Abstract
Artemisinin-based combination therapies (ACTs) are the cornerstone of current strategies for fighting malaria. Over the last decade, ACTs have played a major role in decreasing malaria burden. However, this progress is being jeopardized by the emergence of artemisinin-resistant Plasmodium falciparum parasites. Artemisinin resistance was first detected in western Cambodia in 2008 and has since been observed in neighboring countries in southeast Asia. The problem of antimalarial drug resistance has recently worsened in Cambodia, with reports of parasites resistant to piperaquine, the latest generation of partner drug used in combination with dihydroartemisinin, leading to worrying rates of clinical treatment failure. The monitoring and the comprehension of both types of resistance are crucial to prevent the spread of multidrug-resistant parasites outside southeast Asia, and particularly to Africa, where the public health consequences would be catastrophic. To this end, new tools are required for studying the biological and molecular mechanisms underlying resistance to antimalarial drugs and for monitoring the geographic distribution of the resistant parasites. In this review, we detail the major advances in our understanding of resistance to artemisinin and piperaquine and define the challenges that the malaria community will have to face in the coming years.
Background
In 2016, malaria remains a major human parasitic disease worldwide, affecting almost 3.2 billion people in 108 countries.1 Plasmodium falciparum, the most lethal human malarial pathogen, is responsible annually for 214 million cases and 438,000 deaths.1 Over the last decade, the malaria mortality rate has decreased considerably (by 48% worldwide), mostly due to the reinforcement of malaria control efforts and increases in the financial resources provided by both public and private international organizations. The prevention of malaria transmission, and the early detection and prompt treatment of malaria cases are currently the cornerstones of malaria control and elimination strategies. Malaria prevention is based on vector control measures (e.g., use of nets impregnated with long-lasting insecticide and the spraying of insecticides indoors) to limit contact between humans and vectors, thereby decreasing malaria transmission and morbidity. The management of suspected cases of malaria seen at health facilities or in the community is based on the early biological detection of malaria parasites by microscopy or rapid diagnostic tests, followed by effective and prompt treatment of the confirmed cases. These approaches limit the duration of the disease and prevent severe complications and death. Despite intensive efforts to develop effective vaccines and the promising results reported by vaccine trials (e.g., RTS,S/AS01 vaccine),2 it will be several years before these new preventive weapons can be used in clinical practice.
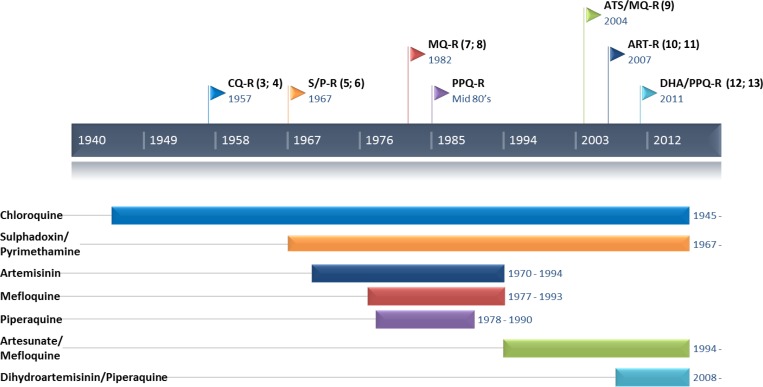
Over the last century, all the antimalarial drugs deployed worldwide have inevitably led to the selection and spread of drug-resistant P. falciparum parasites, particularly in southeast Asia (Figure 1 ).3–13 In the 1950 and 1960s, the World Health Organization (WHO) Global Malaria Eradication Program was based on the massive deployment of chloroquine, along with vector control measures (dichlorodiphenyltrichloroethane [DDT]). This program was initially highly successful, but the progress made was rapidly compromised by the emergence and spread of chloroquine-resistant parasites and DDT-resistant vectors. Later, in the 1970s and 1980s, a similar pattern was observed for alternative antimalarial drugs, such as sulfadoxine–pyrimethamine, which was used in areas in which chloroquine resistance had developed, leading to the selection of parasites resistant to antifolate agents. At the end of the 1990s,14,15 the global health situation, particularly in sub-Saharan Africa, was critical, with more than three million deaths annually due to malaria.16,17 In their attempts to control this alarming situation, stakeholders began to take an interest in derivatives of artemisinin, a sesquiterpene lactone discovered in 1972 by Chinese scientists during a program of research involving the screening of compounds from plants (Military code name: Project 523). Artemisinin derivatives, which are extracted from Artemisia annua, have been used to treat acute malaria in traditional Chinese medicine for more than 2,000 years. Novartis developed and patented the first artemisinin-based combination therapy (ACT), consisting of a combination of artemether and lumefantrine. The massive deployment of this new treatment in South Africa in 2000–2001 along with vector control measures was highly successful, decreasing the number of deaths from malaria by 87.5% in less than 1 year.18 In 2001, the WHO recommended the use of ACT as a first-line treatment of uncomplicated malaria in areas in which the parasites were resistant to monotherapy with other drugs, such as mefloquine.19 This recommendation was extended to the entire world in 2006, and the WHO banned the use of artemisinin in monotherapy to prevent the selection of artemisinin-resistant parasites. In 2012, five different ACTs, including dihydroartemisinin–piperaquine (DHA–PPQ), artesunate–mefloquine (AS–MQ), artemether–lumefantrine, artesunate–sulfadoxine–pyrimethamine, and artesunate–amodiaquine, were deployed in 79 countries with endemic malaria. At present, successful malaria control and, to a greater extent, malaria elimination rely strongly on effective antimalarial treatments. As no alternative to artemisinin derivatives are ready for deployment before 2020, emergence of resistance to artemisinin derivatives and partner drugs represents a major threat to malaria control and elimination efforts worldwide.
Figure 1.
Timeline between the introduction of the main antimalarial drugs and the first case of emergence of resistance.3–5,7–13
Resistance to Artemisinin Derivatives: Past and Present Research Challenges
Clinical phenotype.
Partial resistance to artemisinin in P. falciparum was first reported in 2008, in Battambang Province in western Cambodia.10 It was subsequently confirmed in 2009, in Pailin Province, the well-known epicenter of antimalarial multidrug resistance.11 Artemisinin resistance has since been reported elsewhere in western Cambodia, western Thailand, southern Myanmar (Burma), and southern Vietnam,20–23 and in China.24
Artemisinin resistance is defined clinically as delayed parasite clearance25–27 and a half-life for parasite clearance ≥ 5 hours has been associated with artemisinin resistance in southeast Asia.27 In high-transmission settings, such as sub-Saharan Africa, where the partial immunity of adults plays a significant role in parasite clearance, the threshold need to be defined.28 Currently, this approach is considered to be the gold standard, but suffers from several drawbacks: first, parasite clearance studies are both logistically and financially demanding, requiring the screening of thousands of febrile individuals to identify the small number of patients who will be hospitalized for several days; second, parasite clearance rates depend on many factors other than the intrinsic drug resistance of the parasites, relating to drug administration, immunity, and pharmacodynamics. Alternative in vivo approaches have been proposed. For instance, Beshir and others29 assessed the association between parasite clearance following ACT and transmissibility in mosquitoes by membrane-feeding assays. They found that Kenyan children treated with a standard 3-day ACT course had commonly a persistent residual parasitemia (assessed by quantitative polymerase chain reaction [qPCR]), associated with longer duration of gametocyte carriage and a higher likelihood of infecting mosquitoes. However, even being innovative and elegant, this approach is logistically demanding and not adapted for high-scale application.
Before 2013, the principal alternative approach for studies of artemisinin resistance in P. falciparum was the determination, in classical assays, of IC50 values (the concentration inhibiting parasite growth by 50%) for DHA. However, all in vitro/vivo studies, except one,10 have reported no significant correlation20 between clinical outcome (half-life for parasite clearance) and DHA IC50 values. One potential reason for this observation was later put forward30: the parasites in these assays were exposed to very low concentrations of DHA (≤ 64 nM) for 48–72 hours, whereas parasites in vivo are exposed to higher concentrations of drugs for only 1–2 hours.
Rethinking in vitro phenotype.
A novel assay to overcome this problem was developed in 2012–2013. This assay was based on the exposure of culture-adapted or fresh isolates of P. falciparum parasites to the physiological conditions encountered in humans: P. falciparum parasites were exposed to 700 nM DHA (the active metabolite of all artemisinins) for 6 hours, reflecting typical doses and exposure times for ACTs. This assay (named ring-stage survival assay [RSA]) successfully detected significant differences in susceptibility in vitro between parasites originating from areas in which clinical resistance to artemisinin had and had not been observed.30 In vitro artemisinin susceptibility was found to be stage dependent and strongly linked to the decrease in susceptibility of early ring stages. This further explains the lack of correlation between clinical outcome and DHA IC50 values, during which the parasites are exposed to the drug throughout their cycle. These parasite stages were capable of surviving short periods of drug exposure by entering a state of developmental arrest (dormancy).30–32 This observation was consistent with the findings of previous transcriptome studies showing that artemisinin-resistant parasites had a particularly long ring stage and lower metabolic activities32 and were also consistent with those of modeling studies suggesting a significant decrease in the efficacy of artemisinin derivatives against ring-stage parasites in western Cambodia as early as 2007–2008.33 Survival rates in the RSA were also strongly correlated with clinical phenotype (half-life for parasite clearance).31,34
A molecular marker.
The molecular signatures associated with resistance to artemisinins were then investigated by sequencing the exome of an African strain (F32-Tanzania) that became resistant to artemisinin after repeated exposure to increasing concentrations of the drug over a period of 5 years (F32-ART).35 The exome of F32-ART was compared with that of its sibling clone (F32-TEM) cultured under the same conditions but without the drug. A detailed analysis of the differences between the two exomes identified eight nonsynonymous single nucleotide polymorphisms (SNPs) in seven genes. Analysis of the genomes of three intermediate strains (after 22, 40, or 56 cycles of exposure) and of 49 strains isolated in Cambodia made it possible to determine the chronological order in which these mutations appeared. This analysis revealed a strong association between mutations in the propeller domain of the Kelch gene located on chromosome 13 (K13, PF3D7_1343700) and in vitro susceptibility to artemisinin in the RSA. Previous genome-wide association studies performed by two independent groups identified this region as associated with delayed parasite clearance half-lives.36,37 A study of the temporal (2001–2012) and spatial distribution of K13-mutant parasites showed that they had progressively spread in zones classified as “resistant” (western Cambodia) but not in areas in which artemisinin resistance had never been observed (eastern Cambodia). The detection of parasites with K13 mutations in patients with slow rates of parasite clearance confirmed that this molecular marker was a major determinant of resistance to artemisinin derivatives.38 Definitive evidence was later obtained in site-specific genome editing experiments using zinc-finger nucleases39 or the CRIPSR-Cas9 system.40 This series of experiments showed that the replacement of a wild-type K13 allele in parasites susceptible to artemisinin by different K13 mutant alleles (Y493H, C580Y, M476I, R539T, I543T) conferred artemisinin resistance (survival rates assessed using the RSA increased from ≤ 1–2% to 29% after the insertion of K13 mutations), whereas the removal of K13 mutations in isolates from Cambodia had the opposite effect (survival rates decreased from 13% to 49% to 0.3% to 2.4% after the removal of K13 mutations).
This discovery provided unprecedented opportunities for improving strategies for the monitoring of artemisinin resistance. Several molecular epidemiology studies, mostly in southeast Asia,27,38,41–44 China,45–48 Indian subcontinent49,50 and Africa,27,51–58 investigated the frequency of K13 mutant alleles, their spatial distribution and, in some cases, their association with clinical artemisinin resistance (day 3 positivity rate or parasite clearance half-life). Only 13 independent K13 mutations of about 100 tested were found to be associated with clinical resistance, with evidence of the independent emergence of the same mutation in different areas. Only four of these mutations, all from Asia (C580Y, R539T, I543T, and Y493H), have been validated in vitro. A worldwide map of P. falciparum K13-propeller polymorphism was recently published from a study involving 41 partner institutions and 14,037 samples collected from 163 sites (in 59 countries in which malaria is endemic), providing a global overview of the distribution of artemisinin resistance and fundamental data for drug policy makers and future surveillance activities.59 The main findings of the KARMA study (K13 Artemisinin Resistance Multicenter Assessment) suggest that artemisinin resistance remains confined to southeast Asia (including Cambodia, PDR of Laos, Thailand, Vietnam, and Myanmar) and China. In these two regions, the proportions of parasites with K13 mutations were estimated at 51.8% (95% CI: 48.5–55.3%) and 25.6% (95% CI: 19.3–33.3%), respectively. Two independent foci of emergence were identified—one in Cambodia–Vietnam–PDR of Laos and the other in eastern Thailand–Myanmar–China—with no evidence of parasite flow between the two foci, consistent with other reports that showed that most K13 mutations emerged independently from the same foci in southeast Asia, with the exception of mutations C580Y and Y493H, which probably emerged in Cambodia and subsequently spread to Lao PDR and Vietnam.42 Conversely, no K13 mutant alleles were found in South America, Oceania, the Philippines, and some of the countries in Central Asia. In Africa, the frequency of K13 mutant alleles (< 1.5%) was low. None African K13 mutant alleles were associated with artemisinin resistance and none of the Asian mutants known to be associated with resistance to artemisinin (C580Y, R539T, I543T, and Y493H) were found among the 9,542 African samples tested. The most frequent mutation in Africa, the A578S mutation (3.8%), which was previously detected in India, Bangladesh, and Thailand, was susceptible in the in vitro RSA (0–3 hours) and had no effect on positivity rate on day 3. Thus, not all nonsynonymous K13 mutants are associated with artemisinin resistance. The effective surveillance of artemisinin resistance must therefore be based on a combination of molecular data (genotyping to detect K13 mutants and haplotyping of the neighboring loci to assess the spread of mutant parasites carrying the same K13 mutation) with clinical and in vitro studies (to validate the association of the mutations detected with artemisinin resistance). It is also worth noting that the lack of association between ACT treatment failures and K13 polymorphisms in Africa60 could be a sign of K13-independent mechanisms, which need to be investigated. There is also a possibility that a particular genetic backbone is mandatory to increase the level of resistance conferred by K13 mutations, and this specific genetic background is now only found in Asian parasite populations.61
Insight into resistance mechanisms.
Despite the immense progress of recent years, the mechanisms involved in P. falciparum resistance to artemisinin derivatives remain poorly understood. Such an understanding is essential for the definition of new effective strategies to prevent the spread of resistant strains, and for the development of new detection tools and new drugs targeting these mechanisms. The protein encoded by the K13 gene has a BTB/POZ domain (PDB 4YY8) and a C-terminal helical Kelch domain. The mutations associated with artemisinin resistance affect one of the β sheets of the Kelch domain (Y493H in blade 2, R539T and I543T in blade 3, and C580Y in blade 4). Such domains are frequently involved in protein–protein interactions and the validated mutations may alter the overall structure of the protein, substantially modifying the interactions between K13 and its potential partner proteins. Mbengue and others recently provided new insight into the mechanisms involved in K13-mediated artemisinin resistance.62 They demonstrated that the presence of the C580Y mutation decreased polyubiquitination of the P. falciparum phosphatidylinositol-3-kinase (PfPI3K, which is strongly inhibited by artemisinins) by modifying its binding to K13, thereby limiting PfPI3K proteolysis and increasing production of the phosphatidylinositol-3-phosphate (PI3P). PI3P levels were found to be predictive of artemisinin resistance in both clinically and genetically modified P. falciparum strains, with high PI3P levels associated with an increase in artemisinin resistance in the absence of K13 mutation.
Moreover, the K13 protein displays similarity to the human KEAP1 protein involved in the oxidative stress response mediated by the transcription factor NrF2.63–67 Mutations of the KEAP1 gene impairing the binding of the encoded protein to NrF2 have been found in human cancers resistant to chemotherapy. This resistance is thought to be due to the diffusion of NrF2 in the nucleus and the activation of several genes involved in the antioxidant responses (AREs), promoting cell survival.68 A similar mechanism may account for the resistance of P. falciparum to artemisinin. It is therefore important to screen for proteins interacting with K13, to identify the cellular pathways involving K13 and to understand how these pathways are modified by K13 mutations.
Perspectives.
Major advances have been made since the detection of the first case of artemisinin resistance in 2008, and the identification of the K13 mutation as a major determinant of artemisinin resistance has made it possible to conduct large-scale molecular monitoring programs with simple, inexpensive techniques (e.g., extraction of DNA from finger-prick blood samples on filter paper and its use for the detection of mutations by PCR and sequencing). Such programs can be used to monitor resistance in real time, making it possible to adapt antimalarial treatments according to the epidemiological situation in a given area. The timely implementation of such initiatives should make it possible to avert public health disasters like the spread of chloroquine-resistant parasites in Africa in the 1980s. Studies of the K13 gene may also shed light on the cellular and molecular mechanisms underlying resistance to artemisinin derivatives, thereby making it possible to limit the spread of this resistance phenotype through the development of appropriate new drugs. However, as artemisinin derivatives are systematically administered with a partner drug, it is important to understand the mechanisms involved in the emergence of parasites resistant to partner drugs, particularly in areas of artemisinin resistance, in which the clearance of parasites from the bloodstream of patients depends primarily on the efficacy of the partner drug.
Resistance to PPQ: Research Challenges
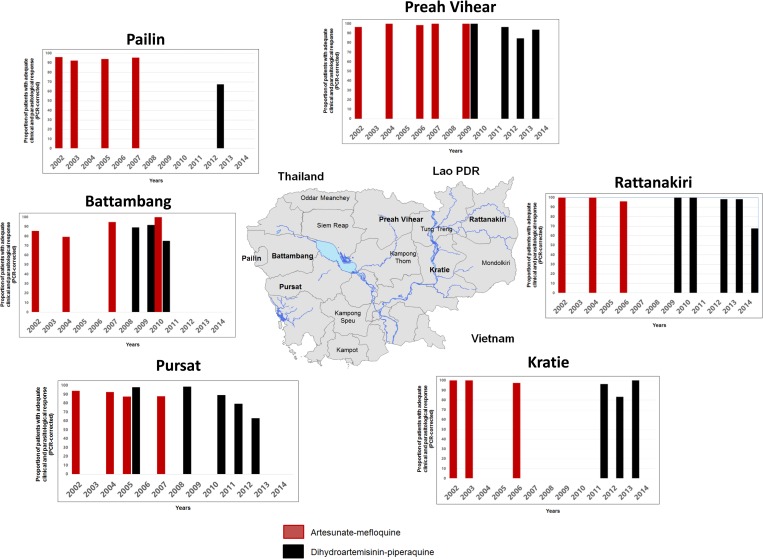
For decades, the Thai–Cambodian border has been the epicenter of multidrug-resistant P. falciparum malaria emergence. This region saw successively the demise of chloroquine (1960s), sulfadoxine–pyrimethamine (1970s), and MQ (1990s) treatments.14 MQ, initially introduced in the 1990s, was replaced, in 2000, by AS–MQ in Cambodia.69 The clinical efficacy of this combination decreased rapidly, from 90.1% in 2001 to 79.4% in 2004,9 probably due to the high proportion of MQ-resistant parasites (high MQ IC50 values and amplified Pfmdr-1 gene copy).70–72 AS–MQ was replaced by DHA–PPQ73,74 in 2008, initially in western Cambodia (Pailin Province) and then throughout the entire country in 2010, as the first-line drug of choice for uncomplicated malaria.75 Compelling data, presented in Table 1 and Figure 2 , clearly illustrate that the decrease of the ACT efficacy is always emerging in western Cambodia before to spread eastward. It is worth to note that AS–MQ regained efficacy promptly after it was replaced by DHA–PPQ (e.g., in Battambang, the reported efficacy of AS–MQ in 2004 was only 79.3% but reaches 100% in 2010, 1 year only after the implementation of DHA–PPQ as a first-line treatment in this area), suggesting that a switch from DHA–PPQ to AS–MQ could be a temporary solution in areas where DHA–PPQ is now failing. Clinical studies assessing artesunate–pyronaridine (ATS–PYRO) efficacy conducted in Cambodia were disappointing: indeed, ATS–PYRO did not meet the WHO efficacy criteria for first-line treatment of P. falciparum malaria in western Cambodia (i.e., > 90% PCR-adjusted adequate clinical and parasitological response at day 42).76 This was largely unexpected, given the high efficacy of this ACT in other Asian countries and in Africa.77–81 The reasons of this disparity remain unclear, but cross-resistance between pyronaridine and other antimalarial drugs like PPQ should be investigated.
Table 1.
Detailed data of the therapeutic efficacy studies performed in different sites in Cambodia between 2001 and 2015 to evaluate the efficacy of 3-day standard course of ACTs (AS–MQ, DHA–PPQ, ATS–PYRO, and A–L)
| Study sites (province) | Start year | End year | Follow-up (days) | Antimalarial drug regimen | ACPR (%) | Reference |
|---|---|---|---|---|---|---|
| Battambang | 2001 | 2001 | 28 | AS–MQ | 96.0 | 9 |
| 2003 | 2003 | 28 | AS–MQ | 92.3 | 9 | |
| 2005 | 2005 | 42 | AS–MQ | 94.1 | # | |
| 2007 | 2007 | 42 | AS–MQ | 95.4 | # | |
| 2012 | 2013 | 42 | DHA–PPQ | 67.5 | 82 | |
| 2014 | 2014 | 42 | ATS–PYRO | 100.0 | # | |
| Kampong Speu | 2003 | 2003 | 28 | AS–MQ | 96.7 | 9 |
| 2010 | 2010 | 42 | AS–MQ | 88.9 | 89 | |
| 2012 | 2013 | 42 | DHA–PPQ | 86.4 | 82 | |
| Kampong Thom | 2012 | 2012 | 42 | DHA–PPQ | 100.0 | # |
| 2012 | 2013 | 42 | DHA–PPQ | 100.0 | 82 | |
| Kampot | 2006 | 2008 | 42 | AS–MQ | 81.2 | 90 |
| 2013 | 2014 | 42 | DHA–PPQ | 94.1 | # | |
| Kratie | 2001 | 2001 | 28 | AS–MQ | 100.0 | 9 |
| 2003 | 2003 | 28 | AS–MQ | 100.0 | 9 | |
| 2006 | 2006 | 42 | AS–MQ | 97.5 | # | |
| 2011 | 2011 | 42 | DHA–PPQ | 96.4 | # | |
| 2011 | 2012 | 42 | DHA–PPQ | 83.3 | 82 | |
| 2013 | 2013 | 42 | DHA–PPQ | 100.0 | # | |
| Mondulkiri | 2014 | 2015 | 42 | DHA–PPQ | 90.0 | # |
| Odder Meanchey | 2003 | 2003 | 28 | AS–MQ | 97.8 | 9 |
| 2012 | 2013 | 42 | DHA–PPQ | 62.5 | 12 | |
| 2012 | 2014 | 42 | DHA–PPQ | 51.1 | 85 | |
| Pailin | 2002 | 2002 | 28 | AS–MQ | 85.7 | 9 |
| 2004 | 2004 | 28 | AS–MQ | 90.1 | 9 | |
| 2004 | 2004 | 42 | AS–MQ | 79.3 | 9 | |
| 2007 | 2008 | 42 | AS–MQ | 95.0 | 11 | |
| 2007 | 2008 | 42 | AS–MQ | 100.0 | 77 | |
| 2007 | 2008 | 42 | ATS–PYRO | 89.8 | 77 | |
| 2008 | 2009 | 42 | DHA–PPQ | 89.4 | 91 | |
| 2009 | 2010 | 42 | DHA–PPQ | 91.9 | 91 | |
| 2010 | 2011 | 42 | DHA–PPQ | 75.0 | 91 | |
| 2011 | 2011 | 42 | AS–MQ | 100.0 | # | |
| 2014 | 2015 | 42 | ATS–PYRO | 82.0 | 76 | |
| Preah Vihear | 2002 | 2002 | 28 | AS–MQ | 96.6 | 9 |
| 2004 | 2004 | 28 | AS–MQ | 100.0 | 9 | |
| 2006 | 2006 | 42 | AS–MQ | 98.6 | # | |
| 2007 | 2007 | 42 | AS–MQ | 100.0 | # | |
| 2009 | 2009 | 42 | DHA–PPQ | 100.0 | 91 | |
| 2009 | 2009 | 42 | AS–MQ | 100.0 | # | |
| 2011 | 2011 | 42 | DHA–PPQ | 96.6 | # | |
| 2011 | 2012 | 42 | DHA–PPQ | 93.4 | 82 | |
| 2012 | 2013 | 63 | DHA–PPQ | 84.6 | 20 | |
| 2013 | 2014 | 42 | DHA–PPQ | 93.7 | # | |
| Pursat | 2002 | 2002 | 28 | AS–MQ | 94.0 | 9 |
| 2004 | 2004 | 28 | AS–MQ | 92.6 | 9 | |
| 2005 | 2005 | 42 | AS–MQ | 87.5 | # | |
| 2005 | 2005 | 28 | A–L | 82.4 | 82 | |
| 2005 | 2005 | 28 | DHA–PPQ | 98.2 | 82 | |
| 2007 | 2007 | 42 | AS–MQ | 87.9 | # | |
| 2008 | 2008 | 42 | DHA–PPQ | 98.7 | 91 | |
| 2010 | 2010 | 42 | DHAPPQ | 89.3 | 91 | |
| 2011 | 2011 | 42 | DHAPPQ | 83.3 | # | |
| 2011 | 2012 | 42 | DHAPPQ | 79.5 | 82 | |
| 2012 | 2013 | 63 | DHAPPQ | 63.2 | 20 | |
| 2012 | 2013 | 42 | DHAPPQ | 97.5 | 82 | |
| 2014 | 2015 | 42 | ATS–PYRO | 89.8 | 76 | |
| Rattanakiri | 2002 | 2002 | 28 | AS–MQ | 100.0 | 9 |
| 2004 | 2004 | 28 | AS–MQ | 100.0 | 9 | |
| 2006 | 2006 | 42 | AS–MQ | 96.2 | # | |
| 2009 | 2010 | 42 | DHA–PPQ | 100.0 | 91 | |
| 2010 | 2010 | 28 | A–L | 95.0 | # | |
| 2010 | 2010 | 42 | DHA–PPQ | 100.0 | 91 | |
| 2010 | 2010 | 42 | DHA–PPQ | 100.0 | # | |
| 2012 | 2013 | 63 | DHA–PPQ | 98.4 | 20 | |
| 2013 | 2013 | 42 | DHA–PPQ | 98.3 | # | |
| 2014 | 2014 | 42 | DHA–PPQ | 67.6 | 83 | |
| Siem Reap | 2014 | 2015 | 42 | DHA–PPQ | 37.5 | # |
| Stung Treng | 2014 | 2015 | 42 | DHA–PPQ | 65.6 | # |
ACPR = adequate clinical and parasitological response (proportion, %); A–L = artemether–lumefantrine; AS–MQ = artesunate–mefloquine; ATS–PYRO = artesunate–pyronaridine; DHA–PPQ: dihydroartemisinin–piperaquine. Follow-up, the last day of follow-up according to the WHO protocol used (28, 42, or 63 days).
National Center for Parasitology Entomology and Malaria Control (CNM) unpublished reports.
Figure 2.
Spatial distribution and evolution of the efficacy of artesunate-mefloquine and dihydroartemisinin-piperaquine 3-day standard course in different sites in Cambodia between 2001 and 2014. Details are provided in Table 1.
DHA–PPQ is failing in Cambodia.
The most recent clinical studies conducted in Cambodia between 2012 and 2015 have reported an alarming increase in the proportion of late clinical treatment failure following a standard 3-day course of DHA–PPQ. Treatment failure rates increased from 25% in 2010 to 46% in 201420,82–85 in western Cambodia, suggesting that DHA–PPQ is becoming ineffective within only a few years of its nationwide deployment. The high treatment failure rates suggest that parasites with reduced susceptibility to both artemisinin and PPQ are now prevalent in western/northern Cambodia. Clinical failures of DHA–PPQ treatment have been linked to the emergence of PPQ resistance, but no strong correlation has been found between clinical outcome (recrudescence 2–4 weeks after the initial treatment) and susceptibility to PPQ in vitro, as assessed by the determination of IC50 for pretreatment (day 0) isolates.83 PPQ resistance remains poorly characterized and there is currently no robust evidence of parasite-dependent resistance to this drug (reliable in vitro phenotype and/or validated genetic molecular marker).
In vitro detection of PPQ resistance: Piperaquine Survival Assay.
Several in vitro studies, conducted in Cambodia, have reported an increase over time in the geometric mean IC50 of PPQ in western Cambodia or a significantly higher geometric mean IC50 of PPQ for isolates from patients with recrudescent infections than for those from cured patients.20 However, no direct association between a high IC50 or 90% inhibitory concentration (IC90) of PPQ for isolates before treatment and the failure of DHA–PPQ treatment has ever been demonstrated. In all published studies, the range of IC50 values for PPQ in patients with recrudescent infections overlapped substantially with those in cured patients, suggesting that the classical in vitro test is not sufficiently reliable for the identification of PPQ-resistant parasites and would therefore probably detect the elimination, in a given area, of the parasites most sensitive to PPQ.20,84 It has been recently shown that the poor performance of the standard isotopic assays for detecting PPQ resistance was associated with a relatively high frequency of non-interpretable curves in assays on PPQ-resistant isolates. Most of the isolates collected from patients with recrudescent infection had non-interpretable IC50 curves, whereas all yielded conventional response curves for the other drugs tested. These anomalous curves presented a paradoxical increase in [3H]-hypoxanthine incorporation at PPQ concentrations above 100–200 nM, the physiological concentration of PPQ observed in the blood of patients treated with a standard 3-day course of DHA–PPQ82 To overcome the limitations of current assays and provide a more robust assessment of PPQ resistance, a new in vitro assay (named “Piperaquine Survival Assay” PSA) based on the same principle as the RSA was developed.82 This assay involves exposing tightly synchronized culture-adapted parasites (in vitro format) or parasites directly collected from patients (ex vivo format) to a pharmacologically relevant dose of PPQ (200 nM for 48 hours), to mimic in vivo conditions. The in vitro PSA was found to discriminate efficiently between PPQ-resistant and sensitive parasites, with resistance defined as a survival rate in the PSA ≥ 10% (defined as the cutoff for resistance on the basis of results for the isolates of patients with recrudescent infection). Patients harboring parasites with K13 mutants with an ex vivo survival rate in the PSA ≥ 10% had a 32 times higher risk of having recrudescent infection. The use of the PSA (in absence of reliable molecular marker) together with K13 genotyping was found to be a suitable method for monitoring resistance to both artemisinin and PPQ, in areas in which DHA–PPQ is currently the recommended first-line treatment and may be a reliable platform for identifying molecular markers associated with PPQ resistance and for evaluating alternative drugs effective against PPQ-resistant parasites.
Toward a molecular marker for PPQ resistance.
Studies conducted in Cambodia and Thailand with the aim of characterizing molecular signatures associated with PPQ resistance have found no association between the in vitro PPQ susceptibility of P. falciparum isolates and copy number variations (CNVs) for the PF3D7_0520500 and PF3D7_0520900 genes located on chromosome 5 previously identified by Eastman and others.85–87 Spring and others recently reported a significant correlation between DHA–PPQ treatment failure and the presence of a triple-mutant haplotype for the propeller domain of the K13 gene and for positions 1718319 and 688956 on chromosomes 13 and 10, respectively (MAL13: 1718319 and MAL10: 688956), previously reported to be associated with delayed parasite clearance in genome-wide association studies.42,84,88 Patients harboring triple-mutant parasites had a 5.4 times higher risk of treatment failure and a higher IC50 for PPQ (33.6 nM for the triple mutant versus 24 nM for other parasites). However, rather than being directly associated with PPQ resistance (no causal impact and no biological rationale), these markers may be associated with a particular artemisinin genetic background favoring the acquisition of resistance to PPQ.89
However, several groups with different in vitro assays showed that parasites with single-copy Pfmdr-1 (and consequently a low IC50 value for MQ) were strongly linked to PPQ resistance.20,82,84,86 For instance, it has been observed that all in vitro PPQ-resistant isolates had a single Pfmdr-1 copy but the converse was not true, as not all isolates with a single copy of Pfmdr-1 were in vitro PPQ resistant. This finding strongly supports recent recommendations for the use of artesunate plus MQ as an alternative first-line treatment in provinces in which DHA–PPQ is failing.20,82,84,86 However, further investigations are required to determine whether the elimination of parasites with multiple copies of Pfmdr-1 has resulted from the removal of MQ pressure or the selective pressure exerted by PPQ. This last finding also suggests that PfMDR1, a pump involved in the intraparasitic influx/efflux of metabolites or drugs, may play a significant role in parasite susceptibility to PPQ. For instance, PfMDR1 may play a significant role in the influx of PPQ into the digestive vacuole, in which PPQ acts on its target to kill parasites. Nonetheless, some parasites sensitive to PPQ harbor a single copy of Pfmdr-1, highlighting the need to find a specific molecular marker.
Several studies have also observed that specific mutations of the Pfcrt gene were significantly associated with PPQ resistance.82,90 For instance, isolates containing parasites with a variant of the Dd2 Pfcrt allele carrying either 97Y, 343L, or 353V have higher median PSA survival rates than those harboring the Dd2 allele.82 In French Guiana, Pelleau and others also found that the acquisition of the C350R substitution in the Pfcrt gene decreased susceptibility to PPQ, suggesting that PPQ pressure drove the expansion of this allele.90 As the 353V mutation (frequently observed in Cambodia) is located close to 350R, further investigations, such as genome editing, should be carried out to assess the role of Pfcrt mutations in the genetic background of Cambodian parasites.
Perspectives.
The development of the PSA as an efficient screening tool for PPQ-resistant parasites is a major advance that may facilitate the identification of a specific molecular marker. Molecular signatures (CNV and SNP) associated with in vitro resistance to PPQ are being investigated by the whole-exome sequencing of PPQ-resistant (PSA survival rates ≥ 10%) and PPQ-susceptible (PSA survival rates < 10%) parasites. In parallel, we repeatedly exposed parasites to 200 nM PPQ, over increasingly long exposure times (from 48 to 96 h), over a period of 1 year (Duru V, Witkowski B, Khim N, and Menard D, unpublished work). This resulted in the generation of a Cambodian PPQ-resistant culture-adapted parasite (PSA survival rate of 21%). The survival rates in the PSA of the initial parasite and its sibling clone cultured under the same conditions but without the drug were estimated at 3% and 0.7%, respectively. The differences between the three exomes is analyzed, and the results obtained should facilitate accurate definition of the molecular signatures associated with PPQ resistance, while this approach has already been successfully used to identify mutations in the propeller domain of the K13 gene as a major determinant of artemisinin resistance.
Concluding Remarks
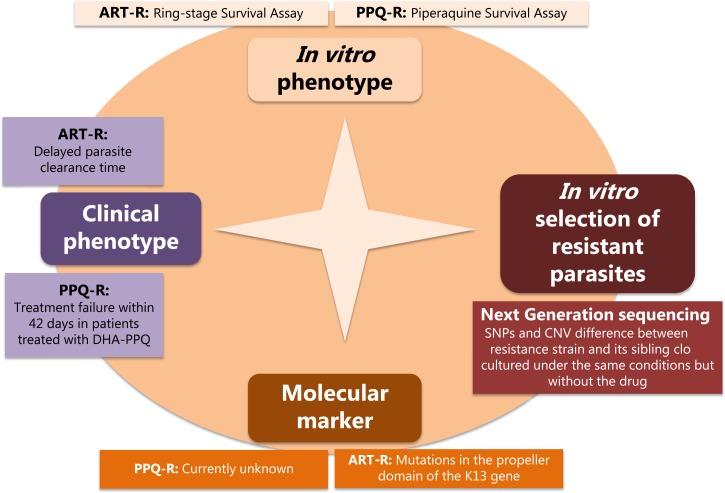
Assessments of antimalarial drug efficacy include clinical drug efficacy studies, in vitro assessments of drug sensitivity, and molecular markers (Figure 3 ). Drug efficacy studies (clinical phenotype) remain the gold standard to evaluate the efficacy of antimalarial drugs. Clinical studies investigate both the parasite clearance time in the first 3 days of treatment to detect artemisinin resistance and the treatment failure rate (evaluated during the follow-up usually until day 42 or day 63 according to the half-life of the partner drugs) to detect partner drug resistance. In vitro phenotype assessing the sensitivity of P. falciparum malaria parasites to antimalarial drugs is also frequently used to complement data from clinical studies. These assays contribute to detect early signs of emergence of drug resistance. In vitro assays are also useful for drug screening and for validating candidate molecular markers associated with drug resistance. Indeed, data obtained from in vitro assay are directly associated to the parasite drug susceptibility and contrarily to in vivo phenotypes, are not affected by host factors, such as acquired immunity, bioavailability, or pharmacokinetics of antimalarial drugs. Classical in vitro assays (assessing IC50 values) have limited sensitivity for detecting resistant parasites to artemisinin and PPQ and the design of such assays needs to be adapted according to the pharmacokinetic/pharmacology profiles of the tested drugs. The RSA and the PSA are excellent examples. Finally, validated molecular markers associated to antimalarial drug resistance are relevant to detect and monitor in real time the geospatial distribution of resistant parasites. They are useful to anticipate risks associated with the emergence and the spread of antimalarial parasite resistance and to inform policy makers. The discovery of the molecular signatures associated to antimalarial drug resistance is challenging. However, at present, next-generation sequencing of well-characterized parasites, which allow to detect SNPs and CNV differences between sensitive and resistant parasites, greatly facilitate the genomic investigations (Figure 3).
Figure 3.
Main tools and strategy required for the detection of the molecular signatures associated with antimalarial drug resistance.
Resistance to both artemisinin and PPQ is now prevalent in Cambodia, as demonstrated by the worrying increase in the number of cases of DHA–PPQ treatment failure. The decrease in ACT efficacy has resulted from adaptation of the parasite to both artemisinin derivatives and the partner drug. Our understanding of artemisinin resistance has progressed considerably, due, in particular, to the discovery of K13 polymorphisms. Evaluations of the global distribution of artemisinin resistance, with the screening of thousands of blood samples for the detection of K13 mutations, are underway. However, greater efforts are required concerning PPQ resistance. There is an urgent need to characterize PPQ resistance in more detail and to identify a reliable and specific molecular marker of this resistance, as this drug is a potential partner drug candidate for inclusion in combination with the next generation of antimalarial drugs, such as ozonides, spiroindolones, and imidozolopiperazines.91
The management of falciparum malaria cases in Cambodia remains challenging. New strategies (even short-term approaches) using existing antimalarial drugs are required. These approaches may include a rotation of different ACTs (DHA–PPQ and AS–MQ), which could be effective due to the contrasting susceptibility profiles of PPQ- and MQ-resistant parasites. However, such strategies would be expected to fail in the long term, with the emergence of parasites resistant to both PPQ and MQ. This strategy is currently implemented in several provinces in Cambodia in which high failure rates have been recorded for DHA–PPQ treatment. Another option is extending the course of treatment to 5 or 7 days, but it may be difficult to ensure compliance with a longer course of treatment. Finally, another alternative approach would involve combining artemisinin derivatives with two slowly eliminated partner drugs in a 3-day triple ACT, in a strategy similar to those already used for HIV and tuberculosis. This approach could involve the use of artemether–lumefantrine–amodiaquine and DHA–PPQ–MQ combinations, which are currently under evaluation for the treatment of uncomplicated falciparum malaria in areas of resistance to both artemisinin and the partner drug in conventional ACT.
ACKNOWLEDGMENTS
We thank all the patients enrolled in the studies, the health center staff and the staff from the National Center for Parasitology, Entomology, and Malaria Control in Cambodia for their support, and the staff of the Institut Pasteur in Cambodia and those from the Malaria Molecular Epidemiology Unit, especially Nimol Khim, Nimol Kloeung, Sopheakvatey Ke, and Anais Domergue. We also want to thank Pascal Ringwald for providing in-depth data of ACT efficacy studies conducted in Cambodia. We also thank the Division Internationale of the Pasteur Institute (Marc Jouan and Eliane Coeffier).
Footnotes
Financial support: This study was supported by the Institut Pasteur in Cambodia, the Institut Pasteur, Paris through a Programme Transversal de Recherche grant (PTR 2015—535) and the KARMA project and the 5% initiative (Vers l'élimination du paludisme ou comment agir efficacement contre la transmission des parasites du paludisme? Prochains défis à relever pour les pays du Sud-est asiatique). Valentine Duru is supported by a doctoral fellowship from the International Division, Institut Pasteur.
Authors' addresses: Valentine Duru, Benoit Witkowski, and Didier Ménard, Malaria Molecular Epidemiology Unit, Institut Pasteur in Cambodia, Phnom Penh, Cambodia, E-mails: vduru@pasteur-kh.org, bwitkowski@pasteur-kh.org, and dmenard@pasteur-kg.org.
References
- 1.World Health Organization . World Malaria Report 2015. Geneva, Switzerland: World Health Organization; 2015. [Google Scholar]
- 2.RTS,S Clinical Trials Partnership Agnandji ST, Lell B, Fernandes JF, Abossolo BP, Methogo BG, Kabwende AL, Adegnika AA, Mordmüller B, Issifou S, Kremsner PG, Sacarlal J, Aide P, Lanaspa M, Aponte JJ, Machevo S, Acacio S, Bulo H, Sigauque B, Macete E, Alonso P, Abdulla S, Salim N, Minja R, Mpina M, Ahmed S, Ali AM, Mtoro AT, Hamad AS, Mutani P, Tanner M, Tinto H, D'Alessandro U, Sorgho H, Valea I, Bihoun B, Guiraud I, Kaboré B, Sombié O, Guiguemdé RT, Ouédraogo JB, Hamel MJ, Kariuki S, Oneko M, Odero C, Otieno K, Awino N, McMorrow M, Muturi-Kioi V, Laserson KF, Slutsker L, Otieno W, Otieno L, Otsyula N, Gondi S, Otieno A, Owira V, Oguk E, Odongo G, Woods JB, Ogutu B, Njuguna P, Chilengi R, Akoo P, Kerubo C, Maingi C, Lang T, Olotu A, Bejon P, Marsh K, Mwambingu G, Owusu-Agyei S, Asante KP, Osei-Kwakye K, Boahen O, Dosoo D, Asante I, Adjei G, Kwara E, Chandramohan D, Greenwood B, Lusingu J, Gesase S, Malabeja A, Abdul O, Mahende C, Liheluka E, Malle L, Lemnge M, Theander TG, Drakeley C, Ansong D, Agbenyega T, Adjei S, Boateng HO, Rettig T, Bawa J, Sylverken J, Sambian D, Sarfo A, Agyekum A, Martinson F, Hoffman I, Mvalo T, Kamthunzi P, Nkomo R, Tembo T, Tegha G, Tsidya M, Kilembe J, Chawinga C, Ballou WR, Cohen J, Guerra Y, Jongert E, Lapierre D, Leach A, Lievens M, Ofori-Anyinam O, Olivier A, Vekemans J, Carter T, Kaslow D, Leboulleux D, Loucq C, Radford A, Savarese B, Schellenberg D, Sillman M, Vansadia P. A phase 3 trial of RTS,S/AS01 malaria vaccine in African infants. N Engl J Med. 2012;367:2284–2295. doi: 10.1056/NEJMoa1208394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Eyles DE, Hoo CC, Warren M, Sandosham AA. Plasmodium falciparum resistant to chloroquine in Cambodia. Am J Trop Med Hyg. 1963;12:840–843. doi: 10.4269/ajtmh.1963.12.840. [DOI] [PubMed] [Google Scholar]
- 4.Young MD, Contacos PG, Stitcher JE, Millar JW. Drug resistance in Plasmodium falciparum from Thailand. Am J Trop Med Hyg. 1963;12:305–314. doi: 10.4269/ajtmh.1963.12.305. [DOI] [PubMed] [Google Scholar]
- 5.Hofler W. Sulfadoxine-pyrimethamine resistant falciparum malaria from Cambodia [in German] Dtsch Med Wochenschr. 1980;105:350–351. [PubMed] [Google Scholar]
- 6.Hurwitz ES, Johnson D, Campbell CC. Resistance of Plasmodium falciparum malaria to sulfadoxine-pyrimethamine (‘Fansidar’) in a refugee camp in Thailand. Lancet. 1981;1:1068–1070. doi: 10.1016/s0140-6736(81)92239-x. [DOI] [PubMed] [Google Scholar]
- 7.Boudreau EF, Webster HK, Pavanand K, Thosingha L. Type II mefloquine resistance in Thailand. Lancet. 1982;2:1335. doi: 10.1016/s0140-6736(82)91532-x. [DOI] [PubMed] [Google Scholar]
- 8.Smithuis FM, van Woensel JB, Nordlander E, Vantha WS, ter Kuile FO. Comparison of two mefloquine regimens for treatment of Plasmodium falciparum malaria on the northeastern Thai-Cambodian border. Antimicrob Agents Chemother. 1993;37:1977–1981. doi: 10.1128/aac.37.9.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Denis MB, Tsuyuoka R, Poravuth Y, Narann TS, Seila S, Lim C, Incardona S, Lim P, Sem R, Socheat D, Christophel EM, Ringwald P. Surveillance of the efficacy of artesunate and mefloquine combination for the treatment of uncomplicated falciparum malaria in Cambodia. Trop Med Int Health. 2006;11:1360–1366. doi: 10.1111/j.1365-3156.2006.01690.x. [DOI] [PubMed] [Google Scholar]
- 10.Noedl H, Se Y, Schaecher K, Smith BL, Socheat D, Fukuda MM. Evidence of artemisinin-resistant malaria in western Cambodia. N Engl J Med. 2008;359:2619–2620. doi: 10.1056/NEJMc0805011. [DOI] [PubMed] [Google Scholar]
- 11.Dondorp AM, Nosten F, Yi P, Das D, Phyo AP, Tarning J, Lwin KM, Ariey F, Hanpithakpong W, Lee SJ, Ringwald P, Silamut K, Imwong M, Chotivanich K, Lim P, Herdman T, An SS, Yeung S, Singhasivanon P, Day NP, Lindegardh N, Socheat D, White NJ. Artemisinin resistance in Plasmodium falciparum malaria. N Engl J Med. 2009;361:455–467. doi: 10.1056/NEJMoa0808859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Saunders DL, Vanachayangkul P, Lon C. Dihydroartemisinin-piperaquine failure in Cambodia. N Engl J Med. 2014;371:484–485. doi: 10.1056/NEJMc1403007. [DOI] [PubMed] [Google Scholar]
- 13.Leang R, Taylor WR, Bouth DM, Song L, Tarning J, Char MC, Kim S, Witkowski B, Duru V, Domergue A, Khim N, Ringwald P, Menard D. Evidence of falciparum malaria multidrug resistance to artemisinin and piperaquine in western Cambodia: dihydroartemisinin-piperaquine open-label multicenter clinical assessment. Antimicrob Agents Chemother. 2015;59:4719–4726. doi: 10.1128/AAC.00835-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mita T, Tanabe K, Kita K. Spread and evolution of Plasmodium falciparum drug resistance. Parasitol Int. 2009;58:201–209. doi: 10.1016/j.parint.2009.04.004. [DOI] [PubMed] [Google Scholar]
- 15.Roper C, Pearce R, Nair S, Sharp B, Nosten F, Anderson T. Intercontinental spread of pyrimethamine-resistant malaria. Science. 2004;305:1124. doi: 10.1126/science.1098876. [DOI] [PubMed] [Google Scholar]
- 16.Trape JF. The public health impact of chloroquine resistance in Africa. Am J Trop Med Hyg. 2001;64:12–17. doi: 10.4269/ajtmh.2001.64.12. [DOI] [PubMed] [Google Scholar]
- 17.Trape JF, Pison G, Preziosi MP, Enel C, Desgrees du Lou A, Delaunay V, Samb B, Lagarde E, Molez JF, Simondon F. Impact of chloroquine resistance on malaria mortality. C R Acad Sci III. 1998;321:689–697. doi: 10.1016/s0764-4469(98)80009-7. [DOI] [PubMed] [Google Scholar]
- 18.Muheki C, McIntyre D, Barnes KI. Artemisinin-based combination therapy reduces expenditure on malaria treatment in KwaZulu Natal, South Africa. Trop Med Int Health. 2004;9:959–966. doi: 10.1111/j.1365-3156.2004.01292.x. [DOI] [PubMed] [Google Scholar]
- 19.World Health Organization . Antimalarial Drug Combination Therapy. Geneva, Switzerland: World Health Organization; 2001. [Google Scholar]
- 20.Amaratunga C, Lim P, Suon S, Sreng S, Mao S, Sopha C, Sam B, Dek D, Try V, Amato R, Blessborn D, Song L, Tullo GS, Fay MP, Anderson JM, Tarning J, Fairhurst RM. Dihydroartemisinin-piperaquine resistance in Plasmodium falciparum malaria in Cambodia: a multisite prospective cohort study. Lancet Infect Dis. 2016;16:357–365. doi: 10.1016/S1473-3099(15)00487-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hien TT, Thuy-Nhien NT, Phu NH, Boni MF, Thanh NV, Nha-Ca NT, Thai le H, Thai CQ, Toi PV, Thuan PD, Long le T, Dong le T, Merson L, Dolecek C, Stepniewska K, Ringwald P, White NJ, Farrar J, Wolbers M. In vivo susceptibility of Plasmodium falciparum to artesunate in Binh Phuoc Province, Vietnam. Malar J. 2012;11:355. doi: 10.1186/1475-2875-11-355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kyaw MP, Nyunt MH, Chit K, Aye MM, Aye KH, Aye MM, Lindegardh N, Tarning J, Imwong M, Jacob CG, Rasmussen C, Perin J, Ringwald P, Nyunt MM. Reduced susceptibility of Plasmodium falciparum to artesunate in southern Myanmar. PLoS One. 2013;8:e57689. doi: 10.1371/journal.pone.0057689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Phyo AP, Nkhoma S, Stepniewska K, Ashley EA, Nair S, McGready R, ler Moo C, Al-Saai S, Dondorp AM, Lwin KM, Singhasivanon P, Day NP, White NJ, Anderson TJ, Nosten F. Emergence of artemisinin-resistant malaria on the western border of Thailand: a longitudinal study. Lancet. 2012;379:1960–1966. doi: 10.1016/S0140-6736(12)60484-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang F, Takala-Harrison S, Jacob CG, Liu H, Sun X, Yang H, Nyunt MM, Adams M, Zhou S, Xia Z, Ringwald P, Bustos MD, Tang L, Plowe CV. A single mutation in K13 predominates in southern China and is associated with delayed clearance of Plasmodium falciparum following artemisinin treatment. J Infect Dis. 2015;212:1629–1635. doi: 10.1093/infdis/jiv249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Flegg JA, Guerin PJ, White NJ, Stepniewska K. Standardizing the measurement of parasite clearance in falciparum malaria: the parasite clearance estimator. Malar J. 2011;10:339. doi: 10.1186/1475-2875-10-339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.White LJ, Flegg JA, Phyo AP, Wiladpai-ngern JH, Bethell D, Plowe C, Anderson T, Nkhoma S, Nair S, Tripura R, Stepniewska K, Pan-Ngum W, Silamut K, Cooper BS, Lubell Y, Ashley EA, Nguon C, Nosten F, White NJ, Dondorp AM. Defining the in vivo phenotype of artemisinin-resistant falciparum malaria: a modelling approach. PLoS Med. 2015;12:e1001823. doi: 10.1371/journal.pmed.1001823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ashley EA, Dhorda M, Fairhurst RM, Amaratunga C, Lim P, Suon S, Sreng S, Anderson JM, Mao S, Sam B, Sopha C, Chuor CM, Nguon C, Sovannaroth S, Pukrittayakamee S, Jittamala P, Chotivanich K, Chutasmit K, Suchatsoonthorn C, Runcharoen R, Hien TT, Thuy-Nhien NT, Thanh NV, Phu NH, Htut Y, Han KT, Aye KH, Mokuolu OA, Olaosebikan RR, Folaranmi OO, Mayxay M, Khanthavong M, Hongvanthong B, Newton PN, Onyamboko MA, Fanello CI, Tshefu AK, Mishra N, Valecha N, Phyo AP, Nosten F, Yi P, Tripura R, Borrmann S, Bashraheil M, Peshu J, Faiz MA, Ghose A, Hossain MA, Samad R, Rahman MR, Hasan MM, Islam A, Miotto O, Amato R, MacInnis B, Stalker J, Kwiatkowski DP, Bozdech Z, Jeeyapant A, Cheah PY, Sakulthaew T, Chalk J, Intharabut B, Silamut K, Lee SJ, Vihokhern B, Kunasol C, Imwong M, Tarning J, Taylor WJ, Yeung S, Woodrow CJ, Flegg JA, Das D, Smith J, Venkatesan M, Plowe CV, Stepniewska K, Guerin PJ, Dondorp AM, Day NP, White NJ. Tracking Resistance to Artemisinin Collaboration Spread of artemisinin resistance in Plasmodium falciparum malaria. N Engl J Med. 2014;371:411–423. doi: 10.1056/NEJMoa1314981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maiga AW, Fofana B, Sagara I, Dembele D, Dara A, Traore OB, Toure S, Sanogo K, Dama S, Sidibe B, Kone A, Thera MA, Plowe CV, Doumbo OK, Djimde AA. No evidence of delayed parasite clearance after oral artesunate treatment of uncomplicated falciparum malaria in Mali. Am J Trop Med Hyg. 2012;87:23–28. doi: 10.4269/ajtmh.2012.12-0058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Beshir KB, Sutherland CJ, Sawa P, Drakeley CJ, Okell L, Mweresa CK, Omar SA, Shekalaghe SA, Kaur H, Ndaro A, Chilongola J, Schallig HD, Sauerwein RW, Hallett RL, Bousema T. Residual Plasmodium falciparum parasitemia in Kenyan children after artemisinin-combination therapy is associated with increased transmission to mosquitoes and parasite recurrence. J Infect Dis. 2013;208:2017–2024. doi: 10.1093/infdis/jit431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Witkowski B, Khim N, Chim P, Kim S, Ke S, Kloeung N, Chy S, Duong S, Leang R, Ringwald P, Dondorp AM, Tripura R, Benoit-Vical F, Berry A, Gorgette O, Ariey F, Barale JC, Mercereau-Puijalon O, Menard D. Reduced artemisinin susceptibility of Plasmodium falciparum ring stages in western Cambodia. Antimicrob Agents Chemother. 2013;57:914–923. doi: 10.1128/AAC.01868-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Klonis N, Xie SC, McCaw JM, Crespo-Ortiz MP, Zaloumis SG, Simpson JA, Tilley L. Altered temporal response of malaria parasites determines differential sensitivity to artemisinin. Proc Natl Acad Sci USA. 2013;110:5157–5162. doi: 10.1073/pnas.1217452110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mok S, Imwong M, Mackinnon MJ, Sim J, Ramadoss R, Yi P, Mayxay M, Chotivanich K, Liong KY, Russell B, Socheat D, Newton PN, Day NP, White NJ, Preiser PR, Nosten F, Dondorp AM, Bozdech Z. Artemisinin resistance in Plasmodium falciparum is associated with an altered temporal pattern of transcription. BMC Genomics. 2011;12:391. doi: 10.1186/1471-2164-12-391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Saralamba S, Pan-Ngum W, Maude RJ, Lee SJ, Tarning J, Lindegardh N, Chotivanich K, Nosten F, Day NP, Socheat D, White NJ, Dondorp AM, White LJ. Intrahost modeling of artemisinin resistance in Plasmodium falciparum. Proc Natl Acad Sci USA. 2011;108:397–402. doi: 10.1073/pnas.1006113108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Amaratunga C, Witkowski B, Dek D, Try V, Khim N, Miotto O, Menard D, Fairhurst RM. Plasmodium falciparum founder populations in western Cambodia have reduced artemisinin sensitivity in vitro. Antimicrob Agents Chemother. 2014;58:4935–4937. doi: 10.1128/AAC.03055-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Witkowski B, Lelievre J, Barragan MJ, Laurent V, Su XZ, Berry A, Benoit-Vical F. Increased tolerance to artemisinin in Plasmodium falciparum is mediated by a quiescence mechanism. Antimicrob Agents Chemother. 2010;54:1872–1877. doi: 10.1128/AAC.01636-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cheeseman IH, Miller BA, Nair S, Nkhoma S, Tan A, Tan JC, Al Saai S, Phyo AP, Moo CL, Lwin KM, McGready R, Ashley E, Imwong M, Stepniewska K, Yi P, Dondorp AM, Mayxay M, Newton PN, White NJ, Nosten F, Ferdig MT, Anderson TJ. A major genome region underlying artemisinin resistance in malaria. Science. 2012;336:79–82. doi: 10.1126/science.1215966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Miotto O, Almagro-Garcia J, Manske M, Macinnis B, Campino S, Rockett KA, Amaratunga C, Lim P, Suon S, Sreng S, Anderson JM, Duong S, Nguon C, Chuor CM, Saunders D, Se Y, Lon C, Fukuda MM, Amenga-Etego L, Hodgson AV, Asoala V, Imwong M, Takala-Harrison S, Nosten F, Su XZ, Ringwald P, Ariey F, Dolecek C, Hien TT, Boni MF, Thai CQ, Amambua-Ngwa A, Conway DJ, Djimdé AA, Doumbo OK, Zongo I, Ouedraogo JB, Alcock D, Drury E, Auburn S, Koch O, Sanders M, Hubbart C, Maslen G, Ruano-Rubio V, Jyothi D, Miles A, O'Brien J, Gamble C, Oyola SO, Rayner JC, Newbold CI, Berriman M, Spencer CC, McVean G, Day NP, White NJ, Bethell D, Dondorp AM, Plowe CV, Fairhurst RM, Kwiatkowski DP. Multiple populations of artemisinin-resistant Plasmodium falciparum in Cambodia. Nat Genet. 2013;45:648–655. doi: 10.1038/ng.2624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ariey F, Witkowski B, Amaratunga C, Beghain J, Langlois AC, Khim N, Kim S, Duru V, Bouchier C, Ma L, Lim P, Leang R, Duong S, Sreng S, Suon S, Chuor CM, Bout DM, Menard S, Rogers WO, Genton B, Fandeur T, Miotto O, Ringwald P, Le Bras J, Berry A, Barale JC, Fairhurst RM, Benoit-Vical F, Mercereau-Puijalon O, Menard D. A molecular marker of artemisinin-resistant Plasmodium falciparum malaria. Nature. 2014;505:50–55. doi: 10.1038/nature12876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Straimer J, Gnadig NF, Witkowski B, Amaratunga C, Duru V, Ramadani AP, Dacheux M, Khim N, Zhang L, Lam S, Gregory PD, Urnov FD, Mercereau-Puijalon O, Benoit-Vical F, Fairhurst RM, Menard D, Fidock DA. Drug resistance. K13-propeller mutations confer artemisinin resistance in Plasmodium falciparum clinical isolates. Science. 2015;347:428–431. doi: 10.1126/science.1260867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ghorbal M, Gorman M, Macpherson CR, Martins RM, Scherf A, Lopez-Rubio JJ. Genome editing in the human malaria parasite Plasmodium falciparum using the CRISPR-Cas9 system. Nat Biotechnol. 2014;32:819–821. doi: 10.1038/nbt.2925. [DOI] [PubMed] [Google Scholar]
- 41.Nyunt MH, Hlaing T, Oo HW, Tin-Oo LL, Phway HP, Wang B, Zaw NN, Han SS, Tun T, San KK, Kyaw MP, Han ET. Molecular assessment of artemisinin resistance markers, polymorphisms in the K13 propeller, and a multidrug-resistance gene in the eastern and western border areas of Myanmar. Clin Infect Dis. 2014;60:1208–1215. doi: 10.1093/cid/ciu1160. [DOI] [PubMed] [Google Scholar]
- 42.Takala-Harrison S, Jacob CG, Arze C, Cummings MP, Silva JC, Dondorp AM, Fukuda MM, Hien TT, Mayxay M, Noedl H, Nosten F, Kyaw MP, Nhien NT, Imwong M, Bethell D, Se Y, Lon C, Tyner SD, Saunders DL, Ariey F, Mercereau-Puijalon O, Menard D, Newton PN, Khanthavong M, Hongvanthong B, Starzengruber P, Fuehrer HP, Swoboda P, Khan WA, Phyo AP, Nyunt MM, Nyunt MH, Brown TS, Adams M, Pepin CS, Bailey J, Tan JC, Ferdig MT, Clark TG, Miotto O, MacInnis B, Kwiatkowski DP, White NJ, Ringwald P, Plowe CV. Independent emergence of artemisinin resistance mutations among plasmodium falciparum in southeast Asia. J Infect Dis. 2015;211:670–679. doi: 10.1093/infdis/jiu491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Talundzic E, Okoth SA, Congpuong K, Plucinski MM, Morton L, Goldman IF, Kachur PS, Wongsrichanalai C, Satimai W, Barnwell JW, Udhayakumar V. Selection and spread of artemisinin-resistant alleles in Thailand prior to the global artemisinin resistance containment campaign. PLoS Pathog. 2015;11:e1004789. doi: 10.1371/journal.ppat.1004789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tun KM, Imwong M, Lwin KM, Win AA, Hlaing TM, Hlaing T, Lin K, Kyaw MP, Plewes K, Faiz MA, Dhorda M, Cheah PY, Pukrittayakamee S, Ashley EA, Anderson TJ, Nair S, McDew-White M, Flegg JA, Grist EP, Guerin P, Maude RJ, Smithuis F, Dondorp AM, Day NP, Nosten F, White NJ, Woodrow CJ. Spread of artemisinin-resistant Plasmodium falciparum in Myanmar: a cross-sectional survey of the K13 molecular marker. Lancet Infect Dis. 2015;15:415–421. doi: 10.1016/S1473-3099(15)70032-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Huang F, Takala-Harrison S, Jacob CG, Liu H, Sun X, Yang H, Nyunt MM, Adams M, Zhou S, Xia Z, Ringwald P, Bustos MD, Tang L, Plowe CV. A single mutation in K13 predominates in southern China and is associated with delayed clearance of Plasmodium falciparum following artemisinin treatment. J Infect Dis. 2015;212:1629–1635. doi: 10.1093/infdis/jiv249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang Z, Shrestha S, Li X, Miao J, Yuan L, Cabrera M, Grube C, Yang Z, Cui L. Prevalence of K13-propeller polymorphisms in Plasmodium falciparum from China-Myanmar border in 2007–2012. Malar J. 2015;14:168. doi: 10.1186/s12936-015-0672-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang Z, Wang Y, Cabrera M, Zhang Y, Gupta B, Wu Y, Kemirembe K, Hu Y, Liang X, Brashear A, Shrestha S, Li X, Miao J, Sun X, Yang Z, Cui L. Artemisinin resistance at the China-Myanmar border and association with mutations in the K13 propeller gene. Antimicrob Agents Chemother. 2015;59:6952–6959. doi: 10.1128/AAC.01255-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ye R, Hu D, Zhang Y, Huang Y, Sun X, Wang J, Chen X, Zhou H, Zhang D, Mungthin M, Pan W. Distinctive origin of artemisinin-resistant Plasmodium falciparum on the China-Myanmar border. Sci Rep. 2016;6:20100. doi: 10.1038/srep20100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mishra N, Prajapati SK, Kaitholia K, Bharti RS, Srivastava B, Phookan S, Anvikar AR, Dev V, Sonal GS, Dhariwal AC, White NJ, Valecha N. Surveillance of artemisinin resistance in Plasmodium falciparum in India using the kelch13 molecular marker. Antimicrob Agents Chemother. 2015;59:2548–2553. doi: 10.1128/AAC.04632-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mohon AN, Alam MS, Bayih AG, Folefoc A, Shahinas D, Haque R, Pillai DR. Mutations in Plasmodium falciparum K13 propeller gene from Bangladesh (2009–2013) Malar J. 2014;13:431. doi: 10.1186/1475-2875-13-431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Conrad MD, Bigira V, Kapisi J, Muhindo M, Kamya MR, Havlir DV, Dorsey G, Rosenthal PJ. Polymorphisms in K13 and falcipain-2 associated with artemisinin resistance are not prevalent in Plasmodium falciparum isolated from Ugandan children. PLoS One. 2014;9:e105690. doi: 10.1371/journal.pone.0105690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cooper RA, Conrad MD, Watson QD, Huezo SJ, Ninsiima H, Tumwebaze P, Nsobya SL, Rosenthal PJ. Lack of artemisinin resistance in Plasmodium falciparum in Uganda based on parasitological and molecular assays. Antimicrob Agents Chemother. 2015;59:5061–5064. doi: 10.1128/AAC.00921-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Escobar C, Pateira S, Lobo E, Lobo L, Teodosio R, Dias F, Fernandes N, Arez AP, Varandas L, Nogueira F. Polymorphisms in Plasmodium falciparum K13-propeller in Angola and Mozambique after the introduction of the ACTs. PLoS One. 2015;10:e0119215. doi: 10.1371/journal.pone.0119215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Isozumi R, Uemura H, Kimata I, Ichinose Y, Logedi J, Omar AH, Kaneko A. Novel mutations in K13 propeller gene of artemisinin-resistant Plasmodium falciparum. Emerg Infect Dis. 2015;21:490–492. doi: 10.3201/eid2103.140898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kamau E, Campino S, Amenga-Etego L, Drury E, Ishengoma D, Johnson K, Mumba D, Kekre M, Yavo W, Mead D, Bouyou-Akotet M, Apinjoh T, Golassa L, Randrianarivelojosia M, Andagalu B, Maiga-Ascofare O, Amambua-Ngwa A, Tindana P, Ghansah A, MacInnis B, Kwiatkowski D, Djimde AA. K13-propeller polymorphisms in Plasmodium falciparum parasites from sub-Saharan Africa. J Infect Dis. 2014;211:1352–1355. doi: 10.1093/infdis/jiu608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ouattara A, Kone A, Adams M, Fofana B, Maiga AW, Hampton S, Coulibaly D, Thera MA, Diallo N, Dara A, Sagara I, Gil JP, Bjorkman A, Takala-Harrison S, Doumbo OK, Plowe CV, Djimde AA. Polymorphisms in the K13-propeller gene in artemisinin-susceptible Plasmodium falciparum parasites from Bougoula-Hameau and Bandiagara, Mali. Am J Trop Med Hyg. 2015;92:1202–1206. doi: 10.4269/ajtmh.14-0605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Taylor SM, Parobek CM, DeConti DK, Kayentao K, Coulibaly SO, Greenwood BM, Tagbor H, Williams J, Bojang K, Njie F, Desai M, Kariuki S, Gutman J, Mathanga DP, Martensson A, Ngasala B, Conrad MD, Rosenthal PJ, Tshefu AK, Moormann AM, Vulule JM, Doumbo OK, Ter Kuile FO, Meshnick SR, Bailey JA, Juliano JJ. Absence of putative artemisinin resistance mutations among Plasmodium falciparum in sub-Saharan Africa: a molecular epidemiologic study. J Infect Dis. 2015;211:680–688. doi: 10.1093/infdis/jiu467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Torrentino-Madamet M, Fall B, Benoit N, Camara C, Amalvict R, Fall M, Dionne P, Ba Fall K, Nakoulima A, Diatta B, Dieme Y, Menard D, Wade B, Pradines B. Limited polymorphisms in k13 gene in Plasmodium falciparum isolates from Dakar, Senegal in 2012–2013. Malar J. 2015;13:472. doi: 10.1186/1475-2875-13-472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ménard D, Khim N, Beghain J, Adegnika AA, Shafiul-Alam M, Amodu O, Rahim-Awab G, Barnadas C, Berry A, Boum Y, Bustos MD, Cao J, Chen JH, Collet L, Cui L, Thakur GD, Dieye A, Djallé D, Dorkenoo MA, Eboumbou-Moukoko CE, Espino FE, Fandeur T, Ferreira-da-Cruz MF, Fola AA, Fuehrer HP, Hassan AM, Herrera S, Hongvanthong B, Houzé S, Ibrahim ML, Jahirul-Karim M, Jiang L, Kano S, Ali-Khan W, Khanthavong M, Kremsner PG, Lacerda M, Leang R, Leelawong M, Li M, Lin K, Mazarati JB, Ménard S, Morlais I, Muhindo-Mavoko H, Musset L, Na-Bangchang K, Nambozi M, Niaré K, Noedl H, Ouédraogo JB, Pillai DR, Pradines B, Quang-Phuc B, Ramharter M, Randrianarivelojosia M, Sattabongkot J, Sheikh-Omar A, Silué KD, Sirima SB, Sutherland C, Syafruddin D, Tahar R, Tang LH, Touré OA, Tshibangu-wa-Tshibangu P, Vigan-Womas I, Warsame M, Wini L, Zakeri S, Kim S, Eam R, Berne L, Khean C, Chy S, Ken M, Loch K, Canier L, Duru V, Legrand E, Barale JC, Stokes B, Straimer J, Witkowski B, Fidock DA, Rogier C, Ringwald P, Ariey F, Mercereau-Puijalon O. KARMA Consortium A worldwide map of plasmodium falciparum K13-propeller polymorphisms. N Engl J Med. 2016;374:2453–2464. doi: 10.1056/NEJMoa1513137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Muwanguzi J, Henriques G, Sawa P, Bousema T, Sutherland CJ, Beshir KB. Lack of K13 mutations in Plasmodium falciparum persisting after artemisinin combination therapy treatment of Kenyan children. Malar J. 2016;15:36. doi: 10.1186/s12936-016-1095-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Miotto O, Amato R, Ashley EA, MacInnis B, Almagro-Garcia J, Amaratunga C, Lim P, Mead D, Oyola SO, Dhorda M, Imwong M, Woodrow C, Manske M, Stalker J, Drury E, Campino S, Amenga-Etego L, Thanh TN, Tran HT, Ringwald P, Bethell D, Nosten F, Phyo AP, Pukrittayakamee S, Chotivanich K, Chuor CM, Nguon C, Suon S, Sreng S, Newton PN, Mayxay M, Khanthavong M, Hongvanthong B, Htut Y, Han KT, Kyaw MP, Faiz MA, Fanello CI, Onyamboko M, Mokuolu OA, Jacob CG, Takala-Harrison S, Plowe CV, Day NP, Dondorp AM, Spencer CC, McVean G, Fairhurst RM, White NJ, Kwiatkowski DP. Genetic architecture of artemisinin-resistant Plasmodium falciparum. Nat Genet. 2015;47:226–234. doi: 10.1038/ng.3189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mbengue A, Bhattacharjee S, Pandharkar T, Liu H, Estiu G, Stahelin RV, Rizk SS, Njimoh DL, Ryan Y, Chotivanich K, Nguon C, Ghorbal M, Lopez-Rubio JJ, Pfrender M, Emrich S, Mohandas N, Dondorp AM, Wiest O, Haldar K. A molecular mechanism of artemisinin resistance in Plasmodium falciparum malaria. Nature. 2015;520:683–687. doi: 10.1038/nature14412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Adams J, Kelso R, Cooley L. The kelch repeat superfamily of proteins: propellers of cell function. Trends Cell Biol. 2000;10:17–24. doi: 10.1016/s0962-8924(99)01673-6. [DOI] [PubMed] [Google Scholar]
- 64.Itoh K, Wakabayashi N, Katoh Y, Ishii T, Igarashi K, Engel JD, Yamamoto M. Keap1 represses nuclear activation of antioxidant responsive elements by Nrf2 through binding to the amino-terminal Neh2 domain. Genes Dev. 1999;13:76–86. doi: 10.1101/gad.13.1.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Li X, Zhang D, Hannink M, Beamer LJ. Crystal structure of the Kelch domain of human Keap1. J Biol Chem. 2004;279:54750–54758. doi: 10.1074/jbc.M410073200. [DOI] [PubMed] [Google Scholar]
- 66.Prag S, Adams JC. Molecular phylogeny of the kelch-repeat superfamily reveals an expansion of BTB/kelch proteins in animals. BMC Bioinformatics. 2003;4:42. doi: 10.1186/1471-2105-4-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhang DD, Hannink M. Distinct cysteine residues in Keap1 are required for Keap1-dependent ubiquitination of Nrf2 and for stabilization of Nrf2 by chemopreventive agents and oxidative stress. Mol Cell Biol. 2003;23:8137–8151. doi: 10.1128/MCB.23.22.8137-8151.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bauer AK, Hill T, 3rd, Alexander CM. The involvement of NRF2 in lung cancer. Oxid Med Cell Longev. 2013;2013:746432. doi: 10.1155/2013/746432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rojanawatsirivej C, Vijaykadga S, Amklad I, Wilairatna P, Looareesuwan S. Monitoring the therapeutic efficacy of antimalarials against uncomplicated falciparum malaria in Thailand. Southeast Asian J Trop Med Public Health. 2003;34:536–541. [PubMed] [Google Scholar]
- 70.Nelson AL, Purfield A, McDaniel P, Uthaimongkol N, Buathong N, Sriwichai S, Miller RS, Wongsrichanalai C, Meshnick SR. pfmdr1 genotyping and in vivo mefloquine resistance on the Thai–Myanmar border. Am J Trop Med Hyg. 2005;72:586–592. [PubMed] [Google Scholar]
- 71.Price RN, Uhlemann AC, Brockman A, McGready R, Ashley E, Phaipun L, Patel R, Laing K, Looareesuwan S, White NJ, Nosten F, Krishna S. Mefloquine resistance in Plasmodium falciparum and increased pfmdr1 gene copy number. Lancet. 2004;364:438–447. doi: 10.1016/S0140-6736(04)16767-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Alker AP, Lim P, Sem R, Shah NK, Yi P, Bouth DM, Tsuyuoka R, Maguire JD, Fandeur T, Ariey F, Wongsrichanalai C, Meshnick SR. Pfmdr1 and in vivo resistance to artesunate-mefloquine in falciparum malaria on the Cambodian-Thai border. Am J Trop Med Hyg. 2007;76:641–647. [PubMed] [Google Scholar]
- 73.Denis MB, Tsuyuoka R, Lim P, Lindegardh N, Yi P, Top SN, Socheat D, Fandeur T, Annerberg A, Christophel EM, Ringwald P. Efficacy of artemether-lumefantrine for the treatment of uncomplicated falciparum malaria in northwest Cambodia. Trop Med Int Health. 2006;11:1800–1807. doi: 10.1111/j.1365-3156.2006.01739.x. [DOI] [PubMed] [Google Scholar]
- 74.Janssens B, van Herp M, Goubert L, Chan S, Uong S, Nong S, Socheat D, Brockman A, Ashley EA, Van Damme W. A randomized open study to assess the efficacy and tolerability of dihydroartemisinin-piperaquine for the treatment of uncomplicated falciparum malaria in Cambodia. Trop Med Int Health. 2007;12:251–259. doi: 10.1111/j.1365-3156.2006.01786.x. [DOI] [PubMed] [Google Scholar]
- 75.WHO . National Treatment Guidelines for Malaria in the Kingdom of Cambodia. Geneva, Switzerland: World Health Organization; 2012. [Google Scholar]
- 76.Leang R, Canavati SE, Khim N, Vestergaard LS, Borghini Fuhrer I, Kim S, Denis MB, Heng P, Tol B, Huy R, Duparc S, Dondorp AM, Menard D, Ringwald P. Efficacy and safety of pyronaridine-artesunate for treatment of uncomplicated Plasmodium falciparum malaria in western Cambodia. Antimicrob Agents Chemother. 2016;60:3884–3890. doi: 10.1128/AAC.00039-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Rueangweerayut R, Phyo AP, Uthaisin C, Poravuth Y, Binh TQ, Tinto H, Penali LK, Valecha N, Tien NT, Abdulla S, Borghini-Fuhrer I, Duparc S, Shin CS, Fleckenstein L. Pyronaridine-Artesunate Study Team Pyronaridine-artesunate versus mefloquine plus artesunate for malaria. N Engl J Med. 2012;366:1298–1309. doi: 10.1056/NEJMoa1007125. [DOI] [PubMed] [Google Scholar]
- 78.Duparc S, Borghini-Fuhrer I, Craft CJ, Arbe-Barnes S, Miller RM, Shin CS, Fleckenstein L. Safety and efficacy of pyronaridine-artesunate in uncomplicated acute malaria: an integrated analysis of individual patient data from six randomized clinical trials. Malar J. 2013;12:70. doi: 10.1186/1475-2875-12-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kayentao K, Doumbo OK, Penali LK, Offianan AT, Bhatt KM, Kimani J, Tshefu AK, Kokolomami JH, Ramharter M, de Salazar PM, Tiono AB, Ouedraogo A, Bustos MD, Quicho F, Borghini-Fuhrer I, Duparc S, Shin CS, Fleckenstein L. Pyronaridine-artesunate granules versus artemether-lumefantrine crushed tablets in children with Plasmodium falciparum malaria: a randomized controlled trial. Malar J. 2012;11:364. doi: 10.1186/1475-2875-11-364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ramharter M, Kurth F, Schreier AC, Nemeth J, Glasenapp I, Belard S, Schlie M, Kammer J, Koumba PK, Cisse B, Mordmuller B, Lell B, Issifou S, Oeuvray C, Fleckenstein L, Kremsner PG. Fixed-dose pyronaridine-artesunate combination for treatment of uncomplicated falciparum malaria in pediatric patients in Gabon. J Infect Dis. 2008;198:911–919. doi: 10.1086/591096. [DOI] [PubMed] [Google Scholar]
- 81.Tshefu AK, Gaye O, Kayentao K, Thompson R, Bhatt KM, Sesay SS, Bustos DG, Tjitra E, Bedu-Addo G, Borghini-Fuhrer I, Duparc S, Shin CS, Fleckenstein L. Pyronaridine-artesunate Study Team Efficacy and safety of a fixed-dose oral combination of pyronaridine-artesunate compared with artemether-lumefantrine in children and adults with uncomplicated Plasmodium falciparum malaria: a randomised non-inferiority trial. Lancet. 2010;375:1457–1467. doi: 10.1016/S0140-6736(10)60322-4. [DOI] [PubMed] [Google Scholar]
- 82.Duru V, Khim N, Leang R, Kim S, Domergue A, Kloeung N, Ke S, Chy S, Eam R, Khean C, Loch K, Ken M, Lek D, Beghain J, Ariey F, Guerin PJ, Huy R, Mercereau-Puijalon O, Witkowski B, Menard D. Plasmodium falciparum dihydroartemisinin-piperaquine failures in Cambodia are associated with mutant K13 parasites presenting high survival rates in novel piperaquine in vitro assays: retrospective and prospective investigations. BMC Med. 2015;13:305. doi: 10.1186/s12916-015-0539-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lon C, Manning JE, Vanachayangkul P, So M, Sea D, Se Y, Gosi P, Lanteri C, Chaorattanakawee S, Sriwichai S, Chann S, Kuntawunginn W, Buathong N, Nou S, Walsh DS, Tyner SD, Juliano JJ, Lin J, Spring M, Bethell D, Kaewkungwal J, Tang D, Chuor CM, Satharath P, Saunders D. Efficacy of two versus three-day regimens of dihydroartemisinin-piperaquine for uncomplicated malaria in military personnel in northern Cambodia: an open-label randomized trial. PLoS One. 2014;9:e93138. doi: 10.1371/journal.pone.0093138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Spring MD, Lin JT, Manning JE, Vanachayangkul P, Somethy S, Bun R, Se Y, Chann S, Ittiverakul M, Sia-Ngam P, Kuntawunginn W, Arsanok M, Buathong N, Chaorattanakawee S, Gosi P, Ta-Aksorn W, Chanarat N, Sundrakes S, Kong N, Heng TK, Nou S, Teja-Isavadharm P, Pichyangkul S, Phann ST, Balasubramanian S, Juliano JJ, Meshnick SR, Chour CM, Prom S, Lanteri CA, Lon C, Saunders DL. Dihydroartemisinin-piperaquine failure associated with a triple mutant including kelch13 C580Y in Cambodia: an observational cohort study. Lancet Infect Dis. 2015;15:683–691. doi: 10.1016/S1473-3099(15)70049-6. [DOI] [PubMed] [Google Scholar]
- 85.Eastman RT, Dharia NV, Winzeler EA, Fidock DA. Piperaquine resistance is associated with a copy number variation on chromosome 5 in drug-pressured Plasmodium falciparum parasites. Antimicrob Agents Chemother. 2011;55:3908–3916. doi: 10.1128/AAC.01793-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lim P, Dek D, Try V, Eastman RT, Chy S, Sreng S, Suon S, Mao S, Sopha C, Sam B, Ashley EA, Miotto O, Dondorp AM, White NJ, Su XZ, Char MC, Anderson JM, Amaratunga C, Menard D, Fairhurst RM. Ex vivo susceptibility of Plasmodium falciparum to antimalarial drugs in western, northern, and eastern Cambodia, 2011–2012: Association with molecular markers. Antimicrob Agents Chemother. 2013;57:5277–5283. doi: 10.1128/AAC.00687-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Veiga MI, Ferreira PE, Malmberg M, Jornhagen L, Bjorkman A, Nosten F, Gil JP. pfmdr1 amplification is related to increased Plasmodium falciparum in vitro sensitivity to the bisquinoline piperaquine. Antimicrob Agents Chemother. 2012;56:3615–3619. doi: 10.1128/AAC.06350-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Takala-Harrison S, Clark TG, Jacob CG, Cummings MP, Miotto O, Dondorp AM, Fukuda MM, Nosten F, Noedl H, Imwong M, Bethell D, Se Y, Lon C, Tyner SD, Saunders DL, Socheat D, Ariey F, Phyo AP, Starzengruber P, Fuehrer HP, Swoboda P, Stepniewska K, Flegg J, Arze C, Cerqueira GC, Silva JC, Ricklefs SM, Porcella SF, Stephens RM, Adams M, Kenefic LJ, Campino S, Auburn S, MacInnis B, Kwiatkowski DP, Su XZ, White NJ, Ringwald P, Plowe CV. Genetic loci associated with delayed clearance of Plasmodium falciparum following artemisinin treatment in southeast Asia. Proc Natl Acad Sci U.S.A. 2013;110:240–245. doi: 10.1073/pnas.1211205110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.van Schalkwyk DA, Sutherland CJ. Malaria resistance to non-artemisinin partner drugs: how to reACT. Lancet Infect Dis. 2015;15:621–623. doi: 10.1016/S1473-3099(15)70080-0. [DOI] [PubMed] [Google Scholar]
- 90.Pelleau S, Moss EL, Dhingra SK, Volney B, Casteras J, Gabryszewski SJ, Volkman SK, Wirth DF, Legrand E, Fidock DA, Neafsey DE, Musset L. Adaptive evolution of malaria parasites in French Guiana: reversal of chloroquine resistance by acquisition of a mutation in pfcrt. Proc Natl Acad Sci USA. 2015;112:11672–11677. doi: 10.1073/pnas.1507142112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Wells TN, Hooft van Huijsduijnen R, Van Voorhis WC. Malaria medicines: a glass half full? Nat Rev Drug Discov. 2015;14:424–442. doi: 10.1038/nrd4573. [DOI] [PubMed] [Google Scholar]