Abstract
This study aimed to compare polymerase chain reaction (PCR) with serum agglutination test (SAT) in the diagnosis of patients before and 6 months after treatment. Peripheral blood specimens from 50 patients with brucellosis (case group) and 30 subjects without brucellosis (control group) were selected and entered into the study. The diagnosis of brucellosis was established using SAT ≥ 1:160 and 2-mercaptoethanol (2-ME) ≥ 1:80 with clinical signs and symptoms compatible with brucellosis. For each case, both before treatment and 6 months after completion of therapy, SAT, 2-ME, and PCR were performed. Subjects in the control group were assessed by the same tests at the initial visit. In the case group, 50 patients (36 males, 14 females) with the mean age of 43.6 ± 14.5 years were evaluated. The mean age of the control group was 40.6 ± 14 years. Among the 50 patients whose nested PCR assays were initially positive, 43 (86%) were negative 6 months after completing treatment. Relapse occurred in five (10%) patients within 6 months after treatment and all were PCR positive. None of the patients in the control group was PCR positive. Results show that PCR seems to be highly sensitive and specific, and therefore is a useful method for both the initial diagnosis and detection of relapse or chronic brucellosis.
Introduction
Human brucellosis is a systemic infectious disease with clinical nonspecific presentation, and phases of the disease may be acute, subacute, and chronic.1,2 Although appropriate regimens of therapy are available for the treatment of the disease, the problem of treating this disease has not been completely resolved, and selection of the best treatment regimen is controversial.3,4 Since the clinical feature of brucellosis has overlapped with an extensive range of infectious and noninfectious diseases, the most reliable technique for the diagnosis of this infection is with the use of laboratory methods, but this goal has not always been successful.5 Proper diagnoses are important, especially in the form of therapeutic failure and relapsed cases. At present, the most commonly used methods for the diagnoses of brucellosis are culture and serological tests. Although the isolation of bacteria is the “gold standard,” microbial culture is often negative and depends on the culture medium, quantity of circulating bacteria, and Brucella species. Therefore, serological testing such as serum agglutination test (SAT) seems to be more effective in the diagnosis of brucellosis. At the early stage of infection, or in the presence of blocking antibodies, the sensitivity of this test is low and false-negative reactions may occur. Sometimes, this test may have a cross-reaction or false-positive reaction in samples obtained from patients with nonspecific symptoms misdiagnosed as having brucellosis.6–8 Because of these limitations, a variety of molecular methods, mainly polymerase chain reaction (PCR), has been developed for the rapid identification of organisms in the clinical setting. PCR method proved to be very simple, quick, sensitive, specific, and very useful in clinical laboratories.9,10 Even some studies showed that standard PCR not only is a useful diagnostic tool for patients with clinical signs and symptoms with negative SAT test, but also a predictive marker for the course of the disease and posttreatment follow-up, which is valuable for the early detection of relapse.11,12
Recently, PCR has been applied for the follow-up of patients with brucellosis treated with doxycycline alone or doxycycline plus gentamicin. It was illustrated that bacterial DNA persisted in the blood of several patients throughout treatment and their follow-up showed a significant clinical recovery.13,14 Different target genes, primer pair PCR techniques, and extraction procedure have been previously developed for the detection of genus Brucella.15,16 Molecular assays targeting the IS711 insertion element, which is found in multiple copies within Brucella chromosomes, also improve analytical sensitivity in clinical applications.17,18 The aim of the present study was to compare PCR with the SAT tests in the diagnosis of brucellosis in patients before and 6 months after treatment.
Materials and Methods
From June 2014 to January 2016, 50 patients with acute brucellosis who were treated and followed up for 6 months at the Department of Infectious Diseases and Tropical Medicine Research Center of Babol University of Medical Sciences, Iran, were entered into the study. The last patient's follow-up was in January 2016. Brucellosis cases (case group) and 30 individuals who were ill and had a disease other than brucellosis with similar mean age, sex, and place of residence (rural or urban) were selected as the control group. For each patient, a record was prepared and the clinical manifestations, demographic features, and outcomes of treatment were noted on it. The exclusion criteria for our research were cases with meningitis, spondylitis, endocarditis, pregnant women, and those who received antibiotics for more than 7 days.
At first, Rose Bengal test (RBT) was done for all cases, whereas SAT and 2-ME tests were performed for those with positive RBT test. For those who had clinical symptoms and signs compatible with brucellosis with SAT < 1:160, the antihuman globulin Coombs test (Coombs Wright test) was done. SAT, 2-ME, and Coombs Wright tests were carried out using the Pasteur protocol kit (Iranian Institute for Health Sciences Research. Co, Tehran, Iran). These tests were also done for the control group. Those who had SAT ≥ 1:160 and 2-ME ≥ 1:80 with clinical symptoms and signs (fever, arthralgia, myalgia, back pain, and sweating) were considered to have brucellosis. For these patients, blood culture was done with 7–10 mL of blood inoculated in Castaneda's biphasic medium and incubated at 37°C for 28 days.
For molecular assessment, 2 mL blood sample was taken from the case and control groups. The treatment regimen was gentamicin for 7 days and doxycycline for 45 days. The gentamicin dosage was 5 mg/kg at most to 240 mg/day intramuscularly and doxycycline 100 mg twice a day. For brucellosis treated cases, an additional 5 mL blood was taken 6 months after completion of the treatment for assessing molecular and serological tests. For the detection of relapse, all patients were advised to refer when clinical symptoms and signs reappeared within 6 months after treatment. Relapse was considered to be the reappearance of clinical symptoms and signs by increasing the previous serological titers, or the appearance of a new focal form highly suggestive of brucellosis.19
Isolation of DNA from clinical blood specimens.
About 2 mL of peripheral blood sample was collected in sodium citrate and was used for PCR analysis. All samples were in aliquot and stored at −20°C until tested. DNA was extracted from whole blood (200 μL) with the QIAamp DNA Blood Mini Kit (Qiagen, Hilden, Germany) in accordance with the manufacturer's instructions.
Isolation of DNA from control positive and negative strains.
To evaluate the specificity of primers and to detect the contamination during the extraction stage of the DNA, we used Brucella melitensis serotype 1 (strain 16M) and Brucella abortus B19 as the positive control (provided by Department of Bacterial Vaccines and Antigens Production, Pasteur Institute of Iran) and the standard strains of Staphylococcus aureus (ATCC 25923), Bacillus cereus (ATCC 9634), Escherichia coli (ATCC 25922), Pseudomonas aeruginosa (ATCC 8821), and Klebsiella pneumoniae (clinical isolate), as the negative control (provided by Persian Type Culture Collection of Iranian Research), which may cause similar clinical symptoms of brucellosis. DNA from these strains was isolated using purification Kit (Roche Applied Science, Mannheim, Germany) according to the manufacturer's instructions. The DNA pellet was suspended in 100 μL of 10 mmol TE buffer and stored at −20°C until required for analysis. DNA concentration and purity were assessed by reading NanoDrop 2000c (Thermo Scientific, Waltham, MA) spectrophotometer A260/A280 values and were confirmed by visualization on 1% agarose gel.
PCR primers.
Three novel specific primers were designed in this study through extensive literature and nucleotide sequence searches in the National Center for Biotechnology Information (NCBI) databases for the detection of IS711 genes. The whole sequence of Brucella chromosome 1 was analyzed and compared with all other chromosomes and the available standard Brucella strains. The primers were designed in a way that the target sequence covers all intraspecies biovars. Primer pairs were studied using the Allele ID6 software (PREMIER Biosoft, Palo Alto, CA). The DNA sequence compared with GenBank database was searched and assessed of species assigned to the genus using BLAST (basic local alignment search tool; NCBI). From solution, 10 pmol primers were used in all experiments.
Thermal cycle reactions PCR.
Subsequently, PCR assays were designed to optimize and evaluate simultaneous detection of Brucella spp., detection for targeting IS711 by detection error tradeoff (Forward: 5′-AGAATAATCCACAGAAGGTAGAG-3′) (Reverse: 5′-ATCCAAGGTCAATCCAACAC-3′). Each PCR reaction mixture contained 2.5 μL 10× amplification buffer (500 mM KCl, 100 mM Tris/HCl [pH 8.5], 1.0% Triton X-100), 0.5 μL 25 mM MgCl2, 0.3 μL each of 2.5 mM deoxynucleotide triphosphates (Fermentas, GmbH, Germany), 0.5 μL forward and reverse primers (20 ng/μL), 0.2 μL Taq DNA polymerase (5 U/μL), and 5 μL extracted DNA. After an initial denaturation step at 94°C for 4 minutes, 30 amplification cycles were performed, each consisting of 1 minute at 94°C, 1 minute at 60°C, and 1 minute at 72°C, followed by a final extension step at 72°C for 12 minutes. Then, the PCR products were analyzed using the electrophoresis technique on 1.5% agarose gel stained by 0.5 μg of ethidium bromide/mL and visualized under ultraviolet transilluminator. The test was considered positive if the signal from the amplified product and 403 base pairs (bp) was clearly visible in both samples (control and clinical strains).
Thermal cycle reactions nested PCR.
The nested PCR assays were designed Nes1 (Forward: 5′-CAAGCCGCTCATATTCAC-3′) (Reverse: 5′-CCAAGGTCAATCCAACAC-3′) and Nes2 (Forward: 5′-CGCTCGCTGCCATACTTGC-3′) (Reverse: 5′-CGCTCGCTGCCATACTTGC-3′) primers. Nested PCR reaction mixture was similar to PCR.
The first PCR amplification consisted of an initial denaturation step at 94°C for 4 minutes, followed by eight cycles of denaturation at 94°C for 1 minute, annealing temperature at 58°C for 1 minute, and elongation at 72°C for 1 minute. Next PCR reaction was carried out similar to the first except that the annealing temperature was 68°C for 27 cycles and final extension at 72°C for 10 minutes. All tests included positive controls of B. melitensis serotype 1 (strain 16M) DNA and negative controls. To detect any possible contamination during the extraction stage of the DNA, all the PCR assays included control samples from a healthy person. Moreover, to ensure the reliability of the results, all of the samples were processed in duplicate. The test was considered positive if the signal from the amplified product 293 bp was clearly visible in samples. Finally, the amplification products (PCR and nested PCR) were extracted from the bands on the gel using Gel Extraction kit (Qiagen, GmbH) and then were evaluated by sequencing.
Statistical analysis.
The collected data were statistically analyzed using SPSS program (software version 17.0). Assay sensitivity was calculated using SAT positive (titer ≥ 1/160) as the reference standard. Sensitivity, specificity, positive and negative predictive values, likelihood ratios, and 95% confidence intervals were calculated using the analyzer program.
Ethical considerations.
This study was approved by the Infectious Diseases Research Center and the Ethics Committee of Babol University of Medical Sciences (protocol no. 4018). Written informed consent was obtained from all participants before involvement in the study.
Results
Fifty patients (36 males, 14 females) with the mean age of 43.6 ± 14.5 years (range, 15–75 years) were evaluated. The mean age of the control group was 40.6 ± 14 years. Patients were mostly from rural areas and 29 (58%) had history of exposure such as contact with animals or consumption of local unpasteurized dairy products. The mean duration of symptoms before the diagnosis of brucellosis was 44 days (range, 7–60 days) in 43 (86%) of the cases. More than 80% of patients presented with fever; they had nonspecific symptoms such as arthralgia and low back pain. The clinical picture in 28 (56.5%) patients was a nonfocal febrile syndrome, and the other 22 (43.5%) had one or more focal forms: sacroiliitis, six (27.3%); knee arthritis, 10 (45.5%); wrist arthritis, two (9.1%); and epididymo-orchitis, four (18.2%). The characteristics of the patients are shown in Table 1.
Table 1.
Epidemiological, clinical, serological, and molecular results of 50 patients with brucellosis
| Characteristics | Values |
|---|---|
| No. of patients (male) | 50 (36 male) |
| Age, mean age ± SD years | 43.6 ± 14.5 years (range 15–75) |
| Rural residence, no. (%) | 29 (58%) |
| Exposure history, no. (%) | 29 (58%) |
| Fever, no. (%) | 37 (80.4%) |
| Arthralgias, no. (%) | 25 (50%) |
| Low back pain, no. (%) | 25 (50%) |
| Complication involvement, no. (%) | 22 (43.5%) |
| Sacroiliitis, no. (%) | 6 (27.3%) |
| Knee arthritis, no. (%) | 10 (45.5%) |
| Wrist arthritis, no. (%) | 2 (9.1%) |
| Epididymo-orchitis, no. (%) | 4 (18.2%) |
| SAT ≥ 1/160, no. (%) | 43 (86%) |
| Antihuman globulin Coombs test, no. (%) | 7 (14%) |
| Patients with positive nested PCR, no. (%) | 50 (100%) |
| Relapsed patients with positive PCR, no. (%) | 5 (10%) |
PCR = polymerase chain reaction; SAT = serum agglutination test; SD = standard deviation.
At the time of diagnosis, the RBT agglutination test, SAT, and 2-ME were significantly positive in 43 patients and seven persons who had SAT < 1:160, their Coombs Wright test was ≥ 1:160. Brucella DNA was detected in 44 (88%) cases and six patients SAT ≥ 1:160 were PCR negative. All 50 (100%) patients were positive by the nested PCR assay. The data from the patients before and 6 months after treatment are shown in Table 2. Blood culture was positive in five (10%) patients. The sensitivity and specificity of our nested PCR assay was 100%, but no PCR amplification product was detected from the negative controls (non-Brucella species or healthy samples) (Figure 1 ). In the control group, RBT, SAT, and Brucella DNA were negative. At the 6-month follow-up after treatment, PCR was negative in 43 (86%) of the 50 patients whose nested PCR was initially positive. Among the seven (14%) patients who continued to have positive posttreatment PCR, five (10%) cases had relapsed (one case at 3 months, two cases at 4 months, and two cases 6 months after treatment), whereas two cases remained asymptomatic. These patients were also positive by both PCR and SAT tests, but their blood cultures were negative. The characteristics of these patients are shown in Table 3. Using nested PCR as the gold standard, the sensitivity and specificity of SAT ≥ 1:160 were 84.2% and 100%, respectively, as shown in Table 4.
Table 2.
Comparison of the PCR evaluated with SAT tests in brucellosis patients before and 6 months after treatment
| SAT titer | No. of cases (%) | First diagnosis | No. of cases (%) | 6 months after treatment | ||
|---|---|---|---|---|---|---|
| PCR (%) | Nested PCR (%) | PCR (%) | Nested PCR (%) | |||
| < 1:160 | 7 (14) | 3 (6) | 7 (14) | 43 (86) | 0 | 0 |
| 1:160 | 14 (28) | 13 (26) | 14 (28) | 2 (4) | 0 | 2 (4) |
| 1:320 | 23 (46) | 22 (44) | 23 (46) | 1 (2) | 1 (2) | 1 (2) |
| 1:640 | 4 (8) | 4 (8) | 4 (8) | 3 (6) | 3 (6) | 3 (6) |
| 1:1,280 | 2 (4) | 2 (4) | 2 (4) | 1 (2) | 1 (2) | 1 (2) |
| Total | 50 (100) | 44 (48) | 50 (100) | 50 (100) | 5 (10) | 7 (14) |
PCR = polymerase chain reaction; SAT = serum agglutination test.
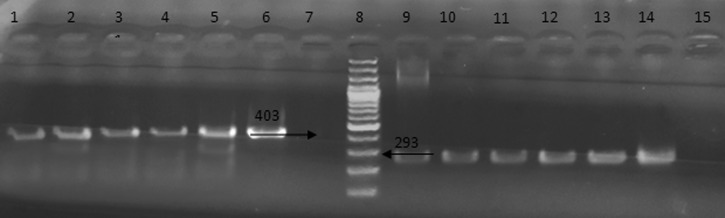
Figure 1.
Agarose gel electrophoresis of polymerase chain reaction products obtained by amplification of human DNA using the primers set. Lanes 1–5 and 9–13 are positive samples; lane 6 and 14, positive control (403 and 293 bp, respectively); lane 7 and 15, negative control; lane 8, DNA ladder.
Table 3.
Patients with positive nested PCR 6 months after treatment (seven cases)
| Patient | Status | Titer of SAT | |
|---|---|---|---|
| Before treatment | After treatment | ||
| 1 | Relapsed | 1:320 | 1:640 |
| 2 | Relapsed | 1:640 | 1:320* |
| 3 | Relapsed | 1:320 | 1:640 |
| 4 | Relapsed | 1:640 | 1:640 |
| 5 | Relapsed | 1:320 | 1:1,280 |
| 6 | Cure | 1:640 | 1:160 |
| 7 | Cure | 1:1,280 | 1:160 |
PCR = polymerase chain reaction; SAT = serum agglutination test.
This patient presented with arthritis of left knee and cured with retreatment.
Table 4.
Diagnostic test parameters of SAT for diagnosis of human brucellosis
| Parameter | Estimate (95% CIs) |
|---|---|
| Sensitivity | 84.21 (72.1–92.5%) |
| Specificity | 100.00 (91.8–100.00%) |
| Positive predictive value | 100.00 (92.6–100.00%) |
| Negative predictive value | 82.69 (69.7–91.8%) |
| Negative likelihood ratio | 0.16 (0.09–0.29) |
CI = confidence interval; SAT = serum agglutination test.
Discussion
In the endemic region, SAT is usually used for the diagnosis of brucellosis. However, the sensitivity and specificity of this test is not significantly high to detect all cases of human brucellosis. In the early stage of the disease, as well as the presence of blocking antibodies, this test may be negative. Thus, another pitfall is that using this test in the follow-up of the treated patients is not appropriate for the selection of relapse and chronic brucellosis.20–22 In this study, we found that only five (10%) patients had positive blood culture. If we consider blood culture as the gold standard of brucellosis, we may lose around 90% of cases of true brucellosis. Other researchers have reported positive blood culture in 10–70% of their cases.6–8,20
With regard to SAT, when we considered SAT for diagnosis of brucellosis, significant titer of SAT was seen in 43 (86%) of our cases. Therefore, another reliable test is needed for the diagnosis and follow-up of all patients with brucellosis.23–25 SAT false-negative results may have occurred in patients with very recent infection or chronic cases.26–28
With regard to the molecular method, we evaluated useful nested PCR assay for the diagnosis of human brucellosis in clinical practice. All 50 (100%) patients were positive by the nested PCR assay. The suitable results obtained 100% sensitivity and specificity. This high sensitivity suggests that the PCR method may replace blood culture as the gold standard for the diagnosis of acute brucellosis.29–31 Our results are similar to the finding of other researchers reporting that the species-specific PCR assay with primers IS711 can detect B. melitensis DNA in both SAT-positive and negative samples. The sensitivity and specificity of PCR-IS711 compared with SAT as the gold standard were found to be between 80% and 98% by other researchers.32–34
In this study, 86% of the patients had a negative PCR 6 months after treatment, a fact showing cure of the patients.29 In our revision, Brucella DNA was detected in two (4%) samples of clinically cured patients with no significant titers of their SAT during a 6-month follow-up after treatment with no evidence of relapse. It is often difficult to decide whether these patients are really cured.
Nevertheless, Brucella DNA was detected in most brucellosis patients during treatment and follow-up, despite the appropriate antibiotic therapy and obvious clinical recovery in some studies.26,28,35
We think that these differences may be due to the short-term follow-up of these studied patients, the high Brucella bacterial loads of their patients at the time of diagnosis or the kind of therapeutic regimens that they used. This issue neither indicates that clinical recovery relating to the eradication of the organism nor stresses the point that the presence of DNA may be representing an active disease that requires treatment.24,26 Hence, bacterial DNA persists in the blood of several patients throughout treatment and follow-up in those with significant clinical recovery.13,14 One of the main characteristics of brucellosis is its noticeable tendency to relapse after treatment.35 Since nearly 90% of relapses happened within 6 months after the completion of the treatment, follow-up of these patients is necessary after treatment to detect any relapse as soon as possible and to provide the sufficient therapy.36,37 In the present study, five (10%) patients had relapse during the follow-up period and were positive by PCR, and had increased titers. Molecular detection of Brucella DNA can be a sign of acute or chronic brucellosis and may also be detected in asymptomatic subjects with a history of brucellosis.22,38
In conclusion, our recommendation for physicians is to consider titer ≥ 1:160 as a diagnostic test in conjunction with a compatible clinical presentation. The weakness of this study is the small number of patients, short follow-up period, and the culture-negative cases that were evaluated. Apart from the disease stage, PCR techniques such as nested PCR are more specific and sensitive than serological tests. Consequently, this could be a useful tool for the diagnosis of the early phases of infection and for posttherapy follow-up of the disease and the early detection of relapse and chronic cases.
ACKNOWLEDGMENTS
We thank the staff of the Department of Infectious Diseases, Babol University of Medical Sciences, Iran, and Evangeline Foronda for the proofreading.
Footnotes
Financial support: This project was fully sponsored by the Infectious Diseases Research Center, Babol University of Medical Sciences, Babol, Iran, with project number 1978.
Authors' addresses: Mohammad Reza Hasanjani Roushan and Zahra Moulana, Infectious Diseases and Tropical Medicine Research Center, Babol University of Medical Sciences, Babol, Iran, E-mails: hagar2q@gmail.com and zmoulana@yahoo.com. Seyed Mahmoud Amin Marashi, Department of Microbiology and Immunology, Alborz University of Medical Sciences, Karaj, Iran, E-mail: parsmicrob@gmail.com.
References
- 1.Christopher S, Umapathy BL, Ravikumar KL. Brucellosis: review on the recent trends in pathogenicity and laboratory diagnosis. J Lab Physicians. 2010;2:55–60. doi: 10.4103/0974-2727.72149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Galinska EM, Zagórski J. Brucellosis in humans: etiology, diagnostics, clinical forms. Ann Agric Environ Med. 2013;20:233–238. [PubMed] [Google Scholar]
- 3.Pappas G. Treatment of brucellosis. BMJ. 2008;336:678–679. doi: 10.1136/bmj.39497.431528.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Solera J. Update on brucellosis: therapeutic challenges. Int J Antimicrob Agents. 2010;36:18–20. doi: 10.1016/j.ijantimicag.2010.06.015. [DOI] [PubMed] [Google Scholar]
- 5.Araj GF. Update on laboratory diagnosis of human brucellosis. Int J Antimicrob Agents. 2010;36:7–12. doi: 10.1016/j.ijantimicag.2010.06.014. [DOI] [PubMed] [Google Scholar]
- 6.Nielsen K, Yu W. Serological diagnosis of brucellosis. Prilozi. 2010;31:65–89. [PubMed] [Google Scholar]
- 7.Pabuccuoglu O, Ecemis T, El S, Coskun A, Akcali S, Sanlidag T. Evaluation of serological tests for diagnosis of brucellosis. Jpn J Infect Dis. 2011;64:272–276. [PubMed] [Google Scholar]
- 8.Al Dahouk S, Nöckler K. Implications of laboratory diagnosis on brucellosis therapy. Expert Rev Anti Infect Ther. 2011;9:833–845. doi: 10.1586/eri.11.55. [DOI] [PubMed] [Google Scholar]
- 9.Zamanian M, Hashemi Tabar GR, Rad M, Haghparast A. Evaluation of different primers for detection of Brucella in human and animal serum samples by using PCR method. Arch Iran Med. 2015;18:44–50. [PubMed] [Google Scholar]
- 10.Yu WL, Nielsen K. Review of detection of Brucella spp. by polymerase chain reaction. Croat Med J. 2010;51:306–313. doi: 10.3325/cmj.2010.51.306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Asaad AM, Alqahtani JM. Serological and molecular diagnosis of human brucellosis in Najran, southwestern Saudi Arabia. J Infect Public Health. 2012;5:189–194. doi: 10.1016/j.jiph.2012.02.001. [DOI] [PubMed] [Google Scholar]
- 12.Gemechu MY, Gill JPS, Arora AK, Ghatak S, Singh DK. Polymerase chain reaction (PCR) assay for rapid diagnosis and its role in prevention of human brucellosis in Punjab, India. Int J Prev Med. 2011;2:170–177. [PMC free article] [PubMed] [Google Scholar]
- 13.Navarro E, Segura JC, Castaño MJ, Solera J. Use of real-time quantitative polymerase chain reaction to monitor the evolution of Brucella melitensis DNA load during therapy and post-therapy follow-up in patients with brucellosis. Clin Infect Dis. 2006;42:1266–1273. doi: 10.1086/503035. [DOI] [PubMed] [Google Scholar]
- 14.Al-Ajlan HH, Ibrahim AS, Al-Salamah AA. Comparison of different PCR methods for detection of Brucella spp. in human blood samples. Pol J Microbiol. 2011;60:27–33. [PubMed] [Google Scholar]
- 15.Navarro E, Escribano J, Fernandez J, Solera J. Comparison of three different PCR methods for detection of Brucella spp. in human blood samples. FEMS Immunol Med Microbiol. 2002;34:147–151. doi: 10.1111/j.1574-695X.2002.tb00616.x. [DOI] [PubMed] [Google Scholar]
- 16.Cloeckaert A, Grayon M, Grepinet O. An IS711 element downstream of the bp26 gene is a specific marker of Brucella spp. isolated from marine mammals. Clin Diagn Lab Immunol. 2000;7:835–839. doi: 10.1128/cdli.7.5.835-839.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Baily GG, Krahn JB, Drasar BS, Stoker NG. Detection of Brucella melitensis and Brucella abortus by DNA amplification. J Trop Med Hyg. 1992;95:271–275. [PubMed] [Google Scholar]
- 18.Bounaadja L, Albert D, Chenais B, Henault S, Zygmunt MS, Poliak S, Garin-Bastuji B. Real-time PCR for identification of Brucella spp. a comparative study of IS711, bcsp31 and per target genes. Vet Microbiol. 2009;28:156–164. doi: 10.1016/j.vetmic.2008.12.023. [DOI] [PubMed] [Google Scholar]
- 19.Hajia M, Fallah F, Angoti G, Karimi A, Rahbar M, Gachkar L, Mokhtar B, Sanaei A, Rastegar Lari A. Comparison of methods for diagnosing brucellosis. Lab Med. 2013;44:29–33. [Google Scholar]
- 20.Alikhani MY, Hashemi SH, Naseri Z, Farajnia S, Peeri-Dogaheh H, Hashemi S. Diagnosis of human brucellosis by blood culture (BACTEC) and PCR method via whole blood and serum. Jundishapur J Microbiol. 2013;6:248–251. [Google Scholar]
- 21.Wang Y, Wang Z, Zhang Y, Bai L, Zhao Y, Liu C, Ma A, Yu H. Ann Clin Microbiol Antimicrob. 2014;13:31. doi: 10.1186/s12941-014-0031-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Castaño MJ, Solera J. Chronic brucellosis and persistence of Brucella melitensis DNA. J Clin Microbiol. 2009;47:2084–2089. doi: 10.1128/JCM.02159-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vrioni G, Bourdakis A, Pappas G, Pitiriga V, Mavrouli M, Pournaras S, Tsakris A. Administration of a triple versus a standard double antimicrobial regimen for human brucellosis more efficiently eliminates bacterial DNA load. Antimicrob Agents Chemother. 2014;58:7541–7544. doi: 10.1128/AAC.03841-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Roushan MR, Amiri MJ, Laly A, Mostafazadeh A, Bijani A. Follow-up standard agglutination and 2-mercaptoethanol tests in 175 clinically cured cases of human brucellosis. Int J Infect Dis. 2010;14:250–253. doi: 10.1016/j.ijid.2009.05.008. [DOI] [PubMed] [Google Scholar]
- 25.Mohammad Hasani S, Mirnejad R, Amani J, Vafadar MJ. Comparing rapid and specific detection of Brucella in clinical samples by PCR-ELISA and multiplex-PCR method. Iran J Pathol. 2016;11:144–150. [PMC free article] [PubMed] [Google Scholar]
- 26.Vrioni G, Pappas G, Priavali E, Gartzonika C, Levidiotou S. An eternal microbe: Brucella DNA load persists for years after clinical cure. Clin Infect Dis. 2008;4612:131–136. doi: 10.1086/588482. [DOI] [PubMed] [Google Scholar]
- 27.Maas KS, Mendez M, Zavaleta M, Manrique J, Franco MP, Mulder M, Bonifacio N, Castaneda ML, Chacaltana J, Yagui E, Guillen A, Blazes DL, Espinosa B, Hall E, Abdoel TH, Smits HL. Evaluation of brucellosis by PCR and persistence after treatment in patients returning to the hospital for follow-up. Am J Trop Med Hyg. 2007;76:698–702. [PubMed] [Google Scholar]
- 28.Al Dahouk S, Sprague LD, Neubauer H. New developments in the diagnostic procedures for zoonotic brucellosis in humans. Rev Sci Tech. 2013;32:177–188. doi: 10.20506/rst.32.1.2204. [DOI] [PubMed] [Google Scholar]
- 29.Kamal IH, Al Gashgari B, Moselhy SS, Kumosani TA, Abulnaja KO. Two-stage PCR assay for detection of human brucellosis in endemic areas. BMC Infect Dis. 2013;13:145. doi: 10.1186/1471-2334-13-145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Al-Nakkas A, Mustafa AS, Wright SG. Large-scale evaluation of a single-tube nested PCR for the laboratory diagnosis of human brucellosis in Kuwait. J Med Microbiol. 2005;54:727–730. doi: 10.1099/jmm.0.45772-0. [DOI] [PubMed] [Google Scholar]
- 31.Al Nakkas A, Wright S, Mustafa A, Wilson S. Single-tube, nested PCR for the diagnosis of human brucellosis in Kuwait. Ann Trop Med Parasitol. 2002;96:397–403. doi: 10.1179/000349802125001203. [DOI] [PubMed] [Google Scholar]
- 32.Hassanain NA, Ahmed WM. Efficacy of serological tests in comparison with PCR for diagnosis of brucellosis. World J Med Sci. 2012;7:243–247. [Google Scholar]
- 33.Masallat D, Moemen D, Eid MI. PCR versus serology for diagnosing of brucellosis in pregnant women: Mansoura University Hospital experience. Egypt J Med Microbiol. 2013;22:133–138. [Google Scholar]
- 34.Khosravi AD, Abassi E, Alavi SM. Isolation of Brucella melitensis and Brucella abortus from brucellosis patients by conventional culture method and polymerase chain reaction technique. Pak J Med Sci. 2006;22:396–400. [Google Scholar]
- 35.Skendros P, Boura P. Immunity to brucellosis. Rev Sci Tech. 2013;32:137–147. doi: 10.20506/rst.32.1.2190. [DOI] [PubMed] [Google Scholar]
- 36.Kim EJ, Lee SJ, Ahn EY, Ryu DG, Choi YH, Kim TH. Relapsed brucellosis presenting as neurobrucellosis with cerebral vasculitis in a patient previously diagnosed with Brucella spondylitis: a case report. Infect Chemother. 2015;47:268–271. doi: 10.3947/ic.2015.47.4.268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Roushan MR, Amiri MS, Janmohammadi N, Hadad MS, Javanian M, Baiani M, Bijani A. Comparison of the efficacy of gentamicin for 5 days plus doxycycline for 8 weeks versus streptomycin for 2 weeks plus doxycycline for 45 days in the treatment of human brucellosis: a randomized clinical trial. J Antimicrob Chemother. 2010;65:1028–1035. doi: 10.1093/jac/dkq064. [DOI] [PubMed] [Google Scholar]
- 38.Mitka S, Anetakis C, Souliou E, Diza E, Kansouzidou A. Evaluation of different PCR assays for early detection of acute and relapsing brucellosis in humans in comparison with conventional methods. J Clin Microbiol. 2007;45:1211–1218. doi: 10.1128/JCM.00010-06. [DOI] [PMC free article] [PubMed] [Google Scholar]