Abstract
Systemic inhibition of miR-21 has proven effective against myocardial fibrosis and dysfunction, while studies in cardiac myocytes suggested a protective role in this cell type. Considering potential implications for therapy, we aimed to determine the cell fraction where miR-21 exerts its pathological activity. We developed a viral vector-based strategy for gene targeting of nonmyocyte cardiac cells in vivo and compared global to cardiac myocyte-specific and nonmyocyte-specific deletion of miR-21 in chronic left ventricular pressure overload. Murine moloney virus and serotype 9 of adeno-associated virus were engineered to encode improved Cre recombinase for genetic deletion in miR-21fl/fl mice. Pericardial injection of murine moloney virus-improved Cre recombinase to neonates achieved highly selective genetic ablation of miR-21 in nonmyocyte cardiac cells, identified as cardiac fibroblasts and endothelial cells. Upon left ventricular pressure overload, cardiac function was only preserved in mice with miR-21 deficiency in nonmyocyte cardiac cells, but not in mice with global or cardiac myocyte-specific ablation. Our data demonstrate that miR-21 exerts its pathologic activity directly in cardiac nonmyocytes and encourage further development of antimiR-21 therapy toward cellular tropism.
Introduction
Myocardial remodeling occurs in response to acute or chronic injuries of the cardiac muscle, e.g., due to aortic valve stenosis or ischemic injury. This process, which may turn into functional deterioration, is characterized by cardiac hypertrophy and fibrosis. Cells that exert either of these morphologic changes may be grouped into cardiac myocytes (CM) and nonmyocyte cells, the latter comprising cardiac fibroblasts (CF), cells of the vasculature, and immune cells. Whereas therapeutic regimes to suppress hypertrophic processes in CM are available, the relatively poor knowledge about pathologic signaling in nonmyocyte cardiac cells and their responsible targets hampers the development of new drug therapies.
MicroRNAs, small noncoding RNAs that regulate gene expression, have been found to be critically involved in cardiac remodeling, either with disease-promoting or protective activity.1 One of the most prominent microRNAs that control cardiac remodeling is miR-21, for which we demonstrated strong upregulation in failing myocardium in humans and in animal disease models. We further showed that synthetic inhibitors for miR-21 (antimiR-21) potently prevent cardiac fibrosis in a mouse model for chronic cardiac pressure overload (induced by transverse aortic constriction, TAC).2,3 In line with this, other labs also showed that miR-21 inhibition reduces pathologic fibrosis in liver, kidney, lung or skeletal muscle.4,5,6,7,8 These findings have made antimiRs against miR-21 an attractive therapeutic option for various fibrotic conditions.4
Although the therapeutic potential of antimiRs is considerable, the fact that these molecules have little to no specificity for tissues or cell types makes it difficult to allocate the (patho-)physiological function of a microRNA. This would be particularly relevant for miR-21, since this microRNA also has beneficial antiapoptotic activity in CM, at least in settings that strongly induce apoptosis, such as ischemia/reperfusion.9 It is thus important to understand where miR-21 exerts its detrimental and possibly also protective activities. To target specific tissues/cells in vivo, typically transgenic “driver” mouse lines are employed for Cre-mediated recombination under the control of tissue-specific promoters. However, the identification of promoters that are specific for cardiac nonmyocytes lags far behind the needs.
Since certain viruses show pronounced tissue tropism and, in some cases, are even specific for cell types, they are increasingly used as vectors for targeted gene expression. For example, serotype 9 of adeno-associated virus (AAV9) efficiently infects myocardium, where it is highly selective for CM.10,11 We have employed this feature to specifically manipulate gene expression in CMs in vivo and thereby to characterize the function of cardiac genes.12,13 However, no viral vector had thus far been shown to infect a larger fraction of the nonmyocytes in myocardial tissue in vivo. Here, we present a viral vector-based strategy for nonmyocyte-targeted genetic deletion or overexpression in vivo. Application of these vectors into the pericardial cavity of neonates allowed us to demonstrate that miR-21 exerts its cardiac profibrotic activity directly in the nonmyocyte fraction of the myocardium.
Results
Vector design for selective manipulation of miR-21 in CM and nonmyocytes
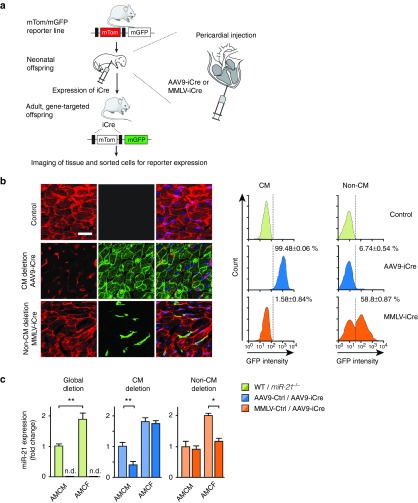
Viral vectors which infect the cardiac muscle include serotypes 2, 6, 8, and 9 of adeno-associated virus,11 adenovirus,10,14 lentivirus,15 and retroviridae.16 Among these vectors, superior tropism for CM has been reported for AAV911 and we have previously corrobated its suitability and cell-type specificity in mice.12,13 For the nonmyocyte pool of cardiac cells, we chose moloney murine leukemia virus (MMLV) based on evidence that it infects proliferating CF in vitro and in vivo,16,17 making it an attractive vector to target fibroblasts also when they are activated by disease-inducing conditions. Since CF proliferation in healthy mice decreases substantially 5 days after birth,18 we further reasoned that, for studies in vivo, administration to the newborn may enhance transduction rate within the heart. We generated AAV9 and MMLV vectors that encode codon-improved Cre recombinase (iCre)19 under the control of the CMV promoter. Following a procedure that had originally been described for adenovirus injection,20 these vectors were administered pericardially into 3–4 days old mice from a line that carries a mtdTomato-mGFPflox/flox construct in the Rosa LacZ locus and allows for high sensitivity of detection with marginal background (Figure 1a). In the presence of iCre, the mtdTomato cassette is deleted, allowing instead the expression of membrane-targeted GFP downstream from it.21 AAV9 was confirmed to target CM with high efficiency and specificity, as noticeable by an exclusive GFP signal in 99.5 % of cells that were morphologically assigned as CM, whereas MMLV-iCre had infected ~60% of the nonmyocyte cell population (Figure 1b). Interestingly, we found by flow cytometric and immunohistochemical analysis that the nonmyocyte cell pool primarily contained endothelial cells (EC) (e.g., 54.3% of cardiac isolates are CD105+), CF (26.2% PDGFRa+) and leukocytes (12.4% CD45+) (see Supplementary Figure S1a), consistent with other reports.22 Among the various cell types, highest endogenous expression of miR-21 was observed in CF (see Supplementary Figure S1c).
Figure 1.
Specificity and efficiency of viral vectors to target cardiac myocytes or nonmyocyte cells. (a) Experimental strategy to test viral vectors for iCre-mediated recombination in mtdTomato-mGFP reporter mice. Viruses encoding iCre were administered pericardially on d3-4 and hearts were collected 3 weeks later for analysis. Each mouse received 5 × 1011 viral genomes (vg) of AAV9 or 5 × 109 vg of MMLV. Controls were carried out with an equivalent titer of empty vector. (b) Left, representative cardiac sections from mtdTomato-mGFP reporter mice after delivery of iCre-endoding AAV9 or MMLV. Green flurorescence indicates successful deletion of the mtdTomato locus. Right, flow cytometric quantification of GFP-positive cardiac myocytes (CM) and nonmyocyte cells (Non-CM) isolated from mouse hearts. Scale bar represents 50 µm. n = 2–3 mice per group. (c) Quantification of miR-21 in adult mouse cardiac myocytes (AMCM) or fibroblasts (AMCF) isolated from global deletion mice (left) or mice treated with either AAV9-iCre (middle) or MMLV-iCre (right). In each experiment, vectors were administered to 3–4 days old neonatal mice and cardiac cells were isolated at 6 weeks of age using Langendorff perfusion method. n = 3–6 mice per group. Data are mean ± SEM and were analyzed using two-way analysis of variance (ANOVA) with Sidak's post-test. *P < 0.05 and **P < 0.01. AAV9, adenoassociated virus of serotype 9; MMLV, moloney murine leukemia virus; PE-Cy7, phycoerythrin-Cy7 conjugated; GFP, Green fluorescent protein.
We then asked how efficient and in which ratio to each other individual cell types are infected by AAV9 or MMLV in vivo. For this, GFP- instead of iCre-encoding AAV9 or MMLV were used to infect wildtype mice, and the number of cells in which GFP colocalized with PDGFRa, CD105, CD45 or Troponin T (the latter for CM) was assessed. About half of the non-CM cells from the MMLV group were GFP-positive and had thus been infected; among these, two out of three were EC (~63% of total EC) and one of three were CF (~60% of total CF). Immune cells had not been infected by MMLV, as judged from quantification of CD45-positive cells after transfection with MMLV-GFP (see Supplementary Figure S1d). In agreement with this, we found minimal to no infection of bone marrow cells and peripheral blood monocytes by MMLV (see Supplementary Figure S1e). Thus, we conclude that pericardial application of MMLV to neonates targets almost exclusively CF and EC in myocardium in vivo.
Next, we assessed the extent to which miR-21 deletion occurs in mice after infection with viral vectors and compared this to mice with global miR-21-deficiency. The latter have been engineered upon breeding a Cre-transgenic mouse line23 with miR-21fl/fl mice24 (kindly provided by S. Orkin and E. Olson, respectively). For targeted manipulation of miR-21, miR-21fl/fl mice received AAV9-iCre or MMLV-iCre vectors on day 3–4 after birth. As controls served AAV9 or MMLV vectors that encode the dsRed cDNA. After 6 weeks, mice were sacrificed and cells were isolated from left ventricles with the Langendorff procedure, providing defined CM and CF isolates in purities >95% each. Quantification of endogenous miR-21 confirmed that AAV9-iCre allowed for a 61% reduction of miR-21 in CM, without affecting its levels in CF. Vice versa, MMLV-iCre led to 58% lower miR-21 levels in CF, but left its expression in CM unchanged (Figure 1c). As an indicator of miR-21 reduction, these mice also showed derepression of Spry1, an established inhibitor of ERK-MAPkinase and target of miR-21 (Spry1 mRNA: MMLV-Dsred TAC 1.0 ± 0.1 versus MMLV-iCre TAC 1.53 ± 0.11,**P < 0.01; SPRY1 protein: MMLV-Dsred TAC 0.71 ± 0.06 versus MMLV-iCre TAC 1.11 ± 0.09, *P < 0.05) and reduction in ERK activity (pERK/tERK: MMLV-Dsred TAC 1.12 ± 0.14 versus MMLV-iCre TAC 0.66 ± 0.06*, P < 0.05).2 Together, these findings provide a solid basis to manipulate miR-21 in vivo in CM and nonmyocytes.
Evaluation of viral tropism for other organs
Despite the effectivity of MMLV-driven deletion of miR-21 in nonmyocytes in myocardium, the possibility that this vector also deregulates miR-21 in cells from other organs must be considered. For this, miR-21 was quantified in kidney, liver, lung and spleen. MMLV-iCre (in miR-21fl/fl mice) did not alter miR-21 expression in any of the other organs tested (see Supplementary Figure S2b). By contrast, AAV9-iCre conferred a 75% reduction of miR-21 in liver and to more moderate, nonsignificant extent in lung (see Supplementary Figure S2a). These findings are consistent with previous reports on the tropism of AAV9.11 However, no morphologic changes in liver or lung were observed. Thus, we conclude that AAV9 and particularly MMLV are well-suited tools to study miR-21 in CM and in CF/EC, respectively, in vivo.
Genetic miR-21 deficiency in cardiac nonmyocyte cells protects from functional impairment
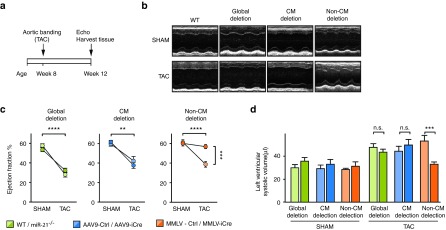
To investigate through which cardiac cell types miR-21 contributes to cardiac function, we applied chronic cardiac pressure overload to mice that had received AAV9-iCre or MMLV-iCre, or to mice with global miR-21 deficiency. Echocardiography was carried out at the beginning of the experiment and 4 weeks after TAC or sham surgery. Expectedly, sham groups that received iCre-encoding AAV9 or MMLV or control vector were healthy and normal, in contrast to the typical impairment of cardiac function that was evident in all TAC-treated control mice where no genetic manipulation of miR-21 had been applied (WT, AAV9-dsRed, MMLV-dsRed) (Figure 2a,b). Importantly, global deficiency of miR-21 or partial reduction in CM did not improve ejection fraction, whereas the nonmyocyte-targeted approach almost completely preserved cardiac function (Figure 2b,c and see Supplementary Table S1). This was also reflected by left ventricular volume (LV volume), which only in TAC-treated, MMLV-iCre-infected mice was similar to sham control (Figure 2d). Together, these findings demonstrate that miR-21 has a crucial role in cardiac nonmyocyte cells and that reducing its expression specifically in these cells protects the myocardium from disease-induced functional impairment.
Figure 2.
Genetic deletion of miR-21 in nonmyocyte cells preserves cardiac function in a cardiac disease model. Echocardiography in mice with global genetic deletion of miR-21 (miR-21-/-) or in mice where deletion was targeted to cardiac myocytes or nonmyocytes upon infecting miR-21fl/fl mice with iCre-encoding AAV9 or MMLV, respectively. Controls for these groups were wildtype (WT) or dsRed-encoding vectors. Mice were either subjected to transverse aortic constriction (TAC) or received sham surgery as a negative control. (a) Experimental strategy. (b) Representative recordings from TAC-treated mice with global or targeted miR-21 deletion. (c) Quantification of the left ventricular ejection fraction in TAC- or sham-operated mice from the above approaches. Green, blue, and orange color codes are used for groups with global, CM- or non-CM-targeted deletion of miR-21, respectively, with darker color for the genetically manipulated mice and brighter color for their genetic controls. (d) Left ventricular volume (LV volume) in the above groups. miR-21 global deletion mice Sham n = 9, TAC n = 16–18. AAV9- Ctrl/-iCre Sham n = 6–7; TAC n = 5–9. MMLV-Ctrl/-iCre Sham n = 6–9; TAC n = 7–8. Data are mean ± SEM. and were analyzed using two-way analysis of variance (ANOVA) with Sidak's post-test. *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001. AAV9, adenoassociated virus of serotype 9; CM, cardiac myocytes; iCre, improved Cre recombinase; MMLV, moloney murine leukemia virus.
Depletion of miR-21 in nonmyocyte cardiac cells reduces pressure overload-induced cardiac remodeling
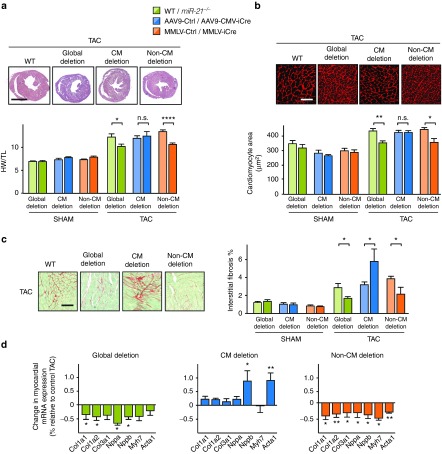
To investigate the cell-specific contribution of miR-21 to cardiac remodeling, we determined the extent of cardiac hypertrophy and fibrosis in mice with global or targeted miR-21 deficiency. Mice carrying a global knockout of the miR-21 gene or recipients of AAV9-iCre, MMLV-iCre or control vectors were further split for subsequent sham or TAC treatment, respectively (Figure 3a). AAV9-iCre-mediated deletion of miR-21 in CM (see Figure 1c) did not alter TAC-induced cardiac hypertrophy (as judged by heart-weight-to-tibia-length ratio, Figure 3a). In line with this, wheat germ agglutinin staining showed that myocyte hypertrophy after TAC was not affected by AAV9-iCre (Figure 3b). Targeting nonmyocyte cardiac cells by MMLV-iCre, instead, prevented cardiac hypertrophy at the morphologic and the cellular level (Figure 3a,b). Interestingly, global miR-21 deficiency also interfered with cardiac hypertrophy, but, as shown above and consistent with other reports,23,24 without functional benefit (Figure 2 and Figure 3a,b). This lack of correlation between cardiac function and hypertrophy seen with the global miR-21 genetic knockout suggests that additional protective mechanisms occur in these mice (see for “Discussion”).
Figure 3.
Protection from TAC-induced cardiac hypertrophy and fibrosis by global or nonmyocyte-targeted deletion of miR-21. (a) Top panel, Representative myocardial cross-sections from TAC-treated miR-21-/- mice, AAV9-iCre or MMLV-iCre mice. Sham- or TAC-treated cardiac tissue from wildtype mice is shown as controls. Diagram, Ratio of heart weight to tibia length (HW/TL) (see Figure 2 for details on color codes). Scale bar represents 2 mm. (b) Wheat germ agglutinin staining in representative myocardial sections. Scale bar represents 50 µm. (c) Left, Representative stainings of myocardial samples by Sirius red/Fast green. Right, Quantification of data. Scale bar represents 50 µm. (d) Quantitative PCR assessment of mRNA levels of collagens (Col1a1, Col1a2, and Col3a1), Nppa, Nppb, Myh7, and Acta1. The dashed line denotes the expression level from controls to which data were normalized. (e). miR-21 global deletion mice Sham n = 9, TAC n = 16–18. AAV9- Ctrl/-iCre Sham n = 6–7; TAC n = 5–9. MMLV- Ctrl/-iCre Sham n = 4–6; TAC n = 4–5. Data are mean ± SEM and were analyzed using two-way analysis of variance (ANOVA) with Sidak's post-test. *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001. AAV9, adenoassociated virus of serotype 9; iCre, improved Cre recombinase; MMLV, moloney murine leukemia virus; PCR, polymerase chain reaction; TAC, transverse aortic constriction.
Staining extracellular matrix proteins in myocardial cross-sections indicated that fibrosis in the TAC/MMLV-iCre group had been significantly prevented compared with TAC-treated control mice (Figure 3c). This was likewise observed in mice with global miR-21 deficiency (Figure 3c). Consistent with these findings, the expression of collagens and of Nppa as biomarkers for cardiac fibrosis and hypertrophy, respectively, was reduced in TAC-treated mice from MMLV-iCre and globally Cre-expressing groups (Figure 3d). In contrast to the results obtained upon miR-21 deletion in nonmyocytes, deficiency of miR-21 in CM did not affect hypertrophy in the TAC model, but increased cardiac fibrosis. This rules out a disadvantageous contribution of CM-based miR-21 to cardiac remodeling, but rather suggests beneficial function(s) in this cell type. In support of this, TAC-treated mice with miR-21 deficiency in CM (AAV9-iCre group) displayed higher numbers of terminal deoxynucleotidyl transferase nick-end labeling (TUNEL)-positive, apoptotic cells in myocardium than controls (see Supplementary Figure S3a). Since TAC typically does not induce extensive cell death, this effect could not be more than moderate (AAV9-Ctrl TAC 0.2 ± 0.05% versus AAV9-iCre TAC 0.35 ± 0.08%), yet it agrees qualitatively with findings made in the ischemia/reperfusion model.9 Similarly, we found that overexpression of miR-21 protected cells from doxorubicin-induced cell death in neonatal rat cardiac myocytes (see Supplementary Figure S3b). Together, these data provide evidence for a direct role of miR-21 in cardiac fibrosis through its activity in cardiac nonmyocytes.
Overexpression in nonmyocyte cells in vivo aggravates the deleterious effects of miR-21 upregulation in disease
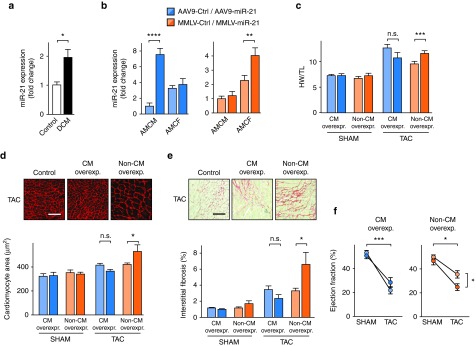
In patients with dilated cardiomyopathy, we observed pronounced upregulation of miR-21 in myocardium (Figure 4a), which is consistent with similar reports from other labs and with our observations.2 Given the potential coexistence of protective and pathological roles of miR-21 in cardiac myocytes and nonmyocytes, we asked whether targeted overexpression of miR-21 in these cell populations would suffice to promote cardiac remodeling and if so, whether this would differ between overexpression in myocytes and nonmyocytes. After pericardial injection of AAV9-miR-21 or MMLV-miR-21 into neonatal mice, quantification in primary cells from the adults revealed a robust increase in CM from the AAV group and in CF from the MMLV group (Figure 4b). Cardiac hypertrophy was largely unchanged in sham groups, but upon TAC, vector-based overexpression of miR-21 caused reciprocal effects on cardiac remodeling: whereas AAV9-miR-21 did not significantly affect cardiac hypertrophy, MMLV-miR-21 promoted cardiac hypertrophy (Figure 4c). Furthermore, TAC-treated mice that overexpressed miR-21 in nonmyocytes (MMLV) showed significantly larger cardiac myocytes than controls or AAV9-miR-21-infected mice (Figure 4d), together with exacerbated cardiac fibrosis (Figure 4e). These morphologic alterations were accompanied by deterioration in cardiac function, as elevation of miR-21 in nonmyocytes significantly impaired ejection fraction, while robust expression of miR-21 in myocytes was not detrimental (Figure 4f). These data further validate the detrimental effects of miR-21 upregulation in the nonmyocyte fraction of the myocardium.
Figure 4.
Elevated expression of miR-21 in cardiac fibroblasts (CF) and endothelial cells exacerbates cardiac remodeling and is detrimental to cardiac function. (a) Quantification of miR-21 in human left ventricular myocardium from healthy and patients suffering from dilated cardiomyopathy (DCM). n = 5 per group. (b) Quantification of miR-21 in adult mouse cardiac myocytes (AMCM) and fibroblasts (AMCF) 5 weeks after infecting mice with miR-21-encoding AAV9 or MMLV, showing specific vector-dependent overexpression. Corresponding control vectors encoded the nonrelated C. elegans miR-39. (c) Ratio of heart weight to tibia length (HW/TL) from mice 4 weeks after TAC or sham surgery. (d) Wheat germ agglutinin staining in representative myocardial sections. (e) Representative Sirius red/Fast green staining of myocardial sections from the above mice (left), and quantitative data (right). (f) Echocardiographic determination of left ventricular ejection fraction in the above mice. Scale bars: 50 µm d and e. AAV9-Ctrl /-miR-21 Sham n = 4; TAC n = 6–7. MMLV-Ctrl/-miR-21 Sham n = 6–7; TAC n = 9. Data are mean ± SEM and were analyzed using two-way analysis of variance (ANOVA) with Sidak's post-test. *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001. AAV9, adenoassociated virus of serotype 9; MMLV, moloney murine leukemia virus; TAC, transverse aortic constriction.
Discussion
MiR-21 has been characterized as an essential promoter of pathologic cardiac remodeling, of which cardiac fibrosis and hypertrophy are key characteristics. We have previously demonstrated that inhibition of miR-21 by synthetic antimiR molecules effectively prevents development and the progression of already established remodeling, highlighting antimiR-21-based therapy as a promising therapeutic approach. However, given that experimental overexpression of miR-21 can reduce the number of apoptotic cardiac myocytes9 and may also affect coupling of cardiac myocytes with other cardiac cells,25 it appears that miR-21 has both disease-promoting as well as protective roles, which display in distinct cardiac cell populations and which depend on the disease condition. A main obstacle to resolve this issue has thus far been the lack of tools to specifically manipulate gene expression in the nonmyocyte pool of cardiac cells, of which EC and CF make up the vast majority.
In this study, we devised strategies to achieve elevated expression or Cre-mediated genetic deficiency of miR-21 in cardiac myocytes and in cardiac nonmyocytes in vivo. Testing of various viruses not only confirmed the specificity of AAV9 for cardiac myocytes, but, using MMLV as a vehicle for Cre expression, we could also manipulate the nonmyocyte cardiac cells in unprecedented specificity and efficiency.
A key observation we made with these targeted approaches was that cardiac dysfunction under disease conditions was only prevented in mice with miR-21 deficiency in nonmyocytes. Myocyte-targeted or global deletion of miR-21 failed to protect from functional impairment, although the latter—like the nonmyocyte-specific approach—allowed for some reduction of cardiac fibrosis and hypertrophy. This finding may have consequences in future therapeutic applications of miR-21-based therapy. MiR-21 thus primarily determines cardiac remodeling and dysfunction through its activity in nonmyocyte cells of the heart. Because MMLV not only infects fibroblasts, but also EC in myocardium (see Supplementary Figure S1d), it is intriguing to further ask in which of the two cell types miR-21 primarily functions as a promoter of cardiac remodeling. For example, it would be plausible to presume that targeting miR-21 in EC alters angiogenesis or antiinflammatory responses.26,27,28,29 In agreement with earlier publications, we observed TAC-induced upregulation of miR-21 in both CF and EC, but not in cardiac myocytes (CM SHAM 1 ± 0.1 versus CM TAC 0.91 ± 0.2; CF SHAM 1 ± 0.07 versus CF TAC 3.87 ± 0.83,**P < 0.01; EC SHAM 1 ± 0.25 versus EC TAC 3.25 ± 0.84,*P < 0.05).2,30 Furthermore, flow cytometry analysis indicated that the number of fibroblasts increased significantly while there was a significant decline in endothelial cell numbers (% of non-CM: CF SHAM 26.7 ± 0.43 versus CF TAC 33. 7 ± 0.46,**P < 0.01; EC SHAM 53.7 ± 1.18 % versus EC TAC 45.5 ± 2.27,*P < 0.05). However, this is apparently not the case, as MMLV-iCre-driven deficiency of miR-21 did not lead to altered Cd31 or Vegfa expression in EC or cardiac vascularization (see Supplementary Figure S4). Taking all evidence together, our data point to a direct and predominant role of miR-21 in CF. MiR-21 thus seems to have dichotomous roles in different cardiac cells and it seems reasonable that their phenotypic expression is determined by type and severity of disease conditions.
How do the data from our viral vector-based approach compare with those obtained upon ubiquitous deletion of the miR-21 gene? Two labs had established such mice by breeding miR-21fl/fl mice with a line that expresses Cre recombinase under the control of an adenovirus type 5 EIIA early (EIIA) promotor23 or a cytomegalovirus early enhancer/chicken ß-actin (CAG) promotor,24 respectively. As described, improved cardiac function was not observed24 (or tested for23), consistent with our findings. Yet, whereas global miR-21 deficiency in the study by Patrick et al. did not alter cardiac remodeling, we detected a reduction of cardiac hypertrophy and fibrosis in this setting (Figure 3). It is unclear whether experimental details such as the extent or duration of TAC (3 weeks versus 4 weeks) or the genetic background and differences in compensation account for this discrepancy. Nonetheless, these considerations do not compromise the most intriguing aspect of our results, that is: no functional benefit by global miR-21 deficiency, but preserved cardiac function upon miR-21 deficiency in non-CM. We rather believe that mice with global miR-21 deficiency are more likely to engage physiological responses that help to mitigate disease severity. In support of the latter, such mice were recently found to have reduced immune response,28 leading to lower inflammation and thus to a compensated phenotype.
It is interesting that the absence of miR-21 in CF and EC not only prevented fibrosis, but also hypertrophy of cardiac myocytes. In agreement to this, overexpression in nonmyocytes forced cardiac myocytes into enlargement. Transferred to the disease scenario mimicked by TAC, this would mean that upregulation of miR-21 in nonmyocyte cells promotes their pathologic signaling to heart muscle cells. It is, at present, unclear what the nature of this signal may be, but several paracrine factors are bona fide suspects.31 How relevant nonmyocyte signalling toward cardiac hypertrophy may be has been suggested by two previous studies32,33; this aspect has also been the focus of a recent review by Kamo et al.34
Another aspect of this study is that the results from targeted miR-21 deletion in nonmyocytes largely phenocopy the effects seen upon pharmacological inhibition of miR-21.2,3 This corroborates the crucial role of nonmyocyte miR-21 in pathomorphologic alterations of the diseased heart and encourages ongoing testing of therapeutic options, wherein targeting ~50% of non-CMs may be sufficient to confer cardioprotection. There is clearly a need to establish reporter systems that help to understand and optimize the routes of such inhibitors in tissue. Likewise, functional characterization of microRNAs in vivo requires cell type-specific approaches. In continuation of these data, it would now be interesting to pinpoint the individual roles of miR-21 in CF and in EC. Promoters that allow for transgenic expression in these cells could be an ideal tool and promising candidates such as the promoters for TCF21, Periostin, or FSP1 have been identified.35 However, their use has limitations with respect to cell- or organospecificity36,37 or promoter strength.38,39 These findings may explain why, thus far, only ubiquitous knockouts of miR-21 in mice have been established and tested.23,24
The intriguing selectivity of engineered MMLV for cardiac cell types in combination with pericardial injection may not only be valuable for the characterization of miR-21 and other microRNAs—it also bears good potential to advance on to gene therapy in patients. Although premature at present, we envisage that treatment of inherited cardiac diseases in infants could benefit from early application of vectors that provide for specific and strong expression in dividing cardiac cells.
Materials and Methods
Animal models. EIIA-CreTg/0; miR-21fl/fl mice (Harvard Medical School, Boston, MA),23 miR-21 floxed mice (Dallas, TX),24 and the transgenic Rosa-tm4 (ACTB-tdTomato,-EGFP) reporter mice21 have been described previously. All mice were on a C57BL/6N genetic background.
TAC was performed on 8–10-week-old male mice as described previously.40 For pre and postoperative analgesia, the mice were injected with buprenorphine (0.08 mg/kg) and metamizole (200 mg/kg) intraperitoneally. Mice were anesthetized in an induction chamber with 4% Isoflurane mixed with 0.5 l/min of 100% O2. During the surgical procedure, anesthesia was maintained at 1.5–2% Isoflurane with 0.5 l/min O2. Partial thoracotomy was performed and a small piece of 6.0 silk sutures was ligated between the innominate and left carotid arteries around 27.5 G needles that was placed parallel to the transverse aorta. In sham surgery, only the chest was opened, but no ligation of the aorta was carried out. Cardiac dimensions and function was analyzed by pulse-wave Doppler Echocardiography before TAC/sham surgery and, at the end of the experiment, before sacrificing the animals. All animal studies were performed in accordance with relevant guidelines and regulations of the responsible authorities.
Construction and production of viral vectors Transgene. A genomic fragment containing 680 nucleotides of the miR-21 (Gene I.D. 262205609) precursor was amplified by polymerase chain reaction (PCR) from mouse genomic DNA and was cloned into self-complementary AAV and MMLV backbone plasmids. For the generation of a miR control virus, the precursor of C. elegans miR-39 (350 nucleotides, Gene I.D. 392893621) was synthesized (GeneArt, Life Technologies, Regensburg, Germany). For knockdown experiments, the open reading frame of codon-improved Cre-recombinase (iCre) was cloned into the respective viral backbone plasmids. Dsred was used as control for knockdown experiments.
AAV9. HEK293-T cells were grown in Cell Factory 10 trays (Thermo Scientific, 140400, Bonn, Germany) for 24 hours before transfection in Dulbecco's modified essential medium (DMEM) containing 10% fetal calf serum (FCS) and 1% penicillin/streptomycin. 420 µg of transgene plasmid and 1.5 mg of helper plasmid (pDP9rs, kindly provided by Roger Hajjar, Icahn School of Medicine at Mount Sinai, New York) were transfected into HEK293-T cells using polyethylenimine (Polysciences, 24765 Warrington, PA). After 72 hours, crude cell lysates were treated with benzonase and the viruses were purified by centrifugation through an iodixanol density gradient (Optiprep, Fresenius Kabi Norge, Oslo, Norway). The virus-containing fraction was then transferred to Vivaspin 20 columns (Sartorius Stedim, VS2042, Goettingen, Germany) and centrifuged to concentrate the viroids and to replace the iodixanol by Ringer lactate buffer (Braun, Melsungen, Germany). The rAAV titers were determined by measuring the copy number of viral genomes which were in the range of 1 × 1013–5 × 1013 genome copies per milliliter (vg/ml).
MMLV. 10 µg of pMXs retroviral vector (kindly provided by Deepak Srivastava, Gladstone Institute of Cardiovascular Diseases, San Francisco) was transfected using polyethylenimine into Plat-E cells which were plated on 15-cm tissue culture dish at a density of 10 × 106 cells per dish, 24 hours before transfection. Viral supernatants were collected from 30 culture dish, each with 15 cm diameter, at 72 and 120 hours after transfection, filtered through a 0.45 µm cellulose filter, and high titer MML viruses were produced by concentrating pooled viral supernatants using ultracentrifugation. The virus pellet was then resuspended in prewarmed Opti-minimum essential medium (MEM) (Gibco, Karlsruhe, Germany) containing 50 mmol/l 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES) buffer (pH 7) (Sigma, Deisenhofen, Germany). The titer was then determined by real time PCR.41
Intrapericardial delivery of viral vectors into neonatal mice. Neonatal mice were injected with viral vectors as described previously.20 Mother was first separated from the litters and kept in a new cage. 3–4 days old mice were anaesthetized with Fentanyl (0.05 mg/kg), Medetomidine (0.5 mg/kg), and Midazolam (5 mg/kg), later a puncture was carried out using a 30-gauge needle at the costoxiphoid angle of the anterior chest (BD Microfine+, 324825, Franklin Lakes, NJ). Injection was below the ribs (subcostal) in cranial direction and ~3 mm deep. Injection depth was ensured by using a microbore tubing (Tygon, I.D: 0.02 in, Saint-Gobain PPL) threaded over the needle to limit its advancement to 3 mm. This approach positions the needle beneath the sternum and anterior to the heart, and also minimizes the risk of pneumothorax caused due to lung injury. About 50 µl of virus was then injected into the pericardial space. Note that MMLV titers above the applied 5 × 109 vg/50 µl induced adverse effects due to high viscosity.
Whole-animal perfusion and fixation. Mice were anaesthetized using isoflurane and the hearts were exposed. A small cut was made in the right atrium and a cannula was inserted into the left ventricular chamber near the apex of the heart. Blood was cleared from the body by perfusing 5 ml of phosphate-buffered saline (PBS) and then the animal was perfused with prewarmed (37°C) 4% para-formaldehyde for 10 minutes. Tissues (heart, liver, lung, kidney, and spleen) were then explanted, fixed in 4% para-formaldehyde for 2 hours at 4°C, dehydrated in 30% sucrose overnight, embedded in O.C.T. (Cryomatrix, Thermo Scientific) embedding medium and sectioned with a microtome (5 µm).
Isolation of primary cells from mouse Primary CM/CF from adult mouse hearts. Hearts were harvested and coronary arteries were perfused briefly with buffer A (113 mmol/l NaCl, 4.7 mmol/l KCl, 0.6 mmol/l KH2PO4, 0.6 mmol/l Na2HPO4, 1.2 mmol/l MgSO4, 12 mmol/l NaHCO3, 10 mmol/l KHCO3, 10 mmol/l HEPES, and 30 mmol/l Taurine) to remove blood in a retrograde manner by cannulating the aorta. Collagenase type II (Worthington, Lakewood, NJ) was added to enzymatically dissociate the ventricular cells. The dissociated cells were then allowed to sediment at 37°C for 10 minutes to allow cardiomyocytes to settle down as a pellet leaving behind a nonmyocyte rich supernatant. The cardiomyocytes were then resuspended in buffer B (47.5 ml buffer A, 2.5 ml FCS and 62.5 µl 10mmol/l CaCl2). CaCl2 was gradually added to the cells to a final concentration of 100 µmol/l and the isolated cardiomyocytes were preplated in MEM containing 5% FCS, 10 mmol/l 2,3-butanedionemonoxime, 2 mmol/l L-glutamine and 1% penicillin/streptomycin for 2 hours at 37°C and 5% CO2. For CFs, nonmyocyte rich supernatant was then centrifuged at 225×g for 5 minutes and the pellets were resuspended in MEM containing 5% FCS, 2 mmol/l L-glutamine and 1% penicillin/streptomycin and kept at 37°C and 5% CO2. CF adhere more rapidly than EC and therefore these plates are enriched predominantly with CF (< 5% EC as contaminating cells). After 2 hours of plating, the plates were washed with prewarmed PBS (37°C) and were stored at −80°C.
Bone marrow cells. Femurs were dissected from the mice and excess tissue was removed using sterile forceps and scissors, keeping the ends of the bone intact. Bones were then disinfected upon immersion in 70% ethanol for 2 minutes and then washed with PBS in a sterile petridish. Tips of the bones were trimmed and the marrow was flushed with 3 ml FACS buffer (0.5% bovine serum albumin) using a 5-ml syringe with 30G × ½ needle into a sterile 15-ml centrifuge tube. The bone marrow cell suspension was then centrifuged at 400×g for 5 minutes. The cells were then treated with 1 ml erythrocyte lysis buffer (nine parts of 155 mmol/l NH4Cl and one part of 0.1mol/l Tris-HCl pH 7.65) for 5 minutes at room temperature before neutralizing with 2 ml of PBS. Cells were washed twice with PBS before they were subjected to FACS sorting and analysis.
Peripheral blood mononuclear cells. 100 µl of blood per mice was collected by retro-orbital bleeding into a heparinized 15-ml sterile centrifuge tube. The sample was then treated with 4 ml of erythrocyte lysis buffer for 10 minutes at room temperature before neutralization with 8 ml of PBS. The cells were then washed two times with PBS as described above.
Flow cytometry/cell sorting. Primary cells were isolated as described above. In the case of nonmyocyte cells, the nonmyocyte-enriched supernatant obtained after sedimentation of collagenase II-digested ventricles was centrifuged at 400×g for 5 minutes (instead of 225×g as in the case for cardiac fibroblast isolation). Primary cells were treated with rat antimouse CD16/CD32 (eBioscience, clone 2.4G2, San Diego, CA) at 4°C for 20 minutes before being incubated with PE-Cy7-conjugated primary antibodies at 4°C for 90 minutes. The following antibodies were used: anti-PDGFRα-PECy7 (clone APA5), anti-CD105-PECy7 (clone MJ7/18), anti-CD45-PECy7 (clone 30-F11), IgG isotype control (each from eBioscience), and anti-TroponinT (Sigma, clone JLT-12). The dye DRAQ7 (Cell Signaling, 7406P, Danvers, MA) was used to control for viability. Cell suspensions were triturated through a 40 µm Nylon cell strainer (Falcon) and then subjected to FACS analysis using a Biorad S3 cell sorter. For FACS sorting, the cardiac cell suspension was separated into cardiomyocyte- (pellet) and nonmyocyte-enriched fractions (supernatant) by centrifugation (900×g for 1 minute). While cardiomyocytes were sorted based on morphology, nonmyocyte fractions were further subjected to antibody-based procedures as described above.
Echocardiography. Transthoracic echocardiography was performed on mice to assess cardiac function. Ultrasound gel was applied to the chest of isoflurane-anaesthetized mice and echocardiographic monitoring was carried out using an ultrasound system with a linear transducer with 32–55 MHz frequency (Visual Sonics, Ontario, Canada) combined with Vevo 770 software. M-mode tracings were recorded and dimensions of myocardium at systole and diastole were measured.
Histochemical and immunohistochemical analyses. For the analysis of collagen deposition, paraffin sections (8 µm) of left ventricular myocardium were stained with Sirius red and Fast green42 or hematoxylin. Whole heart images were taken with a 10× objective using a AxioObserver.Z1 (Zeiss, Jena, Germany) motorized scanning stage microscopy (130 × 85; Märzhäuser, Wetzlar, Germany). Fibrosis was quantified as the ratio of Sirius red-to-Fast green signals in in each section. The cross-sectional area of cardiomyocytes was assessed by staining 6 µm-thick myocardial paraffin sections with Alexa Fluor 647-labeled wheat-germ agglutinin (WGA) (Life Technologies). SYTOX Green (Life Technologies) was applied to detect nuclei (at 488 nm). Images were taken from areas of transversely cut muscle fibers at 20× magnification (Leica TCS SP5 II; laser lines, 488 nm for SYTOX Green and 633 nm for WGA). Metamorph software version 7.7 (Molecular Devices Sunnyvale, CA) was applied for automated cell detection and to determine the average cross-sectional area of cardiomyocytes in one section (n > 50 cells per section; 100–200 cells per heart).12
For the analysis of vascular formation, myocardial sections (6 µm) were fixed with 4% paraformaldehyde for 10 minutes. After permeabilization with 1% TritonX and blocking with 10% goat serum, sections were incubated with primary antibodies against CD31 (Abcam #28364, Cambridge, UK) and TroponinT (Abcam #8295) overnight at 4°C. The sections were then incubated with secondary antibody conjugated to Alexafluor 488 and 4',6-diamidino-2-phenylindole (DAPI) was used to detect nuclei. Images were acquired using confocal microscope and analyzed as described above (n > 4,000 cells per section; 7,000–15,000 cells per heart).
Quantitative real-time PCR. Total RNA was prepared with peqGOLD RNA pure (peqlab) and 1 µg was reverse transcribed with Protoscript reverse transcriptase (NEB), both according to manufacturers' instructions. Quantitative real-time PCR amplification of target mRNAs was performed with primers listed below, using the FastStart universal SYBR Green master mix (Roche, Mannheim, Germany). The specificity of primers was validated by plotting the dissociation curve. The sample volume of 12.5 µl contained 1× SYBR Green master mix, 10 pmoles of each primer and 10 ng of template. PCR was performed using StepOnePlus Real-Time PCR systems (Applied Biosystems, Foster City, CA). Primer sequences were as follows (Gene symbol and species followed by sequences of forward and reverse primers):
Acta1 mouse (5′-CCCAAAGCTAACCGGGAGAAG-3′, 5′-CCAGAATCCAACACGATGCC-3′); Cd31 mouse (5′-ACACCTGCAAAGTGGAATCA-3′, 5′-TCTAACTTCGGCTTGGGAAA-3′); Cd68 mouse (5′-TACATGGCGGTGGAATACAA-3′, 5′-TCAAGGTGAACAGCTGGAGA-3′); Col1a1 mouse (5′-CTGGCAAGAAGGGAGATGA-3′, 5′- CACCATCCAAACCACTGAAA-3′); Col1a2 mouse (5′-AGGTCTTCCTGGAGCTGATG-3′, 5′-ACCCACAGGGCCTTCTTTAC-3′); Col3a1 mouse (5′-ACAGCAAATTCACTTACACAGTTC-3′, 5′-CTCATTGCCTTGCGTGTTT-3′); Myh7 mouse (5′-ACTGTCAACACTAAGAGGGTCA-3′, 5'-TTGGATGATTTGATCTTCCAGGG-3′); Nppa mouse (5′-GCTTCCAGGCCATATTGGAG-3′, 5′-GGGGGCATGACCTCATCTT-3′); Nppb mouse (5'-CCCAAAAAGAGTCCTTCGGTC-3', 5'-CGGTCTATCTTGTGCCCAAAG-3'); Rpl32 mouse (5′-ACATCGGTTATGGGAGCAAC-3′, 5′-GGGATTGGTGACTCTGATGG-3′); Spry1 mouse (5′-GAGCAGCAGCAGTGTGTCTT-3′, 5′-GCCTTCAAGTCTTCCACCAG-3′); Tagln mouse (5′-AGGTGTGGCTGAAGAATGGT-3′, 5′-AGGCACCTTCACTGGCTTC-3'); and Vegfa mouse (5′-CACGACAGAAGGAGAGCAGA-3′, 5′-AGATGTCCACCAGGGTCTCA-3′)
Expression of miR-21 was quantified using LNA-enhanced microRNA assays (Exiqon, Vedbaek, Denmark). 10 ng of total RNA were reverse transcribed using the Universal cDNA Synthesis kit II (Exiqon). The cDNAs were then quantified using the Fast Start Universal SYBR Green master mix (Roche) and LNA-enhanced miRCURY PCR primer sets for miRNAs (Exiqon). Real-time PCRs were performed using thermal-cycling parameters as recommended by Exiqon. U6 snRNA was used as normalization control.
Apoptosis assay. Apoptosis in isolated neonatal rat cardiac myocytes or paraffin-preserved hearts was assessed by TUNEL staining using the In Situ Cell Death Detection Kit, Fluorescein (Roche), according to the manufacturer's instructions. Briefly, sections of paraffin-embedded mouse hearts (6–8 μm) were rehydrated, followed by 5 minutes of microwave irradiation in antigen-retrieval solution (10 mmol/l Citrate/0.05% tween). The samples were incubated with TUNEL reaction mixture for 1 hour at 37°C in the dark. DAPI was added for nuclear counterstaining. Both a negative control (without enzyme solution) and a positive control (pretreatment with DNase for 10 minutes at room temperature) were included. Images of whole-heart sections were acquired by a confocal microscope (×20 objective, laser lines, 358 nm for DAPI and 488 nm for Fluorescein). Green (TUNEL positive) and blue (200,000–300,000 per group) nuclei were automatically counted using an image analysis algorithm (MetaMorph). The fraction of viable cells was determined in vitro, using a colorimetric 3-(4,5-dimethythiazol-2-yl)-2,5-diphenyl tetrazolium bromide (MTT) assay for metabolic activity: Neonatal rat cardiomyocytes were plated in MEM containing 5% FCS 1% BrdU and 1% penicillin/streptomycin for 24 hours. Cells were then transduced with adeno-associated viruses of serotype 6 for further 72 hours. The medium was then changed to 0.1% FCS and after 24 hours, apoptosis was induced by incubating the cells with 500 nmol/l doxorubicin (Doxo) in MEM containing 0.1% FCS for 24 hours. Unstimulated control cells were incubated in neonatal rat cardiac myocytes with 0.1% FCS. The cells were then kept in DMEM containing 0% FCS and 0.5 mg/ml of MTT for 4 hours. The resultant formazan crystals were dissolved in acidified isopropanol (200 μl) and the absorbance intensity was measured by a microplate reader (Tecan Infinite M200) at 570 nm with a reference wavelength of 650 nm.
Statistical analysis. Data are shown as mean ± SEM. Statistical analysis was performed using Graphpad Prism software package (version 6). Data distribution was assessed by Shapiro-Wilk test for normality. Common variance was tested using F-test or Bartlett's test. Differences between two means were tested using two-tailed unpaired Student's t-test. Differences among multiple means were assessed by one-way or two-way analysis of variance followed by Sidak's test. A P-value of <0.05 was considered significant. *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001.
SUPPLEMENTARY MATERIAL Figure S1. Cardiac cell sorting and quantification of endogenous miR-21 and biomarker mRNAs in isolated cardiac cell types. Figure S2. Quantification of miR-21 in heart, kidney, liver, lung and spleen of miR-21fl/fl mice after infection with iCre-encoding AAV9 or MMLV. Figure S3. Survival and viability of cardiac myocytes correlates depends on miR-21 expression. Figure S4. Expression of endothelial cell-specific genes and capillary density after manipulation of miR-21. Table S1. Physiological parameters in pressure-overload induced mice after global, myocyte or non-myocyte targeted deletion of miR-21.
Acknowledgments
We thank Lucia Koblitz for primary cell isolations; Kornelija Sakac and Pascal Baclet for performing mouse surgery and echocardiography; Sabine Brummer for cardiac histology; Stuart Orkin (Harvard Medical School, Boston) for miR-21 knockout mice; Eric Olson (University of Texas Southwestern Medical Center, Dallas, Texas) for miR-21 floxed mice Deepak Srivastava (Gladstone Institute of Cardiovascular Diseases, San Francisco) for providing the MMLV plasmid and Roger Hajjar (Icahn School of Medicine at Mount Sinai, New York) for providing human heart biopsies. This work was supported in part by grants from the Fondation Leducq (to S.E.); the Bavarian Ministry of Sciences, Research and the Arts in the framework of the Bavarian Molecular Biosystems Research Network (to S.E.); and Munich Biotech Cluster m4 (to S.E.). S.E. has filed a patent on the use of miR-21 for therapeutic purposes.
Supplementary Material
References
- Melman, YF, Shah, R and Das, S (2014). MicroRNAs in heart failure: is the picture becoming less miRky? Circ Heart Fail 7: 203–214. [DOI] [PubMed] [Google Scholar]
- Thum, T, Gross, C, Fiedler, J, Fischer, T, Kissler, S, Bussen, M et al. (2008). MicroRNA-21 contributes to myocardial disease by stimulating MAP kinase signalling in fibroblasts. Nature 456: 980–984. [DOI] [PubMed] [Google Scholar]
- Thum, T, Chau, N, Bhat, B, Gupta, SK, Linsley, PS, Bauersachs, J et al. (2011). Comparison of different miR-21 inhibitor chemistries in a cardiac disease model. J Clin Invest 121: 461–2; author reply 462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez, IG, MacKenna, DA, Johnson, BG, Kaimal, V, Roach, AM, Ren, S et al. (2015). Anti-microRNA-21 oligonucleotides prevent Alport nephropathy progression by stimulating metabolic pathways. J Clin Invest 125: 141–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Z, Zha, Y, Hu, W, Huang, Z, Gao, Z, Zang, Y et al. (2013). The autoregulatory feedback loop of microRNA-21/programmed cell death protein 4/activation protein-1 (MiR-21/PDCD4/AP-1) as a driving force for hepatic fibrosis development. J Biol Chem 288: 37082–37093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chau, BN, Xin, C, Hartner, J, Ren, S, Castano, AP, Linn, G et al. (2012). MicroRNA-21 promotes fibrosis of the kidney by silencing metabolic pathways. Sci Transl Med 4: 121ra18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, G, Friggeri, A, Yang, Y, Milosevic, J, Ding, Q, Thannickal, VJ et al. (2010). miR-21 mediates fibrogenic activation of pulmonary fibroblasts and lung fibrosis. J Exp Med 207: 1589–1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ardite, E, Perdiguero, E, Vidal, B, Gutarra, S, Serrano, AL and Muñoz-Cánoves, P (2012). PAI-1-regulated miR-21 defines a novel age-associated fibrogenic pathway in muscular dystrophy. J Cell Biol 196: 163–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayed, D, He, M, Hong, C, Gao, S, Rane, S, Yang, Z et al. (2010). MicroRNA-21 is a downstream effector of AKT that mediates its antiapoptotic effects via suppression of Fas ligand. J Biol Chem 285: 20281–20290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agah, R, Frenkel, PA, French, BA, Michael, LH, Overbeek, PA and Schneider, MD (1997). Gene recombination in postmitotic cells. Targeted expression of Cre recombinase provokes cardiac-restricted, site-specific rearrangement in adult ventricular muscle in vivo. J Clin Invest 100: 169–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bish, LT, Morine, K, Sleeper, MM, Sanmiguel, J, Wu, D, Gao, G et al. (2008). Adeno-associated virus (AAV) serotype 9 provides global cardiac gene transfer superior to AAV1, AAV6, AAV7, and AAV8 in the mouse and rat. Hum Gene Ther 19: 1359–1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganesan, J, Ramanujam, D, Sassi, Y, Ahles, A, Jentzsch, C, Werfel, S et al. (2013). MiR-378 controls cardiac hypertrophy by combined repression of mitogen-activated protein kinase pathway factors. Circulation 127: 2097–2106. [DOI] [PubMed] [Google Scholar]
- Hulot, JS, Fauconnier, J, Ramanujam, D, Chaanine, A, Aubart, F, Sassi, Y et al. (2011). Critical role for stromal interaction molecule 1 in cardiac hypertrophy. Circulation 124: 796–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kass-Eisler, A, Falck-Pedersen, E, Alvira, M, Rivera, J, Buttrick, PM, Wittenberg, BA et al. (1993). Quantitative determination of adenovirus-mediated gene delivery to rat cardiac myocytes in vitro and in vivo. Proc Natl Acad Sci USA 90: 11498–11502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, J, Pettigrew, GJ, Thomas, J, Vandenberg, JI, Delriviere, L, Bolton, EM et al. (2002). Lentiviral vectors for delivery of genes into neonatal and adult ventricular cardiac myocytes in vitro and in vivo. Basic Res Cardiol 97: 348–358. [DOI] [PubMed] [Google Scholar]
- Byun, J, Huh, JE, Park, SJ, Jang, JE, Suh, YL, Lee, JS et al. (2000). Myocardial injury-induced fibroblast proliferation facilitates retroviral-mediated gene transfer to the rat heart in vivo. J Gene Med 2: 2–10. [DOI] [PubMed] [Google Scholar]
- Ieda, M, Fu, JD, Delgado-Olguin, P, Vedantham, V, Hayashi, Y, Bruneau, BG et al. (2010). Direct reprogramming of fibroblasts into functional cardiomyocytes by defined factors. Cell 142: 375–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee, I, Fuseler, JW, Price, RL, Borg, TK and Baudino, TA (2007). Determination of cell types and numbers during cardiac development in the neonatal and adult rat and mouse. Am J Physiol Heart Circ Physiol 293: H1883–H1891. [DOI] [PubMed] [Google Scholar]
- Shimshek, DR, Kim, J, Hübner, MR, Spergel, DJ, Buchholz, F, Casanova, E et al. (2002). Codon-improved Cre recombinase (iCre) expression in the mouse. Genesis 32: 19–26. [DOI] [PubMed] [Google Scholar]
- Zhang, JC, Woo, YJ, Chen, JA, Swain, JL and Sweeney, HL (1999). Efficient transmural cardiac gene transfer by intrapericardial injection in neonatal mice. J Mol Cell Cardiol 31: 721–732. [DOI] [PubMed] [Google Scholar]
- Muzumdar, MD, Tasic, B, Miyamichi, K, Li, L and Luo, L (2007). A global double-fluorescent Cre reporter mouse. Genesis 45: 593–605. [DOI] [PubMed] [Google Scholar]
- Bönner, F, Borg, N, Burghoff, S and Schrader, J (2012). Resident cardiac immune cells and expression of the ectonucleotidase enzymes CD39 and CD73 after ischemic injury. PLoS One 7: e34730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, TX, Hartner, J, Lim, EJ, Fabry, V, Mingler, MK, Cole, ET et al. (2011). MicroRNA-21 limits in vivo immune response-mediated activation of the IL-12/IFN-gamma pathway, Th1 polarization, and the severity of delayed-type hypersensitivity. J Immunol 187: 3362–3373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patrick, DM, Montgomery, RL, Qi, X, Obad, S, Kauppinen, S, Hill, JA et al. (2010). Stress-dependent cardiac remodeling occurs in the absence of microRNA-21 in mice. J Clin Invest 120: 3912–3916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayed, D, Rane, S, Lypowy, J, He, M, Chen, IY, Vashistha, H et al. (2008). MicroRNA-21 targets Sprouty2 and promotes cellular outgrowths. Mol Biol Cell 19: 3272–3282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, LZ, Li, C, Chen, Q, Jing, Y, Carpenter, R, Jiang, Y et al. (2011). MiR-21 induced angiogenesis through AKT and ERK activation and HIF-1α expression. PLoS One 6: e19139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabatel, C, Malvaux, L, Bovy, N, Deroanne, C, Lambert, V, Gonzalez, ML et al. (2011). MicroRNA-21 exhibits antiangiogenic function by targeting RhoB expression in endothelial cells. PLoS One 6: e16979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald, RA, Halliday, CA, Miller, AM, Diver, LA, Dakin, RS, Montgomery, J, et al. (2015). Reducing in-Stent restenosis. J Am Coll Cardiol 65: 2314–2327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheedy, FJ (2015). Turning 21: Induction of miR-21 as a key switch in the inflammatory response. Front Immunol 6: 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumarswamy, R, Volkmann, I, Jazbutyte, V, Dangwal, S, Park, DH and Thum, T (2012). Transforming growth factor-β-induced endothelial-to-mesenchymal transition is partly mediated by microRNA-21. Arterioscler Thromb Vasc Biol 32: 361–369. [DOI] [PubMed] [Google Scholar]
- Kakkar, R and Lee, RT (2010). Intramyocardial fibroblast myocyte communication. Circ Res 106: 47–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oka, T, Xu, J, Kaiser, RA, Melendez, J, Hambleton, M, Sargent, MA et al. (2007). Genetic manipulation of periostin expression reveals a role in cardiac hypertrophy and ventricular remodeling. Circ Res 101: 313–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda, N, Manabe, I, Uchino, Y, Eguchi, K, Matsumoto, S, Nishimura, S et al. (2010). Cardiac fibroblasts are essential for the adaptive response of the murine heart to pressure overload. J Clin Invest 120: 254–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamo, T, Akazawa, H and Komuro, I (2015). Cardiac nonmyocytes in the hub of cardiac hypertrophy. Circ Res 117: 89–98. [DOI] [PubMed] [Google Scholar]
- Song, K, Nam, YJ, Luo, X, Qi, X, Tan, W, Huang, GN et al. (2012). Heart repair by reprogramming non-myocytes with cardiac transcription factors. Nature 485: 599–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong, P, Christia, P, Saxena, A, Su, Y and Frangogiannis, NG (2013). Lack of specificity of fibroblast-specific protein 1 in cardiac remodeling and fibrosis. Am J Physiol Heart Circ Physiol 305: H1363–H1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Acharya, A, Baek, ST, Banfi, S, Eskiocak, B and Tallquist, MD (2011). Efficient inducible Cre-mediated recombination in Tcf21 cell lineages in the heart and kidney. Genesis 49: 870–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsley, A, Snider, P, Zhou, H, Rogers, R, Wang, J, Olaopa, M et al. (2007). Identification and characterization of a novel Schwann and outflow tract endocardial cushion lineage-restricted periostin enhancer. Dev Biol 307: 340–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian, L, Huang, Y, Spencer, CI, Foley, A, Vedantham, V, Liu, L et al. (2012). In vivo reprogramming of murine cardiac fibroblasts into induced cardiomyocytes. Nature 485: 593–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockman, HA, Ross, RS, Harris, AN, Knowlton, KU, Steinhelper, ME, Field, LJ et al. (1991). Segregation of atrial-specific and inducible expression of an atrial natriuretic factor transgene in an in vivo murine model of cardiac hypertrophy. Proc Natl Acad Sci USA 88: 8277–8281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmo, M, Peixoto, C, Coroadinha, AS, Alves, PM, Cruz, PE and Carrondo, MJ (2004). Quantitation of MLV-based retroviral vectors using real-time RT-PCR. J Virol Methods 119: 115–119. [DOI] [PubMed] [Google Scholar]
- Nakajima, H, Nakajima, HO, Salcher, O, Dittiè, AS, Dembowsky, K, Jing, S et al. (2000). Atrial but not ventricular fibrosis in mice expressing a mutant transforming growth factor-beta(1) transgene in the heart. Circ Res 86: 571–579. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.