To the editor:
Muscle wasting occurs with aging and in a wide range of catabolic diseases, such as cancer, diabetes, chronic renal disease, and heart failure, which can lead to a dramatically reduced quality of life and increased disease mortality. Several treatments are currently in development as a result of our understanding of the cellular mechanisms that cause muscle wasting.1 Myostatin, encoded by the MSTN gene, is a well-known cytokine that is activated in most disease conditions associated with muscle wasting, including cancer. Myostatin is primarily secreted by skeletal muscle cells and acts mainly through an autocrine/paracrine mode by activating the SMAD2/SMAD3 signaling pathway that subsequently triggers transcription programs leading to muscle atrophy.1,2 Inhibition of myostatin has been identified as a promising strategy to alleviate muscle wasting, especially given that loss of functional myostatin in humans is not associated with apparent deleterious effects other than muscle hypertrophy.3 Monoclonal antibodies that bind to myostatin to inhibit its function are currently in clinical trials as a potential therapy to treat cachexia syndrome in cancer patients.1,4 However, the nature of antibody-based drugs means that this strategy requires repeated injections chronically.
With the emergence of different genome-editing tools that enable investigators to introduce genetic alterations into specific genomic sites at will, the use of genome editing for human therapeutic applications is being explored in a variety of preclinical models of diseases.5 Recently, in vivo genome editing targeting muscle tissues with clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated (Cas) systems showed efficacy in a mouse model of Duchenne muscular dystrophy.6,7,8 With a view to establishing a novel therapeutic modality toward muscle-wasting syndrome, we used CRISPR/Cas9 to directly target Mstn in vivo in skeletal muscle cells to prevent loss of muscle mass. We reasoned that the ability to disrupt Mstn and block the myostatin pathway in some, if not all, skeletal muscle cells would be sufficient to partially prevent atrophy of targeted cells and its neighbor cells, thus preserving muscle function to some extent.
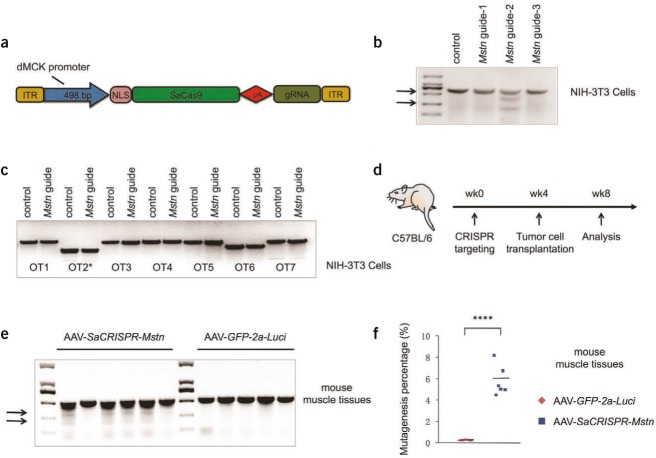
To perform in vivo targeting of skeletal muscle cells, we adapted a Staphylococcus aureus (SaCas9)/adeno-associated virus 8 (AAV8) system9 for specific muscle cell targeting by using a tissue-specific double muscle creatine kinase (dMCK) promoter (~500 bp)10 to drive SaCas9 expression (Figure 1a). Efficient expression of genes driven by the dMCK promoter was confirmed by in vivo luminescence examinations with mice injected with AAV8/dMCK promoter-GFP-2a-luciferase viruses for up to two months (data not shown). We next screened candidate SaCRISPR guide RNAs (gRNAs) targeting the first exon of the mouse Mstn gene in mouse NIH-3T3 cells. Among the three gRNAs we tested, we found that one gRNA (guide-2, targeting sequences: GGGCTGTGTAATGCATGTGCG+TGGAG) displayed a ~50% mutagenesis rate at the on-target site as judged by Surveyor assays (Figure 1b). We further accessed the potential off-target mutagenesis at ten sites that are mostly matched to the guide-2 targeting sequences by genome-wide prediction using the CRISPR Design Server (http://crispr.mit.edu/) (Supplementary Table S1 online). Although one of the sites displayed a relatively high score (5.0) with mismatches by two bases in protospacer matching sequences, lack of protospacer-adjacent motif prevented efficient targeting at this locus. With the limit of detection by the Surveyor assays in NIH-3T3 cells, we found no evidence of significant off-target mutagenesis (Figure 1c). We thus used guide-2 for subsequent in vivo targeting. We made an AAV for efficient delivery of SaCas9 and this gRNA (AAV-SaCRISPR-Mstn) to muscle cells in vivo, using an AAV expressing GFP-2a-luciferase (AAV-GFP-2a-Luci) as a control.
Figure 1.
In vivo mutagenesis of the Mstn gene in skeletal muscle cells with AAV-SaCRISPR/Cas9. (a) A schematic diagram of adapted AAV-SaCRISPR backbone with SaCas9 driven by a skeletal muscle cell-specific dMCK promoter. (b) Surveyor assays performed at the targeting loci with genomic DNA from NIH-3T3 cells transfected with SaCas9 and different guide RNAs (gRNAs) targeting the mouse Mstn gene. (c) Surveyor assays performed at the top genome-wide off-target sites (Supplementary Table S1) with genomic DNA from NIH-3T3 cells transfected with SaCas9 and gRNA-2 targeting the mouse Mstn gene. For duplicated region (OT2*), the first genomic locus listed in the table was analyzed for potential off-target effect. (d) Schematic description of the in vivo targeting experiment. (e) Surveyor assays performed with genomic DNA from mouse gastrocnemius muscle tissues treated with AAV-SaCRISPR-Mstn or AAV-GFP-2a-Luci at the Mstn targeting locus at week 8. (f) Indel rates at the on-target sites from next-generation DNA sequencing of muscle samples from post-treatment animals receiving either AAV-SaCRISPR-Mstn or AAV-GFP-2a-Luci viruses. In b and e, arrows show the cleavage products resulting from the Surveyor assays; the intensity of the cleavage product bands relative to the uncleaved product band corresponds to the mutagenesis rate. Statistical analyses in f were conducted using the two-tailed Student's t-test; ****P < 0.0001. dMCK promoter, double muscle creatine kinase promoter; ITR, inverted terminal repeat; NLS, nuclear localization signal.
To test the hypothesis that genome editing could disrupt Mstn in vivo in muscle tissues and thus prevent muscle loss in catabolic conditions, we administered the AAV-SaCRISPR-Mstn and the AAV-GFP-2a-Luci viruses locally to gastrocnemius muscles (4–5 × 1010 viral particles per injection and six to eight injections per limb) in six-week-old male wild-type C57BL/6 mice, with six mice in each group. We waited around four weeks to allow sufficient expression of SaCas9 in vivo, after which we injected Lewis lung carcinoma cells (3 × 106 cells per mouse) subcutaneously into all mice to induce cancer-associated muscle loss. The mice were then analyzed at the end of week 8 (Figure 1d). Immunostainings of gastrocnemius muscles from AAV-GFP-2a-Luci virus-treated animals showed a mosaic pattern of GFP+ myofibers (Supplementary Figure S1a), indicating efficient but uneven infections by local viral injection. Surveyor analysis in muscle tissues displayed around 5% mutagenesis at the Mstn gene locus in four out of six CRISPR-treated mice, with no evidence of mutagenesis in control mice (Figure 1e). Furthermore, deep sequencing of gRNA targeting sites indicated 4.5 to 8.2% (5.8% on average) indel formation at the target sites in muscle tissues receiving AAV-SaCRISPR-Mstn in all six mice, in contrast to around 0.2% indel formation in muscle tissues receiving AAV-GFP-2a-Luci viruses (Figure 1f). We therefore also included the two mice that showed little editing activity in Surveyor assays (Figure 1e) for all functional analyses.
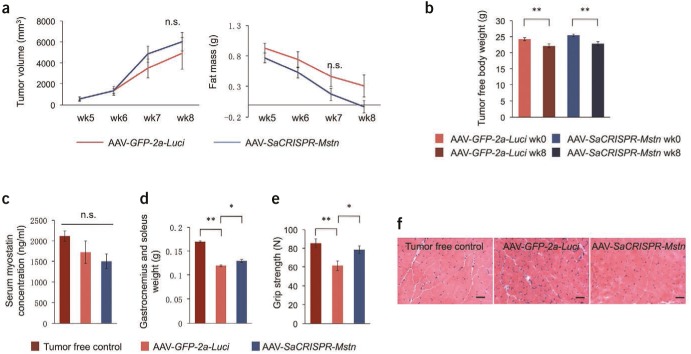
We observed a consistent growth of tumors (as judged by tumor volume measurement) and reduction of fat mass (as judged by NMR analysis) in both groups after tumor cell engraftment (Figure 2a) and a significant decrease of tumor-free body weight (~9%) at the end point (week 8) in comparison to week 0 in both groups (Figure 2b), indicating the development of cancer-associated cachexia; no statistically significant difference was observed in the above measurements between AAV-SaCRISPR-Mstn–treated and control mice.
Figure 2.
Effects of targeting the Mstn gene in skeletal muscle cells in mice with cancer-associated muscle atrophy. (a) Tumor volume (direct measurement) and fat mass (analyzed by NMR) in live animals each week after tumor cell engraftment. (b) Tumor-free body weight at week 0 (starting body weight) and week 8 (body weight at the end point with tumor weight subtracted) in mice treated with AAV-SaCRISPR-Mstn or AAV-GFP-2a-Luci viruses. (c) Serum concentration of myostatin measured by ELISA in animals from indicated groups at week 8. (d) Gastrocnemius and soleus muscle weight in animals from indicated groups measured at week 8. (e) Grip strength of limbs measured in live animals from indicated groups at week 8. (f) Hematoxylin-eosin stains of cross-sectional areas of gastrocnemius muscles of representative mice from indicated groups at week 8. Scale bar = 200 µm. In a–e, n = 6 mice for AAV-SaCRISPR-Mstn or AAV-GFP-2a-Luci group; n = 10 for tumor-free control group. Error bars show standard error of the mean. The unpaired, two-tailed Student's t-test was used for statistical analysis in a and b. One-way analysis of variance, post-hoc Bonferroni multiple-comparison test was used for statistical analysis in c–e. *P < 0.05; **P < 0.01. AAV, adeno-associated virus; ELISA, enzyme-linked immunosorbent assay; n.s., not significant.
Next, we analyzed muscle conditions in mice from different groups, including another group of mice with no tumor engraftment and no virus treatment as healthy controls (designated as “tumor-free control”). No significant differences of myostatin concentration in mouse serum were noticed as a result of the relatively low mutagenesis efficiency (Figure 2c). However, we observed a small but significant ~9% increase of gastrocnemius and soleus muscle weight in the CRISPR-treated group compared to the AAV-GFP-2a-Luci group (Figure 2d). We suspected that this was a result of the major autocrine/paracrine-working mode of myostatin2; disruption of Mstn gene in a few myotubes caused a decrease in local myostatin concentration, leading to attenuated myostatin signaling pathway in CRISPR-targeted cells as well as its neighbor cells, and thus alleviated atrophy of these myotubes. To further test this hypothesis, we also measured the grip strength of virus-treated limbs. Compared to tumor-free mice, mice carrying tumors displayed a dramatic reduction in grip strength as cachexia developed (Figure 2e). However, we observed a significant improvement (>25%) in grip strength in mice treated with AAV-SaCRISPR-Mstn in comparison to AAV-GFP-2a-Luci, indicating partially recovered muscle function in CRISPR-treated mice (Figure 2e). Moreover, CRISPR-treated mice showed clearly ameliorated atrophy of myocytes, as indicated by hematoxylin and eosin staining of muscle fibers (Figure 2f). Interestingly, we found coexistence of myotubes that with severe atrophy (indicated by dotted-line boxes) and with alleviated atrophy (indicated by full-line boxes) in a single slide from CRISPR-treated mice (Supplementary Figure S1b), suggesting that alleviated muscle atrophy is mostly likely a result of decreased local myostatin production instead of a systematic reduction in blood myostatin concentration.
In summary, we demonstrated in this study that AAV8-mediated delivery of SaCas9 driven by a muscle cell–specific promoter can be used to disrupt Mstn in mouse muscle in vivo. Although the overall efficiency of genome editing in muscle tissues was low (around 5% as judged by Surveyor assay and deep sequencing), consistent with previous studies with in vivo muscle targeting,6–8 disruption of myostatin function in a portion of muscle cells was shown to be sufficient to reduce cachexia in targeted cells or neighbor cells due to the major autocrine/paracrine working mode of myostatin, thus preserving muscle function (>25% as judged by grip-strength measurement and ameliorated atrophy) in cancer-induced cachexia syndrome. As a proof of concept, our study raises the possibility that a patient could benefit with alleviated muscle-wasting syndrome induced by cancer or other catabolic conditions through the use of CRISPR/Cas9-mediated targeting of the MSTN gene in skeletal muscle tissues, although continued development of this strategy to improve the targeting efficacy and assessment of safety will be needed before it can be realized in human beings.
SUPPLEMENTARY MATERIAL Table S1. Top ten genome-wide off-target sites used in this study. Figure S1. Effects of targeting the Mstn gene in skeletal muscle cells in mice with cancer-associated muscle atrophy.
Acknowledgments
We thank Kiran Musunuru (University of Pennsylvania) and Yan Chen (Institute for Nutritional Sciences, SIBS, China) for helpful comments, suggestions, and critical reading of this manuscript. We thank Xianfeng Chen for assistance in data analysis and Junjie Xiao (Shanghai University, China) for help in measuring mouse-grip strength. This work was supported by grants from Ministry of Science and Technology of the People's Republic of China (2016YFC1304900), the Shanghai Institutes for Biological Sciences Fellowship (Y5Y1X41491) (Y.Z.), the National Natural Science Foundation of China (81500614 to Y.Z.), the Hundred Talents Program of the Chinese Academy of Sciences (Q.D.), the National Youth 1000 Talents Program (Q.D.), the Shanghai Pujiang Program (15PJ1409200 to Q.D.), and the National Natural Science Foundation of China (31670829 to Q.D.).
Supplementary Material
References
- Cohen, S, Nathan, JA and Goldberg, AL (2015). Muscle wasting in disease: molecular mechanisms and promising therapies. Nat Rev Drug Discov 14: 58–74. [DOI] [PubMed] [Google Scholar]
- Lee, YS, Huynh, TV and Lee, SJ (2016). Paracrine and endocrine modes of myostatin actin. J Appl Physiol (1985) 120: 592–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuelke, M, Wagner, KR, Stolz, LE, Hubner, C, Riebel, T, Komen, W et al. (2014). Myostatin mutation associated with gross muscle hypertrophy in a child. N Engl J Med 350: 2682–2688. [DOI] [PubMed] [Google Scholar]
- Ebner, N, Steinbeck, L, Doehner, W, Anker, SD and von Heahling, S (2014). Highlights from the 7th Cachexia Conference: muscle wasting pathophysiological detection and novel treatment strategies. J Cachexia Sarcopenia Muscle 5: 27–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeder, ML and Gersbach, CA (2016). Genome-editing technologies for gene and cell therapy. Mol Ther 24: 475–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long, C, Amoasii, L, Mireault, AA, McAnally, JR, Li, H, Sanchez-Ortiz, E et al. (2016). Postnatal genome editing partially restores dystrophin expression in a mouse model of muscular dystrophy. Science 351: 400–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson, CE, Hakim, CH, Ousterout, DG, Thakore, PI, Moreb, EA, Castellanos Rivera, RM et al. (2016). In vivo genome editing improves muscle function in a mouse model of Duchenne muscular dystrophy. Science 351: 403–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabebordbar, M, Zhu, K, Cheng, JK, Chew, WL, Widrick, JJ, Yan, WX et al. (2016). In vivo gene editing in dystrophic mouse muscle and muscle stem cells. Science 351: 407–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran, FA, Cong, L, Yan, WX, Scott, DA, Gootenberg, JS, Kriz, AJ et al. (2014). In vivo genome editing using Staphylococcus aureus Cas9. Nature 520: 186–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, B, Li, J, Fu, FH, Chen, C, Zhu, X, Zhou, L et al. (2008). Construction and analysis of compact muscle-specific promoters for AAV vectors. Gene Ther 15: 1489–1499. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.