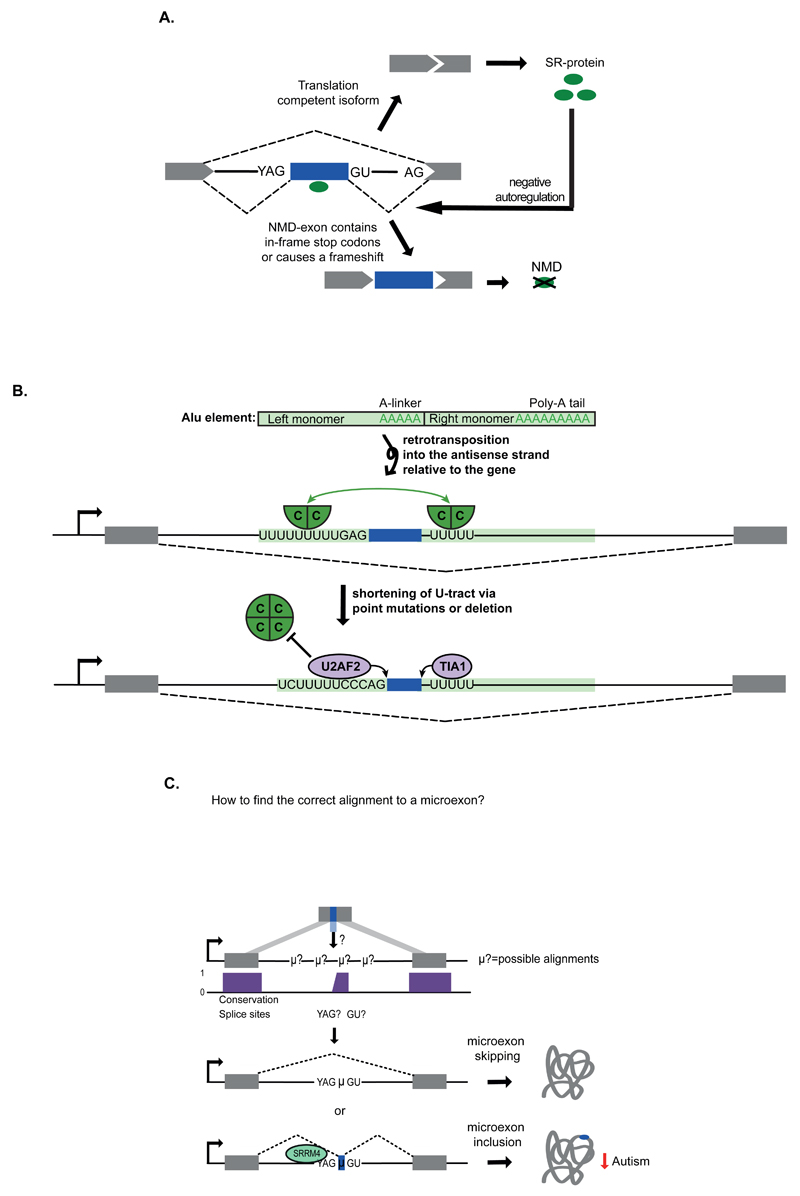
Figure 1. Cryptic exons and microexons.
a) Many introns contain proximally spaced sequences that resemble splice sites, which can in some cases lead to splicing of ‘cryptic’ exons. Cryptic exons often introduce premature termination codons (PTCs), which may target the resulting transcripts for nonsense-mediated decay (NMD). Such NMD-exons are common within transcripts that encode splicing activators, where they function as part of autoregulatory mechanisms33–35. In this example, the SR protein enhances inclusion of an NMD-exon within its own mRNA as part of a negative autoregulatory feedback that maintains appropriate steady-state abundance. b) An Alu element is normally composed of two arms, which contain an A-linker and polyA tail. The Alu can become retrotransposed into the antisense strand relative to the gene, so that transcription of the gene produces antisense Alu sequence that contains two U-tracts at the beginning of each arm. Many such antisense Alu elements are capable of forming cryptic exons owing to the presence of splice site-like motifs37. However, they are normally repressed by a hnRNP C tetramer (green circle), possibly because each U-tract can bind the two RNA Recognition Motif domains that are present on the opposite surfaces of the tetramer (as indicated by the green arrow)8,186. The example provided here shows the U-tracts around the Alu exon from the CD55 gene (encoding CD55 molecule). Below, mutations in the U-tracts are shown that decrease binding of hnRNP C, allowing binding of U2 small nuclear RNA auxillary factor (U2AF2) and TIA1 cytotoxic granule-associated RNA binding protein (TIA1), which initiate splicing of a cryptic Alu exon8,37,39. C = hnRNP C protein. C) Microexons can be detected from gapped regions in sequencing reads11,13,44. After mapping of multiple parts of the sequence read to flanking exons, unmapped intervening sequences are aligned to the intronic sequence present between the two exons, with preference given to those that are flanked by conserved splice site motifs. Inclusion of microexons can be enhanced by RNA binding proteins (RBPs) such as Serine/Arginine Repetitive Matrix 4 (SRRM4), an SR protein that binds upstream of microexons and promotes microexon splicing. Inclusion of microexons typically leads to modulation of overlapping or adjacent protein domains to change protein activity. SRRM4 is reduced in autism patients leading to decreased inclusion of microexons12. YAG, 3' splice site; GU, 5' splice site; NMD, Nonsense-mediated decay; μ?, possible microexon; μ, microexon.
Cryptic exons often emerge from transposable elements (TEs). In primates, antisense Alu sequences are the best substrates for the exonisation process, as they require a low number of mutations to form potent splice sites37. The evolution of the Alu family consisted of two phases. The original Alu monomers arose from a fusion of the 5′ and 3′ ends of the 7SL RNA gene, which encodes an RNA component of the signal recognition particle (SRP). A further fusion of these monomers led to the modern Alu elements that are composed of left and right arms joined by an A-rich linker and followed by an A-tail (Figure 1B)38. Notably, more than 330,000 Alu elements (annotated by RepeatMasker) are present in introns of protein-coding genes in an antisense orientation, where they are transcribed in a reverse orientation, thus containing two U-tracts instead of the A-tail and linker. These U-tracts can function as binding sites for splicing factors, especially U2 small nuclear RNA auxillary factor (U2AF2) and T-cell intracellular antigen (TIA) proteins, which can induce the formation of cryptic or alternative Alu exons8,37,39. Mechanisms regulating splicing of Alu exons and other cryptic exons have been uncovered with studies that map the binding sites of RNA binding proteins (RBPs) with cross-linking and immunoprecipitation (CLIP), individual nucleotide CLIP (iCLIP) and related techniques40. The Alu-derived exons were found to be tightly repressed by heterogeneous nuclear ribonucleoprotein C (hnRNP C), which can displace U2AF2 from the long U-tracts (Figure 1B)8, and other cryptic exons were found to be repressed by NOVA7, RBP fox-1 homologue (C. elegans) 2 (Rbfox2)6 and TAR DNA binding protein 43 (TDP-43)9. Together with deep sequencing of RNA from tissues, these studies revealed thousands of previously unknown cryptic exons, and some of these are becoming recognised as regulated alternative exons10 (Figure 1A). Interestingly, repressive sequences were found to be more common at cryptic exons compared to the established alternative exons that emerged from transposable elements, indicating that loss of repression may have a role in the formation of new exons41.
Microexons
Exons that are shorter than 30 nt have traditionally been referred to as microexons42–44. New computational methods for analysis of sequencing data revealed hundreds of previously unidentified microexons, 60% of which are preferentially included in neuronal tissues11–13,44. Interestingly, microexons tend to be flanked by intronic motifs that are required for their inclusion, which bind to RBPs such as Serine/Arginine Repetitive Matrix 4 (SRRM4; also known as nSR100)12, RBP Fox (RBFOX) or Polypyrimidine Tract Binding Protein 1 (PTBP1)13. SRRM4 is an SR-related protein that acts in an unusual way. Unlike other SR proteins that bind to exonic enhancers, SRRM4 binds to enhancers that are embedded within the unusually long polypyrimidine tract present upstream of microexons, thereby compensating for the limited space available for enhancer sequences within the microexons (Figure 1C).
Recursive splice sites
RS sites, also referred to as ‘zero-length exons’, are defined by a sequence that combines the 3' and 5' splice site consensus motifs. This allows an intron to be spliced in multiple consecutive steps: the 3' splice site is used to splice the preceding part of the intron, which reconstitutes a full 5' splice site that is then used to splice the remaining part of the intron (Figure 2). First discovered in the long introns of three Drosophila melanogaster genes45,46, analyses using total RNA-seq and iCLIP identified 197 RS sites in D. melanogaster15 and 11 in human14,15. In addition to detection of splice-junction reads bearing the RS site motif, these studies also required the presence of co-transcriptional splicing patterns (Figure 2A), which can only be reliably evaluated in long introns with high read coverage14,47,48 (Box 1). Accordingly, these numbers are probably underestimates. For instance, 419 cryptic splicing events were found at putative RS junctions in human samples prior to considering the co-transcriptional splicing patterns14. Notably, intrasplicing is another mechanism that can affect alternative splicing by using non-annotated splice sites. However, here the first splicing reaction reconstitutes a new 3' splice site, which can then be used by an upstream exon to remove the remaining intron49.
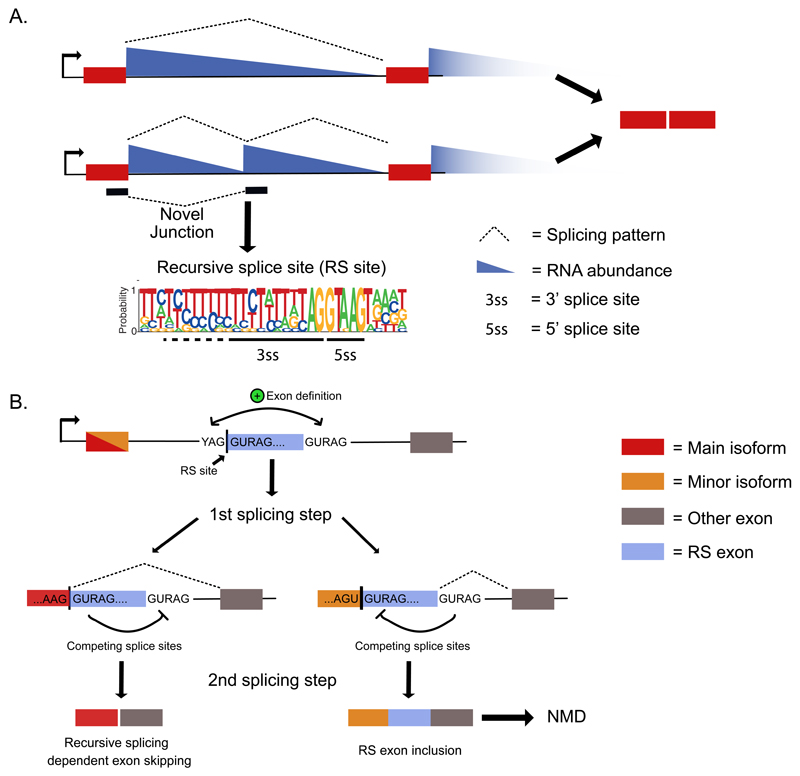
Figure 2. Recursive splicing of long introns.
a) Total RNA-seq read counts display a characteristic pattern of depletion from the start to the ends of long introns, which can be used to infer exon positions and splicing events47,48. “Sawtooth” patterns that overlap novel junction reads indicate splicing at deep intronic loci and are candidates for recursive splicing14,15. Here, the upstream exon first uses a 3' splice site to remove the first part of the intron. This process reconstitutes a 5' splice site that can then be used to remove the next section of the intron. This special type of splice site that is shown in the weblogo is referred to as a recursive splicing site (RS site). b) Recursive splicing in vertebrates requires the RS site to overlap a cryptic ‘RS exon’, which initiates the exon definition mechanism, required for recognition of the 3' splice site of the RS site14. After the first splicing step, the 5' splice site of the RS site competes with the 5' splice site of the RS exon. In the second step of splicing, the outcome of this competition decides whether the RS exon is skipped owing to recursive splicing, or included as an NMD-exon. While the preceding exons from major isoforms end in sequences that favour RS exon skipping, the minor isoforms and cryptic elements end in sequences that favour RS exon inclusion. RS site, Recursive splice site; RS exon, Recursive splicing exon; YAG, 3' splice site; GURAG, 5' splice site.
Even though RS sites do not normally lead to splicing of an exon, they employ the exon definition mechanism in vertebrates (Figure 2B). These RS sites are present at the start of cryptic exons, referred to as RS exons14. Both the RS sites and the downstream 5' splice site that is required for exon definition are highly conserved. Definition of the RS exon is essential to initiate splicing at the 3' splice site. After splicing of the preceding intron, the RS site reconstitutes a strong 5' splice site, which leads to skipping of the RS exon via recursive splicing. Whereas the exon definition mechanism is required for recursive splicing in human and zebrafish, it remains unclear how RS sites are defined in the fruit fly. The first RS sites discovered in the fruit fly overlapped with the start of annotated exons, indicating that an exon definition mechanism might be involved45,46. Alternatively, the 3' splice site of intronic RS sites is often strongly conserved across Drosophila, which is consistent with the sensitivity of fruit fly recursive splicing to depletion of U2AF215. It is also possible that RS sites are preceded by additional enhancer elements similar to microexons, which are flanked by binding motifs of multiple regulators, including SRRM412, RBFOX or PTBP113.
Retained introns
Even though the precision and efficiency of splicing is very high, it is not perfect. Both in plants and animals, decreased efficiency of splicing at some introns can lead to their retention within polyadenylated transcripts21,25,50–52. In fact, comparison of RNA from human and mouse tissues detected retained introns in alternative transcripts of most genes21–24. Intron retention can be a result of various trans- and cis-acting mechanisms (Figure 3A). Most often, it is caused by an inefficient recognition of canonical splice sites53. Under conditions of limiting spliceosome availability, such as upon downregulation of spliceosomal components, deficient splice site recognition can affect hundreds of introns in this way24. Moreover, inclusion of shorter introns in mammalian cells can be more dependent on intron definition, a mechanism that brings the splice sites at both ends of the intron into closer proximity27. This mechanism can be regulated by RBPs that bind at both ends of the intron and interact with each other54. These proposed mechanisms agree with the generally weaker 5' and 3' splice sites, and shorter length of the retained introns compared to other introns21,53,55. Moreover, retained introns have higher GC content compared to average introns, which might make them more sensitive to RNA polymerase II stalling21,53. Certain RBPs can also promote specific intron retention events22,56,57. For example, PTBP1 can repress recognition of a canonical splice site in an intron of the FosB gene56, while Poly(A) Binding Protein, Nuclear 1 (PABPN1) promotes retention of the last intron within its own transcript by binding to an adenosinerich region in the 3' UTR57. Finally, depletion of exon junction complex (EJC) components also leads to retention of long introns in D. melanogaster58,59, although apparently not in human cells60.
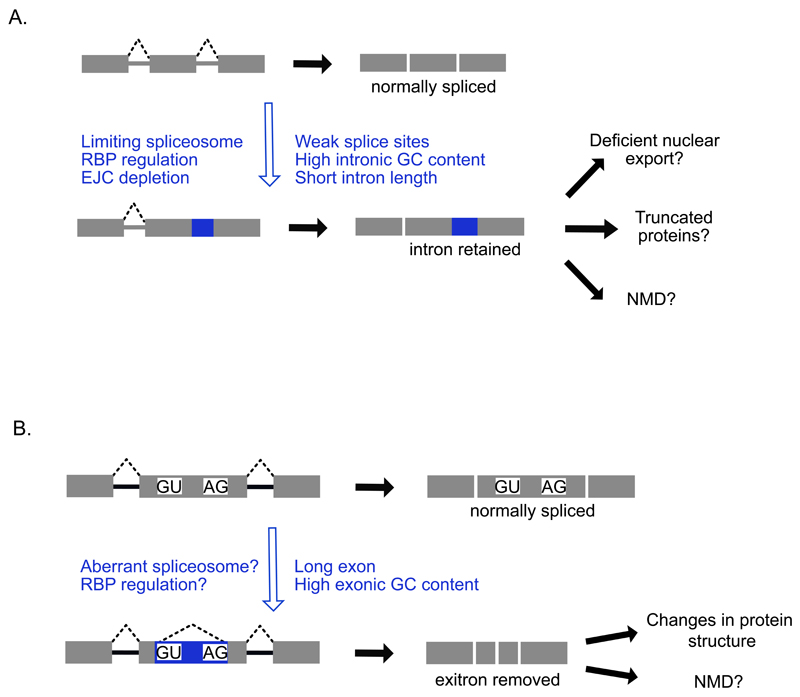
Figure 3. Intron retention and exitrons.
a) Intron retention events are detected as an accumulation of reads across intronic regions, or increases in the ratio of exon-intron reads to exon-exon reads21–25. Intron retention events are characterised by numerous features including weak splice sites, high GC content and short intron lengths. Trans-acting factors such as RBPs, the spliceosome and the EJC can also regulate specific intron retention events. The resulting transcripts are typically either retained in the nucleus or targeted for NMD in the cytoplasm or may result in truncated proteins21,53,55. Other intron retention events might be translated into truncated proteins. b) Exitrons are introns within annotated protein-coding exons that can be removed owing to the presence of internal splice site motifs within the exon25,26. Exitron-containing exons are longer than typical exons, and removal of the exitron can lead to changes in protein structure or degradation via NMD. NMD, Nonsense-mediated decay; AG, 3' splice site; GU, 5' splice site.
Exitrons
Some alternatively spliced introns are also present within regions annotated as exons. These introns are rarely spliced, and therefore they are referred to as cryptic introns, or also as ‘exitrons’25,26 (Figure 3B). A total of 923 exitrons have been discovered within regions that are normally annotated as exons26. Similar to retained introns, exitrons are shorter than average introns and have weak splice sites, which can explain why they are retained under normal conditions. Exitrons are formed from exons that are amongst the longest known in humans, and have higher GC content than typical exons. They are formed when cryptic splice sites within an exon go on to pair with the canonical splice sites that flank the same exon, thereby leading to definition of two smaller exons26. Unlike retained introns, exitrons don’t normally contain PTCs. Instead, their removal can change protein structure or lead to frame-shifts that introduce PTCs, which can target the resulting transcripts to NMD (Figure 3B).
Circular RNAs (CircRNAs)
CircRNAs are formed as a result of pre-mRNA splicing that doesn’t follow its canonical 5' to 3' order20,61. The mechanism responsible for this is referred to as back splicing, or head-to-tail splicing, where a branch point upstream of an exon attacks a downstream splice donor62–64. In some cases this happens with a single exon, whereas in others the start of an upstream exon splices to the end of a downstream exon, producing multi-exonic circRNAs17 (Figure 4A). In these multi-exonic circRNAs, the intervening intron can be spliced out. We refer to such single- or multi-exonic circular transcripts that lack introns as ‘exonic circRNAs’. Alternatively, if the intron between the exons remains retained, the resulting circular transcript is referred to as ‘exon-intron circRNA’20 (Figure 4A). Finally, ‘intronic circRNAs’ can be produced from intron lariats that are resistant to de-branching due to presence of C-rich motifs near the branch point20. These diverse types of circRNAs have been discovered in all domains of life 16–20,65,66. While most are quite rare, some are highly abundant in a specific tissue due to their resistance to exonucleases (Figure 4B)67. Many have tissue-specific expression patterns16,17,20,66, and in the central nervous system they tend to be enriched within neuronal dendrites68.
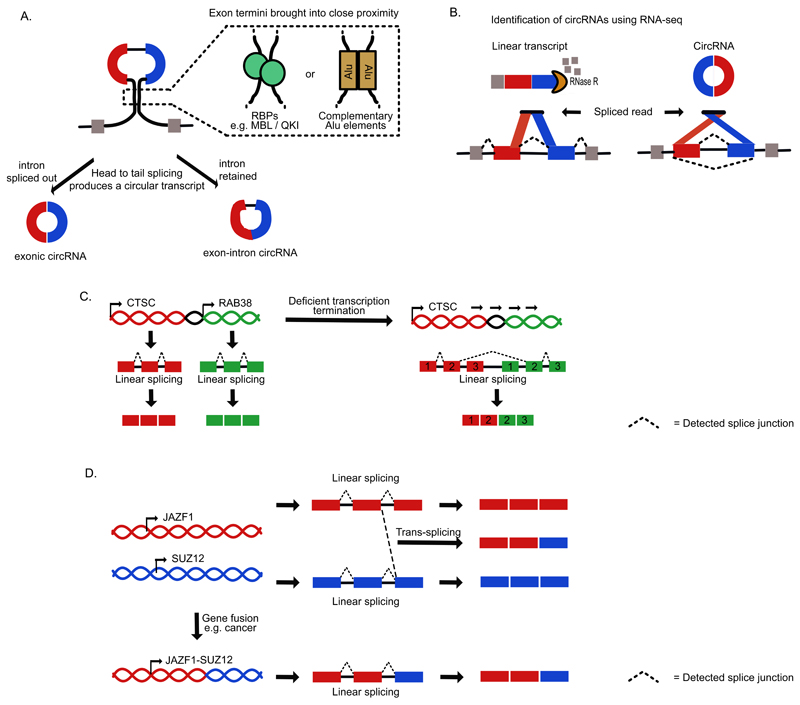
Figure 4. Formation of circRNAs and chimeric transcripts.
a) CircRNAs are produced by head to tail splicing and can be both mono- or multi-exonic. In this multi-exonic example the 3' splice site of an upstream exon becomes spliced to the 5' splice site of a downstream exon to generate a circular transcript that that either has the intervening intron removed (exonic circRNA) or retained between the two circularized exons (intron-exon circRNA) 20. Their formation is promoted when the pre-mRNA regions flanking the exon termini are brought in proximity. This can be due to the action of RNA-binding proteins such as Quaking (QKI) or muscleblind-like (MBNL), which bind to flanking regions74,75. Alternatively, this can be due to RNA hybridisation of the flanking regions, which can be caused by Alu elements in primates70. b) Circular RNAs are resistant to RNase R, which can be used for their enrichment during preparation of cDNA libraries. They can then be detected in sequencing data by junction reads that are in a head-to-tail orientation16–19. c) Chimeric RNA products can also be produced by cis-splicing when transcript termination is deficient76. This process results in read-through of one gene into its neighbouring gene, before splicing occurs between the penultimate exon of gene 1 and the second exon of gene 2, which is seen in the CTSC-RAB38 genes in cancer. d) Trans-splicing occurs when exons of two different transcripts become spliced together80–87. Alternatively, the same chimeric transcripts can be produced when genes become fused at the level of the DNA, such as in JAZF1-SUZ12 genes in some cancer, which leads to the same chimeric transcript being produced by a linear splicing reaction. RBP, RNA-binding protein.
The head-to-tail splicing can be promoted by the presence of intronic inverted repeat sequences, which hybridise and thereby bring the ends of the relevant exons in proximity69–72 (Figure 4A). In primates, hybridisation can be directed by inverted Alu repeats in flanking introns70. As inverted Alu repeats are known to be a target for RNA editing, it is thus possible that formation of circRNAs could be regulated by editing. Indeed, dsRNA hybridisation sites that are edited by adenosine deaminase acting on RNA (ADAR) are seen in introns that flank circRNAs in C. elegans73. However, formation of a dsRNA structure is not always required for circRNA formation69. RBPs such as Quaking (QKI) and muscleblind-like (MBNL) proteins are also able to regulate circRNA biogenesis via binding sites in the flanking introns20,71,74,75 (Figure 4A).
Chimeric RNAs
Modified algorithms for analysis of RNA-seq data can identify chimeric RNAs, which are produced when splicing joins the exons of different genes (Box 1). Cis-splicing was proposed to result from deficient transcriptional termination, which allows proximal genes to be transcribed as a single unit, thereby resulting in splicing of the penultimate exon of the upstream gene to the second exon of the downstream gene76. Such chimeric transcripts that combine exons of adjacent genes have been detected in several human tissues77–79 (Figure 4C). In contrast, trans-splicing joins exons derived from distant genomic locations (Figure 4D). The resulting chimeric transcripts have been best documented in trypanosomes, C.elegans and insects80–85, and to a lesser extent also in humans86,87.
Atypical splice sites
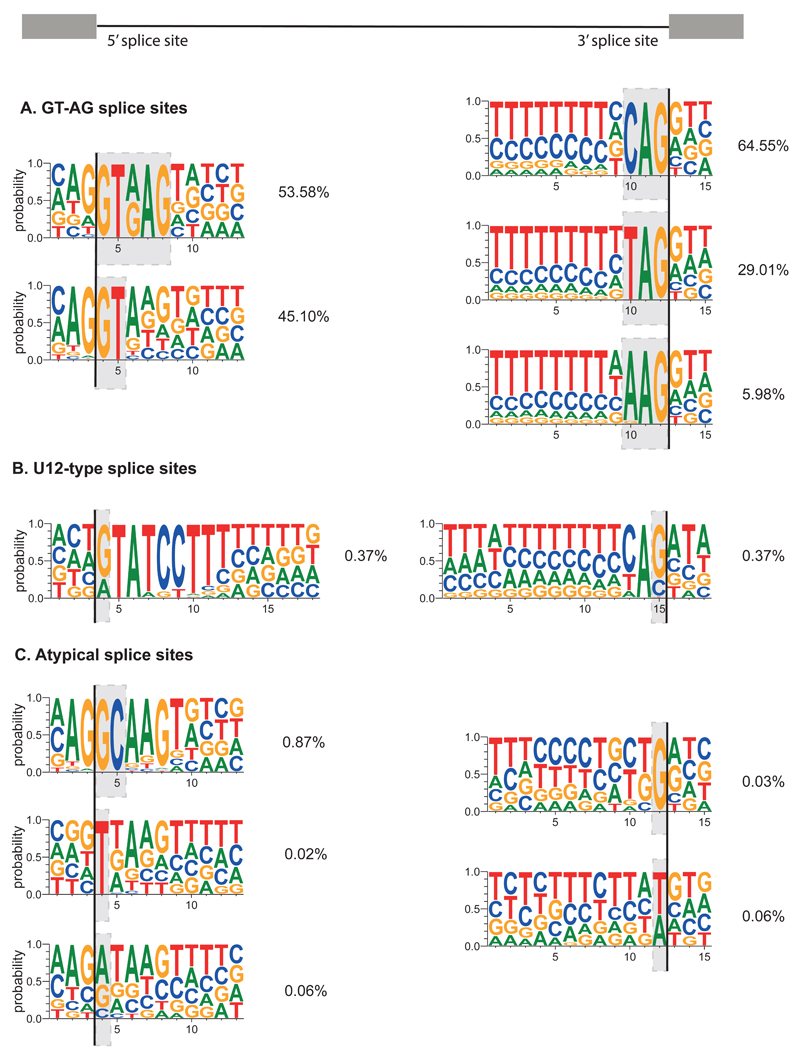
More than 99% of human introns are spliced by the major U2-dependent spliceosome. Most 5’ splice sites start with GTRAG, and remaining ones have a stronger preference for AG at the end of the exon, while 3’ splice sites end with CAG, TAG or more rarely, AAG (Figure 5A). The introns spliced by the minor U12-dependent spliceosome can be distinguished by the longer consensus sequence at the 5' splice site and at the branch point88,89(Figure 5B). Meanwhile, 5' splice sites that start with a GC are the most common atypical U2-type splice sites90,91 (Figure 5C). Each 3' splice site is normally preceded by a branch point that contains an adenine nucleotide. It is common that multiple branch points are present92, and this can affect the choice of alternative 3' splice sites. This was recently demonstrated genome-wide following mutation of SF3B1 splicing factor93–95.
Figure 5. A summary of human splice site consensus motifs.
Summarised splice site sequences are classified using the nucleotides marked by the grey boxes. All borders of human exons within Ensembl v83 multi-exon transcripts that overlap with RefSeq mRNA IDs were used. Identical coordinates from overlapping transcripts were collapsed into a single occurrence such that junctions were not counted multiple times. First exons had only their exon-intron junction evaluated, whilst terminal exons had only their intron-exon junction evaluated. This led to a total of 189,255 5' splice sites (shown on the left, with the line showing exon-intron border) and 187,091 of 3' splice sites (shown on the right, with the line showing intron-exon border). U12-type splice site sequences were obtained from U12DB187. After identifying the 5' and 3' sites overlapping with the U12-type splice sites, respectively, the remaining U2-type splice site sequences were examined. 5' and 3' splice sites were classified independently and sequentially based on the indicated nucleotides. For example, 53.58% of unique U1-type exon-intron junctions contain GTRAG, and the remaining U1-type junctions were classified based on the first two intronic nucleotides. The percentage of unique junctions containing each motif are indicated. Weblogo 3 was used to show the relative frequency of nucleotides at each position188. a) The U1-type 5' splice sites with GT at the border, and U2-type 3' splice sites with AG at the border, b) The U11-type 5' splice sites and U12-type 3' splice sites, c) The U1-type 5' splice sites with GC at the border, remaining U1-type 5' splice sites with TN at the border, where N stands for any nucleotide, U1-type 5' splice sites with VN at the border, where V stands for any nucleotide except T, U2-type 3' splice sites with BG at the border, where B stands for any non-A nucleotide, the U2-type 3' splice sites with W at the border, where W stands for T or A.
Multiple mechanisms could explain recognition of these atypical splice sites, including shifted base-pairing of small nuclear RNAs (snRNAs)96 and bulged nucleotides that retain base-pairing to snRNAs97. Some sites were also found to be modified by A-to-I RNA editing, in which inosine is effectively read as a guanosine91,98. An example of this mechanism is the ADAR2-dependent editing of an AA-3' dinucleotide within its own pre-mRNA, which then functions as a strong AG-3' splice site to change splicing of its transcript as part of an auto-regulatory mechanism98. Finally, the unconventional cytoplasmic splicing of XBP1 and other mRNAs during unfolded protein response, which employs the RNase Inositol-requiring enzyme 1 (IRE1) and RNA ligase RtcB, can create new exon-exon junctions that don’t contain the standard consensus sequences99–101.
The functions of non-canonical splicing
Non-canonical splicing events contribute to a great diversity of cellular mechanisms and biological functions. Perhaps the best understood of these functions is that of microexons, which are enriched in genes associated with synapse biology and axonogenesis12. Microexons are highly conserved, and their length generally comes in multiples of three, thereby preserving the open reading frame (ORF)11–13. Microexons are enriched within modular interaction domains, where they tend to encode charged residues that are accessible at the surface, and often overlap lipid or peptide binding domains12,13,102 (Figure 1B). It has been shown that inclusion of microexons alters the interactomes of several proteins12. Owing to their common brain-specific splicing patterns, microexons have a major role in increasing proteome diversity in the brain. Therefore it is not surprising that widespread skipping of microexons upon loss of SRRM4 in mice leads to neurodevelopmental defects103. Another type of newly discovered events that lead to new protein variants are exitrons, as most of these preserve the reading frame, and are enriched within disordered regions of the encoded proteins26,104 (Figure 3B).
Other forms of non-canonical splicing most often have a role in regulating gene expression. One of the best studied examples is the neuronal-expressed circRNA CDR1as/ciRS-7, which contains at least 63 conserved miR-7 binding sites that sequester this miRNA and thereby increases translation of its mRNA targets16,17. Moreover, circRNAs can contribute to mechanisms that regulate transcription or splicing20. For example, by enhancing production of circRNAs in its own transcript, the MBNL1 RBP decreases the amount of translation-competent transcripts produced from its own gene74. In fact, many splicing factors regulate splicing of cryptic exons, introns or circRNAs in their own transcripts or those of other RBPs, as part of auto- or cross-regulatory mechanisms6,7,20,21,24,33–35,55,105 (Figure 1A). Retained introns often lead to retention of the host mRNA in the nucleus, where it undergoes exosome-mediated degradation22. If exported to the cytoplasm, most retained introns introduce PTCs, and may thereby promote NMD of the resulting transcript or lead to production of truncated proteins22,23,106,107 (Figure 3A). Intron retention was found to coordinate expression of related genes in granulocyte differentiation24, at certain stages of the cell cycle55 and across tissues21. Interestingly, intron retention is more common in transcripts that are less required for the physiology of a particular tissue21.
The function of recursive splicing in regulating gene expression remains to be fully understood. In human, RS sites are found in the extremely long introns of genes that are expressed mainly in the brain, and function in neuronal axon guidance and cell adhesion14,15. It is tempting to speculate that recursive splicing could be important for splicing integrity of these introns. However, steric blocking of recursive splicing failed to reduce the overall splicing of the long intron in two human genes14. An alternative regulatory role was proposed for human RS sites14. These RS sites are followed by RS exons, which are spliced out of dominant isoforms, but included in minor isoforms that arise from use of upstream cryptic exons or rare alternative promoters (Figure 2B). The reason for inclusion of RS exons in minor isoforms is that the preceding cryptic exons end with suboptimal sequences, and therefore they do not reconstitute a sufficiently strong 5' splice site at the RS site. Interestingly, most RS exons contain PTCs, and therefore their inclusion prevents translation of full-length proteins and targets the resulting transcripts to NMD (Figure 2B). It remains to be seen how many RS sites are involved in the regulation of alternative splicing.
Evolutionary perspectives on non-canonical splicing
Even though exemplary functions of individual non-canonical splicing events have been discussed in this review, the same function cannot be ascribed to all events of the same type. For example, even though a few circRNAs can sequester a miRNA, most of them are not abundant enough to have such a function108. It is likely that many transcripts produced by non-canonical splicing have no function, and their presence reflects the capacity of cellular quality control mechanisms to protect from potential damaging effects of these transcripts. Most newly-emerging exons contain PTCs, and their initial emergence is likely to produce truncated or misfolded proteins that are likely to be deleterious for the organism. It is therefore not surprising that quality control mechanisms minimise the deleterious effects of such events. These include RBPs or snRNP complexes that have secondary activities aside from their usual roles in spliceosome function or regulation of canonical exons. These RBPs or snRNPs can repress splicing of cryptic exons8,74,75,109,110, edit the nascent RNA to represses splicing111,112, prevent mRNA export, or decrease the stability of aberrant mRNAs8,60,107,113,114.
Many non-canonical events are introduced by transposable elements (TEs), which make up as much as two-thirds of the human genome115. For example, over 1.5 million degenerated long interspersed elements (LINE) sequences are annotated in human genome (http://repeatmasker.org), and while many are transcribed as parts of other genes, fewer than 100 of them are capable of retrotransposition116. This indicates that evolution constantly puts the degenerated TEs to new uses, and when present in transcribed regions, they are a rich source of new exons and other elements for post-transcriptional control37,117,118. The newly-emerging Alu exons are controlled by an antagonistic interplay between two RBPs, hnRNP C and U2AF2, which compete for binding to U-tracts, thereby affecting the splicing outcome8 (Figure 1B). While mutations creating a splice site can cause a major increase in the inclusion of an Alu exon, mutations that change a single uridine within the U-tract are likely to only slightly modify the inclusion of Alu exons. Thus, repression by hnRNP C might ensure that new Alu exons emerge gradually, rather than in discrete steps119. Notably, hnRNP C is a conserved protein in vertebrates, and therefore it has preceded the insertion of Alu elements into primate genomes. It remains unknown how such conserved RBPs controlled the transition of diverse classes of TEs within vertebrate genomes from a state of repressed TE-derived cryptic exons into functionally regulated alternative exons.
One way to explain the evolutionary functions of emerging non-canonical splicing events is the multilevel selection theory120. According to this theory, even if only a small number of individual species- or clade-specific TEs were beneficial at the level of organisms, the prevalence of TEs could have adaptive value for the species or clade by promoting speciation or preventing extinction. Similarly, the prevalence of cryptic splicing might increase the probability for emergence of a few species-specific splicing events that can reset the gene regulatory networks. Such evolutionary tinkering is particularly important for complex organisms, in which it is linked to the increased size of the non-coding genomic regions121. Notably, the genes with the longest introns tend to be most highly expressed in the brain14, and these long introns produce the highest number of non-canonical splicing events122,123. It remains to be seen if and how such events may have contributed to the evolution of regulatory networks in the vertebrate brains.
By decreasing the expression of new transcript variants produced by non-canonical splicing, the cellular quality control pathways not only protect our cells from their potentially toxic effects, but also decrease the negative selection against these variants during evolution. Thus, the low expression level of transcripts produced by non-canonical splicing provides an opportunity for evolution to test the newly emerging variants and to select against toxic protein isoforms before expressing them at higher levels. It remains to be seen if established transcripts that generate functional protein isoforms created by alternative splicing in our cells might have initially emerged as cryptic splicing events in an ancestral species, and were then gradually co-opted by evolution for new functions.
Non-canonical splicing and disease
Disease-associated variations
Approximately a third of disease-causing mutations are presently estimated to disrupt pre-mRNA splicing124–126. This effect can occur either via mutations in cis-elements within pre-mRNAs, or via mutations or misregulation of trans-regulatory factors that bind to pre-mRNAs5. This figure may be an underestimate, since it does not include the disease-linked synonymous variants within exons that can affect splicing127. Moreover, even though standard computational models focus on positions close to canonical splice sites to identify variants that might affect RNA splicing, new models are being developed that can predict variants at other positions128. These models were successful in the analysis of variants in spinal muscular atrophy (SMA), colorectal cancer and autism spectrum disorder (ASD).
Mutations that are located far from canonical splice sites can activate non-canonical splicing to cause disease12,86,129–139. For example, the core spliceosomal component U1 snRNP can repress cryptic exons when it binds in a non-productive conformation (Figure 6A). Deletion of the repressive U1 snRNP binding sites was found to activate splicing of cryptic exons in both ataxia telangiectasia and Laron syndrome110,140. Generally, most studied mutations that induce splicing of cryptic exons achieve this by inactivating repressive sequences or secondary structures133–135 or increasing the strength of a cryptic splice site (Figure 6B). Moreover, disruption of canonical splicing can activate distal cryptic polyadenylation sites136,137 (Figure 6B). For example, triplet repeats within the first exon of the HTT transcript inhibit splicing of the following intron, thereby activating a cryptic polyadenylation site within the intron138. The resulting transcript can be translated into short toxic peptides that contribute to the molecular pathogenesis of Huntington disease.
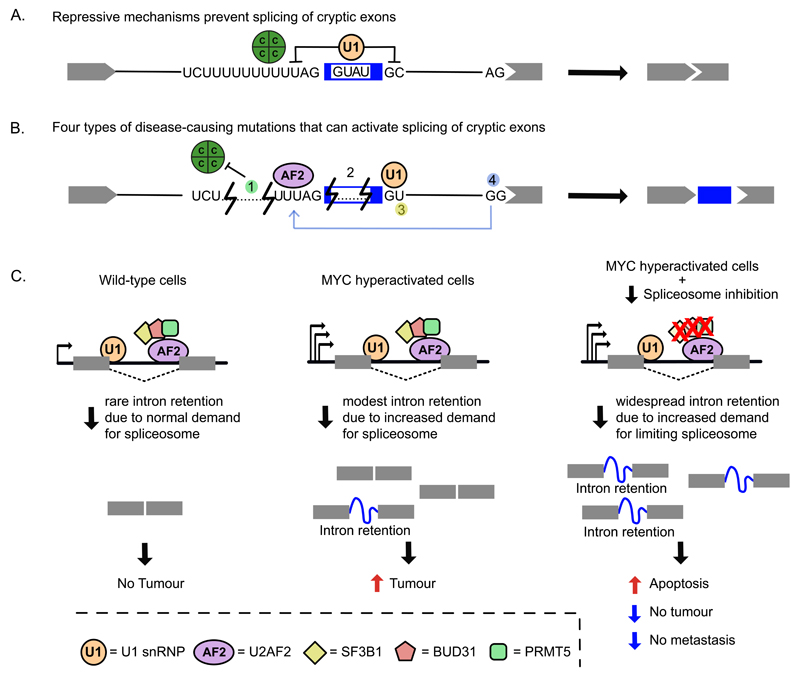
Figure 6. Cryptic splicing in disease and therapeutic strategies.
a) Cryptic exons are normally repressed by RBPs such as hnRNPC (green circle) or by U1 snRNP. b) Examples of mutations (numbered) in deep intronic regions that can activate cryptic splicing events in disease-associated genes. (1) hnRNPC (green circle) binding to a U-tract upstream of an antisense Alu element represses recognition of the cryptic 3' splice site within the element. Intronic deletions or point mutations that shorten U-tract can impede hnRNPC recruitment but allow U2AF2 (shown in purple) binding, leading to Alu exonisation. A deletion within an Alu in the PTS gene (encoding 6-Pyruvoyltetrahydropterin Synthase) leads to splicing of an Alu exon that introduces a frameshift, thereby causing the neurologic disease hyperphenylalaninaemia8,141. (2) In the ATM gene, U1snRNP (orange circle) binding to an intronic element within a cryptic exon inhibits its recognition as a splicing competent exon. Patients with ataxia telangiectasia present a 4 nt deletion that abolishes U1snRNP interaction, causing cryptic exon activation110. (3) A point mutation within a deep intronic sequence of the CFTR gene generates an active 5' splice site that allows insertion of a cryptic exon within the CFTR transcripts, which causes cystic fibrosis135. (4) In the BRCA2 gene, a point mutation that disrupts a canonical 3' splice site activates (depicted by a grey arrow) an upstream cryptic exon136. Disrupted BRCA2 expression causes breast, ovarian and other cancer types. c) New therapeutic strategies in cancer involve spliceosome targeting156,162,163. In MYC-driven tumours, oncogenic MYC causes transcriptional amplification, which overloads the splicing machinery and makes these cells more sensitive to alterations in splicing fidelity. Genetic knockdown or pharmacological inhibition of spliceosomal components leads to accumulation of retained introns that results in increased apoptosis and reduced tumorigenic and metastatic potential of MYC-driven tumours. C, hnRNP C protein; U1, U1 snRNP; AF2, U2AF2 protein.
About a half of the cryptic exons that are linked to disease are derived from TEs, particularly Alu elements, which have diverged from their original sequence by accumulating mutations that create splice sites37,141. For example, a cryptic Alu-derived exon can disrupt expression of the DMD gene, thereby causing the Duchenne muscular dystrophy (DMD) phenotype130. A particularly rich source of variation within the antisense Alu elements are the U-tracts, which can control the formation of an Alu exon by affecting the competition between hnRNP C and U2AF28,129. This mechanism has been seen in the PTS gene, in which a >50nt deletion containing the U-tract leads to splicing of a cryptic Alu exon (Figure 3C, 6A), which disrupts PTS gene expression and leads to the neurological condition hyperphenylalaninaemia129.
In addition to cis-acting mutations, a changed activity of a trans-acting factor can perturb non-canonical splicing in a manner that leads to disease12,142–146. A link between the reduced expression of SRRM4 mRNA and decreased splicing of microexons was also observed in individuals with ASD12 (Figure 1B). Moreover, TDP-43, a major component of aggregates in ~50% of cases of frontotemporal dementia and ~98% of amyotrophic lateral sclerosis cases, was found to repress splicing of a large number of cryptic exons with potential relevance for disease mechanisms9. The resulting disease-associated inclusion of a cryptic exon into the autophagy-associated gene, ATG4B, might lead to the defects in autophagy that are commonly linked to these diseases147. In addition, mutations in components of the minor spliceosome can lead to specific diseases by causing retention of U12-type introns, without affecting U2-type introns142–146,148.
The disease associations of other recently described non-canonical splicing events remain to be examined. While potential roles of circRNAs in disease have been suggested139,149,150, they might also be candidates for disease biomarkers owing to their high levels of stability151,152. Genes that undergo recursive splicing have been linked to neurodevelopmental disorders14, but it remains to be seen whether variations in RS sites are involved in these diseases.
Non-canonical splicing in cancer
A prevalent feature of most cancer types is widespread intron retention131,153–155. This could relate to competition for the spliceosome due to the high transcriptional activity in tumours, as limiting spliceosomal activity is a known cause of intron retention156 (Figure 6C). Intron retention more often occurs in genes encoding RNA splicing and export factors, and therefore it may perturb the autoregulatory mechanisms of these genes in cancers. Moreover, enrichment in intron retention is associated with the presence of somatic single nucleotide variants in cancer, particularly in tumour-suppressor genes154. Cancer-associated mutations in splicing factors such as U2 Small Nuclear RNA Auxiliary Factor 1 (U2AF1) or Splicing Factor 3b, Subunit 1 (SF3B1) can also promote intron retention or use of alternative 3’ splice sites157,158. It was proposed that mutations in SF3B1 induce selection of cryptic 3’ splice sites through use of a different branch point95,157, which leads to partial inclusion of the 3’ end of the intron. Importantly, half of these aberrantly spliced transcripts are NMD-sensitive and lead to downregulation of corresponding mRNAs and proteins157. Finally, differential splicing of several exitrons has been observed in breast cancer26,159.
Genomic deletion breakpoints and chromosomal rearrangements that generate chimeric transcripts are another common feature of cancer. Notably, the same chimeric transcripts can also be detected at a lower level in non-cancer cells that do not contain the chromosomal rearrangement. In this case, trans-splicing generates the chimeric transcript. For example, JAZF1-SUZ12 and PAX3-FOXO1 chimeric transcripts are normally generated by trans-splicing of independent transcripts in both endometrial and mesenchymal stem cells, whereas chromosomal rearrangements result in fusions of these genes in endometrial stromal tumours and rhabdomyosarcomas, respectively86,132 (Figure 4D). This could reflect that parental genes have properties, such as spatial gene proximity or sequence features, that facilitate the trans-splicing of individual transcripts in wild-type cells and homologous recombination to cause chromosomal rearrangements in cancer. Alternatively, constitutive generation of trans-spliced molecules in wild type cells might in some way facilitate the long-term chromosomal rearrangements observed in cancer by unknown mechanisms. Furthermore, cis-splicing between adjacent genes also commonly produces chimeric transcripts in cancer, such as the SLC45A3–ELK4 transcript in prostate cancer160,161. Taken together, these observations suggest that non-canonical splicing events such as intron retention and chimeric transcripts could have important roles in cancer.
Therapeutic opportunities
Splicing can be exploited for three types of therapeutic strategies: those that modify activity of splicing factors, those that change specific splicing events, and those that exploit non-canonical splicing mechanisms. The first holds particularly great potential in certain types of cancer, where genetic knockdown or pharmacological inhibition of spliceosomal components can prevent the growth and metastasis of MYC-driven tumours156,162,163. In spite of these components being required in all cells, the increased demand for spliceosomal components induces accumulation of retained introns and increases apoptosis specifically in tumours (Figure 6C).
In cases in which specific splicing events need to be corrected, the pioneer studies restored normal splicing of β-globin in β-thalasaemia through the use of chemically modified antisense oligonucleotides (ASOs) that sterically block binding of the splicing machinery while avoiding RNAse H-mediated degradation of the target RNA164. This approach was successful in correcting splicing in SMA and many other diseases165,166. Antisense sequences can also be delivered as modified U-snRNA molecules, using viral vectors that efficiently transfer a modified U-snRNA gene into the affected tissue, which allows continuous expression without the need of repetitive administration167,168. Both ASOs and modified U-snRNAs can be directed either to splice sites, to branch points or to other regulatory elements, such as splicing enhancers or silencers, and therefore they were successful in preventing splicing of cryptic exons in a variety of diseases that are caused by deep-intronic mutations155,166–170. To increase their efficiency, bifunctional ASOs or U-snRNAs can be designed, which contain an RNA binding domain and an effector domain, which recruits splicing factors that either enhance or silence splicing171,172. Finally, therapeutic strategies based on CRISPR-Cas9 genome editing have recently been successful to induce exon-skipping in vivo in adult mice173–175, indicating that this tool is likely to prove valuable as therapy to correct various types of canonical and cryptic splicing events in human diseases.
Several non-canonical splicing mechanisms can also be exploited as therapies. Trans-splicing has been applied to correct genetic mutations in monogenic disorders. In this technique, known as Spliceosome-Mediated RNA Trans-splicing (SMaRT), specific regions within the mutated mRNA are replaced using engineered RNA trans-splicing molecules as templates176,177. These template molecules contain the wild-type mRNA sequence to be replaced, a domain with the essential splicing elements and a domain that binds the target region. This strategy has been applied to many diseases, such as muscular dystrophies, haemophilia and cancer177,178. Other types of non-canonical events might also prove useful for therapies, such as for example the designed artificial circRNAs that could serve as aptamers, trans-cleaving ribozymes, small interfering RNAS (siRNAs), or as sponges to sequester micro RNAS (miRNAs) or RBPs151,179.
Future perspectives
In this Review we have seen how new methods have led to the discovery of various types of splicing events. The next challenge will be to systematically examine non-canonical splicing events that occur as a result of genetic variation, as this would clarify their importance from the perspective of evolution and disease. So far, many mutations affecting splicing have been identified by exome sequencing, which can only identify intronic mutations within a limited region around the annotated exon-intron boundaries. Therefore, genome-wide sequencing will be required to reveal the full range of intronic variation that can activate cryptic splicing, perturb distal branch points or disturb regulatory regions32. It is also important to bear in mind that abnormally processed pre-mRNAs can interfere with transcription or cause co-transcriptional decay36,180. Dedicated genomic and transcriptomic experiments and computational approaches will therefore be needed to detect the full range of mutations that cause disease via non-canonical splicing.
Even though it is clear that many non-canonical splicing events take place in human transcripts, our understanding of their roles in disease and physiology remains limited. We first need to better understand their roles in generating alternative transcripts with modified stability, translation or localisation, production of new protein isoforms, or sequestration of specific RBPs and miRNAs. We also need to uncover their roles in diversifying tissue-specific or cell-specific patterns of gene expression across populations181,182. Many non-canonical splicing events are enriched in the central nervous system, including cryptic exons10, microexons12, RS sites14,15 and circRNAs183. Much remains to be learnt about how these mechanisms contribute to the complexity of gene regulation and the diversity of protein isoforms produced in the brain.
As the next round of ENCODE data on protein-RNA interactions becomes available, understanding of non-canonical splicing events that are hidden deep within introns will be crucial to help explain those interactions for which a function has not yet been identified184. Chromatin structure, DNA methylation, histone marks, nucleosome positioning and the kinetics of transcriptional elongation all contribute to splicing regulation in coordination with RBPs and the spliceosome185. It remains to be seen if these factors cooperate in the control of non-canonical splicing. It is likely that diverse regulatory interactions within intronic regions contribute to the quality control that prevents aberrant cryptic splicing from causing disease27. Nevertheless, it is clear that many non-canonical splicing events escape this quality control, and their role as a source for new molecular functions during evolution will remain a fascinating subject of research for many years.
Acknowledgment
We thank Dr. K. Zarnack for helpful comments on the manuscript. This work was supported by European Research Council (617837-Translate) and Marie Curie Post-doctoral Research Fellowship (627783-NeuroCRYSP) to LB, and an Edmond and Lily Safra fellowship to CRS.
Glossary
- β-thalasaemia
A genetic blood disorder characterized by a defective synthesis of the β-globin chains of hemoglobin, thus causing abnormal erythropoiesis and anemia.
- Alu element
A retrotransposon belonging to the family of short interspersed elements (SINE), consisting of an ~300 nt sequence, which originally derived from the 7SL RNA.
- Aptamers
Oligonucleotide (or peptide) molecules that have secondary and tertiary structures that strongly bind to specific proteins or other cellular targets.
- Ataxia telangiectasia
Autosomal recessive disorder involving cerebellar degeneration, immunodeficiency, chromosomal instability, radiosensitivity and cancer predisposition. It is caused by mutations in ATM gene.
- Autophagy
Intracellular pathway responsible for regulated disassembly of unnecessary or dysfunctional cellular components after their targeting to lysosomes.
- Axonogenesis
Generation and outgrowth of axons during neuronal development.
- CLIP
A method used to identify the RNA targets bound by an RNA-binding protein-of-interest that employs crosslinking, immunoprecipitation and stringent purification of protein-RNA complexes by SDS-PAGE.
- Chimeric transcript
Transcript formed when sections of two or more different genes are joined together in a new transcript either via splicing or as a result of chromosomal fusions.
- CircRNA
RNA that has become circularised owing to intramolecular ligation of its 5' and 3' ends.
- Co-transcriptional decay
RNA surveillance mechanism that acts in the nucleus while transcripts are still associated with the chromatin template.
- Cryptic exon
An exon that is not annotated by the current genomic databases, such as ENSEMBL, and are often only revealed after removing a repressive RBP or after a genomic mutation that increases its splicing efficiency.
- Duchenne muscular dystrophy
A progressive proximal muscular dystrophy caused by mutations in the dystrophin (DMD) gene.
- Exitron
An intron located within an annotated exon.
- Exon definition
The process by which exons are recognised and defined as functional units via interactions between multiple snRNPs and RBPs, especially U1 and U2 snRNPs and SR proteins.
- Hyperphenylalaninaemia
A neurologic disorder caused by autosomal recessive mutations in the genes encoding enzymes involved in the synthesis or regeneration of BH4 cofactor. The most common form is caused by mutations in the PTS gene.
- Intrasplicing
An unconventional splicing mechanism in which splicing to a 3' splice site reconstitutes a new 3' splice to be used in a subsequent splicing step.
- Laron syndrome
Autosomal recessive disorder characterized by short stature that results from mutations in growth hormone (GH) receptor gene.
- Microexon
Exon that is shorter than 30 nts.
- NMD
Nonsense mediated decay, a pathway that initiates decay of certain transcripts, especially those containing a PTC.
- NMD-exon
Exon that contains a PTC, and is therefore targeted for NMD.
- NOVA
A joint name for RBPs encoded by two partially redundant genes that are expressed in the brain; neuro-oncological ventral antigen 1 and 2 (NOVA1 and NOVA2).
- PTC
premature termination codon
- RBP
RNA-binding protein
- RS
recursive splicing, a mechanism that which allows an intron to be spliced in two or more steps.
- RS exon
An exon that follows an RS site and which is required for the exon definition mechanism that initiates splicing at the RS site.
- RS site
The site of recursive splicing, which consists of a 3' splice site that is followed by a sequence that reconstitutes a 5' splice site after the first splicing event.
- Seed sequence
The section of a sequencing read that is used to align the read to the genome or transcriptome.
- snRNPs
Ribonucleoprotein complexes assembled around the small nuclear RNAs (snRNAs) that interact with splice sites or the branch point on pre-mRNA and thereby coordinate and catalyse the splicing reaction.
- Splice sites
Sequences at the boundary of exons and introns, which contain motifs that recruit snRNPs and RBPs to initiate the splicing reaction. 3' and 5' splice sites are located upstream and downstream of exons, respectively.
- Spliceosome
A macromolecular machine consisting of snRNPs and additional RBPs that coordinate and catalyse the splicing reaction.
- SR proteins
A family of RBPs containing a protein domain with long repeats of serine and arginine that generally promote exon definition when binding to exons.
- U2AF complex
Complex of two U2 auxiliary factor RBPs that bind the 3' splice site and facilitate the recruitment of the U2 snRNP to the branch point.
References
- 1.Raj B, Blencowe BJ. Alternative Splicing in the Mammalian Nervous System: Recent Insights into Mechanisms and Functional Roles. Neuron. 2015;87:14–27. doi: 10.1016/j.neuron.2015.05.004. [DOI] [PubMed] [Google Scholar]
- 2.Fu XD, Ares M., Jr Context-dependent control of alternative splicing by RNA-binding proteins. Nat Rev Genet. 2014;15:689–701. doi: 10.1038/nrg3778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Derrien T, et al. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome research. 2012;22:1775–1789. doi: 10.1101/gr.132159.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Matera AG, Wang Z. A day in the life of the spliceosome. Nat Rev Mol Cell Biol. 2014;15:108–121. doi: 10.1038/nrm3742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Scotti MM, Swanson MS. RNA mis-splicing in disease. Nat Rev Genet. 2015 doi: 10.1038/nrg.2015.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jangi M, Boutz PL, Paul P, Sharp PA. Rbfox2 controls autoregulation in RNA-binding protein networks. Genes & development. 2014;28:637–651. doi: 10.1101/gad.235770.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eom T, et al. NOVA-dependent regulation of cryptic NMD exons controls synaptic protein levels after seizure. eLife. 2013;2:e00178. doi: 10.7554/eLife.00178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zarnack K, et al. Direct competition between hnRNP C and U2AF65 protects the transcriptome from the exonization of Alu elements. Cell. 2013;152:453–466. doi: 10.1016/j.cell.2012.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ling JP, Pletnikova O, Troncoso JC, Wong PC. TDP-43 repression of nonconserved cryptic exons is compromised in ALS-FTD. Science. 2015;349:650–655. doi: 10.1126/science.aab0983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yan Q, et al. Systematic discovery of regulated and conserved alternative exons in the mammalian brain reveals NMD modulating chromatin regulators. Proc Natl Acad Sci U S A. 2015;112:3445–3450. doi: 10.1073/pnas.1502849112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wu J, Anczukow O, Krainer AR, Zhang MQ, Zhang C. OLego: fast and sensitive mapping of spliced mRNA-Seq reads using small seeds. Nucleic acids research. 2013;41:5149–5163. doi: 10.1093/nar/gkt216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Irimia M, et al. A highly conserved program of neuronal microexons is misregulated in autistic brains. Cell. 2014;159:1511–1523. doi: 10.1016/j.cell.2014.11.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li YI, Sanchez-Pulido L, Haerty W, Ponting CP. RBFOX and PTBP1 proteins regulate the alternative splicing of micro-exons in human brain transcripts. Genome research. 2015;25:1–13. doi: 10.1101/gr.181990.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sibley CR, et al. Recursive splicing in long vertebrate genes. Nature. 2015;521:371–375. doi: 10.1038/nature14466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Duff MO, et al. Genome-wide identification of zero nucleotide recursive splicing in Drosophila. Nature. 2015;521:376–379. doi: 10.1038/nature14475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hansen TB, et al. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384–388. doi: 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- 17.Memczak S, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333–338. doi: 10.1038/nature11928. [DOI] [PubMed] [Google Scholar]
- 18.Salzman J, Gawad C, Wang PL, Lacayo N, Brown PO. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS One. 2012;7:e30733. doi: 10.1371/journal.pone.0030733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Danan M, Schwartz S, Edelheit S, Sorek R. Transcriptome-wide discovery of circular RNAs in Archaea. Nucleic acids research. 2012;40:3131–3142. doi: 10.1093/nar/gkr1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen LL. The biogenesis and emerging roles of circular RNAs. Nat Rev Mol Cell Biol. 2016 doi: 10.1038/nrm.2015.32. [DOI] [PubMed] [Google Scholar]
- 21.Braunschweig U, et al. Widespread intron retention in mammals functionally tunes transcriptomes. Genome research. 2014;24:1774–1786. doi: 10.1101/gr.177790.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yap K, Lim ZQ, Khandelia P, Friedman B, Makeyev EV. Coordinated regulation of neuronal mRNA steady-state levels through developmentally controlled intron retention. Genes & development. 2012;26:1209–1223. doi: 10.1101/gad.188037.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Boutz PL, Bhutkar A, Sharp PA. Detained introns are a novel, widespread class of post-transcriptionally spliced introns. Genes & development. 2015;29:63–80. doi: 10.1101/gad.247361.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wong JJ, et al. Orchestrated intron retention regulates normal granulocyte differentiation. Cell. 2013;154:583–595. doi: 10.1016/j.cell.2013.06.052. [DOI] [PubMed] [Google Scholar]
- 25.Marquez Y, Brown JW, Simpson C, Barta A, Kalyna M. Transcriptome survey reveals increased complexity of the alternative splicing landscape in Arabidopsis. Genome research. 2012;22:1184–1195. doi: 10.1101/gr.134106.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Marquez Y, Hopfler M, Ayatollahi Z, Barta A, Kalyna M. Unmasking alternative splicing inside protein-coding exons defines exitrons and their role in proteome plasticity. Genome research. 2015;25:995–1007. doi: 10.1101/gr.186585.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.De Conti L, Baralle M, Buratti E. Exon and intron definition in pre-mRNA splicing. Wiley Interdiscip Rev RNA. 2013;4:49–60. doi: 10.1002/wrna.1140. [DOI] [PubMed] [Google Scholar]
- 28.Robberson BL, Cote GJ, Berget SM. Exon definition may facilitate splice site selection in RNAs with multiple exons. Molecular and cellular biology. 1990;10:84–94. doi: 10.1128/mcb.10.1.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dobin A, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kelly S, et al. Splicing of many human genes involves sites embedded within introns. Nucleic acids research. 2015;43:4721–4732. doi: 10.1093/nar/gkv386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kapustin Y, et al. Cryptic splice sites and split genes. Nucleic acids research. 2011;39:5837–5844. doi: 10.1093/nar/gkr203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ni JZ, et al. Ultraconserved elements are associated with homeostatic control of splicing regulators by alternative splicing and nonsense-mediated decay. Genes & development. 2007;21:708–718. doi: 10.1101/gad.1525507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lareau LF, Inada M, Green RE, Wengrod JC, Brenner SE. Unproductive splicing of SR genes associated with highly conserved and ultraconserved DNA elements. Nature. 2007;446:926–929. doi: 10.1038/nature05676. [DOI] [PubMed] [Google Scholar]
- 35.Jangi M, Sharp PA. Building robust transcriptomes with master splicing factors. Cell. 2014;159:487–498. doi: 10.1016/j.cell.2014.09.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vaz-Drago R, et al. Transcription-coupled RNA surveillance in human genetic diseases caused by splice site mutations. Hum Mol Genet. 2015;24:2784–2795. doi: 10.1093/hmg/ddv039. [DOI] [PubMed] [Google Scholar]
- 37.Keren H, Lev-Maor G, Ast G. Alternative splicing and evolution: diversification, exon definition and function. Nat Rev Genet. 2010;11:345–355. doi: 10.1038/nrg2776. [DOI] [PubMed] [Google Scholar]
- 38.Quentin Y. Origin of the Alu family: a family of Alu-like monomers gave birth to the left and the right arms of the Alu elements. Nucleic acids research. 1992;20:3397–3401. doi: 10.1093/nar/20.13.3397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gal-Mark N, Schwartz S, Ram O, Eyras E, Ast G. The pivotal roles of TIA proteins in 5' splice-site selection of alu exons and across evolution. PLoS genetics. 2009;5:e1000717. doi: 10.1371/journal.pgen.1000717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Konig J, Zarnack K, Luscombe NM, Ule J. Protein-RNA interactions: new genomic technologies and perspectives. Nat Rev Genet. 2011;13:77–83. doi: 10.1038/nrg3141. [DOI] [PubMed] [Google Scholar]
- 41.Corvelo A, Eyras E. Exon creation and establishment in human genes. Genome biology. 2008;9:R141. doi: 10.1186/gb-2008-9-9-r141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dominski Z, Kole R. Selection of splice sites in pre-mRNAs with short internal exons. Molecular and cellular biology. 1991;11:6075–6083. doi: 10.1128/mcb.11.12.6075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Black DL. Does steric interference between splice sites block the splicing of a short c-src neuron-specific exon in non-neuronal cells? Genes & development. 1991;5:389–402. doi: 10.1101/gad.5.3.389. [DOI] [PubMed] [Google Scholar]
- 44.Volfovsky N, Haas BJ, Salzberg SL. Computational discovery of internal micro-exons. Genome research. 2003;13:1216–1221. doi: 10.1101/gr.677503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Burnette JM, Miyamoto-Sato E, Schaub MA, Conklin J, Lopez AJ. Subdivision of large introns in Drosophila by recursive splicing at nonexonic elements. Genetics. 2005;170:661–674. doi: 10.1534/genetics.104.039701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hatton AR, Subramaniam V, Lopez AJ. Generation of alternative Ultrabithorax isoforms and stepwise removal of a large intron by resplicing at exon-exon junctions. Molecular cell. 1998;2:787–796. doi: 10.1016/s1097-2765(00)80293-2. [DOI] [PubMed] [Google Scholar]
- 47.Herzel L, Neugebauer KM. Quantification of co-transcriptional splicing from RNA-Seq data. Methods. 2015 doi: 10.1016/j.ymeth.2015.04.024. [DOI] [PubMed] [Google Scholar]
- 48.Ameur A, et al. Total RNA sequencing reveals nascent transcription and widespread co-transcriptional splicing in the human brain. Nature structural & molecular biology. 2011;18:1435–1440. doi: 10.1038/nsmb.2143. [DOI] [PubMed] [Google Scholar]
- 49.Parra MK, Tan JS, Mohandas N, Conboy JG. Intrasplicing coordinates alternative first exons with alternative splicing in the protein 4.1R gene. The EMBO journal. 2008;27:122–131. doi: 10.1038/sj.emboj.7601957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ner-Gaon H, et al. Intron retention is a major phenomenon in alternative splicing in Arabidopsis. Plant J. 2004;39:877–885. doi: 10.1111/j.1365-313X.2004.02172.x. [DOI] [PubMed] [Google Scholar]
- 51.Galante PA, Sakabe NJ, Kirschbaum-Slager N, de Souza SJ. Detection and evaluation of intron retention events in the human transcriptome. Rna. 2004;10:757–765. doi: 10.1261/rna.5123504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kan Z, States D, Gish W. Selecting for functional alternative splices in ESTs. Genome research. 2002;12:1837–1845. doi: 10.1101/gr.764102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sakabe NJ, de Souza SJ. Sequence features responsible for intron retention in human. BMC genomics. 2007;8:59. doi: 10.1186/1471-2164-8-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Martinez-Contreras R, et al. Intronic binding sites for hnRNP A/B and hnRNP F/H proteins stimulate pre-mRNA splicing. PLoS Biol. 2006;4:e21. doi: 10.1371/journal.pbio.0040021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wickramasinghe VO, et al. Regulation of constitutive and alternative mRNA splicing across the human transcriptome by PRPF8 is determined by 5' splice site strength. Genome biology. 2015;16:201. doi: 10.1186/s13059-015-0749-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Marinescu V, Loomis PA, Ehmann S, Beales M, Potashkin JA. Regulation of retention of FosB intron 4 by PTB. PLoS One. 2007;2:e828. doi: 10.1371/journal.pone.0000828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bergeron D, Pal G, Beaulieu YB, Chabot B, Bachand F. Regulated Intron Retention and Nuclear Pre-mRNA Decay Contribute to PABPN1 Autoregulation. Molecular and cellular biology. 2015;35:2503–2517. doi: 10.1128/MCB.00070-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Malone CD, et al. The exon junction complex controls transposable element activity by ensuring faithful splicing of the piwi transcript. Genes & development. 2014;28:1786–1799. doi: 10.1101/gad.245829.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hayashi R, Handler D, Ish-Horowicz D, Brennecke J. The exon junction complex is required for definition and excision of neighboring introns in Drosophila. Genes & development. 2014;28:1772–1785. doi: 10.1101/gad.245738.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang Z, Murigneux V, Le Hir H. Transcriptome-wide modulation of splicing by the exon junction complex. Genome biology. 2014;15:551. doi: 10.1186/s13059-014-0551-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nigro JM, et al. Scrambled exons. Cell. 1991;64:607–613. doi: 10.1016/0092-8674(91)90244-s. [DOI] [PubMed] [Google Scholar]
- 62.Schindewolf C, Braun S, Domdey H. In vitro generation of a circular exon from a linear pre-mRNA transcript. Nucleic acids research. 1996;24:1260–1266. doi: 10.1093/nar/24.7.1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pasman Z, Been MD, Garcia-Blanco MA. Exon circularization in mammalian nuclear extracts. Rna. 1996;2:603–610. [PMC free article] [PubMed] [Google Scholar]
- 64.Braun S, Domdey H, Wiebauer K. Inverse splicing of a discontinuous pre-mRNA intron generates a circular exon in a HeLa cell nuclear extract. Nucleic acids research. 1996;24:4152–4157. doi: 10.1093/nar/24.21.4152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Suzuki H, et al. Characterization of RNase R-digested cellular RNA source that consists of lariat and circular RNAs from pre-mRNA splicing. Nucleic acids research. 2006;34:e63. doi: 10.1093/nar/gkl151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Guo JU, Agarwal V, Guo H, Bartel DP. Expanded identification and characterization of mammalian circular RNAs. Genome biology. 2014;15:409. doi: 10.1186/s13059-014-0409-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Jeck WR, Sharpless NE. Detecting and characterizing circular RNAs. Nature biotechnology. 2014;32:453–461. doi: 10.1038/nbt.2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.You X, et al. Neural circular RNAs are derived from synaptic genes and regulated by development and plasticity. Nat Neurosci. 2015;18:603–610. doi: 10.1038/nn.3975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Liang D, Wilusz JE. Short intronic repeat sequences facilitate circular RNA production. Genes & development. 2014;28:2233–2247. doi: 10.1101/gad.251926.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Jeck WR, et al. Circular RNAs are abundant, conserved, and associated with ALU repeats. Rna. 2013;19:141–157. doi: 10.1261/rna.035667.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kramer MC, et al. Combinatorial control of Drosophila circular RNA expression by intronic repeats, hnRNPs, and SR proteins. Genes & development. 2015;29:2168–2182. doi: 10.1101/gad.270421.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhang XO, et al. Complementary sequence-mediated exon circularization. Cell. 2014;159:134–147. doi: 10.1016/j.cell.2014.09.001. [DOI] [PubMed] [Google Scholar]
- 73.Ivanov A, et al. Analysis of intron sequences reveals hallmarks of circular RNA biogenesis in animals. Cell reports. 2015;10:170–177. doi: 10.1016/j.celrep.2014.12.019. [DOI] [PubMed] [Google Scholar]
- 74.Ashwal-Fluss R, et al. circRNA biogenesis competes with pre-mRNA splicing. Molecular cell. 2014;56:55–66. doi: 10.1016/j.molcel.2014.08.019. [DOI] [PubMed] [Google Scholar]
- 75.Conn SJ, et al. The RNA binding protein quaking regulates formation of circRNAs. Cell. 2015;160:1125–1134. doi: 10.1016/j.cell.2015.02.014. [DOI] [PubMed] [Google Scholar]
- 76.Grosso AR, et al. Pervasive transcription read-through promotes aberrant expression of oncogenes and RNA chimeras in renal carcinoma. eLife. 2015;4 doi: 10.7554/eLife.09214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Akiva P, et al. Transcription-mediated gene fusion in the human genome. Genome research. 2006;16:30–36. doi: 10.1101/gr.4137606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Qin F, et al. Discovery of CTCF-sensitive Cis-spliced fusion RNAs between adjacent genes in human prostate cells. PLoS genetics. 2015;11:e1005001. doi: 10.1371/journal.pgen.1005001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Jividen K, Li H. Chimeric RNAs generated by intergenic splicing in normal and cancer cells. Genes Chromosomes Cancer. 2014;53:963–971. doi: 10.1002/gcc.22207. [DOI] [PubMed] [Google Scholar]
- 80.Sutton RE, Boothroyd JC. Evidence for trans splicing in trypanosomes. Cell. 1986;47:527–535. doi: 10.1016/0092-8674(86)90617-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Allen MA, Hillier LW, Waterston RH, Blumenthal T. A global analysis of C. elegans trans-splicing. Genome research. 2011;21:255–264. doi: 10.1101/gr.113811.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.McManus CJ, Duff MO, Eipper-Mains J, Graveley BR. Global analysis of trans-splicing in Drosophila. Proc Natl Acad Sci U S A. 2010;107:12975–12979. doi: 10.1073/pnas.1007586107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Dorn R, Reuter G, Loewendorf A. Transgene analysis proves mRNA trans-splicing at the complex mod(mdg4) locus in Drosophila. Proc Natl Acad Sci U S A. 2001;98:9724–9729. doi: 10.1073/pnas.151268698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Gabler M, et al. Trans-splicing of the mod(mdg4) complex locus is conserved between the distantly related species Drosophila melanogaster and D. virilis. Genetics. 2005;169:723–736. doi: 10.1534/genetics.103.020842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kong Y, et al. The evolutionary landscape of intergenic trans-splicing events in insects. Nat Commun. 2015;6:8734. doi: 10.1038/ncomms9734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Li H, Wang J, Mor G, Sklar J. A neoplastic gene fusion mimics trans-splicing of RNAs in normal human cells. Science. 2008;321:1357–1361. doi: 10.1126/science.1156725. [DOI] [PubMed] [Google Scholar]
- 87.Wu CS, et al. Integrative transcriptome sequencing identifies trans-splicing events with important roles in human embryonic stem cell pluripotency. Genome research. 2014;24:25–36. doi: 10.1101/gr.159483.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Dietrich RC, Incorvaia R, Padgett RA. Terminal intron dinucleotide sequences do not distinguish between U2- and U12-dependent introns. Molecular cell. 1997;1:151–160. doi: 10.1016/s1097-2765(00)80016-7. [DOI] [PubMed] [Google Scholar]
- 89.Wu Q, Krainer AR. Splicing of a divergent subclass of AT-AC introns requires the major spliceosomal snRNAs. Rna. 1997;3:586–601. [PMC free article] [PubMed] [Google Scholar]
- 90.Sheth N, et al. Comprehensive splice-site analysis using comparative genomics. Nucleic acids research. 2006;34:3955–3967. doi: 10.1093/nar/gkl556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Parada GE, Munita R, Cerda CA, Gysling K. A comprehensive survey of non-canonical splice sites in the human transcriptome. Nucleic acids research. 2014;42:10564–10578. doi: 10.1093/nar/gku744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Mercer TR, et al. Genome-wide discovery of human splicing branchpoints. Genome research. 2015;25:290–303. doi: 10.1101/gr.182899.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.DeBoever C, et al. Transcriptome sequencing reveals potential mechanism of cryptic 3' splice site selection in SF3B1-mutated cancers. PLoS Comput Biol. 2015;11:e1004105. doi: 10.1371/journal.pcbi.1004105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Darman RB, et al. Cancer-Associated SF3B1 Hotspot Mutations Induce Cryptic 3' Splice Site Selection through Use of a Different Branch Point. Cell reports. 2015;13:1033–1045. doi: 10.1016/j.celrep.2015.09.053. [DOI] [PubMed] [Google Scholar]
- 95.Alsafadi S, et al. Cancer-associated SF3B1 mutations affect alternative splicing by promoting alternative branchpoint usage. Nat Commun. 2016;7:10615. doi: 10.1038/ncomms10615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Roca X, Krainer AR. Recognition of atypical 5' splice sites by shifted base-pairing to U1 snRNA. Nature structural & molecular biology. 2009;16:176–182. doi: 10.1038/nsmb.1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Roca X, et al. Widespread recognition of 5' splice sites by noncanonical base-pairing to U1 snRNA involving bulged nucleotides. Genes & development. 2012;26:1098–1109. doi: 10.1101/gad.190173.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Rueter SM, Dawson TR, Emeson RB. Regulation of alternative splicing by RNA editing. Nature. 1999;399:75–80. doi: 10.1038/19992. [DOI] [PubMed] [Google Scholar]
- 99.Shen X, et al. Complementary signaling pathways regulate the unfolded protein response and are required for C. elegans development. Cell. 2001;107:893–903. doi: 10.1016/s0092-8674(01)00612-2. [DOI] [PubMed] [Google Scholar]
- 100.Yoshida H, Matsui T, Yamamoto A, Okada T, Mori K. XBP1 mRNA is induced by ATF6 and spliced by IRE1 in response to ER stress to produce a highly active transcription factor. Cell. 2001;107:881–891. doi: 10.1016/s0092-8674(01)00611-0. [DOI] [PubMed] [Google Scholar]
- 101.Filipowicz W. Making ends meet: a role of RNA ligase RTCB in unfolded protein response. The EMBO journal. 2014;33:2887–2889. doi: 10.15252/embj.201490425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Dergai M, et al. Microexon-based regulation of ITSN1 and Src SH3 domains specificity relies on introduction of charged amino acids into the interaction interface. Biochem Biophys Res Commun. 2010;399:307–312. doi: 10.1016/j.bbrc.2010.07.080. [DOI] [PubMed] [Google Scholar]
- 103.Quesnel-Vallieres M, Irimia M, Cordes SP, Blencowe BJ. Essential roles for the splicing regulator nSR100/SRRM4 during nervous system development. Genes & development. 2015;29:746–759. doi: 10.1101/gad.256115.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Wright PE, Dyson HJ. Intrinsically disordered proteins in cellular signalling and regulation. Nat Rev Mol Cell Biol. 2015;16:18–29. doi: 10.1038/nrm3920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Rossbach O, et al. Auto- and cross-regulation of the hnRNP L proteins by alternative splicing. Molecular and cellular biology. 2009;29:1442–1451. doi: 10.1128/MCB.01689-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Buckley PT, Khaladkar M, Kim J, Eberwine J. Cytoplasmic intron retention, function, splicing, and the sentinel RNA hypothesis. Wiley Interdiscip Rev RNA. 2014;5:223–230. doi: 10.1002/wrna.1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Sibley CR. Regulation of gene expression through production of unstable mRNA isoforms. Biochemical Society transactions. 2014;42:1196–1205. doi: 10.1042/BST20140102. [DOI] [PubMed] [Google Scholar]
- 108.Jens M, Rajewsky N. Competition between target sites of regulators shapes post-transcriptional gene regulation. Nat Rev Genet. 2015;16:113–126. doi: 10.1038/nrg3853. [DOI] [PubMed] [Google Scholar]
- 109.Dhir A, Buratti E, van Santen MA, Luhrmann R, Baralle FE. The intronic splicing code: multiple factors involved in ATM pseudoexon definition. The EMBO journal. 2010;29:749–760. doi: 10.1038/emboj.2009.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Pagani F, et al. A new type of mutation causes a splicing defect in ATM. Nature genetics. 2002;30:426–429. doi: 10.1038/ng858. [DOI] [PubMed] [Google Scholar]
- 111.Liu N, et al. N(6)-methyladenosine-dependent RNA structural switches regulate RNA-protein interactions. Nature. 2015;518:560–564. doi: 10.1038/nature14234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Solomon O, et al. Global regulation of alternative splicing by adenosine deaminase acting on RNA (ADAR) Rna. 2013;19:591–604. doi: 10.1261/rna.038042.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Lovci MT, et al. Rbfox proteins regulate alternative mRNA splicing through evolutionarily conserved RNA bridges. Nature structural & molecular biology. 2013;20:1434–1442. doi: 10.1038/nsmb.2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Bitton DA, et al. Widespread exon skipping triggers degradation by nuclear RNA surveillance in fission yeast. Genome research. 2015;25:884–896. doi: 10.1101/gr.185371.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.de Koning AP, Gu W, Castoe TA, Batzer MA, Pollock DD. Repetitive elements may comprise over two-thirds of the human genome. PLoS genetics. 2011;7:e1002384. doi: 10.1371/journal.pgen.1002384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Brouha B, et al. Hot L1s account for the bulk of retrotransposition in the human population. Proc Natl Acad Sci U S A. 2003;100:5280–5285. doi: 10.1073/pnas.0831042100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Jacob F. Evolution and tinkering. Science. 1977;196:1161–1166. doi: 10.1126/science.860134. [DOI] [PubMed] [Google Scholar]
- 118.Cowley M, Oakey RJ. Transposable elements re-wire and fine-tune the transcriptome. PLoS genetics. 2013;9:e1003234. doi: 10.1371/journal.pgen.1003234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Ule J. Alu elements: at the crossroads between disease and evolution. Biochemical Society transactions. 2013;41:1532–1535. doi: 10.1042/BST20130157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Brunet TD, Doolittle WF. Multilevel Selection Theory and the Evolutionary Functions of Transposable Elements. Genome Biol Evol. 2015;7:2445–2457. doi: 10.1093/gbe/evv152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Feschotte C. Transposable elements and the evolution of regulatory networks. Nat Rev Genet. 2008;9:397–405. doi: 10.1038/nrg2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Roy M, Kim N, Xing Y, Lee C. The effect of intron length on exon creation ratios during the evolution of mammalian genomes. Rna. 2008;14:2261–2273. doi: 10.1261/rna.1024908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Pickrell JK, Pai AA, Gilad Y, Pritchard JK. Noisy splicing drives mRNA isoform diversity in human cells. PLoS genetics. 2010;6:e1001236. doi: 10.1371/journal.pgen.1001236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Lopez-Bigas N, Audit B, Ouzounis C, Parra G, Guigo R. Are splicing mutations the most frequent cause of hereditary disease? FEBS Lett. 2005;579:1900–1903. doi: 10.1016/j.febslet.2005.02.047. [DOI] [PubMed] [Google Scholar]
- 125.Daguenet E, Dujardin G, Valcarcel J. The pathogenicity of splicing defects: mechanistic insights into pre-mRNA processing inform novel therapeutic approaches. EMBO Rep. 2015 doi: 10.15252/embr.201541116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Singh RK, Cooper TA. Pre-mRNA splicing in disease and therapeutics. Trends Mol Med. 2012;18:472–482. doi: 10.1016/j.molmed.2012.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Supek F, Minana B, Valcarcel J, Gabaldon T, Lehner B. Synonymous mutations frequently act as driver mutations in human cancers. Cell. 2014;156:1324–1335. doi: 10.1016/j.cell.2014.01.051. [DOI] [PubMed] [Google Scholar]
- 128.Xiong HY, et al. RNA splicing. The human splicing code reveals new insights into the genetic determinants of disease. Science. 2015;347:1254806. doi: 10.1126/science.1254806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Meili D, et al. Disease-causing mutations improving the branch site and polypyrimidine tract: pseudoexon activation of LINE-2 and antisense Alu lacking the poly(T)-tail. Human mutation. 2009;30:823–831. doi: 10.1002/humu.20969. [DOI] [PubMed] [Google Scholar]
- 130.Ferlini A, et al. A novel Alu-like element rearranged in the dystrophin gene causes a splicing mutation in a family with X-linked dilated cardiomyopathy. Am J Hum Genet. 1998;63:436–446. doi: 10.1086/301952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Sowalsky AG, et al. Whole transcriptome sequencing reveals extensive unspliced mRNA in metastatic castration-resistant prostate cancer. Mol Cancer Res. 2015;13:98–106. doi: 10.1158/1541-7786.MCR-14-0273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Yuan H, et al. A chimeric RNA characteristic of rhabdomyosarcoma in normal myogenesis process. Cancer Discov. 2013;3:1394–1403. doi: 10.1158/2159-8290.CD-13-0186. [DOI] [PubMed] [Google Scholar]
- 133.Greer K, et al. Pseudoexon activation increases phenotype severity in a Becker muscular dystrophy patient. Molecular genetics & genomic medicine. 2015;3:320–326. doi: 10.1002/mgg3.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Buratti E, Dhir A, Lewandowska MA, Baralle FE. RNA structure is a key regulatory element in pathological ATM and CFTR pseudoexon inclusion events. Nucleic acids research. 2007;35:4369–4383. doi: 10.1093/nar/gkm447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Highsmith WE, et al. A novel mutation in the cystic fibrosis gene in patients with pulmonary disease but normal sweat chloride concentrations. N Engl J Med. 1994;331:974–980. doi: 10.1056/NEJM199410133311503. [DOI] [PubMed] [Google Scholar]
- 136.Chen X, et al. Intronic alterations in BRCA1 and BRCA2: effect on mRNA splicing fidelity and expression. Human mutation. 2006;27:427–435. doi: 10.1002/humu.20319. [DOI] [PubMed] [Google Scholar]
- 137.Lualdi S, et al. Multiple cryptic splice sites can be activated by IDS point mutations generating misspliced transcripts. J Mol Med (Berl) 2006;84:692–700. doi: 10.1007/s00109-006-0057-1. [DOI] [PubMed] [Google Scholar]
- 138.Sathasivam K, et al. Aberrant splicing of HTT generates the pathogenic exon 1 protein in Huntington disease. Proc Natl Acad Sci U S A. 2013;110:2366–2370. doi: 10.1073/pnas.1221891110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Ghosal S, Das S, Sen R, Basak P, Chakrabarti J. Circ2Traits: a comprehensive database for circular RNA potentially associated with disease and traits. Frontiers in genetics. 2013;4:283. doi: 10.3389/fgene.2013.00283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Akker SA, et al. Pre-spliceosomal binding of U1 small nuclear ribonucleoprotein (RNP) and heterogenous nuclear RNP E1 is associated with suppression of a growth hormone receptor pseudoexon. Mol Endocrinol. 2007;21:2529–2540. doi: 10.1210/me.2007-0038. [DOI] [PubMed] [Google Scholar]
- 141.Vorechovsky I. Transposable elements in disease-associated cryptic exons. Human genetics. 2010;127:135–154. doi: 10.1007/s00439-009-0752-4. [DOI] [PubMed] [Google Scholar]
- 142.Madan V, et al. Aberrant splicing of U12-type introns is the hallmark of ZRSR2 mutant myelodysplastic syndrome. Nat Commun. 2015;6:6042. doi: 10.1038/ncomms7042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Edery P, et al. Association of TALS developmental disorder with defect in minor splicing component U4atac snRNA. Science. 2011;332:240–243. doi: 10.1126/science.1202205. [DOI] [PubMed] [Google Scholar]
- 144.He H, et al. Mutations in U4atac snRNA, a component of the minor spliceosome, in the developmental disorder MOPD I. Science. 2011;332:238–240. doi: 10.1126/science.1200587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Merico D, et al. Compound heterozygous mutations in the noncoding RNU4ATAC cause Roifman Syndrome by disrupting minor intron splicing. Nat Commun. 2015;6:8718. doi: 10.1038/ncomms9718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Yoshida K, et al. Frequent pathway mutations of splicing machinery in myelodysplasia. Nature. 2011;478:64–69. doi: 10.1038/nature10496. [DOI] [PubMed] [Google Scholar]
- 147.Menzies FM, Fleming A, Rubinsztein DC. Compromised autophagy and neurodegenerative diseases. Nat Rev Neurosci. 2015;16:345–357. doi: 10.1038/nrn3961. [DOI] [PubMed] [Google Scholar]
- 148.Argente J, et al. Defective minor spliceosome mRNA processing results in isolated familial growth hormone deficiency. EMBO Mol Med. 2014;6:299–306. doi: 10.1002/emmm.201303573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Bachmayr-Heyda A, et al. Correlation of circular RNA abundance with proliferation--exemplified with colorectal and ovarian cancer, idiopathic lung fibrosis, and normal human tissues. Scientific reports. 2015;5:8057. doi: 10.1038/srep08057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Wang YH, Yu XH, Luo SS, Han H. Comprehensive circular RNA profiling reveals that circular RNA100783 is involved in chronic CD28-associated CD8(+)T cell ageing. Immun Ageing. 2015;12:17. doi: 10.1186/s12979-015-0042-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Li J, et al. Circular RNAs in cancer: novel insights into origins, properties, functions and implications. Am J Cancer Res. 2015;5:472–480. [PMC free article] [PubMed] [Google Scholar]
- 152.Memczak S, Papavasileiou P, Peters O, Rajewsky N. Identification and Characterization of Circular RNAs As a New Class of Putative Biomarkers in Human Blood. PLoS One. 2015;10:e0141214. doi: 10.1371/journal.pone.0141214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Dvinge H, Bradley RK. Widespread intron retention diversifies most cancer transcriptomes. Genome Med. 2015;7:45. doi: 10.1186/s13073-015-0168-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Jung H, et al. Intron retention is a widespread mechanism of tumor-suppressor inactivation. Nature genetics. 2015;47:1242–1248. doi: 10.1038/ng.3414. [DOI] [PubMed] [Google Scholar]
- 155.Romano M, Buratti E, Baralle D. Role of pseudoexons and pseudointrons in human cancer. Int J Cell Biol. 2013;2013:810572. doi: 10.1155/2013/810572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Hsu TY, et al. The spliceosome is a therapeutic vulnerability in MYC-driven cancer. Nature. 2015;525:384–388. doi: 10.1038/nature14985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Darman RB, et al. Cancer-Associated SF3B1 Hotspot Mutations Induce Cryptic 3? Splice Site Selection through Use of a Different Branch Point. Cell reports. 2015;13:1033–1045. doi: 10.1016/j.celrep.2015.09.053. [DOI] [PubMed] [Google Scholar]
- 158.Ilagan JO, et al. U2AF1 mutations alter splice site recognition in hematological malignancies. Genome research. 2015;25:14–26. doi: 10.1101/gr.181016.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Milde-Langosch K, Kappes H, Riethdorf S, Loning T, Bamberger AM. FosB is highly expressed in normal mammary epithelia, but down-regulated in poorly differentiated breast carcinomas. Breast Cancer Res Treat. 2003;77:265–275. doi: 10.1023/a:1021887100216. [DOI] [PubMed] [Google Scholar]
- 160.Rickman DS, et al. SLC45A3-ELK4 is a novel and frequent erythroblast transformation-specific fusion transcript in prostate cancer. Cancer Res. 2009;69:2734–2738. doi: 10.1158/0008-5472.CAN-08-4926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Zhang Y, et al. Chimeric transcript generated by cis-splicing of adjacent genes regulates prostate cancer cell proliferation. Cancer Discov. 2012;2:598–607. doi: 10.1158/2159-8290.CD-12-0042. [DOI] [PubMed] [Google Scholar]
- 162.Bonnal S, Vigevani L, Valcarcel J. The spliceosome as a target of novel antitumour drugs. Nat Rev Drug Discov. 2012;11:847–859. doi: 10.1038/nrd3823. [DOI] [PubMed] [Google Scholar]
- 163.Koh CM, et al. MYC regulates the core pre-mRNA splicing machinery as an essential step in lymphomagenesis. Nature. 2015;523:96–100. doi: 10.1038/nature14351. [DOI] [PubMed] [Google Scholar]
- 164.Dominski Z, Kole R. Restoration of correct splicing in thalassemic pre-mRNA by antisense oligonucleotides. Proc Natl Acad Sci U S A. 1993;90:8673–8677. doi: 10.1073/pnas.90.18.8673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Hua Y, et al. Peripheral SMN restoration is essential for long-term rescue of a severe spinal muscular atrophy mouse model. Nature. 2011;478:123–126. doi: 10.1038/nature10485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.McClorey G, Wood MJ. An overview of the clinical application of antisense oligonucleotides for RNA-targeting therapies. Curr Opin Pharmacol. 2015;24:52–58. doi: 10.1016/j.coph.2015.07.005. [DOI] [PubMed] [Google Scholar]
- 167.Goyenvalle A, et al. Rescue of dystrophic muscle through U7 snRNA-mediated exon skipping. Science. 2004;306:1796–1799. doi: 10.1126/science.1104297. [DOI] [PubMed] [Google Scholar]
- 168.Gorman L, Suter D, Emerick V, Schumperli D, Kole R. Stable alteration of pre-mRNA splicing patterns by modified U7 small nuclear RNAs. Proc Natl Acad Sci U S A. 1998;95:4929–4934. doi: 10.1073/pnas.95.9.4929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Uchikawa H, et al. U7 snRNA-mediated correction of aberrant splicing caused by activation of cryptic splice sites. J Hum Genet. 2007;52:891–897. doi: 10.1007/s10038-007-0192-8. [DOI] [PubMed] [Google Scholar]
- 170.Blazquez L, et al. In vitro correction of a pseudoexon-generating deep intronic mutation in LGMD2A by antisense oligonucleotides and modified small nuclear RNAs. Human mutation. 2013;34:1387–1395. doi: 10.1002/humu.22379. [DOI] [PubMed] [Google Scholar]
- 171.Goyenvalle A, Babbs A, van Ommen GJ, Garcia L, Davies KE. Enhanced exon-skipping induced by U7 snRNA carrying a splicing silencer sequence: Promising tool for DMD therapy. Molecular therapy : the journal of the American Society of Gene Therapy. 2009;17:1234–1240. doi: 10.1038/mt.2009.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Garcia-Blanco MA, Baraniak AP, Lasda EL. Alternative splicing in disease and therapy. Nature biotechnology. 2004;22:535–546. doi: 10.1038/nbt964. [DOI] [PubMed] [Google Scholar]
- 173.Xu L, et al. CRISPR-mediated Genome Editing Restores Dystrophin Expression and Function in mdx Mice. Molecular therapy : the journal of the American Society of Gene Therapy. 2015 doi: 10.1038/mt.2015.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Nelson CE, et al. In vivo genome editing improves muscle function in a mouse model of Duchenne muscular dystrophy. Science. 2015 doi: 10.1126/science.aad5143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Tabebordbar M, et al. In vivo gene editing in dystrophic mouse muscle and muscle stem cells. Science. 2015 doi: 10.1126/science.aad5177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Puttaraju M, DiPasquale J, Baker CC, Mitchell LG, Garcia-Blanco M. Messenger RNA repair and restoration of protein function by spliceosome-mediated RNA trans-splicing. Molecular therapy : the journal of the American Society of Gene Therapy. 2001;4:105–114. doi: 10.1006/mthe.2001.0426. [DOI] [PubMed] [Google Scholar]
- 177.Koller U, Wally V, Bauer JW, Murauer EM. Considerations for a Successful RNA Trans-splicing Repair of Genetic Disorders. Mol Ther Nucleic Acids. 2014;3:e157. doi: 10.1038/mtna.2014.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Chao H, et al. Phenotype correction of hemophilia A mice by spliceosome-mediated RNA trans-splicing. Nat Med. 2003;9:1015–1019. doi: 10.1038/nm900. [DOI] [PubMed] [Google Scholar]
- 179.Petkovic S, Muller S. RNA circularization strategies in vivo and in vitro. Nucleic acids research. 2015;43:2454–2465. doi: 10.1093/nar/gkv045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Davidson L, Kerr A, West S. Co-transcriptional degradation of aberrant pre-mRNA by Xrn2. The EMBO journal. 2012;31:2566–2578. doi: 10.1038/emboj.2012.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Shen S, et al. Widespread establishment and regulatory impact of Alu exons in human genes. Proc Natl Acad Sci U S A. 2011;108:2837–2842. doi: 10.1073/pnas.1012834108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Tajnik M, et al. Intergenic Alu exonisation facilitates the evolution of tissue-specific transcript ends. Nucleic acids research. 2015;43:10492–10505. doi: 10.1093/nar/gkv956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Rybak-Wolf A, et al. Circular RNAs in the Mammalian Brain Are Highly Abundant, Conserved, and Dynamically Expressed. Molecular cell. 2015;58:870–885. doi: 10.1016/j.molcel.2015.03.027. [DOI] [PubMed] [Google Scholar]
- 184.Kellis M, et al. Defining functional DNA elements in the human genome. Proc Natl Acad Sci U S A. 2014;111:6131–6138. doi: 10.1073/pnas.1318948111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Naftelberg S, Schor IE, Ast G, Kornblihtt AR. Regulation of alternative splicing through coupling with transcription and chromatin structure. Annu Rev Biochem. 2015;84:165–198. doi: 10.1146/annurev-biochem-060614-034242. [DOI] [PubMed] [Google Scholar]
- 186.Konig J, et al. iCLIP reveals the function of hnRNP particles in splicing at individual nucleotide resolution. Nature structural & molecular biology. 2010;17:909–915. doi: 10.1038/nsmb.1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Alioto TS. U12DB: a database of orthologous U12-type spliceosomal introns. Nucleic acids research. 2007;35:D110–115. doi: 10.1093/nar/gkl796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: a sequence logo generator. Genome research. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Pulyakhina I, et al. SplicePie: a novel analytical approach for the detection of alternative, non-sequential and recursive splicing. Nucleic acids research. 2015;43:e80. doi: 10.1093/nar/gkv242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Chuang TJ, et al. NCLscan: accurate identification of non-co-linear transcripts (fusion, trans-splicing and circular RNA) with a good balance between sensitivity and precision. Nucleic acids research. 2015 doi: 10.1093/nar/gkv1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Szabo L, et al. Statistically based splicing detection reveals neural enrichment and tissue-specific induction of circular RNA during human fetal development. Genome biology. 2015;16:126. doi: 10.1186/s13059-015-0690-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.McPherson A, et al. deFuse: an algorithm for gene fusion discovery in tumor RNA-Seq data. PLoS Comput Biol. 2011;7:e1001138. doi: 10.1371/journal.pcbi.1001138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Maher CA, et al. Transcriptome sequencing to detect gene fusions in cancer. Nature. 2009;458:97–101. doi: 10.1038/nature07638. [DOI] [PMC free article] [PubMed] [Google Scholar]