Abstract
Babesiosis is an emerging tick-borne disease caused by apicomplexan parasites of the genus Babesia. Most human infections in the United States are caused by Babesia microti, but other infection-causing Babesia parasites have been documented as well. Polymerase chain reaction (PCR)–based methods can be used to identify this parasite to the species level. In this study, published real-time PCR assays for the specific detection of B. microti were evaluated against conventional PCR for their analytical performance. All evaluated real-time PCR assays had comparable dynamic range and amplification efficiency, but the sensitivity and specificity varied. The best performing test, a TaqMan assay targeting the 18S ribosomal RNA gene, was further evaluated for diagnostic performance using blood specimens submitted to the Centers for Disease Control and Prevention for parasite detection and was found to have 100% sensitivity and specificity. In conclusion, the 18S TaqMan real-time PCR assay is a sensitive, specific, and rapid method for identification of B. microti among cases of babesiosis in the United States.
Introduction
Babesiosis is a zoonosis caused by intraerythrocytic apicomplexan parasites of the genus Babesia. These parasites are generally transmitted by tick bites, but blood transfusion and congenital transmissions have been reported as well.1,2 Babesia infection may be asymptomatic in otherwise healthy people, but can be severe in immunocompromised or asplenic individuals. Common clinical manifestations include nonspecific flu-like symptoms (e.g., fever) and hemolytic anemia.
The Centers for Disease Control and Prevention (CDC) has supported health departments in their investigations of cases of babesiosis in the United States since the first documented case was identified in 1966.3 Babesiosis is considered an emerging infectious disease, with a majority of cases reported in the northeast and upper midwest. Babesiosis became a nationally notifiable disease in January 2011; that year, CDC was notified of 1,124 cases that fulfilled the established case definition.4
The current gold standard for the laboratory diagnosis of babesiosis is microscopic examination of Giemsa-stained blood smears.5 However, microscopy can identify Babesia only to the genus level, and morphological features alone are not always sufficient to distinguish Babesia from Plasmodium. Molecular methods or serology can be used to confirm the microscopic findings and in addition provide a species-specific diagnosis.6,7
In the United States, babesiosis is most often caused by Babesia microti, but sporadic human cases caused by other Babesia parasites have been documented.8,9 For many years, CDC relied on a nested polymerase chain reaction (PCR) assay for the species-specific identification of B. microti in human clinical specimens10; however, this assay is time consuming and requires handling of amplified DNA, which is a risk factor for false-positive results due to contamination. To facilitate a faster diagnosis in a more practical format, we evaluated different real-time PCR assays specific for B. microti, aiming to identify an alternative for the detection of this species in human blood specimens.
Materials and Methods
Specimens.
Seventy-eight ethylenediaminetetraacetic acid–whole blood human specimens from different areas of the United States received at CDC between 2001 and 2016 were included in this study. The specimens had originally been submitted for confirmatory diagnosis of parasitic infections and were used in accordance with the CDC Human Subjects Research Protocol titled “Use of residual diagnostic specimens from humans for laboratory methods research.” As part of CDCs reference diagnostic services, these specimens were examined by microscopy of Giemsa-stained blood smears upon reception at CDC. Following the diagnostic algorithm for babesiosis at CDC, specimens that were microscopy positive for Babesia spp. (N = 44) were subjected to a B. microti–specific nested PCR.10 Thirty-six specimens were positive for B. microti by this PCR assay. Specimens positive for Babesia by microscopy but negative for B. microti by PCR (N = 8) were subjected to a generic PCR assay followed by DNA sequencing analysis of the amplicon to determine the infecting species11: two were positive for Babesia duncani (of which one case was described previously),12 one for a Babesia CA1-type parasite, four for Babesia MO1-type parasites, and one for another Babesia divergens–like parasite.13 Specimens negative for Babesia spp. by microscopy but positive for other blood-borne pathogens were confirmed with species-specific PCR and included as specificity controls.14,15 Fifteen specimens were included as parasite-free negative controls. In addition to the specificity controls described above, the specificity of the real-time PCR assay was evaluated with four laboratory isolates of B. divergens s.s. (of which one was the Purnell strain),16 originally cultured from cattle in Europe. Genomic DNA was extracted from 200 μL of each blood specimen using QIAamp DNA Blood Mini Kit using the QIAcube automated system (QIAGEN, Valencia, CA) according to the manufacturer's instructions.
Nested PCR assay.
All specimens positive for Babesia spp. by microscopy were subjected to a nested PCR specific for B. microti using primers and thermal cycling structure described elsewhere.10 The PCR reactions contained AmpliTaq Gold PCR Master Mix (Applied Biosystems, Foster City, CA), 50 pmol of each primer (Bab1 and Bab4 for the first step reaction, and Bab2 and Bab3 for the nested reaction), and 1 μL template DNA (genomic DNA or the product from the first step reaction) in a 50 μL total volume. The nested PCR products were visualized by 1.5% agarose gel electrophoresis stained with ethidium bromide.
Real-time PCR assays.
Nine published real-time PCR assays specific for B. microti were identified by a literature search in March 2014.17–24 In silico analysis of the oligonucleotides were performed by comparing the primer and probe sequences with the target gene; if more than one unique target gene sequence was available in GenBank, a pairwise or multiple alignment was made. The oligonucleotides were mapped to the target genes using Geneious v.8.1 (Biomatters Inc., Newark, NJ). If any mismatches were identified between the primer and probe sequences and the target gene sequences, the assay was not evaluated further since such mismatches can lead to false-negative results due to suboptimal annealing.
The four real-time PCR assays selected for laboratory evaluation were performed as described in the original publications, but with the following adjustments: 1 μL of DNA template was combined with QuantiTect Multiplex PCR Master Mix (QIAGEN) for the probe-based assays, or the QuantiTect SYBR Green PCR Master Mix (QIAGEN) for the SYBR Green assay; all assays were performed in Mx3000 thermal cyclers and the fluorescence data was analyzed using MxPro (Version 4.10; Agilent, 2007, Santa Clara, CA). For all assays, specimens with a Ct value < 40 (plus correct melting curve results for the SYBR Green assay) were considered positive for B. microti.
The B. microti Gray strain was propagated in male Golden Syrian hamsters (Mesocricetus auratus) to obtain enough parasites for the creation of broad dynamic range standard curves. Golden hamsters have been shown to be suitable animal models for human babesiosis.25 The hamsters were handled in accordance with CDC animal use protocol 2641BISMULC-A2. Standard curves were made with parasite-infected blood (versus genomic DNA) to account for possible variability in DNA extraction efficiency at different levels of parasitemia, and to get a realistic limit of detection. Blood samples from three experimentally infected hamsters were serially diluted in parasite-free hamster blood down to 0.1 parasites/μL. The three hamsters had parasitemia levels of 4.4, 18.7, and 25.6%, respectively, which were calculated to correspond to 220,000, 935,000, and 1,280,000 parasites/μL, respectively.5
Data analysis.
PCR test performance measures were calculated according to the Clinical Laboratory Improvement Amendment of 1988. Confidence intervals were calculated based on the Wilson procedure for a single proportion.26
Results and Discussion
Selection of real-time PCR assays for evaluation.
Upon literature review, nine published real-time PCR assays were identified and included in this evaluation. Seven of them target the 18S ribosomal RNA (rRNA) gene, one targets the surface antigen 1 gene (sa1), and one targets the thiamine pyrophosphokinase gene (tpk). Five assays targeting the 18S rRNA gene had at least one oligonucleotide with one or more mismatches when aligned to the target gene (Supplemental Figure 1); these assays were eliminated from further evaluation to avoid potential problems with false-negative results. Four assays had correct oligonucleotide designs according to this in silico test and were therefore selected to be evaluated in the laboratory: the SA1 TaqMan assay, the TPK molecular beacon assay, the 18S TaqMan assay, and the 18S SYBR Green assay.17,19,21
Comparison of four real-time PCR assays.
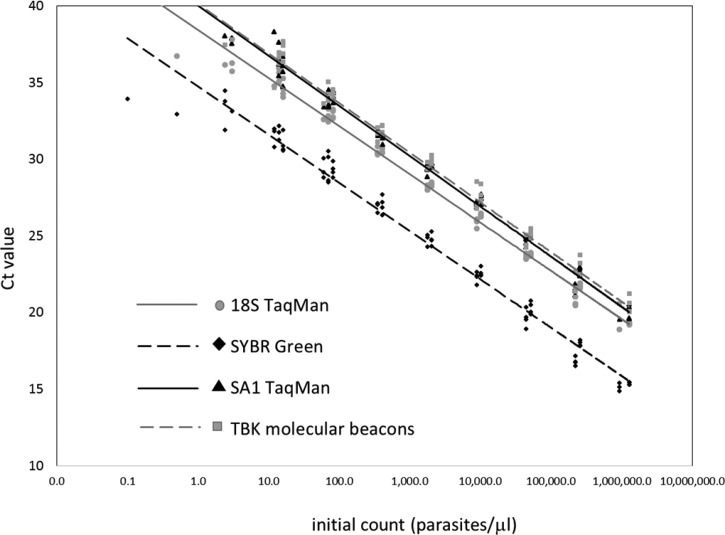
The detection limits, amplification efficiencies, and dynamic ranges of the four real-time PCR assays were determined using serial dilutions of three samples with known parasite densities. Results are summarized in Table 1 and Figure 1 . All real-time PCR assays had comparable dynamic range and amplification efficiency, but the analytical sensitivity (limit of detection) varied. The most sensitive assay was the SYBR Green assay with 2.4 parasites/μL, which is the same level of sensitivity as for the nested PCR. The 18S TaqMan assay had a detection limit of 12 parasites/μL, but it also produced positive results on some samples with fewer parasites, although inconsistently (positive results for two of three replicates of 3 parasites/μL and one of three replicates of 0.5 parasites/μL). The SA1 and TBK assays were both less sensitive, with a detection limit of 14 parasites/μL. The multicopy nature of the 18S rRNA gene probably contributed to the superior detection limit for the nested, SYBR Green, and 18S TaqMan assays.
Table 1.
Comparison of analytical performance of five PCR-based assays for Babesia microti detection
| Assay | Target gene | Amplicon size (base pairs) | Detection mode | Amplification efficiency (%) | Detection limit (parasites/μL) | Lowest detected (parasites/μL) | Analytical specificity (95% CI) | Reference |
|---|---|---|---|---|---|---|---|---|
| Nested | 18S rRNA | 154 | Agarose gel | NA | 2.4 | 0.5 | 31/31 × 100 = 100% (89.0–100%) | 10 |
| 18S TaqMan | 18S rRNA | 98 | TaqMan probe | 108 | 12 | 0.5 | 31/31 × 100 = 100% (89.0–100%) | 21 |
| SYBR Green | 18S rRNA | 154 | SYBR Green | 108 | 2.4 | 0.1 | 22/31 × 100 = 71% (53.4–83.9%) | 17 |
| SA1 TaqMan | sa1 | 114 | TaqMan probe | 102 | 14 | 2.4 | 31/31 × 100 = 100% (89.0–100%) | 21 |
| TBK | tbk | 141 | Molecular beacon | 104 | 14 | 2.4 | 31/31 × 100 = 100% (89.0–100%) | 19 |
CI = confidence interval; NA = not applicable; PCR = polymerase chain reaction; rRNA = ribosomal RNA.
Figure 1.
Standard curves based on serial dilutions of blood samples with calculated parasitemia from experimentally infected hamsters. The Ct values obtained from each dilution were plotted against the initial parasite count. The data points: 18S rRNA assay, gray circles; SYBR Green assay, black diamonds; SA1 TaqMan assay, black triangles; and TBK assay, gray squares. Logarithmic trend lines were calculated for each real-time PCR assay: 18S rRNA assay, solid gray line; SYBR Green assay, dashed black line; SA1 TaqMan assay, solid black line; and TBK assay, dashed gray line.
The analytical specificity of the assays was determined with a panel of 27 clinical specimens positive for parasites other than B. microti, plus four B. divergens isolates (see Materials and Methods section for details). Only one test, the SYBR Green assay, displayed evidence of nonspecific amplification with nine false-positive results. The Ct values for the false positives were all 33 or above (mean = 36), but the melting curve analysis did not separate the false positives from the weak B. microti specimens that were detected with similar Ct values. A BLAST similarity search of the SYBR Green primers in the GenBank database revealed that these primers have the capability to bind efficiently to other parasites besides B. microti (data not shown). Thus, the SYBR Green assay was considered not specific for B. microti. Taken together, these data indicated that the 18S TaqMan assay was the most accurate real-time PCR assay for B. microti.
Diagnostic validation of the 18S TaqMan assay.
The 18S TaqMan assay was validated for sensitivity, specificity, accuracy, and reproducibility as defined by the Clinical Laboratory Improvement Amendments of 1988. The assay was performed with 78 clinical specimens (of which 36 were positive for B. microti, 27 were positive for other parasites, and 15 were parasite free; see Materials and Methods section for details). We found the sensitivity and specificity for B. microti to be 100% (Table 2). To assess reproducibility, the 78 specimens were retested in six separate runs on four different days, totaling 468 individual result points. All positive specimens remained positive in all runs, and among the negative specimens, only one borderline positive result (Ct = 39) was obtained with one malaria specimen in one single run. These findings led to a reproducibility of almost 100% (Table 2). Since the false-positive result was so weak and not repeatable, it was most likely due to a random contamination event. Positive and negative predictive values were not calculated since the real prevalence of babesiosis in the test population is unknown and the numbers of positives and negatives included in this validation most likely did not reflect the real prevalence.
Table 2.
Diagnostic performance of the 18S TaqMan real-time PCR assay
| Characteristic | Calculations | Value | 95% CI |
|---|---|---|---|
| Sensitivity | (true positives)/(true positives + false negatives) | 36/(36 + 0) × 100 = 100% | 90.3–100.0% |
| Specificity | (true negatives)/(true negatives + false positives) | 42/(42 + 0) × 100 = 100% | 91.6–100.0% |
| Accuracy | (true positives + true negatives)/(true positives + true negatives + false negatives + false positives) | (36 + 42)/(36 + 42 + 0 + 0) × 100 = 100% | 95.3–100.0% |
| Reproducibility | (results in agreement)/(total no. of results) | 467/468 × 100 = 99.8% | 98.8–100.0% |
CI = confidence interval; PCR = polymerase chain reaction.
Concluding remarks.
Real-time PCR assays can provide a faster diagnosis compared with conventional PCR assays, especially nested assays. This study compared the performance of published real-time PCR assays against a conventional nested PCR assay for the specific detection of B. microti in human blood specimens and identified a TaqMan assay targeting the 18S rRNA gene to be the best performing assay. The 18S rRNA gene is highly conserved within each Babesia species and is present in high copy number in the genome. PCR-based assays targeting this gene therefore have the potential to be very sensitive and species specific. Although not quite as analytically sensitive as the nested PCR assay, the 18S TaqMan showed a 100% diagnostic sensitivity and did not yield any false-negative results for the actual clinical specimens analyzed in this study.
This study only evaluated the use of real-time PCR as a qualitative method to detect B. microti. The possibility to use this as a quantitative assay to estimate parasitemia remains to be explored. Although parasitemia calculations are not part of the diagnostic algorithm at CDC, the parasitologists sometimes noted an estimate of the number of infected blood cells when reading the blood smears. Based on these estimates, 18 of the B. microti–positive specimens were classified as having few parasites; the mean Ct value for these specimens in the 18S TaqMan assay was 26 (range, 17–37). Four specimens were classified as having high parasite density, with a mean Ct value of 22 (range, 20–23). More studies are needed to clarify the usefulness of this real-time PCR assay for quantification of parasitemia in babesiosis cases.
In conclusion, the 18S TaqMan real-time PCR assay is a rapid, sensitive, and specific method for detection of B. microti in human blood specimens. This assay has now replaced the nested PCR as the method of choice for the identification of B. microti among cases of babesiosis at the CDC.
Supplementary Material
ACKNOWLEDGMENTS
We thank Maniphet Xayavong (former laboratory technician at CDC) for technical assistance, and Barbara Herwaldt and Elizabeth Gray (epidemiologists at CDC) for critical reviews of the manuscript.
Disclaimer: The findings and conclusions in this manuscript are those of the author(s) and do not necessarily represent the official position of the U.S. Centers for Disease Control and Prevention.
Footnotes
Authors' addresses: Samaly S. Souza, Henry S. Bishop, Patrick Sprinkle, and Yvonne Qvarnstrom, Division of Parasitic Diseases and Malaria, Center for Global Health, Centers for Disease Control and Prevention, Atlanta, GA, E-mails: samalysouza@gmail.com, hsb2@cdc.gov, grc4@cdc.gov, and bvp2@cdc.gov.
References
- 1.Aderinboye O, Syed SS. Congenital babesiosis in a four-week-old female infant. Pediatr Infect Dis J. 2010;29:188. doi: 10.1097/INF.0b013e3181c3c971. [DOI] [PubMed] [Google Scholar]
- 2.Herwaldt BL, Linden JV, Bosserman E, Young C, Olkowska D, Wilson M. Transfusion-associated babesiosis in the United States: a description of cases. Ann Intern Med. 2011;155:509–519. doi: 10.7326/0003-4819-155-8-201110180-00362. [DOI] [PubMed] [Google Scholar]
- 3.Scholtens RG, Braff EH, Healey GA, Gleason N. A case of babesiosis in man in the United States. Am J Trop Med Hyg. 1968;17:810–813. doi: 10.4269/ajtmh.1968.17.810. [DOI] [PubMed] [Google Scholar]
- 4.Centers for Disease Control and Prevention Babesiosis surveillance: 18 States, 2011. Morb Mortal Wkly Rep. 2012;61:505–509. [PubMed] [Google Scholar]
- 5.Garcia LS. Procedures for Detecting Blood Parasites. Diagnostic Medical Parasitology. Washington, DC: ASM Press; 2001. pp. 829–849. [Google Scholar]
- 6.Quick RE, Herwaldt BL, Thomford JW, Garnett ME, Eberhard ML, Wilson M, Spach DH, Dickerson JW, Telford SR, 3rd, Steingart KR, Pollock R, Persing DH, Kobayashi JM, Juranek DD, Conrad PA. Babesiosis in Washington State: a new species of Babesia? Ann Intern Med. 1993;119:284–290. doi: 10.7326/0003-4819-119-4-199308150-00006. [DOI] [PubMed] [Google Scholar]
- 7.Vannier E, Gewurz BE, Krause PJ. Human babesiosis. Infect Dis Clin North Am. 2008;22:469–488. doi: 10.1016/j.idc.2008.03.010. viii–ix. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Conrad PA, Kjemtrup AM, Carreno RA, Thomford J, Wainwright K, Eberhard M, Quick R, Telford SR, 3rd, Herwaldt BL. Description of Babesia duncani n.sp. (Apicomplexa: Babesiidae) from humans and its differentiation from other piroplasms. Int J Parasitol. 2006;36:779–789. doi: 10.1016/j.ijpara.2006.03.008. [DOI] [PubMed] [Google Scholar]
- 9.Herwaldt B, Persing DH, Precigout EA, Goff WL, Mathiesen DA, Taylor PW, Eberhard ML, Gorenflot AF. A fatal case of babesiosis in Missouri: identification of another piroplasm that infects humans. Ann Intern Med. 1996;124:643–650. doi: 10.7326/0003-4819-124-7-199604010-00004. [DOI] [PubMed] [Google Scholar]
- 10.Persing DH, Mathiesen D, Marshall WF, Telford SR, Spielman A, Thomford JW, Conrad PA. Detection of Babesia microti by polymerase chain reaction. J Clin Microbiol. 1992;30:2097–2103. doi: 10.1128/jcm.30.8.2097-2103.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bonnet S, Jouglin M. Babesia sp. EU1 from roe deer and transmission within Ixodes ricinus. Emerg Infect Dis. 2007;13:1208–1210. doi: 10.3201/eid1308.061560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bloch EM, Herwaldt BL, Leiby DA, Shaieb A, Herron RM, Chervenak M, Reed W, Hunter R, Ryals R, Hagar W, Xayavong MV, Slemenda SB, Pieniazek NJ, Wilkins PP, Kjemtrup AM. The third described case of transfusion-transmitted Babesia duncani. Transfusion. 2012;52:1517–1522. doi: 10.1111/j.1537-2995.2011.03467.x. [DOI] [PubMed] [Google Scholar]
- 13.Herwaldt BL, de Bruyn G, Pieniazek NJ, Homer M, Lofy KH, Slemenda SB, Fritsche TR, Persing DH, Limaye AP. Babesia divergens-like infection, Washington State. Emerg Infect Dis. 2004;10:622–629. doi: 10.3201/eid1004.030377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Qvarnstrom Y, Schijman AG, Veron V, Aznar C, Steurer F, da Silva AJ. Sensitive and specific detection of Trypanosoma cruzi DNA in clinical specimens using a multi-target real-time PCR approach. PLoS Negl Trop Dis. 2012;6:e1689. doi: 10.1371/journal.pntd.0001689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rougemont M, Van Saanen M, Sahli R, Hinrikson HP, Bille J, Jaton K. Detection of four Plasmodium species in blood from humans by 18S rRNA gene subunit-based and species-specific real-time PCR assays. J Clin Microbiol. 2004;42:5636–5643. doi: 10.1128/JCM.42.12.5636-5643.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Purnell RE, Brocklesby DW, Hendry DJ, Young ER. Separation and recombination of Babesia divergens and Ehrlichia phagocytophila from a field case of redwater from Eire. Vet Rec. 1976;99:415–417. doi: 10.1136/vr.99.21.415. [DOI] [PubMed] [Google Scholar]
- 17.Bloch EM, Lee TH, Krause PJ, Telford SR, 3rd, Montalvo L, Chafets D, Usmani-Brown S, Lepore TJ, Busch MP. Development of a real-time polymerase chain reaction assay for sensitive detection and quantitation of Babesia microti infection. Transfusion. 2013;53:2299–2306. doi: 10.1111/trf.12098. [DOI] [PubMed] [Google Scholar]
- 18.Bown KJ, Lambin X, Telford GR, Ogden NH, Telfer S, Woldehiwet Z, Birtles RJ. Relative importance of Ixodes ricinus and Ixodes trianguliceps as vectors for Anaplasma phagocytophilum and Babesia microti in field vole (Microtus agrestis) populations. Appl Environ Microbiol. 2008;74:7118–7125. doi: 10.1128/AEM.00625-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chan K, Marras SA, Parveen N. Sensitive multiplex PCR assay to differentiate Lyme spirochetes and emerging pathogens Anaplasma phagocytophilum and Babesia microti. BMC Microbiol. 2013;13:295. doi: 10.1186/1471-2180-13-295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hersh MH, Tibbetts M, Strauss M, Ostfeld RS, Keesing F. Reservoir competence of wildlife host species for Babesia microti. Emerg Infect Dis. 2012;18:1951–1957. doi: 10.3201/eid1812.111392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hojgaard A, Lukacik G, Piesman J. Detection of Borrelia burgdorferi, Anaplasma phagocytophilum and Babesia microti, with two different multiplex PCR assays. Ticks Tick Borne Dis. 2014;5:349–351. doi: 10.1016/j.ttbdis.2013.12.001. [DOI] [PubMed] [Google Scholar]
- 22.Rollend L, Bent SJ, Krause PJ, Usmani-Brown S, Steeves TK, States SL, Lepore T, Ryan R, Dias F, Ben Mamoun C, Fish D, Diuk-Wasser MA. Quantitative PCR for detection of Babesia microti in Ixodes scapularis ticks and in human blood. Vector Borne Zoonotic Dis. 2013;13:784–790. doi: 10.1089/vbz.2011.0935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Teal AE, Habura A, Ennis J, Keithly JS, Madison-Antenucci S. A new real-time PCR assay for improved detection of the parasite Babesia microti. J Clin Microbiol. 2012;50:903–908. doi: 10.1128/JCM.05848-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tonnetti L, Eder AF, Dy B, Kennedy J, Pisciotto P, Benjamin RJ, Leiby DA. Transfusion-transmitted Babesia microti identified through hemovigilance. Transfusion. 2009;49:2557–2563. doi: 10.1111/j.1537-2995.2009.02317.x. [DOI] [PubMed] [Google Scholar]
- 25.Cullen JM, Levine JF. Pathology of experimental Babesia microti infection in the Syrian hamster. Lab Anim Sci. 1987;37:640–643. [PubMed] [Google Scholar]
- 26.Newcombe RG. Two-sided confidence intervals for the single proportion: comparison of seven methods. Stat Med. 1998;17:857–872. doi: 10.1002/(sici)1097-0258(19980430)17:8<857::aid-sim777>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.